The Comoros Show the Earliest Austronesian Gene Flow into the Swahili Corridor (original) (raw)
Abstract
At the dawn of the second millennium, the expansion of the Indian Ocean trading network aligned with the emergence of an outward-oriented community along the East African coast to create a cosmopolitan cultural and trading zone known as the Swahili Corridor. On the basis of analyses of new genome-wide genotyping data and uniparental data in 276 individuals from coastal Kenya and the Comoros islands, along with large-scale genetic datasets from the Indian Ocean rim, we reconstruct historical population dynamics to show that the Swahili Corridor is largely an eastern Bantu genetic continuum. Limited gene flows from the Middle East can be seen in Swahili and Comorian populations at dates corresponding to historically documented contacts. However, the main admixture event in southern insular populations, particularly Comorian and Malagasy groups, occurred with individuals from Island Southeast Asia as early as the 8th century, reflecting an earlier dispersal from this region. Remarkably, our results support recent archaeological and linguistic evidence-based suggestions that the Comoros archipelago was the earliest location of contact between Austronesian and African populations in the Swahili Corridor.
Keywords: Comoros, Madagascar, East Africa, population genetics, admixture, migration
Introduction
The economic development of the Indian Ocean trading network led to the emergence of regional geopolitical powers, and the Swahili Corridor represented its African pole.1, 2, 3 Eventually comprising a string of city-states along the East African coast and offshore islands, from southern Somalia in the north to the Comoros archipelago, Madagascar, and Central Mozambique in the south, this was a major zone of proto-globalization that, at various times, featured direct contact between populations from Africa, the Middle East, South Asia, and Island Southeast Asia. During cyclical periods of intense trading, this dense network of interactions drove the flow of goods, ideas, and crucially, genes.4, 5, 6
The Swahili culture began to emerge during the first millennium CE from small-scale coastal and island Bantu communities engaged in farming, fishing, hunting, pottery, iron production, and local as well as Indian Ocean trade.1, 7, 8 As a result of their unique geographical position at a cross-road between Africa and the Indian Ocean world and their exploitation of maritime resources, the Swahili became merchant middlemen, providing inland African markets with iron, textiles, shell beads, and agricultural produce in return for gold, ivory, and slaves for inter-regional trade. With the intensification of the Indian Ocean trading network at the beginning of the second millennium, together with maritime technological developments, notably the emergence of larger boats, these coastal dwellers began sailing farther afield to far-distant locations in the Middle East and western India.7 Urban centers were accordingly restructured with the extension of ports, the building of coral-rag “stonehouses,” and the movement of mosques closer to the shore.7 The intensity of exchanges between Swahili villages and trading towns over many centuries was a major driver of the cultural unity of the Swahili Corridor. Major entrepôts established on the East African coast and islands rapidly became cosmopolitan centers in which the Swahili cohabited at various times with merchants from the Middle East and Asia.1, 2, 3, 6
While trade links to the Middle East played a key role in the emergence of the Swahili Corridor, the southern part of the region, particularly Madagascar, was shaped by contact with the Austronesian world far to the east.1 Originating in Island Southeast Asia, the Austronesian dispersal across the Indian Ocean was mostly mediated by traders linked to Southeast Asian maritime polities. Dominating maritime routes, Island Southeast Asian traders were one of the main geopolitical powers of the region, notably during the reign of the Srivijaya Empire (6th–13th centuries).4 Despite being centered on the Malaysian Peninsula, Sumatra, and Java islands, the empire established several trading posts around the Indonesian archipelago. In Southeast Borneo, they controlled the important strategic entrepôt of Banjarmasin, and their long-standing presence in this area put the Malay in contact with local groups such as the Ma’anyan, creating new population dynamics marked by long-distance migrations.5, 9 We have previously shown that the group who descended from this contact, the Banjar, are currently the closest population to the ancestors of the Asian genetic background found in the Malagasy, 7,500 km away.10 This migration is also reflected in the Malagasy language, which is closely related to the Ma’anyan language but also borrows from Malay11, 12 and includes minor contributions from African languages (mainly Sabaki, a branch of Bantu).13 This contact appears to have occurred during a time of peak activity in the Indian Ocean trading network, most likely after a migration following a direct route across the Indian Ocean.5, 14 However, the broader history of Austronesian settlement in East Africa remains unclear. The presence of Island Southeast Asians in Madagascar and the Comoros archipelago is supported by archaeological analyses of ancient crop remains, which reveal that Asian species, such as rice, dominated agricultural subsistence from the early stages of settlement on these islands.15 On the continent and nearby coastal islands, Asian crops were only identified in minor proportions at a small number of sites (mainly trading ports) and only became dominant in rare cases several centuries later. This pattern suggests that long-term Austronesian settlement could have been limited to these two insular territories. Present-day populations on Madagascar and the Comoros have genetic inheritance from Island Southeast Asia, pointing to commonality in the genetic ancestry of Malagasy and Comorian groups.16, 17, 18, 19, 20, 21 Nonetheless, the two regions also possess important differences, notably linguistic ones: an Austronesian language, Malagasy, is spoken today on Madagascar, and a Bantu Sabaki language, Comorian, is spoken in the Comoros.22 In addition, uniparental markers have shown higher Middle Eastern gene flow and more limited Austronesian inheritance in Comorians than in Malagasy.18 Recent archaeological data suggest that Austronesian settlement might have occurred earlier in the Comoros (8th–11th centuries) than in Madagascar (11th–13th centuries), though data is limited.15 All of these studies highlight the complex dynamics of the Austronesian dispersal into the Swahili Corridor.
By analyzing uniparental and genome-wide genetic diversities in several modern Kenyan Swahili communities together with Comorian groups, we have dual objectives: (1) to characterize the historical genetic interactions that took place along the Swahili Corridor and (2) to reconstruct the phases of Austronesian dispersal into East Africa.
Material and Methods
Sample Collection and Ethics
A total of 276 DNA samples were sampled from six groups in Kenya and the Comoros islands: Swahili communities from Mombasa (n = 31), Kilifi (n = 93), and the Lamu archipelago (n = 104) in Kenya, and Comorian communities from Anjouan (n = 16), Grande Comore (n = 18), and Moheli (n = 15) in the Comoros archipelago (Figure S1). All samples were collected from healthy unrelated adult donors, all of whom provided written informed consent. DNA from Comorians was extracted from blood samples with a standard salting-out procedure. DNA from Swahilis was extracted from saliva samples with the Oragene sampling kit according to the manufacturer’s instructions. In each location, after a full presentation of the project to a wide audience, a discussion with each individual willing to participate ensured that the project was fully understood. Once participants had signed informed-consent forms, we interviewed them to obtain information on their date and place of birth, their spoken language(s), and similar data related to their genealogy (up to 2nd or 3rd generation) to establish local ancestry. This sampling strategy, for which we have long-standing experience, allows a relatively random sampling of anthropological interest. This study was approved by the French Ethics Committees (Committees of Protection of Persons) and the Lamu Council of Elders (Lamu County, Kenya). A subset of 140 individuals from our sampling was used for genome-wide genotyping: Swahili communities from Mombasa (n = 23), Kilifi (n = 37), and the Lamu archipelago (n = 31); and Comorian communities from Anjouan (n = 16), Grande Comore (n = 18), and Moheli (n = 15). Genome-wide SNP genotyping was performed with the Illumina Human Omni5 Bead Chip (Illumina), which surveys 4,284,426 single-nucleotide markers regularly spaced across the genome. See Accession Numbers.
Paternal lineages of the Swahili groups were characterized via a method described previously23 (n = 109). DNA quantity for the Comorian samples prevented us from performing analyses on the non-recombining region of the Y chromosome (NRY). In brief, 96 binary markers on the NRY were analyzed with a nanofluidic dynamic array and the BioMark HD system (Fluidigm, USA). Haplogroups were assigned on the basis of the updated ISOGG Y-DNA haplogroup tree24 and the Y-Phylotree25 (Table S1). The full list of markers is described by Kusuma and colleagues.23 We characterized maternal lineages for all samples by sequencing the complete mtDNA (n = 276). In brief, double bar-coded libraries were prepared and enriched for mtDNA as described previously.26, 27 Base calling, quality filtering, and further steps aimed at obtaining consensus sequences were carried out as described previously.28 Sequences (see Accession Numbers) were then analyzed and aligned against the revised Cambridge Reference Sequence (rCRS)29 with MAFFT aligner v7.30 Mitochondrial haplogroups were determined with the HaploGrep program based on Phylotree build 1731 (Table S2).
Dataset
We gathered genome-wide data from previously published studies of populations from Africa, Madagascar, the Middle East, Southeast Asia, South Asia, East Asia, and Europe (Table S3). Two datasets were compiled respective to their analytical use: a low-SNP-density dataset of populations covering a large geographical area and a high-SNP-density dataset of populations comprising a more limited subset of the populations of the low-SNP-density dataset. To avoid any statistical bias that might result from the over-representation of some populations in the datasets, we randomly selected a maximum of 25 individuals for each group, such that each population had between 4 and 25 individuals. We applied quality controls by using Plink v1.932 to filter for and exclude (1) close relatives by using an identity-by-descent (IBD) estimation with an upper threshold of 0.25 (second-degree relatives); (2) SNPs that failed the Hardy-Weinberg exact test (p < 10−6) within each group; (3) samples with a call rate < 0.99 and displaying missing rates > 0.05 across all samples in each population; and (4) variants in high linkage disequilibrium (r2 > 0.2). After this filtering, the low-SNP-density dataset included 3,477 individuals from 193 populations genotyped for 171,728 SNPs, and the high-SNP-density dataset included 1,664 individuals from 83 populations genotyped for 411,432 SNPs. All genotypes in the high-SNP-density dataset were then phased with SHAPEIT v2.r79033 with the 1000Genomes phased data34 as a reference panel and the HapMap phase 2 genetic map.35
Comparative datasets of uniparental haplogroup data were compiled from published data. The NRY dataset comprises 4,831 individuals from 72 populations, and the mtDNA dataset comprises 4,281 individuals from 59 populations (Tables S4 and S5).
Statistical Analyses
The genetic diversity of our dataset pruned for LD was first analyzed by Principal Components Analysis (PCA) computed with EIGENSOFT v6.0.1.36 We used EEMS v137 to define genetic barriers and corridors on a geographic map. We used approximate geographic coordinates and a genetic dissimilarity matrix between population and a map of the East African coast defining a grid of 500 modeled demes. Depending on their location, several populations can be included in one deme, whose size increases accordingly. We ran 3 × 106 MCMC iterations before checking for convergence of the MCMC chain (Figure S2). Plots were generated in R v3.2.2 according to instructions in the EEMS v1 manual.37 Runs of homozygosity and inbreeding coefficient analyses were performed in PLINK v1.9. Uniparental genetic diversity of our datasets was analyzed by PCA with SPSS v20.038 and FST genetic distances were analyzed with ARLEQUIN v3.5.2.2.39
Three-population (_f_3) statistics40 were computed for each trio of populations, comprising two populations from the low-SNP density dataset and either a Swahili or a Comorian group, so that groups showing potential admixture events could be identified. Genetic ancestries of the pruned dataset were estimated by ADMIXTURE v1.341, with default settings, for components K = 2 to K = 30 on both autosomal and X chromosome data. Ten iterations with randomized seeds were run and compiled with CLUMPAK v1.42 We used the minimum average cross-validation value to define the most informative K components (here, K = 26 for autosomal data and K = 3 for the X chromosome data), and the major modes defined by CLUMPAK v142 are reported. We calculated sex-biased admixture by performing t tests between ancestries estimated for autosomes and the X chromosome at K = 3, which defines an Asian component, an African component, and a Middle Eastern component. Plots were obtained with Genesis v0.2.5.43 TreeMix v1.12 analysis44 was performed with all South African, East African, and Island Southeast Asian populations in the low-SNP-density dataset (npop = 88); setting blocks to 200 SNPs accounted for linkage disequilibrium, and migration edges were added sequentially until the model explained 99% of the variance.
Population structure of the phased high-density dataset was evaluated with the fineSTRUCTURE v2.07 package.45 This package performs a model-based Bayesian clustering of genotypes based on the shared IBD fragments between each pair of individuals, without self-copying, calculated with Chromopainter v2.0.45 From the results, a coancestry heatmap and a dendrogram were inferred to visualize the number of statistically defined clusters that describe the data (Figure S3). This procedure, along with PCA, is commonly used to identify individuals as potential genetic outliers. Genetic clustering, based on shared ancestry, reduces the noise that can result from having individuals with different ancestries in the same geographic population, such as in the case of recent migrants, in analyses regarding past demographic events.45, 46 Most of these correspond to anthropologically defined populations, such as those on each Comorian island. Each of these groups was analyzed individually in the IBD-based analyses. The one exception is the Swahili populations, which show no substructure between the three communities (Figure S3), leading us to include all Swahili individuals in a single cluster. However, for anthropological reasons, we also perform these analyses with their location as a clustering factor. Haplotype sharing between pairs of individuals was estimated from the phased high-SNP-density dataset by the Refined IBD algorithm in Beagle v4.0,47, 48 filtering for detected fragments with a logarithm of odds ratio greater than 3. Detected fragments between the same pairs of populations were summed, normalized by the number of individuals, and visualized with Cytoscape v3.2.1.49 All maps used in the present study were generated with Global Mapper v15. Local ancestry analysis in Comorian and Malagasy individuals was performed via PCAdmix v1.050 with three parental metapopulations of 100 individuals with African ancestry (randomly selected from Yoruba, South African Bantu from Soweto, Baka Pygmy from Cameroon, Kenyan Luhya, and Swahili groups), Middle Eastern ancestry (randomly selected from UAE Dubai Arab, Saudi, Yemeni, and Iranian groups), and Asian ancestry (randomly selected from Chinese Han, Indian Brahmin, Indonesian Banjar, Bajo, Ma’anyan, Singaporean Malay, and Filipino Kankanaey “Igorot” groups). The phased Comorian and Malagasy data were screened with linkage disequilibrium information so that the probability of common ancestry of each haplotype with each “parental” metapopulation could be defined. The Viterbi algorithm was then used for masking all haplotypes according to Asian and Middle Eastern ancestries in the Comorian and Malagasy individuals. _f_3 statistics,40 FST calculations with EIGENSOFT v6.0.1.,36 and TreeMix v1.12 analyses44 were then performed on this masked dataset (12,283 SNPs for 3477 individuals). Haplotype “painting” with Chromopainter v245 was performed on the high-density SNP dataset, and each cluster of populations was defined as a target or as either a donor or a surrogate according to the anthropological question addressed. Running an estimation-maximization algorithm in Chromopainter v2 on all 22 autosomes for the entire dataset with 10 iterations provided estimates of mutational rates and effective population size parameters.45 The weighted average of these parameters, according to the SNP coverage of each chromosome and the number of individuals, was then used for computing the chromosomal painting. Each cluster of Swahili, Comorian, and Malagasy populations was successively identified as a target, and the others were identified as surrogates. Because common ancestry between Comorian and Malagasy individuals was a possibility, we ran two chromosomal paintings for each group and allowed one to be a surrogate of the other (unidirectionally only). For all Comorian and Malagasy population, both outputs were analyzed in parallel. We used the painted chromosomes obtained for each cluster in GLOBETROTTER v1.051 to estimate the ratios and dates of the potential admixture events characterizing them. We estimated coancestry curves with and without standardization by using a “NULL” individual, and we checked consistency between each estimated parameter. We performed 100 bootstrap resamplings to estimate the probability value of the admixture events (if we considered the “NULL” individual) and the 95% confidence interval for the obtained dates (if we did not consider the “NULL” individual). The “best-guess” scenario given by GLOBETROTTER v1.051 was considered for each target population. By using the parental populations given by GLOBETROTTER v1.0,51 we also estimated dates of admixture with MALDER v1,52 a modified version of the ALDER v1.352 software designed to allow observation of multiple admixture events. The estimated dates of genetic admixture most likely reflects the midpoint or endpoint of noticeable admixture, rather than the start of this contact.53 Dates of admixture, given in generations, were converted to chronological time with a generation interval of 29 years.54, 55 A correlation test and t tests were performed with SPSS v20.0.38
Results
Population Structure
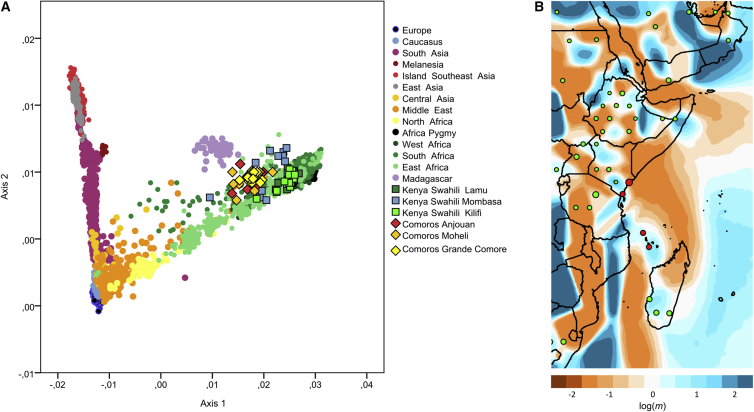
The genome-wide genetic diversity of the Comorian and Kenyan Swahili communities together with populations from a low-SNP-density comparative dataset (3,477 individuals genotyped for 171,728 SNPs) can be represented by a PCA (Figure 1A; Table S3). This analysis shows that the Kenyan Swahili individuals overlap with other mainland Sub-Saharan groups and that the Swahili communities cannot be distinguished from one another, except for a few outlying individuals. Comorian individuals fall between the Swahili and Malagasy, the latter group being pulled away from the African pole by their Asian ancestry. In analyses of African individuals alone, both the Swahili and Comorian individuals cluster together with Bantu-speakers and Malagasy (Figure S4). The same pattern is reflected on PCAs and FST genetic distances with both mtDNA and Y chromosome (NRY) data (Figure S5; Tables S2, S3, S6, and S7). Uniparental data show that the vast majority of lineages in these six groups belong to haplogroups frequent in Africa (mtDNA: L∗ haplogroup: 95.4%; NRY: E∗ and B∗ haplogroups: 99.1%), and there is only a limited presence of Asian lineages (mtDNA F3b1b in one individual from Grande Comore and NRY O2a in one Swahili individual from Kilifi) (Tables S1 and S2). Furthermore, Comorian and Kenyan Swahili groups show genome-wide diversity values (runs of homozygosity; Figure S6) similar to those observed in neighboring populations, but we note that Comorian groups have relatively less diversity than Malagasy groups, which could occur as a result of their smaller island environment or a lower level of admixture.
Figure 1.
Genetic Diversity and Differentiation of Swahili and Comoros Populations Relative to Comparative Populations in the Low-SNP-Density Dataset
(A) Principal-component analysis of the low-SNP-density dataset with EIGENSOFT.36 Swahili individuals are represented by squares, Comoros individuals are represented by diamonds, and all other individuals in the comparative dataset are represented by circles.
(B) An EEMS37 gradient map is centered on populations along the East African coast (convergence of 3 × 106 MCMC iterations). The color scale reveals low (blue) to high (orange) genetic barriers between populations localized on a grid of 500 demes. Each dot is proportional to the number of populations included. Red demes represent Swahili and Comoros populations; and green demes represent Middle Eastern, East African, and South African populations in the low SNP-density dataset.
These results converge with a fineSTRUCTURE45 analysis that characterizes the genetic structure of these individuals, together with samples from a high-SNP-density comparative dataset (1,664 individuals genotyped for 411,432 SNPs; Table S3; Figure S3). This analysis identifies 70 groups of individuals that can be statistically defined as genetically separated populations45 according to their shared genetic profiles (IBD) (Figure S3). Whereas Comorians cluster together genetically according to their respective islands, the three Kenyan Swahili communities are undifferentiated and form a single cluster. Nine Swahili individuals fall outside this main cluster and instead group with Somali individuals and perhaps reflecting recent migrants (Figure S3). These individuals are also outliers in the PCA plot and were excluded from subsequent analyses of the Swahili because these data would bias analyses on past demographic processes. However, we note that they represent a non-negligible number of individuals in our sampling (9 out of 91), suggesting a relatively important number of recently integrated individuals of Somali origin, many of whom are present in Kenya,56 into Swahili groups. Kenyan Swahili individuals have shared IBD fragments mainly with other Bantu speakers, notably the eastern and southern Bantu. This indicates that the three Swahili communities form a homogeneous genetic cluster that is differentiated from other sub-Saharan individuals and, notably, from other East Africans, thus emphasizing the uniqueness of the Swahili genome. The Kenyan Swahili show limited IBD sharing with individuals from Ethiopia and Somalia and non-Bantu-speakers in Kenya. Beyond continental Bantu speakers, Kenyan Swahili individuals share a substantial proportion of IBD fragments with Comorian and Malagasy individuals and thus cluster together with those groups (Figure S3). Comorian individuals also cluster into homogeneous genetic groups according to their respective island of origin: Anjouan, Moheli, or Grande Comore.
The shared genetic ancestry between Swahili, Comorian, and Malagasy groups was visualized by an EEMS37 plot, which shows genetic differentiation between East African populations in the low-SNP-density comparative dataset (Figure 1B). Strong genetic barriers were identified between the Swahili communities and other continental African populations, particularly Ethiopian groups. The plot also reveals a striking genetic continuum from the Kenyan Swahili groups to the Comorians and the Malagasy. This pattern is reflected in FST genetic distances as well, and Comorian groups are as genetically close to the Swahili as they are to the Malagasy (Table S8). On the continent, Swahili groups show lower genetic differentiation (i.e., low FST values) with Bantu-speaking groups in Kenya and South Africa.
These observations agree with a RefinedIBD47, 48 analysis (Figure S7) that shows high IBD sharing between Swahili, Comorians, and Malagasy. It is particularly striking that Swahili individuals share more genetic material with Comorians than with other continental African groups such as the Luhya, another Bantu-speaking group from Kenya. The genetic continuity is also shown by the high level of mtDNA haplotype sharing between Comorian and Swahili groups (Figure S8; Table S2). All of these analyses emphasize the sea-oriented focus of Swahili populations, formalizing the Swahili Corridor as a genetic continuum.
Admixture Scenario
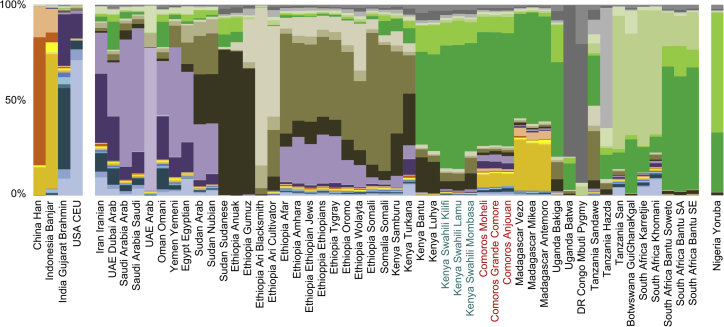
Having characterized the genetic continuum of the Swahili Corridor, we then analyzed the genetic ancestries that define it. All Swahili and Comorian communities result from admixture processes (_f_3-statistics Z score < −2; Table S9). The lowest significant _f_3 statistical results for Swahili communities are obtained for an admixture scenario between a South African Bantu group and a Eurasian group, and the most significant scenario for the Comorians, as well as for the Malagasy, involves a South African Bantu group and a Southeast Asian group. The genetic ancestries present in the low-SNP-density dataset were then decomposed with ADMIXTURE41 (Figure 2; Figure S9). The lowest cross-validation values are obtained when 26 ancestries are defined (Figure S10). The Swahili populations have one major component (dark green in Figure 2) that is also present in other Bantu-speaking groups, but in higher percentages, similar those in South Africa. Other minor ancestries are shared with populations from the Horn of Africa (brown gradient) and western Bantu speakers (light green). No Asian or Middle Eastern genetic ancestry is distinguishable in the Kenyan Swahili, highlighting the indigenous African origins of the Swahili people. This absence is the main difference between the Kenyan Swahili and Comorians, who do share genetic ancestry with Middle Eastern (purple gradient: 6%–7%) and Island Southeast Asian individuals (yellow gradient: 8%–9%). Comorians show larger Middle Eastern and smaller Southeast Asian ancestral components than do the Malagasy. The main Swahili ancestry (dark green) is shared by both Comorian and Malagasy populations, confirming the previously outlined Swahili genetic continuum, albeit with increased complexity as a result of specific ancestries from the Middle East and Southeast Asia.
Figure 2.
ADMIXTURE Plot of the Low-SNP-Density Comparative Dataset for the Major Mode of K = 26, as Defined by CLUMPAK
For clarity, only Near Eastern, Middle Eastern, East African, and South African populations as well as five representative populations from neighboring regions are represented (see Figure S9 for the full plot). Each colored line represents a sampled population whose genetic background can be decomposed into 26 genetic components.
This pattern is confirmed by a PCAdmix50 analysis in which the African ancestry of the Comorians and Malagasy was extracted and analyzed separately (Figure S11). In a TREEMIX44 analysis on this African ancestry, alongside _f_3 outgroup statistics and FST distances, the Comorians and Malagasy are shown to be genetically linked to the African continent by the major ancestry components present in Bantu speakers, notably the Kenyan Swahili (Figure S11; Table S10). This African ancestry component differentiates Bantu-speaking groups from Cushitic-speaking groups, emphasizing the Bantu genetic origin of the Swahili Corridor (Figure S12).
As observed in the ADMIXTURE41 plot, the Bantu genetic continuum of the Swahili Corridor is not defined by an IBD model but rather by differential gene flows, notably from the Horn of Africa, the Middle East, and Southeast Asia (Figure 2). A TREEMIX44 analysis shows that the Kenyan Swahili, Malagasy, and Comorian groups are all closely related (Figure S13). As expected, significant gene flow from island Southeast Asian populations into the Malagasy (33%–39%) was detected.
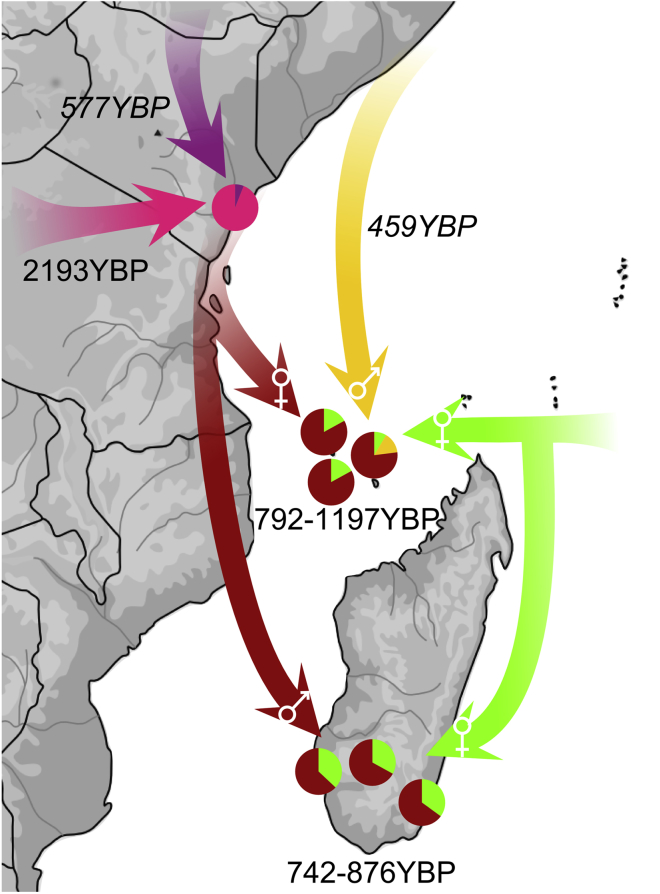
Admixture scenarios were then estimated with GLOBETROTTER51 and MALDER,52 giving highly correlated dates of admixture (r2 = 0.78; p = 0.002). All of the estimated dates reflect the most recent detectable admixture events.53 We computed the admixture scenario for the Kenyan Swahili by treating all three communities analyzed as single group. The best-fit scenario was obtained for two admixture events between, first, a South African Bantu population (South Africa from Soweto: 38%) and a Central African Bantu population (Namibia Kwangali: 62%) around 2193 YBP (95% CI: 1070–3312 YBP). This was followed by a second wave of gene flows from a South African Bantu population (South Africa from Soweto: 88%) and a population from the Horn of Africa (Ethiopian Oromo: 12%) around 577 YBP (95% CI: 200–682 YBP) (Figure 3; Table S11). These two waves of admixture are also inferred by MALDER52 (1535 ± 235 YBP and 247 ± 61 YBP; Table S12). When each Swahili community is analyzed separately, only the most recent event—that is, between a South African Bantu group (87%–89%) and a population from the Horn of Africa or Middle East around 691–732 YBP (Table S11)—is inferred, although two waves of admixture are still inferred with MALDER52 for the Swahili from Kilifi (1262 ± 197 YBP and 220 ± 67 YBP; Table S12).
Figure 3.
Admixture Scenario for Populations along the Swahili Corridor as Estimated by GLOBETROTTER and ADMIXTURE
Dark red arrows represent Swahili gene flow; light green arrows represent Island Southeast Asian Banjar or Malay; a yellow arrow represents Middle Eastern gene flow; and the purple arrow represents gene flow from the Horn of Africa. The pink arrow represents gene flow from central and southern Bantu speakers. Dates refer to the last detectable admixture event; dates below pie charts refer to the admixture event between the Swahili and Banjar or Malay; dates in italics represent secondary gene flow. Sex-biased gene flows are represented by male and female symbols in the tip of the arrows; note that they are not present in Malagasy Antemoro and Comorian Moheli.
Admixture scenarios were also estimated for the Comorian groups. When the Malagasy are excluded from being surrogates (so there is no potential bias because of their close ancestry), all Comorian communities have a best-fit scenario between a Swahili group (Swahili from Mombasa: 80%–87%) and an Island Southeast Asian group (Indonesian Banjar or Singaporean Malay: 17%–20%) around 792–1197 YBP (Table S11). These scenarios are confirmed by MALDER52 (Table S12). However, Comorian communities from Anjouan and Moheli both show significant secondary gene flows (Figure 3; Table S11). Comorians from Anjouan result from a second admixture event between a Swahili group (Swahili from Mombasa: 74%) and a Middle Eastern group (UAE Dubai Arab: 26%) around 459 YBP (95% CI: 35–673 YBP). Comorians from Moheli have a second genetic input from a Swahili group (Swahili from Mombasa: 85%) and an Island Southeast Asian group (Indonesian Banjar or Singaporean Malay: 15%) around 145 YBP (95% CI: 41–258 YBP). We note that if Malagasy groups are allowed as surrogates, then this second admixture event in Comorians from Moheli appears to come from a Swahili group (Swahili from Mombasa: 74%) and a Malagasy group (Malagasy Antemoro: 26%) around 90 YBP (95% CI: 34–319 YBP) (Table S11), suggesting that this secondary gene flow could come from Madagascar.
All Malagasy groups result from a one-date admixture scenario between a Swahili group (Swahili from Mombasa: 63%–67%) and an Island Southeast Asian group (Indonesian Banjar: 33%–37%) around 742–876 YBP (Tables S11 and S12). If Comorians are allowed as surrogates, the scenario obtained is statistically unreliable, most likely as a result of Malagasy gene flow into Comorians from Moheli (Tables S11).
Finally, we tested for sex-biased admixture events by performing t tests based on the three genetic ancestries estimated by the ADMIXTURE41 analyses (Figure 3; Figure S14; Table S13). Although, consistent with uniparental data (Tables S1 and S2), no sex bias was observed in Kenyan Swahili populations, Comorian communities all show significant female-biased gene flow from Africa (p_c_ < 0.05) and Asia (p < 0.05). This contrasts with strong male-biased gene flow from the Middle East, significant in Anjouan and Grande Comore islands (p_c_ < 0.01), as previously reported from uniparental markers.18 This pattern was not observed in the Malagasy, but male-biased African ancestry and female-biased Asian ancestry was observed in Malagasy Mikea (p_c_ < 0.01; Table S13) and nominally observed in Malagasy Vezo, as previously observed across Madagascar.19 Although the main ancestral groups (Swahili and Banjar) are common to both Comorian and Malagasy populations, the timing and sex-bias are different, suggesting different population dynamics.
Discussion
The Swahili Corridor Is a Bantu Genetic Continuum
In concert with the expansion and intensification of the Indian Ocean trading network, the Swahili established multiple influential urban centers along the East African coast and offshore islands, from Somalia to Mozambique and the Comoros, along what is known as the Swahili Corridor.2 We show that this exceptional cultural and historical entity represents a relatively homogeneous genetic group. Despite the cosmopolitan nature of these cities, which at various times might have incorporated merchants and migrants from the Middle East, South Asia, East Asia, and Island Southeast Asia, African genetic ancestry is the main characteristic of Kenyan Swahili groups, Comorians, and the Malagasy (Table S11; Figures S3, S7, and S11). These data suggest constant interaction between the Swahili communities and these African islands and thus define the Swahili Corridor as a genetic continuum (Figure 1; Figure S7). Although our sampling does not include Swahili communities from Tanzania or Mozambique, which could bring some complexity to the presently drawn genetic landscape, the high proportion of specific African genetic ancestries shared by populations from both ends of the Swahili Corridor supports the idea of a genetic continuum. This African genetic diversity has a major input from Bantu-speaking populations (Figure S12), especially those currently living in Central Africa, and most likely corresponds to the results of previous archaeological and genetic studies that identify a Bantu migration from the Congo River toward the eastern African coast at the end of the first millennium BCE, as identified in previous archaeological and genetic studies (Table S11 and S12).57, 58 The ancestors of the Swahili established prosperous fisher-farmer-hunter communities and increasingly engaged with the growing trading network of the Indian Ocean. Archaeological studies indicate that groups from the Lamu archipelago were key players in the early stages of the Indian Ocean trading network.2, 59, 60 Contacts with the other populations brought new goods and social practices to East Africa.7 However, in Kenya, this interaction did not drive intense gene flows, contrary to what is observed in many other groups around the Indian Ocean rim.5 For example, our sampling from a Kenyan Swahili community of Lamu, whose tradition evokes marriages with Chinese sailors from Zheng He’s expedition to Africa (1405–1433 CE), shows no significant Asian gene flow (Figure 2; Table S11).61 The most noticeable genetic input in Kenya comes from the Middle East.
Middle Eastern gene flow is rather limited in the Swahili Corridor (Figure 2). It is detected in some Swahili and Comorian groups after admixture events dated from the mid-13th century in Mombasa and from the 15th century on Anjouan Island (Tables S11 and S12). Middle Eastern ancestry dating to around the 15th century has also been found previously in a Muslim group from Madagascar, the Antemoro.3, 62 This contact was detected at a low level and only on the Y chromosome,17 perhaps explaining its limited appearance in this study. Our analyses also show this sex-biased admixture, which is particularly significant in Anjouan (p_c_ < 0.05; Table S13). This male-driven gene flow from Middle Eastern groups, albeit limited, could coincide with their cultural influence in the Swahili Corridor, initiated since the end of the first millennium.8 Particularly, the period around the 13th–14th centuries CE is characterized by migrations of peasants, merchants, and religious teachers, known as sharifs, mostly from Hadramawt (south Arabia) to East Africa and the Comoros.1, 63 They established culturally, politically, and economically influential families that integrated into the Swahili communities. Interestingly some of these family lineages were even present in both Comoros and Lamu archipelagos.1, 63 Overall, the Swahili genetic characteristics correspond well to the known history of their culture, and archaeological and linguistic evidence supports its emergence as an indigenous African phenomenon that experienced long-term contacts and exchanges with Middle Eastern groups.1, 64
The Comoros Are the Earliest Meeting Point between African and Asian Genomes
The mechanism of Austronesian settlement in East Africa has long been unclear, and questions concerning the exact origin of Austronesian-speaking groups, route(s) taken, relative dates of migration, and admixture with local populations remain. We find here that the Comoros archipelago shows the earliest genetic contact between Austronesian and East African peoples and that this contact results from the dispersal of a group genetically close to the present Banjar population from southeast Borneo around the end of the first millennium.10 Both Comorian and Malagasy groups derive from an admixture event between a Swahili community and a Malay or Banjar group (Figure 3; Tables S11 and S12). Remarkably, this Austronesian gene flow is not detected on the African continent or elsewhere in the Indian Ocean rim west of Southeast Asia (e.g., in southern Eurasia), confirming previous results that suggest this dispersal followed a relatively direct route toward the Comoros archipelago and Madagascar.5, 10 However, the dynamics of admixture strongly differ in these two insular territories. Whereas the proportion of the Banjar or Malay genetic ancestry reaches 37% in the studied Malagasy populations and can reach up to 64% in the Highlands,19, 65, 66 its highest frequency in Comorians is 20% (Figures 2 and 3; Tables S11 and S12). This difference can be explained either by limited Asian gene flow into Comorian groups as compared to Malagasy groups or by a higher Swahili genetic input into Comorian populations (Figure S13). The latter hypothesis seems more likely, because the Comoros archipelago was host to major Swahili settlements and Comorian groups currently speak a Bantu language, as opposed to the Austronesian language spoken in Madagascar.
Another major difference lies in the timing of these admixture events. These dates do not track migration events per se but do illuminate strong anthropological interactions between individuals of Swahili and Austronesian ancestries. Confirming previous results,10, 16, 19 the earliest detected admixture event in Madagascar occurred during the late 11th century in groups located on the easternmost coast of the island. This postdates the earliest date of admixture in the Comoros, which is estimated to be in the 8th century for the communities of Anjouan, the eastern island of the archipelago in our dataset (Tables S11 and S12). These dates coincide broadly with the time frame of Austronesian settlement in each territory as inferred from archaeological data. For example, analyses of ancient crop remains suggest older dates for Asian crops in the Comoros (8th–11th centuries) than in Madagascar (11th–13th centuries).15 More archaeological and genetic data from the north of Madagascar, where Austronesians are thought to have first settled on the island, are currently lacking to ground this conclusion, but it is remarkable that genetic and archaeological analyses broadly converge chronologically. We cannot precisely determine whether Malagasy and Comorian populations are descendants of two separate admixture events involving similar ancestral populations, or whether they are instead daughter groups from an already admixed common ancestor. Some linguistic analyses of the Malagasy language suggest contact with African culture prior to its dispersal to Madagascar and thus support the latter option.67, 68, 69 However, our analyses, which were based on testing of different admixture scenarios, do not show that populations currently present in the Comoros and Madagascar arose from a single admixed source population. On the other hand, we did find evidence that significant Malagasy gene flow occurred specifically into the Comorian community on Moheli Island as late as the 19th century (Table S11). This almost certainly reflects recent interactions between the two areas, rather than direct early ancestry, though, given that Moheli island, which belonged to the Ndzuwani Sultanate from Anjouan for centuries, was partially depopulated by the slave trade and only became independent in 1830 after the arrival of Malagasy migrants.3 Because of the genetic similarity between populations from Madagascar and Comoros, other gene flows between the two cannot be ruled out, but our result suggests that, if there were any gene flows, they were limited. Overall, our analyses point to a shared genetic history between modern Comorians and Malagasy and suggest that this shared history is linked to the exceptional Austronesian migration to East Africa, but they also emphasize independent population dynamics that led to genetic diversification on each set of islands. Although more genetic data from northern Madagascar and mainland East Africa would help to test these hypotheses, the broad convergence of our results with recent findings from archaeology15 point to the Comoros archipelago as the primary gateway for Austronesian gene flow into the Swahili Corridor at the dawn of the second millennium.
Acknowledgments
We acknowledge support from the GenoToul bioinformatics facility of Genopole Toulouse Midi-Pyrénées, France. This research was supported by French Ministry of Research grant ANR-14-CE31-0013-01 (OCEOADAPTO) to F.-X.R, a Rutherford Fellowship from the Royal Society of New Zealand (RDF-10-MAU-001), a fellowship from the Alexander von Humboldt Foundation to M.P.C., and grants from COMPETE 2020 and a Fundação para a Ciência e a Tecnologia-funded project (POCI-01-0145-FEDER-016609) to V.F. We thank Kamil Merito for his help collecting East African samples. We thank all the local Swahili and Comorian communities who participated in this study.
Published: January 4, 2018
Footnotes
Contributor Information
Nicolas Brucato, Email: nicolas.brucato@univ-tlse3.fr.
François-Xavier Ricaut, Email: francois-xavier.ricaut@univ-tlse3.fr.
Accession Numbers
The accession numbers for the 140 genotyping array data reported in this paper are EGA: S00001002565 and S00001002569. The accession numbers for the 276 complete mtDNA sequences reported in this paper are GenBank: MF695863–MF696138.
Web Resources
- European Genome-phenome Archive, https://www.ebi.ac.uk/ega/home
- Genesis. http://www.bioinf.wits.ac.za/software/genesis
- Global Mapper v15, http://www.bluemarblegeo.com/products/global-mapper.php
- Haplogrep, http://haplogrep.uibk.ac.at
- ISOGG 2014 Y-DNA Haplogroup Tree, http://www.isogg.org/tree
- SPSS, http://www.ibm.com/analytics/us/en/technology/spss/
Supplemental Data
Document S1. Figures S1–S14 and Tables S1–S13
Document S2. Article plus Supplemental Data
References
- 1.Horton M., Middleton J. Blackwell; 2000. The Swahili: The social landscape of a mercantile society. [Google Scholar]
- 2.Horton M. The Swahili Corridor. Sci. Am. 1987;255:86–93. [Google Scholar]
- 3.Beaujard, P. (2012). L’océan Indien, au cœur des globalisations de l’Ancien Monde (7e-15e siècles). In Volume 2, Les Mondes de l’Ocean Indien, Armand Collin, ed.
- 4.Beaujard P. The Indian Ocean in Eurasian and African World-Systems before the Sixteenth Century. J. World Hist. 2005;16:441–465. [Google Scholar]
- 5.Brucato N., Kusuma P., Beaujard P., Sudoyo H., Cox M.P., Ricaut F.-X. Genomic admixture tracks pulses of economic activity over 2,000 years in the Indian Ocean trading network. Sci. Rep. 2017;7:2919. doi: 10.1038/s41598-017-03204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boivin N., Crowther A., Helm R., Fuller D.Q. East Africa and Madagascar in the Indian Ocean world. J. World Prehist. 2013;26:213–281. [Google Scholar]
- 7.Fleisher J., Lane P., LaViolette A., Horton M., Pollard E., Quintana Morales E., Vernet T., Christie A., Wynne-Jones S. When Did the Swahili Become Maritime? Am. Anthropol. 2015;117:100–115. doi: 10.1111/aman.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaViolette A. Swahili Cosmopolitanism in Africa and the Indian Ocean World, A.D. 600–1500. Archaeologies. 2008;4:24–49. [Google Scholar]
- 9.Kusuma P., Brucato N., Cox M.P., Letellier T., Manan A., Nuraini C., Grangé P., Sudoyo H., Ricaut F.X. The last sea nomads of the Indonesian archipelago: genomic origins and dispersal. Eur. J. Hum. Genet. 2017;25:1004–1010. doi: 10.1038/ejhg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brucato N., Kusuma P., Cox M.P., Pierron D., Purnomo G.A., Adelaar A., Kivisild T., Letellier T., Sudoyo H., Ricaut F.X. Malagasy Genetic Ancestry Comes from an Historical Malay Trading Post in Southeast Borneo. Mol. Biol. Evol. 2016;33:2396–2400. doi: 10.1093/molbev/msw117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adelaar K.A. Malay influence on Malagasy: linguistic and culture-historical implications. Oceanic Linguistics. 1989;28:1–46. [Google Scholar]
- 12.Adelaar A. Who were the first Malagasy, and what did they speak? In: Acri A., Blench R., Landmann A., editors. Spirit and Ships: Cultural Transfer in Early Monsoon Asia. Institute of Southeast Asian Studies; 2017. pp. 441–469. [Google Scholar]
- 13.Blench R. The Austronesians in Madagascar and their interaction with the Bantu of East African coast: Surveying the linguistic evidence for domestic and translocated animals. Philippines Journal of Linguistics. 2009;18:18–43. [Google Scholar]
- 14.Fitzpatrick S.M., Callaghan R. Islands of inquiry: Colonisation, seafaring and the archaeology of maritime landscapes. ANU Press; 2008. Seafaring simulations and the origin of prehistoric settlers to Madagascar; pp. 47–58. [Google Scholar]
- 15.Crowther A., Lucas L., Helm R., Horton M., Shipton C., Wright H.T., Walshaw S., Pawlowicz M., Radimilahy C., Douka K. Ancient crops provide first archaeological signature of the westward Austronesian expansion. Proc. Natl. Acad. Sci. USA. 2016;113:6635–6640. doi: 10.1073/pnas.1522714113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierron D., Razafindrazaka H., Pagani L., Ricaut F.-X., Antao T., Capredon M., Sambo C., Radimilahy C., Rakotoarisoa J.-A., Blench R.M. Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc. Natl. Acad. Sci. USA. 2014;111:936–941. doi: 10.1073/pnas.1321860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capredon M., Brucato N., Tonasso L., Choesmel-Cadamuro V., Ricaut F.-X., Razafindrazaka H., Rakotondrabe A.B., Ratolojanahary M.A., Randriamarolaza L.-P., Champion B., Dugoujon J.M. Tracing Arab-Islamic inheritance in Madagascar: study of the Y-chromosome and mitochondrial DNA in the Antemoro. PLoS ONE. 2013;8:e80932. doi: 10.1371/journal.pone.0080932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Msaidie S., Ducourneau A., Boetsch G., Longepied G., Papa K., Allibert C., Yahaya A.A., Chiaroni J., Mitchell M.J. Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean. Eur. J. Hum. Genet. 2011;19:89–94. doi: 10.1038/ejhg.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierron D., Heiske M., Razafindrazaka H., Rakoto I., Rabetokotany N., Ravololomanga B., Rakotozafy L.M., Rakotomalala M.M., Razafiarivony M., Rasoarifetra B. Genomic landscape of human diversity across Madagascar. Proc. Natl. Acad. Sci. USA. 2017;114:E6498–E6506. doi: 10.1073/pnas.1704906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurles M.E., Sykes B.C., Jobling M.A., Forster P. The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am. J. Hum. Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tofanelli S., Bertoncini S., Castrì L., Luiselli D., Calafell F., Donati G., Paoli G. On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages. Mol. Biol. Evol. 2009;26:2109–2124. doi: 10.1093/molbev/msp120. [DOI] [PubMed] [Google Scholar]
- 22.Nurse D., Hinnebusch T.J. University of California Press; 1993. Swahili and Sabaki. [Google Scholar]
- 23.Kusuma P., Cox M.P., Pierron D., Razafindrazaka H., Brucato N., Tonasso L., Suryadi H.L., Letellier T., Sudoyo H., Ricaut F.-X. Mitochondrial DNA and the Y chromosome suggest the settlement of Madagascar by Indonesian sea nomad populations. BMC Genomics. 2015;16:191. doi: 10.1186/s12864-015-1394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International Society of Genetic. (2014). ISOGG 2014 Y-DNA Haplogroup Tree, Version: 9.70.
- 25.van Oven M., Van Geystelen A., Kayser M., Decorte R., Larmuseau M.H.D. Seeing the wood for the trees: a minimal reference phylogeny for the human Y chromosome. Hum. Mutat. 2014;35:187–191. doi: 10.1002/humu.22468. [DOI] [PubMed] [Google Scholar]
- 26.Kircher M., Sawyer S., Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maricic T., Whitten M., Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arias L., Barbieri C., Barreto G., Stoneking M., Pakendorf B. High resolution mitochondrial DNA analysis sheds light on human diversity, cultural interactions, and population mobility in Northwest Amazonia. bioRxiv. 2017 doi: 10.1002/ajpa.23345. 101101/160218. [DOI] [PubMed] [Google Scholar]
- 29.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 32.Chang C.C., Chow C.C., Tellier L.C.A.M., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 34.Delaneau O., Marchini J., 1000 Genomes Project Consortium. 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petkova D., Novembre J., Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 2016;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.IBM. (2011). SPSS Statistics.
- 39.Excoffier L., Lischer H.E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 40.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopelman N.M., Mayzel J., Jakobsson M., Rosenberg N.A., Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchmann, R., and Hazelhurst, S. (2014). Genesis.
- 44.Pickrell J.K., Pritchard J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawson D.J., Hellenthal G., Myers S., Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montinaro F., Busby G.B., Pascali V.L., Myers S., Hellenthal G., Capelli C. Unravelling the hidden ancestry of American admixed populations. Nat. Commun. 2015;6:6596. doi: 10.1038/ncomms7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browning B.L., Browning S.R. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brisbin A., Bryc K., Byrnes J., Zakharia F., Omberg L., Degenhardt J., Reynolds A., Ostrer H., Mezey J.G., Bustamante C.D. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum. Biol. 2012;84:343–364. doi: 10.3378/027.084.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hellenthal G., Busby G.B.J., Band G., Wilson J.F., Capelli C., Falush D., Myers S. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loh P.R., Lipson M., Patterson N., Moorjani P., Pickrell J.K., Reich D., Berger B. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hodgson J.A., Mulligan C.J., Al-Meeri A., Raaum R.L. Early back-to-Africa migration into the Horn of Africa. PLoS Genet. 2014;10:e1004393. doi: 10.1371/journal.pgen.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fenner J.N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 55.Langergraber K.E., Prüfer K., Rowney C., Boesch C., Crockford C., Fawcett K., Inoue E., Inoue-Muruyama M., Mitani J.C., Muller M.N. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl. Acad. Sci. USA. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliver R.A. Clarendon Press; 1963. History of East Africa. [Google Scholar]
- 57.Patin E., Lopez M., Grollemund R., Verdu P., Harmant C., Quach H., Laval G., Perry G.H., Barreiro L.B., Froment A. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science. 2017;356:543–546. doi: 10.1126/science.aal1988. [DOI] [PubMed] [Google Scholar]
- 58.Bostoen K., Clist B., Doumenge C., Grollemund R., Hombert J.-M., Muluwa J.K., Maley J., Blench R., Di Carlo P., Good J. Middle to late holocene paleoclimatic change and the early bantu expansion in the rain forests of Western Central Africa. Curr. Anthropol. 2015;56:367–368. [Google Scholar]
- 59.Horton M. British Institute in Eastern Africa; 1996. Shanga: The archaeology of a Muslim trading community on the coast of East Africa. [Google Scholar]
- 60.Chittick N. British Institute in Eastern Africa; 1984. Manda: Excavations at an island port on the Kenya coast. [Google Scholar]
- 61.Viviano F. In National Geographic; 2005. China’s Great Armada, Admiral Zheng He; pp. 28–53. [Google Scholar]
- 62.Ferrand G. E. Leroux; 1891. Les musulmans à Madagascar et aux iles Comores. [Google Scholar]
- 63.Martin B.G. Arab Migrations to East Africa in Medieval Times. Int. J. Afr. Hist. Stud. 1974;7:367–390. [Google Scholar]
- 64.Nurse D., Spear T. University of Pennsylvania Press; 1985. The Swahili: Reconstructing the history and language of an African society, 800–1500. [Google Scholar]
- 65.Hodgson J.A., Pickrell J.K., Pearson L.N., Quillen E.E., Prista A., Rocha J., Soodyall H., Shriver M.D., Perry G.H. Natural selection for the Duffy-null allele in the recently admixed people of Madagascar. Proc. Biol. Sci. 2014;281:20140930. doi: 10.1098/rspb.2014.0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hodgson J.A. A Genomic Investigation of the Malagasy Confirms the Highland-Coastal Divide, and the Lack of Middle Eastern Gene Flow. In: Campbell G., editor. Early Exchange between Africa and the Wider Indian Ocean World. S.I. Publishing; 2016. pp. 231–254. [Google Scholar]
- 67.Adelaar A. Malagasy Phonological History and Bantu Influence. Oceanic Linguistics. 2012;51:123–159. [Google Scholar]
- 68.Adelaar A. Towards an Integrated Theory about the lndonesian Migrations to Madagascar. In: Peregrine P.N., Peiros I., F.M., editors. Ancient human migrations: a multidisciplinary approach. University of Utah Press; 2009. pp. 149–172. [Google Scholar]
- 69.Deschamps H. Histoire de Madagascar. Revue française d'histoire d'outre-mer. 1960;170:136–138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S14 and Tables S1–S13
Document S2. Article plus Supplemental Data