The hippocampal network model: A transdiagnostic metaconnectomic approach (original) (raw)
Abstract
Purpose
The hippocampus plays a central role in cognitive and affective processes and is commonly implicated in neurodegenerative diseases. Our study aimed to identify and describe a hippocampal network model (HNM) using trans-diagnostic MRI data from the BrainMap® database. We used meta-analysis to test the network degeneration hypothesis (NDH) (Seeley et al., 2009) by identifying structural and functional covariance in this hippocampal network.
Methods
To generate our network model, we used BrainMap's VBM database to perform a region-to-whole-brain (RtWB) meta-analysis of 269 VBM experiments from 165 published studies across a range of 38 psychiatric and neurological diseases reporting hippocampal gray matter density alterations. This step identified 11 significant gray matter foci, or nodes. We subsequently used meta-analytic connectivity modeling (MACM) to define edges of structural covariance between nodes from VBM data as well as functional covariance using the functional task-activation database, also from BrainMap. Finally, we applied a correlation analysis using Pearson's r to assess the similarities and differences between the structural and functional covariance models.
Key findings
Our hippocampal RtWB meta-analysis reported consistent and significant structural covariance in 11 key regions. The subsequent structural and functional MACMs showed a strong correlation between HNM nodes with a significant structural-functional covariance correlation of r = .377 (p = .000049).
Significance
This novel method of studying network covariance using VBM and functional meta-analytic techniques allows for the identification of generalizable patterns of functional and structural abnormalities pertaining to the hippocampus. In accordance with the NDH, this framework could have major implications in studying and predicting spatial disease patterns using network-based assays.
Keywords: Anatomic likelihood estimation, ALE, BrainMap, Functional covariance, Functional MRI, Gray matter density, Hippocampal network model, Hippocampus, Magnetic resonance imaging, MRI, Meta-analysis, Meta-analytic connectivity modeling, MACM, Structural covariance, Structural MRI, Voxel-based morphometry, VBM
Highlights
- •
We derived regions that structurally co-vary with the hippocampus in a network model using a transdiagnostic meta-analytic approach. - •
We used meta-analytic connectivity mapping to assess inter-regional connectivity from BrainMap's structural and functional databases. - •
We tested the network degeneration hypothesis by identifying network correlations between structural and functional networks.
1. Introduction
The hippocampus is arguably the most well studied sub-cortical region in both humans and animals due in large part to its functional role in the cognition of learning and memory. When compromised, symptoms of hippocampal dysfunction are distinguished by impairments in memory, attention, emotion, spatial navigation, and executive function. Pathologically, hippocampal disease is characterized by neuronal degeneration progressing to structural atrophy detectable by magnetic resonance imaging (MRI), as in the cases of Alzheimer's disease (AD) (Schröder and Pantel, 2016), mild cognitive impairment (MCI) (Huijbers et al., 2015), mesial temporal lobe epilepsy (MTLE) (Mumoli et al., 2013), and schizophrenia (Radulescu et al., 2014). In terms of network topology, the hippocampus has been characterized as a hub, exhibiting structural covariance with multiple other brain regions, in keeping with its central role in multiple cognitive processes. It has been suggested that metabolic demands of hubs make such structures selectively vulnerable to neuropsychiatric diseases (Goodkind et al., 2015; Crossley et al., 2014), arguing for converging structural and functional network abnormalities. Advances in resting state functional MRI (fMRI) and sophisticated meta-analytic methodologies have allowed researchers to exhaustively map and quantify functional covariance networks (Fox and Raichle, 2007; Smith et al., 2009; Crossley et al., 2013). Nevertheless, our understanding of the relationships between functional and structural networks in the human brain, as can be studied using MRI, remains in its infancy. In 2009, Seeley et al., postulated the “network degeneration hypothesis” (NDH), which predicts that disease atrophy patterns should recapitulate healthy functional network architecture with recent studies beginning to address this question in the context of disease-specific network changes (Lefort-Besnard et al., 2018). Our aim in this study was to test the NDH meta-analytically and trans-diagnostically using the hippocampus as the central hub, thereby creating a hippocampal network model (HNM) that should be applicable in multiple neuropsychiatric and systemic disorders.
Structural and functional connectivity are two procedurally distinct, but conceptually linked constructs, both exhibiting network properties. Functional connectivity networks can be extracted by functional covariances, either at rest or during task performance, and exhibit interconnected sets of brain regions that interact to perform specific perceptual, motor, cognitive, and affective functions (Bressler and Menon, 2010). Structural covariance networks are inferred from inter-regional morphometric covariances across subjects (e.g. gray matter atrophy, cortical thinning). The notion that structural covariance is fundamental to functional network connectivity is based on the expectation that functional processes can exert trophic influences that, over time, modify gray matter volume enough to be detected as structural covariance patterns (Seeley et al., 2009; Gong et al., 2012). Multiple competing and converging hypotheses have been put forward to explain disease-related structural covariance, these include: 1) transneuronal spread, 2) nodal stress, 3) trophic failure, and 4) shared vulnerability (Zhou et al., 2012). For this reason, structural covariance has been extensively studied in the characterization of neurocognitive development (Alexander-Bloch et al., 2013; Lerch et al., 2006), aging (Marstaller et al., 2015; Montembeault et al., 2016), and in neurodegenerative disorders (Seeley et al., 2009; Spreng and Turner, 2013). The NDH suggests a close relationship between structure and function whereby abnormalities in structural covariance, functional covariance and behavior will be mutually predictive in brain disorders.
Meta-analytic computation of functional covariance is well-established (Smith et al., 2013; Crossley et al., 2013). In assessing functional connectivity networks, activation likelihood estimation (ALE) is an extensively validated technique used in meta-analyses that pools 3D coordinates in stereotactic space from a number of like studies applied to functional task-activation studies (Laird et al., 2005; Turkeltaub et al., 2011). It achieves this by analyzing voxel-wise, univariate effects across experiments and generates a probability distribution centered at the respective coordinates. A natural extension of ALE, meta-analytic connectivity modeling (MACM) is a study-wise multivariate approach used to generate functional covariance networks from activation patterns reported across a range of experimental neuroimaging tasks and paradigms (Laird et al., 2009a; Robinson et al., 2010; Eickhoff et al., 2010a). In MACM, an ALE score is generated for every voxel, which are then converted into p values to identify areas of significance with scores higher than empirically-derived null distributions (Turkeltaub et al., 2011; Laird et al., 2005; Eickhoff et al., 2012). This technique has been found to correspond well with numerous mathematical computational formalisms including seed-based resting state (Jiang et al., 2015), independent components analyses (Smith et al., 2009), and graph theory (Crossley et al., 2013). Although MACM and ALE have been validated and replicated considerably in functional studies and, recently, using structural data (Langner et al., 2014; Reid et al., 2016), the present study aims to adopt these methods for investigating structural covariance using a meta-analytical and trans-diagnostic approach with VBM.
Voxel-based morphometry (VBM) is a widely-used technique to identify subtle, disease-related structural changes that cannot be easily observed on visual analysis. VBM achieves this by standard space brain registration and group-averaging, comparing gray matter densities between patients and controls, producing disease-specific atrophy patterns computed in a univariate, voxel-wise manner. VBM studies have reported regional gray matter atrophy and hypertrophy patterns in thousands of peer-reviewed publications spanning over one hundred diseases. Indeed, VBM studies constitute a large body of quantitative literature reporting a vast number of areas of focal structural change in the brain (Ashburner and Friston, 2000). Coordinate-based reporting and whole-brain coverage are sine qua non for meta-analysis; VBM meets both criteria. Using VBM, then, meta-analytic methods 1) can compute convergent patterns of atrophy and hypertrophy in a cross-study manner; 2) can compute structural covariance network, using between-study, co-atrophy patterns. The multiple reports of VBM studies conducted within diseases and across diseases using a standardized coordinate space makes it well suited for coordinate-based meta-analytic structural covariance analyses studies (Glahn et al., 2008; Fox et al., 2014; Crossley et al., 2015).
A transdiagnostic approach, as opposed to disease-specific, for studying structural covariance networks can most readily be achieved through meta-analysis. In recent years, a growing literature of transdiagnostic neuroimaging has been used in the meta-analytic investigation of neuropsychiatric pathology. Numerous reports indicate that diseases of the brain tend to exhibit patterns of convergence, both structurally and functionally (Seeley et al., 2009; Crossley et al., 2013; Goodkind et al., 2015; McTeague et al., 2016). This new approach – grouping studies by neurobiological effects rather than by diagnostic category – corresponds closely to the Research Domain Criteria (RDoC) initiative set forth in 2010 to “create a new kind of taxonomy for mental disorders by bringing the power of modern research and approaches in genetics, neuroscience, and behavioral science to the problems of mental illness.” (Insel et al., 2010). Clinical and neurobiological data has also suggested that psychiatric disorders are more comorbid than previously thought. They share common imaging and genetic markers, and demonstrate alterations across neural networks that mediate cognition and other mental processes (Etkin and Cuthbert, 2014). Moreover, there is much interest and support within the neuroscience community in leveraging large brain imaging databases for the purpose gaining deeper insight into neuroscientific phenomena (Crossley et al., 2016; Bzdok and Yeo, 2017).
BrainMap (Fox et al., 1994; Fox and Lancaster, 2002) is a neuroimaging database of published neuroimaging experiments with coordinate-based results in standard space. Its collection of functional task-based activation and structural gray matter atrophy enables investigators to study human brain function and structure in healthy and disease subjects meta-analytically. Its functional database currently contains 3197 functional publications spanning over one hundred paradigms with 15,834 experiments and 69,727 subjects. Additionally, the VBM database contains 993 publications spanning over one hundred diseases with 3150 experiments and 75,709 subjects (Laird et al., 2012; Laird et al., 2009b). In our study, we utilized MACM, a study-wise multivariate method to assess covariance across structural networks using BrianMap's VBM transdiagnostic literature (Eickhoff et al., 2010b). Previously, MACM has been used to investigate functional covariance between brain regions that are functionally connected across task-activation experiments (Fox et al., 2005a; Robinson et al., 2010; Laird et al., 2013). To our knowledge this paper is the first to employ MACM to generate a structural covariance model using BrainMap's transdiagnostic VBM database. In our endeavor to test the NDH, we believe that using the MACM method is well suited to investigate anatomic/activation likelihood estimate correlations found between structural and functional covariance models.
In this study, we computed a structural covariance model of the hippocampus – the HNM – beginning with a region-to-whole brain trans-diagnostic VBM meta-analysis (also known as a single-seed MACM analysis). We identified 11 significant nodes using a rigorous voxel-level family-wise error (FWE) of 0.01. These 11 nodes of interest were then re-seeded into the BrainMap database as standardized spherical ROIs to generate two MACM models: 1) structural covariance (significant regions of gray matter density changes), from the VBM database, and 2) coactivation (significant blood-oxygen level dependency signal changes in task studies), from the functional database in the form of ALE values and p statistics. We hypothesized that using MACM, nodes derived from our RtWB meta-analysis will reveal covariance patterns in the form of edges for both structural and functional covariance analyses. Finally, in accordance with the NDH, we postulated that our data-driven meta-analytic structural and functional covariance models generated from the HNM will show a significant degree of concordance.
2. Materials & methods
2.1. Region-to-whole brain (RtWB) meta-analysis
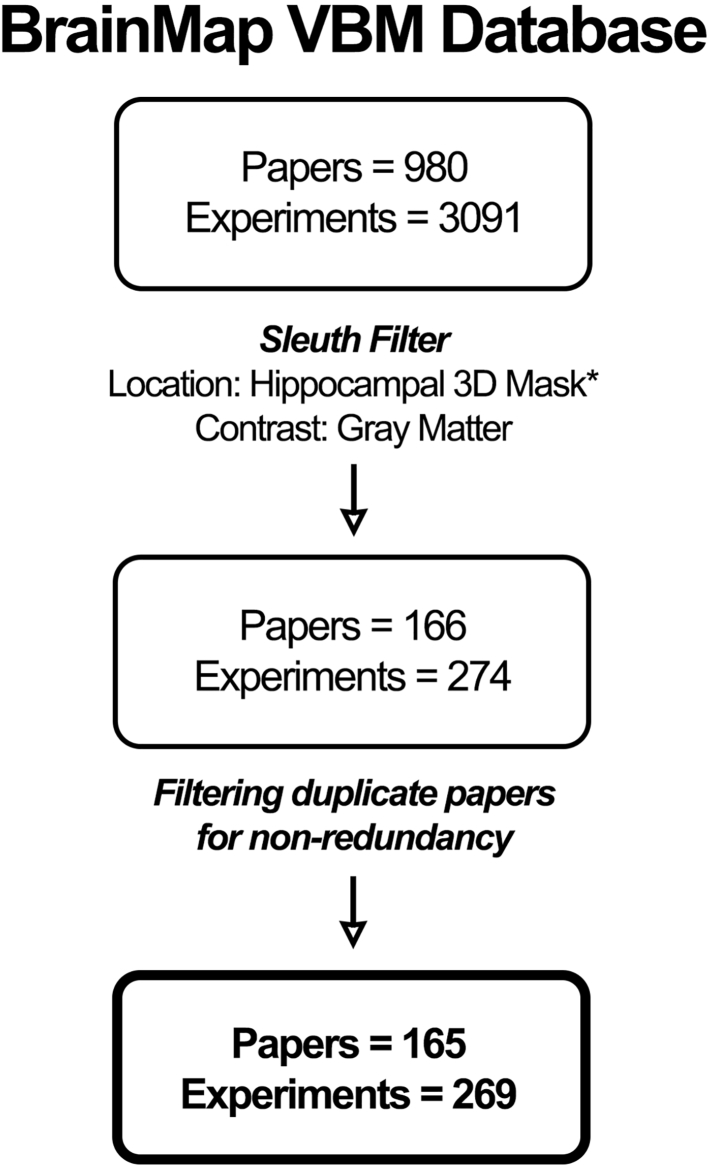
The BrainMap VBM database included, at the time the experiment was conducted, a total of 980 VBM studies comprising 3091 experiments and 74,013 subjects and spanning a total of 143 different ICD-10 codable neurological and psychiatric diseases (Supplementary Table 1) as well as 23 non-ICD-10 classifications. We searched this database for papers reporting hippocampal gray matter alterations using a region-based approach, with the hippocampus as the region of interest. Hippocampal volumetric masks, including both anterior and posterior sections of the hippocampus, were derived from the Harvard-Oxford Structural Probability Atlas (Jenkinson et al., 2012) and subsequently binarized and transformed into Talairach space for further analysis. The VBM database was searched using BrainMap's Sleuth 2.4 software (Fox et al., 2005b) to identify voxel-wise associated clusters with the hippocampal volumetric seed across all VBM studies (search performed on 15 April 2017). Sleuth search: Locations → Talairach Image is Binarized Hippocampus Volume Mask; Experiments → Contrast is Gray Matter (Fig. 1). The search criteria included experiments with gray matter contrast and locations found to co-occur with the binarized hippocampal volumes. Although the vast majority of VBM studies address changes in the form of gray matter atrophy (Controls > Patients), we chose to include in our analysis all studies involved hippocampal structural change, these include studies demonstrating both atrophy and hypertrophy. Using a broad filter encompassing all structural covariance studies present in the database serves to provide a more robust and statistically powerful sampling of studies addressing hippocampal structural covariance. The initial search yielded a total of 166 papers comprising 274 experiments, 10,695 subjects, and 3156 locations. We used additional manual filtering for data non-redundancy to avoid within-lab and sample-specific bias, this brought down our samples to 165 papers, 269 experiments, 10,653 subjects, and 3123 locations. According to Eickhoff et al. (2016), a cluster- or voxel-level FWE correction presents the most appropriate method of statistical inference. In addition, Eickhoff recommends at least 20 experiments be included in an ALE meta-analysis to achieve sufficient statistical power for reporting moderate effects. Our experiment exemplifies some of the most conservative (voxel-level FWE) and robust (269 experiments) parameters set forth by Eickhoff's recommendations.
Fig. 1.
Prisma table: Pipeline demonstrating inclusion criteria for VBM studies incorporating hippocampal gray matter density changes across BrainMap's VBM database.
* Binarized hippocampal 3D mask in Talairach space was derived from the Harvard-Oxford Structural Probability Atlas (Jenkinson et al., 2012).
An ALE map of the filtered workspace containing 269 experiments, 10,653 subjects, and 38 diseases was performed using GingerALE 2.3.6 (brainmap.org/ale/index.html) (Eickhoff et al., 2009) with the following parameters: voxel-level family-wise error (FWE) of 0.01, threshold of 1000 permutations, and minimum cluster volume of 50 mm3 (Table 1). Spatial concordance among the reported VBM foci was computed with the modified anatomic likelihood estimation (ALE) algorithm (Eickhoff et al., 2012). Previous methods papers (Eickhoff et al., 2009; Turkeltaub et al., 2011) have described how to report spatial uncertainty in VBM foci where ALE describes each VBM focus as a Gaussian probability distribution. Three-dimensional Gaussian distributions were pooled in a voxel-wise manner first within experimental contrasts and then across contrasts within a group to create a whole-brain ALE probability cluster map. Each voxel was essentially assigned a unique ALE value that represents the likelihood of experimental effects, such as gray matter structural change in the case of VBM, present within that voxel. ALE maps were also tested against a null distribution to reflect random spatial associations between different experiments at a set threshold of permutations. Voxels that survived this statistical threshold were then reported as ALE clusters reflecting significance in meta-analytic convergence of peak voxels. These voxels reflect significant changes in gray matter within and across experiments. ALE also represents an analytic, data-driven technique in creating the null-distribution used in statistical inference, and uses a Monte-Carlo based approach to permit more accurate cluster-level inference (Eickhoff et al., 2012).
Table 1.
RtWB meta-analysis of BrainMap VBM database peak foci: voxel-level FWE 0.01, 1000 permutations, 50 mm3 minimum cluster volume.
| Description | Size mm3 | Weighted center (x, y, z) | Hemisphere | Lobe | Region | Sub-region | ||
|---|---|---|---|---|---|---|---|---|
| Node 1 - left anterior hippocampus (LAH) | 13065a | −26 | −14 | −14 | Left cerebrum | Limbic lobe | Parahippocampus | Hippocampus |
| Node 2 - left posterior hippocampus (LPH) | 13065a | −26 | −34 | −4 | Left cerebrum | Limbic lobe | Parahippocampus | Hippocampus |
| Node 3 - right anterior hippocampus (RAH) | 10672a | 24 | −12 | −14 | Right cerebrum | Limbic lobe | Parahippocampus | Hippocampus |
| Node 4 - right posterior hippocampus (RPH) | 10672a | 28 | −32 | −4 | Right cerebrum | Limbic lobe | Parahippocampus | Hippocampus |
| Node 5 - right MDN of thalamus (RMDN) | 2659a | 4 | −18 | 12 | Right cerebrum | Sub-lobar | Thalamus | Med dorsal nucleus |
| Node 6 - left midline nucleus of thalamus (LMN) | 2659a | −6 | −16 | 14 | Left cerebrum | Sub-lobar | Thalamus | Midline nucleus |
| Node 7 - right claustrum (RClaus) | 464 | 36 | 6 | 4 | Right cerebrum | Sub-lobar | Claustrum | |
| Node 8 - left claustrum (LClaus) | 292 | −34 | −8 | 0 | Left cerebrum | Sub-lobar | Claustrum | |
| Node 9 - left insula (LIns) | 280 | −34 | 16 | 2 | Left cerebrum | Sub-lobar | Insula | BA13 |
| Node 10 - right caudate body (RCB) | 173 | 10 | 12 | 8 | Right cerebrum | Sub-lobar | Caudate | Caudate body |
| Node 11 - left caudate head (LCH) | 64 | −6 | 6 | −2 | Left cerebrum | Sub-lobar | Caudate | Caudate head |
2.2. Meta-analytic connectivity modeling
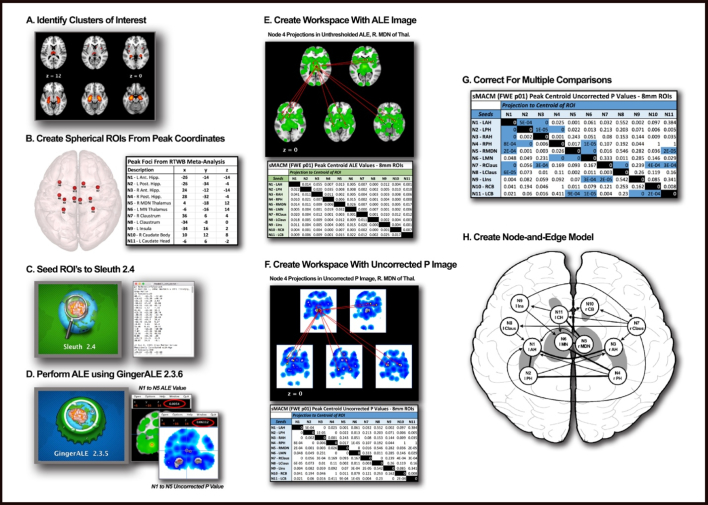
MACM investigates whole-brain connectivity patterns by assessing the covariance between two or more nodes quantitatively using ALE, with ALE values being defined as the probability estimate that an activation (or “atrophy”) exists within a particular coordinate across experiments (Robinson et al., 2010; Robinson et al., 2012). MACM analyses resulting in ALE maps have been validated with diffusion tensor imaging (DTI) and connectivity atlases (CocoMac) (Robinson et al., 2010) and have been demonstrated to be the meta-analytic equivalent of resting-state functional connectivity maps (Laird et al., 2009b; Smith et al., 2009). Further, MACM has also been validated as a method for functional connectivity by recovering known, microscopically distinguishable compartments within a same brain region in a process known as connectivity-based parcellation (Bzdok et al., 2013; Barron et al., 2015). A step-by-step visualization of the MACM method is provided in Fig. 2.
Fig. 2.
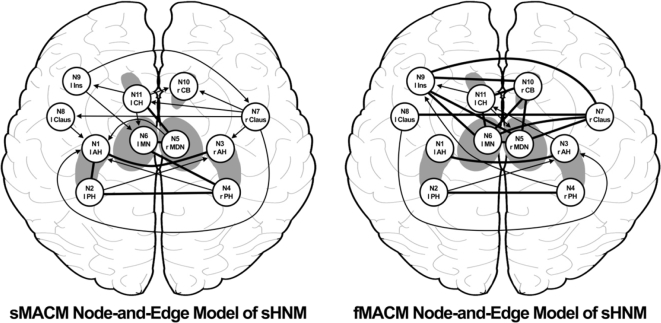
MACM pipeline: Pipeline showing a step-by-step process of using BrainMap and its corresponding software, Sleuth and GingerALE, to conduct a MACM analysis leading to the development of a node-and-edge model. A) Derive nodes from peak foci using region-to-whole brain meta-analysis. (Other methods for identifying nodes are also eligible for MACM analysis). B) Create spherical ROIs of nodes using peak foci coordinates. 8 mm diameter for VBM studies, 12 mm diameter for functional studies and save as ROIs. C) Seed individual ROIs in BrainMap's Sleuth 2.4 to search database and develop a workspace. Filter appropriately for VBM or functional database searches. Use consistent brain space (i.e. Talairach, MNI). D) Perform ALE using GingerALE 2.3.6 using appropriate and consistent thresholds. E) Create workspace using Mango of spherical ROIs. Overlay raw ALE images derived from GingerALE unto workspace and record ALE values from the centroid of each node to create a table. F) Repeat process using Uncorrected P output file from GingerALE and record P values from the centroid of each node to create a table. G) Define appropriate statistical thresholds to correct the raw Uncorrected P value table. Presently, we used Bonferroni correction for multiple comparisons threshold and to define the most robust paths. H) Create a node-and-edge model where nodes are spherical representations of peak structural variance or activation foci (VBM or functional respectively). Dark edges represent bidirectional covariance, arrows represent unidirectional covariance.
Covariance between nodes can be measured as structural covariance, using gray matter VBM contrasts, or as functional covariance, using hemodynamic response contrasts from task-activation paradigms in normal subjects. From our RtWB meta-analysis of the hippocampus, which yielded an ALE map of 11 significant foci, or nodes, located within 7 clusters reporting structural covariance transdiagnostically (Table 1, Fig. 3), we proceeded to conduct a structural and a functional MACM experiment. We first performed a structural MACM (sMACM) for each of the 11 reported nodes identified as the peak foci from the RtWB meta-analysis (Table 1). Spherical 8 mm diameter ROIs were created and centered on the coordinates of the peak ALE voxel at each cluster. Each of the 11 ROIs was then seeded into the VBM database (a separate analysis for each ROI) in a seed-to-whole brain (StWB) meta-analysis (also known as a network MACM analysis), similar to how we derived our nodes using a RtWB meta-analysis of the hippocampus. We used the same criteria as the RtWB analysis for our seeds in Sleuth 2.4 whereby only gray matter contrasts and defined regions (spherical ROIs) were used in the search. Manual filtering for data non-redundancy was also performed for each ROI to avoid within-lab and sample-specific bias for each search. The newly filtered workspace containing the experiments and coordinates of studies reporting StWB structural covariance (Table 2) were processed using GingerALE 2.3.6 (voxel-level FWE of 0.01, thresholded to 1000 permutations, and minimum cluster volume of 50 mm3). ALE values were derived for each node in the HNM from each ROI's StWB meta-analysis and reported in a seed-to-projection table of ALE values (Table 3). A seed-to-projection coefficient (i.e. edge) is the ALE value obtained from the center voxel, or center coordinate, of each HNM node from a seed node's ALE analysis (e.g. If node 1 is the seed for an ALE analysis, a seed-to-projection coefficient is obtained from the ALE value representing the center voxel of nodes 2 through 11). Fig. 4 shows Bonferroni-corrected (.01) statistical p values reported from the seed-to-projection Uncorrected P output file from GingerALE. If reciprocal significance was present (i.e. N1 → N2 and N2 → N1) then co-directionality of edges was determined (Fig. 5).
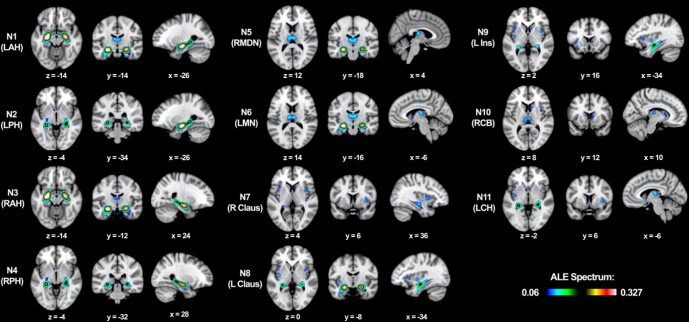
Fig. 3.
Hippocampi region to whole brain meta-analysis: Regions with specified peak foci showing significant structural covariance among trans-diagnostic VBM gray matter contrast studies (voxel-level FEW 0.01, thresholded at 1000 permutations and minimum reported cluster volume of 50 mm3). LAH = left anterior hippocampus, LPH = left posterior hippocampus, RAH = right anterior hippocampus, RPH = right posterior hippocampus, RMDN = right medial dorsal nucleus of the thalamus, LMN = left midline nucleus of the thalamus, R Claus = right claustrum, L Claus = left claustrum, L Ins = left insula, RCB = right caudate body, LCH = left caudate head.
Table 2.
Structural MACM workspace for 8 mm nodes.
| Description | Papers | Subjects | Experiments | Locations |
|---|---|---|---|---|
| Node 1 - left anterior hippocampus (LAH) | 21 | 1311 | 24 | 307 |
| Node 2 - left posterior hippocampus (LPH) | 13 | 1123 | 18 | 314 |
| Node 3 - right anterior hippocampus (RAH) | 20 | 1453 | 24 | 259 |
| Node 4 - right posterior hippocampus (RPH) | 13 | 935 | 14 | 141 |
| Node 5 - right MDN of thalamus (RDMN) | 9 | 835 | 12 | 177 |
| Node 6 - left midline nucleus of thalamus (LMN) | 13 | 800 | 15 | 181 |
| Node 7 - right claustrum (RClaus) | 12 | 943 | 13 | 153 |
| Node 8 - left claustrum (LClaus) | 9 | 836 | 13 | 301 |
| Node 9 - left insula (LIns) | 17 | 1594 | 20 | 278 |
| Node 10 - right caudate body (RCB) | 12 | 1140 | 12 | 137 |
| Node 11 - left caudate head (LCH) | 11 | 1168 | 14 | 382 |
Table 3.
Structural MACM seed-to-whole brain (FWE p01) peak centroid ALE values – 8 mm ROIs.
Fig. 4.
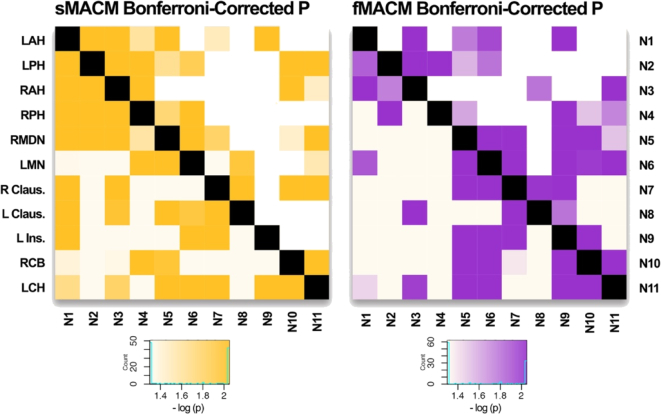
Heat map of Bonferroni-corrected P values in sMACM and fMACM of sHNM: Histogram color values are reported in −log10 (p). Values greater than or equal to 2.04 are darkest and represent regions of covariance with a Bonferroni-Corrected P ≤ .01 (≤.00091) for 11 nodes. Values less than or equal to 1.30 are lightest and represent regions of least covariance (p > .05).
Fig. 5.
Node-and-Edge Diagrams of MACMs: Each diagram delineates paths from the Bonferroni-Corrected P values used in assessing inter-nodal covariance. Bolded lines represent bi-directionality, indicating that variance in two nodes are predictive of variance in each other. Arrows represent uni-directionality, indicating that variance in one node is predictive of variance in another, but not vice versa.
This MACM strategy was replicated in the functional database to produce a functional MACM of the 11 nodes derived from the VBM hippocampal RtWB meta-analysis. One noteworthy difference between the two MACMs is the sizes of the seeds used in the analysis. For the functional MACM, 12 mm diameter ROIs were used. The reason for this is because a larger ROI is required to obtain comparable numbers of coactivation clusters from the functional database in BrainMap. The selection of 8 mm diameter ROIs for sMACM and 12 mm diameter ROIs for fMACM were chosen to account for differences in cluster number and size between the structural and functional database, allowing for the balancing of ALE clusters between datasets using spatial filtering. ROI diameters were thus obtained iteratively within lab through evaluating the number and location of the pertinent voxels against the VBM and functional databases. As in the structural MACM, a non-redundancy filtered table of StWB studies from BrainMap's functional dataset was generated (Table 4), with its corresponding seed-to-projection ALE value table (Table 5). Our ability to perform both structural and functional MACMs allows us to integrate whole-brain structural covariance and whole-brain functional coactivation patterns from BrainMap in a common numerical form of anatomic likelihood estimates. This allows us to run statistics on the ALE matrices (Table 3, Table 5) comparing structure and function.
Table 4.
Functional MACM workspace for 12 mm nodes.
| Description | Papers | Subjects | Experiments | Conditions | Locations |
|---|---|---|---|---|---|
| Node 1 - left anterior hippocampus (LAH) | 67 | 1071 | 76 | 207 | 1049 |
| Node 2 - left posterior hippocampus (LPH) | 35 | 486 | 38 | 105 | 517 |
| Node 3 - right anterior hippocampus (RAH) | 65 | 1133 | 80 | 164 | 1144 |
| Node 4 - right posterior hippocampus (RPH) | 36 | 607 | 41 | 91 | 685 |
| Node 5 - right MDN of thalamus (RMDN) | 120 | 2102 | 148 | 328 | 2635 |
| Node 6 - left midline nucleus of thalamus (LMN) | 94 | 1358 | 103 | 254 | 2012 |
| Node 7 - right claustrum (RClaus) | 75 | 1068 | 85 | 192 | 1444 |
| Node 8 - left claustrum (LCLaus) | 48 | 699 | 55 | 122 | 819 |
| Node 9 - left insula (LIns) | 214 | 3331 | 267 | 633 | 4235 |
| Node 10 - right caudate body (RCB) | 73 | 1092 | 80 | 186 | 1256 |
| Node 11 - left caudate head (LCH) | 66 | 1203 | 79 | 182 | 1045 |
Table 5.
Functional MACM seed-to-whole brain (FWE p01) peak centroid ALE values – 12 mm ROIs.
2.3. Statistical analysis of functional and structural HNMs
A Pearson's correlation was used to evaluate the ALE seed-to-projection coefficients derived from the fMACM and sMACM of the 11 individual nodes from the hippocampal RtWB brain VBM meta-analysis. We use a node-and-edge model as a framework to illustrate the degree of covariance between the HNM nodes as determined by structural and functional MACM analyses. A Pearson's r and _r_2 was calculated for each seed-to-projection ALE values comparing functional and structural values (e.g. ALE coefficients from the node 1 seed to nodes 2–11 projections in sMACM analysis were correlated to ALE coefficients from the node 1 seed to nodes 2–11 projections in fMACM analysis). In addition, a Pearson's correlation was generated for the entire ALE coefficient matrix comparing all projections from all nodes in the sMACM analysis to all projections from all nodes in the fMACM analysis. 110 candidate paths, or edges, were identified with p values from the original ALE FWE-corrected analysis. Only paths meeting the significance threshold, a Bonferroni-adjusted p of .01 were deemed significant. Paths that met the Bonferroni correction threshold are thus represented as edges in the node-and-edge models in Fig. 5.
2.4. Disease, behavior, and paradigm analysis
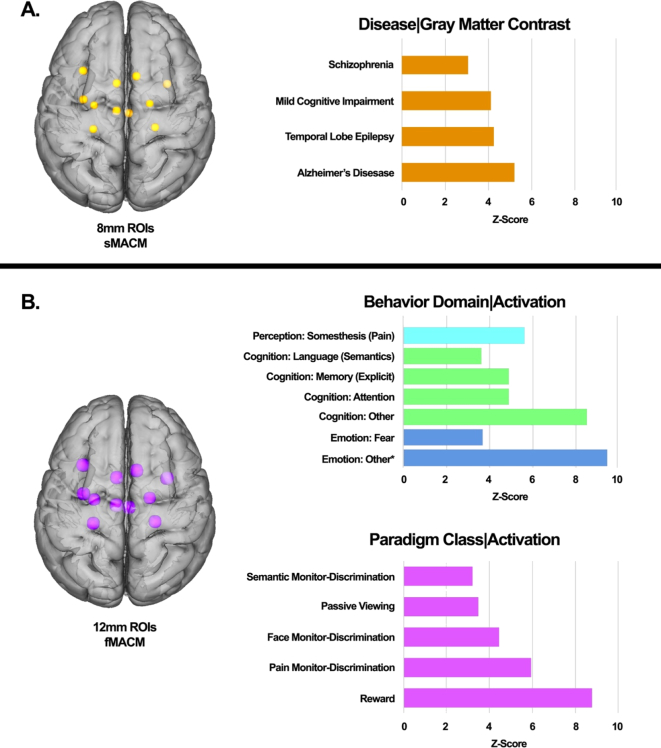
Disease, behavior, and paradigm class were analyzed using the 11 nodes from the hippocampal analysis (Fig. 6). We used Lancaster et al.'s (2012) paradigm class analysis, which utilizes the BrainMap database to isolate activation foci for each behavioral sub-domain. This technique compares activations from the functional database, or atrophies from the VBM database, from within the ROI to the fraction expected without regional behavior, as if activations were uniformly distributed throughout the brain. Z-scores are generated for observed-minus-expected values for each behavior sub-domain, or disease sub-domain in the case of VBM, with the operational threshold z-score of 3.0 as comparable to a group p-value of .05. This analysis has been previously validated in both the behavior domain and paradigm class analyses (Lancaster et al., 2012). This study represents the first application of a disease paradigm analysis using VBM to identify diseases that are likely to be expressed in the specified regions of interest (Fig. 6).
Fig. 6.
Specific behavioral, paradigm class, and disease weightings on the structural hippocampal network model.
A. Structural characterization of sHNM based on Disease Classification meta-data of the BrainMap VBM structural database representing peak structural MACM disease classes.
B. Functional characterization of sHNM based on the Behavioral Domain and Paradigm Class meta-data of the BrainMap functional database representing peak functional MACM behavior domains and task paradigms.
Only paradigms fulfilling the minimal threshold of z ≥ 3.0 are reported.
*The designation “other” indicates an emotion experimental contrast that does not meet the eligibility criteria for pre-selected emotions in the BrainMap database: anger, anxiety, disgust, fear, happiness, humor, sadness.
3. Results
3.1. Hippocampus region-to-whole brain meta-analysis
The ALE results from the hippocampus RtWB meta-analysis (Table 1) shows 11 significant foci located within clusters representing extra-hippocampal gray matter density alterations across diagnoses as well as two distinct foci within each hippocampus (Fig. 3). For each focus, we reported location of maximum ALE values as (x, y, z) coordinates in Talairach space. The list of all 165 papers included in this meta-analysis is presented in a Supplementary table.
3.2. Meta-analytic connectivity modeling
We obtained significant results using structural (Table 2) and functional (Table 4) MACM of the resulting 11 ROIs from the HNM, reported as regions from the Talairach-Daemon (Lancaster et al., 2000). Two separate tables are reported for each MACM: ALE values from BrainMap's structural VBM database (Table 2) and task-activation functional database (Table 4) and Bonferroni corrected P .01 (Fig. 4). These corrected _p_-values, which represent covariance statistics between nodes and projections in Fig. 4, were used to generate the edges in our node-and-edge model. Edges were deemed significant with a Bonferroni correction threshold, if only one edge between two nodes was significant, the connection was assigned as unidirectional (i.e. N1 → N2 but not N2 → N1); if both edges between two nodes were significant, then the connection was deemed bidirectional (i.e. N1 → N2 and N2 → N1) (Fig. 5). Fig. 5 illustrates the presence of a number of significant nodes bearing structural bidirectional covariance properties. Among these are between the left anterior hippocampus (LAH, N1) and left posterior hippocampus (LPH, N2), LAH (N1) and right anterior hippocampus (RAH, N3), LPH (N2) and right posterior hippocampus (RPH, N4), left midline nucleus of the thalamus (LMN, N6) and RPH (N4), and left caudate head (LCH, N11) and right medial dorsal nucleus of the thalamus (RMDN, N5). Similarly, significant nodes bearing functional covariance bi-directional coactivation properties can be found between the LAH (N1) and RAH (N3), LPH (N2) and RPH (N4), left claustrum (LClaus, N8) and right claustrum (RClaus, N7), left insula (LIns, N9) and RClaus, LIns and right caudate body (RCB, N10), LIns (N9) and RMDN (N5), LIns (N9) and LMN (N6), LCH (N11) and RCB (N10), LCH (N11) and LMN (N6), LMN (N6) and RCB (N10), LMN (N6) and RClaus (N7), LMN (N6) and RMDN (N5), and RMDN (N5) and RClaus (N7). Significant unidirectional paths are also reported in both structural and functional MACM figures (Fig. 4, Fig. 5).
3.3. Structural and functional correlation of HNM
The matrix correlation between sMACM and fMACM found a moderate but significant correlation of r = .377 (p = .00005) as defined by Cohen (1988) using Pearson Correlation. A similar finding was generated using the Mantel (1967) Test for matrix correlations whereby a moderate and significant correlation of r = .397 (p = .0013) was identified. We also reported _r_2 to describe the variance explained (14.2%) by the association of structure and function between the nodes of the sHNM. The nodes, whose seed-to-projection ALE values demonstrate the greatest and most significant correlations between structural and functional covariance include: nodes 1 (left anterior hippocampus, r = .942 and p = 4.83 × 10−5), 2 (left posterior hippocampus, r = .830 and p = 4.83 × 10−5), and 3 (right anterior hippocampus, r = .935 and p = 7.34 × 10−5) (Table 6).
Table 6.
sMACM to fMACM statistical comparison of node-to-whole-brain (node to all other nodes) covariance using ALE values and sMACM to fMACM ALE matrix correlation.
| sMACM to fMACM statistical comparisons | |||
|---|---|---|---|
| Seed-to-projection | r | _r_2 | p |
| N1 (LAH) | .942 | .887 | 4.83e-5⁎ |
| N2 (LPH) | .830 | .688 | 4.83e-5⁎ |
| N3 (RAH) | .935 | .874 | 7.34e-5⁎ |
| N4 (RPH) | .379 | .144 | .280 |
| N5 (RMDN) | .347 | .120 | .326 |
| N6 (LMN) | .618 | .382 | .057 |
| N7 (RClaus) | −.096 | .009 | .792 |
| N8 (LClaus) | .143 | .020 | .694 |
| N9 (LIns) | .580 | .336 | .079 |
| N10 (RCB) | .579 | .335 | .080 |
| N11 (LCB) | .687 | .472 | .028 |
| sMACM and fMACM ALE matrix statistical comparisons | |||
|---|---|---|---|
| Statistical test | r | r 2 | p |
| Pearson correlation | .377 | .142 | 4.89e-5⁎ |
| Mantel test | .397 | .158 | .001 |
3.4. Disease, behavior, and paradigm analysis
The 4 most significant diseases identified in the nodes from the RtWB meta-analysis include Alzheimer's Disease (AD), Mesial Temporal Lobe Epilepsy (MTLE), Mild Cognitive Impairment (MCI), and Schizophrenia. Likewise, an evaluation of these same 11 nodes using the behavioral domain paradigm from BrainMap's functional cohort of healthy subjects identified the emotional subdomains of fear and other emotion, the cognition subdomains of attention, explicit memory, semantic language, and other cognition, and the perception subdomain of pain somesthesis (Fig. 6).
4. Discussion
The purpose of our study was to identify and define consistent gray matter structural abnormalities that co-exist with hippocampal atrophy, and address the network degeneration hypothesis (NDH) by testing and quantifying the correlation between structural and functional covariance. Our analysis found that indeed, a moderate correlation exists between meta-analytically derived structural and functional covariance models of nodes derived from a structurally-driven hippocampal network model (HNM). By applying the meta-analytic connectivity modeling (MACM) pipeline procedure (Fig. 2) to our HNM using BrainMap's voxel-based morphometry (VBM) and functional database, we were able to validate our hypothesis, that MACM can be applied to the VBM literature and generate structural covariance models using the trans-diagnostic literature. Additionally, our resulting Pearson's correlation analysis using ALE values obtained from the structural and functional covariance networks of the HNM was found to support the NDH, establishing that the structural-functional relationship of interconnected brain regions is topographically correlated.
We interpret these structural covariances as related to four of the primary trophic influences outlined by Zhou et al., 2012. The transneuronal spread mechanism begins in an epicenter, where neuronal inclusions in the form of neurofibrillary tangles, cytoplasmic Lewy bodies, iron accumulation, prion diseases, viral inclusions, a diverse array of protein aggregates, etc. (Fornai et al., 2005) spread along axonal pathways to other groups of neurons located in more distant regions (Zhou et al., 2012; Ahmed et al., 2016). The nodal stress mechanism suggests that brain hubs (i.e., regions with high levels of connectivity across the brain, more metabolic demand, and higher blood flow) are selectively susceptible to damage from oxidative stress and endothelial damage leading to impairments, such as a leaky blood brain barrier (Crossley et al., 2014). The mechanism of trophic failure involves the sub-cellular dysfunction of trophic factors, e.g. brain-derived neurotrophic factor (BDNF), which is involved in dendritic and axonal branching (Cohen-Cory et al., 2010). When trophic factor expression and regulation is impaired, trophic failure in the form of poor cellular and synaptic maintenance impairs structural connectivity (Fornito et al., 2015). Lastly, shared vulnerability is a mechanism whereby similar neuronal cell types across the brain are susceptible to disease-specific changes due to shared genetic and metabolic profiles (Cioli et al., 2014). Because our study is trans-diagnostic, it is impossible to identify which mechanisms predominate as different diseases exhibit differing and perhaps overlapping trophic influences.
4.1. HNM connectivity
The functional and structural association of the hippocampus to multiple brain regions has been and continues to be an important area of research interest. The identification of hippocampal atrophy in normal aging as well as in diseases that differ in their symptomatology has generated interest in identifying network models explaining the associations between the hippocampus and other cortical and subcortical structures. The goal of developing a comprehensive hippocampal network model has its roots in the use of structural and functional biomarkers in specified brain regions that can be utilized to investigate disease progression, treatment efficacy, and differences in normal aging (Marstaller et al., 2015).
Hippocampal network connectivity has been studied in other established networks, most notably the reward circuit (anterior cingulate cortex, orbital prefrontal cortex, ventral striatum, ventral pallidum, midbrain dopamine neurons, dorsal prefrontal cortex, amygdala, hippocampus, thalamus, lateral habenular nucleus, pedunculopontine nucleus, and raphe nucleus) (Haber and Knutson, 2010) and the default mode network (posterior parietal cortex, precuneus, lateral temporal gyrus, posterior cingulate cortex, hippocampus, and cerebellum) (Laird et al., 2009a; Chang et al., 2015). Both networks provide empirically based models for the study of diseases that alter these networks' properties. Network modeling and structural equation modeling has also been used to predict behavioral and metabolic metrics using the reward circuit and DMN respectively (Laird et al., 2009a; Park et al., 2015; Lin et al., 2015).
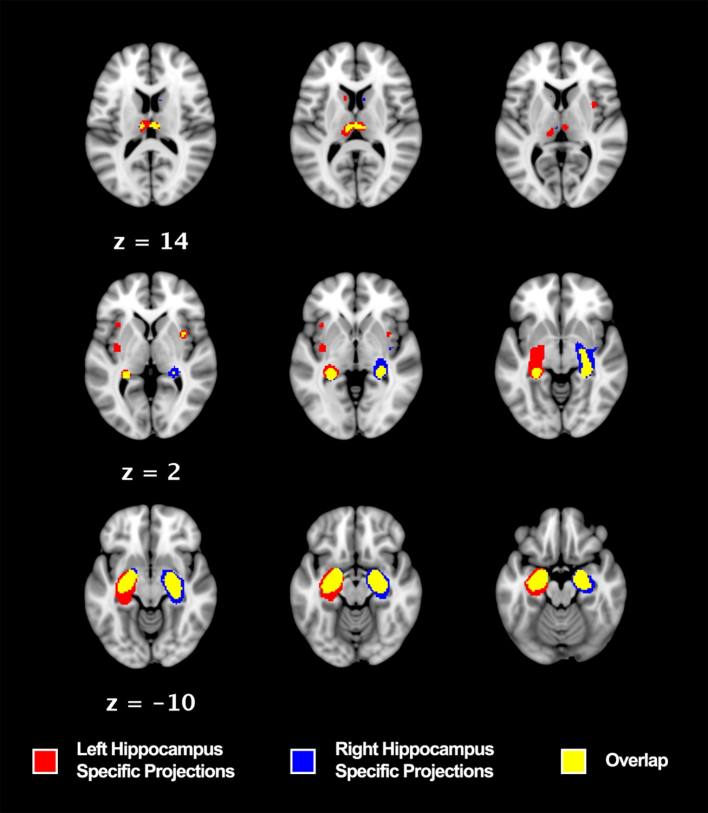
Importantly, the most robust and well-described hippocampal connectivity profile is that which occurs between the contralateral hippocampi, validated in this present study both structurally and functionally (Fig. 5, Table 6). This association has long been established as both hippocampi share direct inter-hemispheric connections via the hippocampal commissure (Spencer et al., 1987). This contralateral structural connectivity can be observed using a visual sMACM of each hippocampus when seeded in the VBM database independently showing key points of difference as well as overlap (Fig. 7). Moreover, there is ample evidence that the hippocampus is a key component in networks relating to both normal functioning and pathology. In our analysis, we used a novel approach to answering the question of pathological significance relating to hippocampal structural covariance.
Fig. 7.
Laterality of structural covariance between right and left hippocampi: Regions in red are those that co-vary significantly with the left hippocampus seeded in a RtWB meta-analysis. Regions in blue are those that co-vary significantly with the right hippocampus seeded in a RtWB meta-analysis. Regions in yellow represent the overlapping structures of the right and left hippocampus RtWB meta-analyses. All analyses were conducted using a voxel-level FEW 0.01, thresholded to 1000 permutations, and reporting only minimal cluster volumes of 50mm3. Left Hippocampus Specific Projections: left caudate head, left insula, left claustrum. Right Hippocampus Specific Projections: right caudate body, right claustrum (different focus than overlap right claustrum). Overlap: left anterior hippocampus, left posterior hippocampus, right anterior hippocampus, right posterior hippocampus, right MDN of thalamus, left MDN of thalamus, right claustrum (different focus than right hippocampus specific projection).
The hippocampus itself is not a monolithic structure. It is subdivided into histological regions (dentate gyrus, CA1-4, subiculum) and has been shown to differ in structural and functional properties along the longitudinal axis. Connectivity-based parcellation experiments of the hippocampus have recently defined this anterior-posterior differentiation of the hippocampus more precisely (Robinson et al., 2015). Other meta-analytic methods have shown functional differences between anterior and posterior hippocampal activation patterns (Chase et al., 2015). Importantly, seeding the hippocampi as volumetric binarized regions in the present VBM database yields not only extra-hippocampal foci, but foci within the hippocampus itself. This lends credence to the aforementioned studies identifying distinct anterior–posterior functional and structural differentiation of the hippocampus in the form of focal atrophy. We identified two distinct ALE foci (Fig. 3) in each hippocampus that were subsequently regarded as independent nodes.
Indeed, our study also identifies several well-studied connections between the hippocampus and other brain structures. We found, unsurprisingly, that specific regions known to be structurally and functionally connected to the hippocampus also appeared in our meta-analysis: the claustrum, insula, MDN of the thalamus, and caudate. Tracer studies in the macaque brain have previously identified claustral projections to other subcortical structures found in our sHNM, specifically the dorsal thalamic nuclei, hippocampus, and caudate nucleus (Mathur, 2014). Crossley et al. (2014) found through DTI meta-analytic lesion mapping that there are a number of imaging connectome hubs relating to anatomical locations of MRI lesions, to which the bilateral hippocampi, thalamus, and insula belong. It has also been demonstrated that blood-brain barrier (BBB) breakdown in the human hippocampus, found to be correlated with normal aging as well as MCI, is also strongly associated with breakdown of the BBB in the caudate nucleus specifically (Montagne et al., 2015). The caudate-hippocampal connection has also been validated with a VBM-ALE meta-analysis study of gray matter atrophy in schizophrenia (Li et al., 2017). Thalamo-hippocampal connectivity is another well-established association in both animal and human models (Vertes et al., 2007; Barron et al., 2013). With regards to the thalamus, specific thalamic nuclei have been shown to be significantly associated with hippocampal pathology, namely the pulvinar, anterior nucleus, and medial dorsal nucleus (Rosenberg et al., 2009). This suggests that different subcortical structures may be differentially connected to the hippocampus by distinct structurally-related mechanisms across a range of diseases.
Our sMACM analysis demonstrate that the most consistent structural change in gray matter density across diseases implicating the hippocampus follows a distinct pattern involving regions previously identified as being implicitly associated with the hippocampus. Furthermore, our fMACM analysis supports the notion that structural and functional network properties are significantly correlated (Sui et al., 2014) (Table 6). This validation can serve the purpose of using the present HNM and its nodes as biomarkers for the study of relevant pathologies (e.g. AD, MTLE, MCI, Schizophrenia, aging, etc.).
4.2. Hippocampal pathology
Hippocampal pathology in Alzheimer's disease, mild cognitive impairment, epilepsy, and schizophrenia has been long established. Hippocampal volume loss is a common finding in imaging studies of individuals with AD, MCI and, normal aging (Apostolova et al., 2012), MTLE (Wei et al., 2016), and Schizophrenia (Koolschijn et al., 2010). Moreover, other studies have demonstrated that systemic pathologies like metabolic syndrome (Anand and Dhikav, 2012), type 2 diabetes mellitus and obesity (Wu et al., 2008; Wang et al., 2014; Stranahan, 2015), and normal aging (Apostolova et al., 2012) also report hippocampal atrophy. Recent studies have ascertained that volume loss in the hippocampus corresponds to neuronal cell death due in part to oxidative stress introduced by a leaky blood brain barrier (Montagne et al., 2015), along with sclerosis (Wei et al., 2016) and amyloid plaques and Tau protein tangles (Apostolova et al., 2010), etc. It is for this reason that a trans-diagnostically-driven HNM is justified as a tool to understand structural network abnormalities in the presence of hippocampal dysfunction.
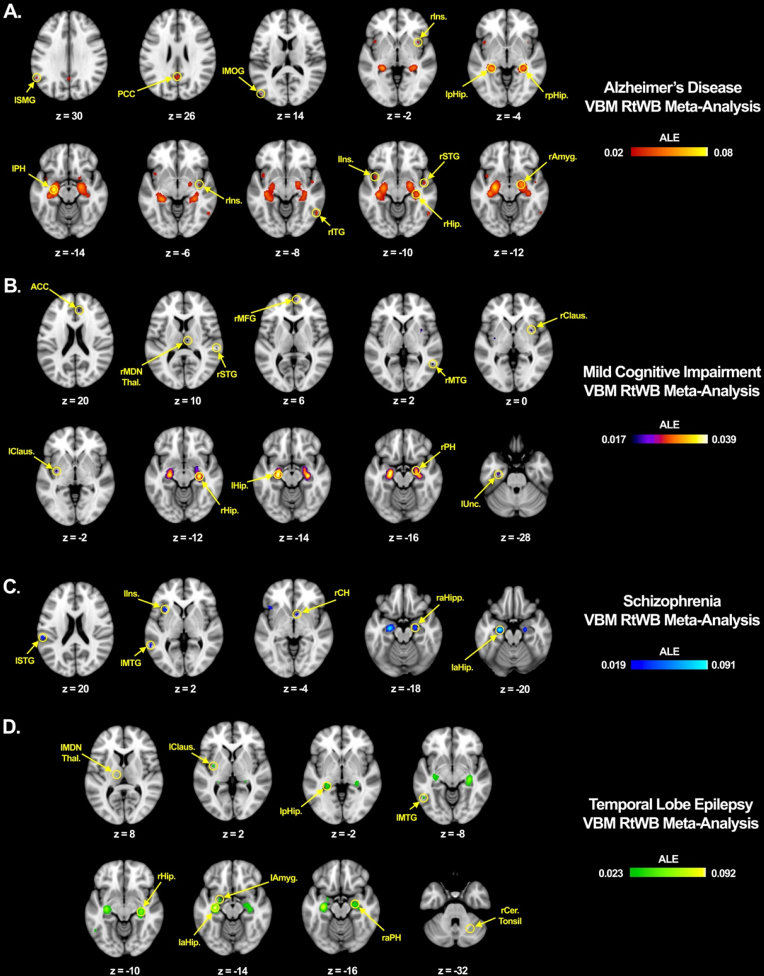
This raises the question: what would the pattern of gray matter atrophy look like if the hippocampus were seeded and constrained only to specific diseases? We tested this by conducting a very similar RtWB analysis as the original trans-diagnostic study. By constraining our regional search to only the diseases that appear to be most strongly correlated to the sHNM based on the disease paradigm analysis (Fig. 6) and using a less stringent statistical threshold (Cluster-level 0.05, FDR pN 0.05, with 1000 threshold permutations) we were able to identify a number of significant regions of hippocampal structural covariance within these top four diseases. Fig. 8 shows a visual comparison of the hippocampal structural covariance patterns for these diseases in order to illustrate that there are important similarities and differences across diseases, workspaces are available as Supplementary data. We can conclude from this analysis that there are certain network pathways that are equally atrophogenic across diseases and co-vary significantly with damage to the hippocampus, whether that is due to sclerosis, plaques and tangles, or inflammatory infiltrates via a leaky BBB. Whatever the disease pathogenic onset that leads to hippocampal cell loss, our sHNM model clearly shows that hippocampal gray matter density is markedly associated with at least 11 brain regions, which together are known to be associated with attention, emotion, reward, and memory, among others (Fig. 6).
Fig. 8.
Hippocampal region-to-whole brain within-disease meta-analysis: All RtWB within-disease meta-analyses were conducted using GingerALE's Cluster-Level 0.05 analysis with FDR pN 0.05 at 1000 threshold permutations.
A. The hippocampus was seeded BrainMap's VBM database constrained only to AD VBM studies. 11 significant foci were identified: left supramarginal gyrus (lSMG), posterior cingulate cortex (PCC), left middle occipital gyrus (lMOG), right insula (rIns.) in two locations, left posterior hippocampus (lpHip.), right posterior hippocampus (rpHip.), left parahippocampus (lPH), right inferior temporal gyrus (rITG), left insula (lIns.), rSTG (right superior temporal gyrus), right hippocampus (rHip.), and right amygdala (rAmyg.). Study consisted of 23 papers, 1350 subjects, 39 experiments, and 443 locations.
B. The hippocampus was seeded BrainMap's VBM database constrained only to MCI VBM studies. 11 significant foci were identified: Anterior Cingulate Cortex (ACC), right medial dorsal nucleus of the thalamus (rMDN Thal.), right superior temporal gyrus (rSTG), right medial frontal gyrus (rMFG), right middle temporal gyrus (rMTG), right claustrum (rClaus.), left claustrum (lClaus.), right hippocampus (rHip.), left hippocampus (lHip.), right parahippocampus (rPH), and left uncus (lUnc.). Study consisted of 14 papers, 995 subjects, 23 experiments, and 287 locations.
C. The hippocampus was seeded BrainMap's VBM database constrained only to Schizophrenia VBM studies. 6 significant foci were identified: left superior temporal gyrus (lSTG), left middle temporal gyrus (lMTG), left insula (lIns.), right caudate head (rCH), right anterior hippocampus (raHip.), and left anterior hippocampus (laHip.). Study consisted of 15 papers, 1119 subjects, 20 experiments, and 269 locations.
D. The hippocampus was seeded BrainMap's VBM database constrained only to temporal lobe epilepsy and mesial temporal lobe epilepsy VBM studies. 9 significant foci were identified: left medial dorsal nucleus of the thalamus (lMDN Thal.), left claustrum (lClaus.), left posterior hippocampus (lpHip.), left middle temporal gyrus (lMTG), right hippocampus (rHip.), left anterior hippocampus (laHip.), left amygdala (lAmyg.), right anterior parahippocampus (raPH), and right cerebellar tonsil (rCer. Tonsil). Study consisted of 18 papers, 1571 subjects, 44 experiments, and 461 locations.
4.3. Methodological considerations
Meta-analysis has been defined as the post hoc combination of independently performed studies to better estimate a parameter of interest (Fox et al., 2005a). Several meta-analytic methods have since been developed. One of the earliest was the use of label-based meta-analysis whereby an investigator would assign labels from multiple studies and tally them without performing any statistical analysis (Keller and Roberts, 2008). This method was heavily criticized for being spatially imprecise and producing misleading results. With the advent of coordinate-based meta-analysis, studies began reporting peak activation or atrophy regions using an x, y, z coordinate system in a standardized brain space (e.g. Talairach and MNI). This method offers a statistically rigorous, and spatially precise alternative to the label-based and image-based methods. Therefore, coordinate-based meta-analysis makes it possible to use published peer-reviewed literature to conduct large scale, multi-laboratory, multi-contrast, and multi-study meta-analysis, as was done presently.
MACM is a tool that takes specific advantage of large volume of data from multiple labs, contrasts, subjects, and paradigms. Although the MACM technique has existed since Laird et al. (2009a), its use has been limited to the study functional covariance. In recent years, it has been used to study functional covariance of specific brain regions such as the caudate (Robinson et al., 2012), nucleus accumbens (Cauda et al., 2011), insula (Cauda et al., 2012), left inferior parietal cortex (Müller et al., 2013), and amygdala (Robinson et al., 2010); specific brain processes such as emotion regulation (Kohn et al., 2014); and specific diseases such as spinocerebellar ataxia (Reetz et al., 2012), MTLE (Barron et al., 2013) and PTSD (Ramage et al., 2012) among others. To our knowledge, this study is the first to use BrainMap's VBM database to directly compare structural covariance with functional covariance using the same methodology.
Our study takes advantage of seven years of new and refined data uploaded into BrainMap since Robinson et al. (2010), providing greater statistical power to MACM analyses. We applied inclusion filters for whole-brain analyses and data non-redundancy. We conducted a RtWB meta-analysis by seeding the hippocampus and searching through the entire BrainMap VBM database to identify the most significant extra and intra-hippocampal foci associated with change in hippocampal gray matter density (Table 1 & Fig. 3). We used the Eickhoff et al. (2012) algorithm, which employs a data-driven approach to create the null-distribution used in statistical inference as well as a Monte-Carlo based approach permitting more accurate cluster-level and voxel-wise inferencing (Eickhoff et al., 2012). We then performed two separate MACM analyses (Table 2, Table 3, Table 4, Table 5, Fig. 4, Fig. 5), following the most up-to-date approach as delineated in Langner et al. (2014) with minor improvements to account for differences in VBM and functional data (Fig. 2). These improvements to the meta-analytic process allows for a more statistically robust method of performing MACM analyses to determine both structural and functional covariance using coordinate-based meta-analytic methods. Using this method, we have also demonstrated that indeed, structural covariance and functional covariance are significantly correlated (Table 6). Future studies will no doubt benefit from using coordinate-based databases such as BrainMap to perform MACM analyses to derive preliminary biomarkers and network models as a strategy to inform studies using clinically-relevant resting state and T1 primary data (Biswal et al., 2010).
4.4. Clinical purpose and dividends
Behavioral characteristics (Fig. 6) have been shown to correspond to hippocampal function and dysfunction based on clinical behavior profiles of individuals with diseases of the hippocampus such as in Alzheimer's disease (Ballard et al., 2011) and schizophrenia (Tregellas et al., 2014). The paradigm class activation studies identified in the sHNM also validates the hippocampus' role in the reward circuit (Haber and Knutson, 2010) and its involvement in other tasks involving memory and attention (pain monitor-discrimination, face monitor-discrimination, semantic monitor-discrimination, and passive viewing). Furthermore, behavior characteristics are important features in the evaluation and analysis of individuals with hippocampal pathology.
Future analyses will aim to apply the sHNM to both structural (T1 weighted images) and functional (resting state fMRI) primary data and use statistical methods such as structural equation modeling, to define path coefficients with greater accuracy. We further hypothesize that structural equation modeling applied to primary data and restricted to nodes of the sHNM will increase our understanding of hippocampal pathology and introduce a novel approach to assessing its relationship to cognition, behavior, metabolism, and aging. We believe that a targeted analysis of the sHNM with primary data could subsequently bear prospective clinical dividends on the per-patient level by giving investigators and clinicians an objective anatomically derived method of assessing cognitive decline and improvement as it relates to disease progression and treatment.
5. Conclusion
In our region-to-whole brain meta-analysis of the hippocampi, we found consistent gray matter structural change across diseases in BrainMap's VBM database in 11 specific brain regions, termed the sHNM. These structures were found to show distinct patterns of structural and functional covariance using meta-analytic connectivity modeling. Functional and structural ALE values derived from MACM were found to be significantly correlated. Likewise, a paradigm analysis of behavior domains and paradigm class activation studies show that the sHNM corresponds to known hippocampal function in normal subjects (emotion, memory, attention) and known hippocampal atrophy in diseases subjects (AD, MCI, MTLE, schizophrenia). Our large sample size of VBM studies allowed us to use more conservative statistical thresholding in reporting the most significant ALE values in our structural and functional covariance studies. Hippocampal pathology is commonly observed in animals and humans, suggesting that a comprehensive structural hippocampal network model could serve as a clinically useful biomarker in the evaluation disease effects on cognitive function. These questions await further investigations.
The following are the supplementary data related to this article.
Hippocampal Network Model: Structural MACM BrainMap Workspace.
Temporal Lobe Epilepsy Hippocampal Network Structural MACM BrainMap Workspace.
Schizophrenia Hippocampal Network Structural MACM BrainMap Workspace.
Alzheimer's Disease Hippocampal Network Structural MACM BrainMap Workspace.
Mild Cognitive Impairment Hippocampal Network Structural MACM BrainMap Workspace.
Hippocampal Network Model Masks.
Supplementary Table 1
BrainMap's VBM database arranged by ICD-10 codes, total number of papers per codable disease, total number of experiments per codable disease, and total number of foci reported per codable disease.
Acknowledgments
We thank Dr. Amy Garrett for her valuable edits and Cheasequah J. Blevins for her neuroscience input and emotional support. This study was supported by funds from the National Institute of Health (NIMH: R01 MH074457, NIMH: 3R01MH074457-11S1) and Congressionally Directed Medical Research Programs (CDMRP: W81xWH-14-1-0316).
References
- Ahmed R.M., Devenney E.M., Irish M., Ittner A., Naismith S., Ittner L.M., Rohrer J.D., Halliday G.M., Eisen A., Hodges J.R., Kiernan M.C. Neuronal network disintegration: common pathways linking neurodegenerative diseases. J. Neurol. Neurosurg. Psychiatry. 2016;87(11):1234–1241. doi: 10.1136/jnnp-2014-308350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A., Giedd J.N., Bullmore E. Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci. 2013;14(5):322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K.S., Dhikav V. Hippocampus in health and disease: an overview. Ann. Indian Acad. Neurol. 2012;15(4):239. doi: 10.4103/0972-2327.104323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Hwang K.S., Andrawis J.P., Green A.E., Babakchanian S., Morra J.H. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol. Aging. 2010;31(8):1284–1303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Green A.E., Babakchanian S., Hwang K.S., Chou Y.-Y., Toga A.W., Thompson P.M. Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment (MCI), and Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2012;26(1):17–27. doi: 10.1097/WAD.0b013e3182163b62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Voxel-based morphometry—the methods. NeuroImage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ballard C., Gauthier S., Corbett A., Brayne C., Aarsland D., Jones E. Alzheimer's disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- Barron D.S., Fox P.M., Laird A.R., Robinson J.L., Fox P.T. Thalamic medial dorsal nucleus atrophy in medial temporal lobe epilepsy: a VBM meta-analysis. NeuroImage. 2013;2(C):25–32. doi: 10.1016/j.nicl.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron D.S., Eickhoff S.B., Clos M., Fox P.T. Human pulvinar functional organization and connectivity. Hum. Brain Mapp. 2015;36(7):2417–2431. doi: 10.1002/hbm.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B.B., Mennes M., Zuo X.N., Gohel S., Kelly C., Smith S.M. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. 2010;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler S.L., Menon V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn. Sci. 2010;14(6):277–290. doi: 10.1016/j.tics.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Bzdok D., Yeo B.T.T. Inference in the age of big data: future perspectives on neuroscience. NeuroImage. 2017;155:549–564. doi: 10.1016/j.neuroimage.2017.04.061. [DOI] [PubMed] [Google Scholar]
- Bzdok D., Laird A.R., Zilles K., Fox P.T., Eickhoff S.B. An investigation of the structural, connectional, and functional subspecialization in the human amygdala. Hum. Brain Mapp. 2013;34:3247–3266. doi: 10.1002/hbm.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauda F., Cavanna A.E., D'Agata F., Sacco K., Duca S., Geminiani G.C. Functional connectivity and coactivation of the nucleus accumbens: a combined functional connectivity and structure-based meta-analysis. J. Cogn. Neurosci. 2011;23(10):2864–2877. doi: 10.1162/jocn.2011.21624. [DOI] [PubMed] [Google Scholar]
- Cauda F., Costa T., Torta D.M.E., Sacco K., D'Agata F., Duca S. Meta-analytic clustering of the insular cortex. NeuroImage. 2012;62(1):343–355. doi: 10.1016/j.neuroimage.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-T., Huang C.-W., Chang Y.-H., Chen N.-C., Lin K.-J., Yan T.-C. Amyloid burden in the hippocampus and default mode network. Medicine. 2015;94(16):e763–8. doi: 10.1097/MD.0000000000000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase H.W., Clos M., Dibble S., Fox P., Grace A.A., Phillips M.L., Eickhoff S.B. Evidence for an anterior–posterior differentiation in the human hippocampal formation revealed by meta-analytic parcellation of fMRI coordinate maps: focus on the subiculum. NeuroImage. 2015;113:44–60. doi: 10.1016/j.neuroimage.2015.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioli C., Abdi H., Beaton D., Burnod Y., Mesmoudi S. Differences in human cortical gene expression match the temporal properties of large-scale functional networks. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Lawrence Erlbaum Associates; Hove: 1988. Statistical Power Analysis for the Behavioral Sciences. [Google Scholar]
- Cohen-Cory S., Kidane A.H., Shirkey N.J., Marshak S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Dev. Neurobiol. 2010;85 doi: 10.1002/dneu.20774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Vértes P.E., Winton-Brown T.T., Patel A.X., Ginestet C.E. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc. Natl. Acad. Sci. 2013;110(38):15502. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Scott J., Carletti F., Fox P.T., McGuire P., Bullmore E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137(8):2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Scott J., Ellison-Wright I., Mechelli A. Neuroimaging distinction between neurological and psychiatric disorders. Br. J. Psychiatry. 2015;207(5):429–434. doi: 10.1192/bjp.bp.114.154393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Fox P.T., Bullmore E.T. Meta-connectomics: human brain network and connectivity meta-analysis. Psychol. Med. 2016;46(5):897–907. doi: 10.1017/S0033291715002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S.B., Laird A.R., Grefkes C., Wang L.E., Zilles K., Fox P.T. Coordinate-based activation likelihood estimation meta-analysis of neuroimaging data: a random-effects approach based on empirical estimates of spatial uncertainty. Hum. Brain Mapp. 2009;30(9):2907–2926. doi: 10.1002/hbm.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S.B., Jbabdi S., Caspers S., Laird A.R., Fox P.T., Zilles K., Behrens T.E.J. Anatomical and functional connectivity of cytoarchitectonic areas within the human parietal operculum. J. Neurosci. 2010;30(18):6409–6421. doi: 10.1523/JNEUROSCI.5664-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S.B., Nickl-Jockschat T., Kurth F. Meta-analyses in clinical brain research. Nervenarzt. 2010;81(1):32–38. doi: 10.1007/s00115-009-2826-x. [DOI] [PubMed] [Google Scholar]
- Eickhoff S.B., Bzdok D., Laird A.R., Kurth F., Fox P.T. Activation likelihood estimation meta-analysis revisited. NeuroImage. 2012;59(3):2349–2361. doi: 10.1016/j.neuroimage.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S.B., Nichols T.E., Laird A.R., Hoffstaedter F., Amunts K., Fox P.T., Bzdok D., Eickhoff C.R. Behavior, sensitivity, and power of activation likelihood estimation characterized by massive empirical simulation. NeuroImage. 2016;137:70–85. doi: 10.1016/j.neuroimage.2016.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A., Cuthbert B. Beyond the DSM: development of a transdiagnostic psychiatric neuroscience course. Acad. Psychiatry. 2014;38(2):145–150. doi: 10.1007/s40596-013-0032-4. [DOI] [PubMed] [Google Scholar]
- Fornai F., Soldani P., Lazzeri G., Bandettini di Poggio A., Biagioni F., Fulceri F., Batini S., Ruggieri S., Paparelli A. Neuronal inclusions in degenerative disorders, do they represent static features or a key to understand the dynamics of the disease? Brain Res. Bull. 2005;65(4):275–290. doi: 10.1016/j.brainresbull.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Fornito A., Zalesky A., Breakspear M. The connectomics of brain disorders. Nat. Rev. Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- Fox P.T., Lancaster J.L. Mapping context and content: the BrainMap model. Nat. Rev. Neurosci. 2002;3:319–321. doi: 10.1038/nrn789. [DOI] [PubMed] [Google Scholar]
- Fox M.D., Raichle M.E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007;8(9):700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox P.T., Mikiten S., Davis G., Lancaster J.L. BrainMap: a database of human functional brain mapping. In: Thatcher R.W., Hallet M., Zeffiro T., John E.R., Huerta M., editors. Functional Neuroimaging. Academic Press; San Diego: 1994. [Google Scholar]
- Fox P.T., Laird A.R., Lancaster J.L. Coordinate-based voxel-wise meta-analysis: dividends of spatial normalization. Report of a virtual workshop. Hum. Brain Mapp. 2005;25(1):1–5. doi: 10.1002/hbm.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P.T., Laird A.R., Fox S.P., Fox P.M., Uecker A.M., Crank M. Brainmap taxonomy of experimental design: description and evaluation. Hum. Brain Mapp. 2005;25(1):185–198. doi: 10.1002/hbm.20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P.T., Lancaster J.L., Laird A.R., Eickhoff S.B. Meta-analysis in human neuroimaging: computational modeling of large-scale databases. Annu. Rev. Neurosci. 2014;37(1):409–434. doi: 10.1146/annurev-neuro-062012-170320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D.C., Laird A.R., Ellison-Wright I., Thelen S.M., Robinson J.L., Lancaster J.L. Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol. Psychiatry. 2008;64(9):774–781. doi: 10.1016/j.biopsych.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G., He Y., Chen Z.J., Evans A.C. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. NeuroImage. 2012;59(2):1239–1248. doi: 10.1016/j.neuroimage.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Goodkind M., Eickhoff S.B., Oathes D.J., Jiang Y., Chang A., Jones-Hagata L.B. Identification of a common neurobiological substrate for mental illness. JAMA Psychiat. 2015;72(4):305–323. doi: 10.1001/jamapsychiatry.2014.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber S.N., Knutson B. The reward circuit: linking primate anatomy and human imaging. Neuropsychopharmacology. 2010;35(1):4–26. doi: 10.1038/npp.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers W., Mormino E.C., Schultz A.P., Wigman S., Ward A.M., Larvie M. Amyloid-β deposition in mild cognitive impairment is associated with increased hippocampal activity, atrophy and clinical progression. Brain. 2015;138(4):1023–1035. doi: 10.1093/brain/awv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T., Cuthbert B., Garvey M., Heinssen R., Pine D.S., Quinn K. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatr. 2010;167(7):748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E.J., Woolrich M.W., Smith S.M. FSL. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jiang L., Xu T., He Y., Hou X.-H., Wang J., Cao X.-Y. Toward neurobiological characterization of functional homogeneity in the human cortex: regional variation, morphological association and functional covariance network organization. Brain Struct. Funct. 2015:1–23. doi: 10.1007/s00429-014-0795-8. [DOI] [PubMed] [Google Scholar]
- Keller S.S., Roberts N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia. 2008;49(5):741–757. doi: 10.1111/j.1528-1167.2007.01485.x. [DOI] [PubMed] [Google Scholar]
- Kohn N., Eickhoff S.B., Scheller M., Laird A.R., Fox P.T., Habel U. Neural network of cognitive emotion regulationean ALE meta-analysis and MACM analysis. Neuroimage. 2014;87 doi: 10.1016/j.neuroimage.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolschijn P.C.M.P., van Haren N.E.M., Cahn W., Schnack H.G., Janssen J., Klumpers F. Hippocampal volume change in schizophrenia. J. Clin. Psychiatry. 2010;71(06):737–744. doi: 10.4088/JCP.08m04574yel. [DOI] [PubMed] [Google Scholar]
- Laird A.R., Fox P.M., Price C.J., Glahn D.C., Uecker A.M., Lancaster J.L. ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Hum. Brain Mapp. 2005;25(1):155–164. doi: 10.1002/hbm.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A.R., Eickhoff S.B., Li K., Robin D.A., Glahn D.C., Fox P.T. Investigating the functional heterogeneity of the default mode network using coordinate-based meta-analytic modeling. J. Neurosci. 2009;29(46):14496–14505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A.R., Eickhoff S.B., Kurth F., Fox P.M., Uecker A.M., Turner J.A. ALE meta-analysis workflows via the BrainMap database: progress towards a probabilistic functional brain atlas. Front. Neuroinform. 2009;3:1–11. doi: 10.3389/neuro.11.023.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A.R., Fox P.M., Eickhoff S.B., Turner J.A., Ray K.L., McKay D.R. Behavioral interpretations of intrinsic connectivity networks. J. Cogn. Neurosci. 2012;23(12):4022–4037. doi: 10.1162/jocn_a_00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A.R., Eickhoff S.B., Rottschy C., Bzdok D., Ray K.L., Fox P.T. Networks of task co-activations. NeuroImage. 2013;80(C):505–514. doi: 10.1016/j.neuroimage.2013.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J.L., Woldorff M.G., Parsons L.M., Liotti M., Freitas C.S., Rainey L. Automated Talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J.L., Laird A.R., Eickhoff S.B., Martinez M.J., Fox M.P., Fox P.T. Automated regional behavioral analysis for human brain images. Front. Neuroinform. 2012;6(23):1–12. doi: 10.3389/fninf.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner R., Rottschy C., Laird A.R., Fox P.T., Eickhoff S.B. Meta-analytic connectivity modeling revisited: controlling for activation base rates. NeuroImage. 2014;99:559–570. doi: 10.1016/j.neuroimage.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort-Besnard J., Bassett D.S., Smallwood J., Margulies D.S., Derntl B., Gruber O., Aleman A., Jardri R., Varoquaux G., Thirion B., Eickhoff S.B., Bzdok D. Different shades of default mode disturbance in schizophrenia: subnodal covariance estimation in structure and function. Hum. Brain Mapp. 2018;39(2):644–661. doi: 10.1002/hbm.23870. (Epub 2017 Nov 3. PubMed ID: 29105239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch J.P., Worsley K., Shaw W.P., Greenstein D.K., Lenroot R.K., Giedd J., Evans A.C. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. NeuroImage. 2006;31(3):993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Li Y., Li W.-X., Xie D.-J., Wang Y., Cheung E.F.C., Chan R.C.K. Grey matter reduction in the caudate nucleus in patients with persistent negative symptoms: an ALE meta-analysis. Schizophr. Res. 2017:1–7. doi: 10.1016/j.schres.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Lin P., Yang Y., Jovicich J., De Pisapia N., Wang X., Zuo C.S., Levitt J.J. Static and dynamic posterior cingulate cortex nodal topology of default mode network predicts attention task performance. Brain Imaging Behav. 2015;10(1):212–225. doi: 10.1007/s11682-015-9384-6. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and generalized regression approach. Cancer Res. 1967;27(2):209–220. [PubMed] [Google Scholar]
- Marstaller L., Williams M., Rich A., Savage G., Burianová H. Aging and large-scale functional networks: white matter integrity, gray matter volume, and functional connectivity in the resting state. Neuroscience. 2015;290(C):369–378. doi: 10.1016/j.neuroscience.2015.01.049. [DOI] [PubMed] [Google Scholar]
- Mathur B.N. The claustrum in review. Front. Syst. Neurosci. 2014:1–11. doi: 10.3389/fnsys.2014.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTeague L.M., Goodkind M.S., Etkin A. Transdiagnostic impairment of cognitive control in mental illness. J. Psychiatr. Res. 2016;83(C):37–46. doi: 10.1016/j.jpsychires.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne A., Barnes S.R., Sweeney M.D., Halliday M.R., Sagare A.P., Zhao Z. Blood-brain barrier breakdown in the aging human hippocampus. Neuron. 2015;85(2):296–302. doi: 10.1016/j.neuron.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montembeault M., Rouleau I., Provost J.-S., Brambati S.M. Altered gray matter structural covariance networks in early stages of Alzheimer's disease. Cereb. Cortex. 2016;26(6):2650–2662. doi: 10.1093/cercor/bhv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V.I., Cieslik E.C., Laird A.R., Fox P.T., Eickhoff S.B. Dysregulated left inferior parietal activity in schizophrenia and depression: functional connectivity and characterization. Front. Hum. Neurosci. 2013:1–11. doi: 10.3389/fnhum.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumoli L., Labate A., Vasta R., Cherubini A., Ferlazzo E., Aguglia U. Detection of hippocampal atrophy in patients with temporal lobe epilepsy: a 3-Tesla MRI shape. Epilepsy Behav. 2013;28(3):489–493. doi: 10.1016/j.yebeh.2013.05.035. [DOI] [PubMed] [Google Scholar]
- Park B.-Y., Seo J., Yi J., Park H. Structural and functional brain connectivity of people with obesity and prediction of body mass index using connectivity. PLoS One. 2015;10(11):e0141376–14. doi: 10.1371/journal.pone.0141376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu E., Ganeshan B., Shergill S.S., Medford N., Chatwin C., Young R.C.D., Critchley H.D. Grey-matter texture abnormalities and reduced hippocampal volume are distinguishing features of schizophrenia. Psychiatry Res. Neuroimaging. 2014;223(3):179–186. doi: 10.1016/j.pscychresns.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Ramage A.E., Laird A.R., Eickhoff S.B., Acheson A., Peterson A.L., Williamson D.E. A coordinate-based meta-analytic model of trauma processing in posttraumatic stress disorder. Hum. Brain Mapp. 2012;34(12):3392–3399. doi: 10.1002/hbm.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetz K., Dogan I., Rolfs A., Binkofski F., Schulz J.B., Laird A.R. Investigating function and connectivity of morphometric findings — exemplified on cerebellar atrophy in spinocerebellar ataxia 17 (SCA17) NeuroImage. 2012;62(3):1354–1366. doi: 10.1016/j.neuroimage.2012.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A.T., Hoffstaedter F., Gong G., Laird A.R., Fox P., Evans A.C. A seed-based cross-modal comparison of brain connectivity measures. Brain Struct. Funct. 2016;222(3):1131–1151. doi: 10.1007/s00429-016-1264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.L., Laird A.R., Glahn D.C., Lovallo W.R., Fox P.T. Metaanalytic connectivity modeling: delineating the functional connectivity of the human amygdala. Hum. Brain Mapp. 2010;9(3, Part 1) doi: 10.1002/hbm.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.L., Laird A.R., Glahn D.C., Blangero J., Sanghera M.K., Pessoa L. The functional connectivity of the human caudate: an application of meta-analytic connectivity modeling with behavioral filtering. NeuroImage. 2012;60(1):117–129. doi: 10.1016/j.neuroimage.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.L., Barron D.S., Kirby L.A.J., Bottenhorn K.L., Hill A.C., Murphy J.E. Neurofunctional topography of the human hippocampus. Hum. Brain Mapp. 2015;36(12):5018–5037. doi: 10.1002/hbm.22987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg D.S., Mauguiere F., Catenoix H., Faillenot I., Magnin M. Reciprocal thalamocortical connectivity of the medial pulvinar: a depth stimulation and evoked potential study in human brain. Cereb. Cortex. 2009;19(6):1462–1473. doi: 10.1093/cercor/bhn185. [DOI] [PubMed] [Google Scholar]
- Schröder J., Pantel J. Neuroimaging of hippocampal atrophy in early recognition of Alzheimer's disease – a critical appraisal after two decades of research. Psychiatry Res. Neuroimaging. 2016;247(c):71–78. doi: 10.1016/j.pscychresns.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Seeley W.W., Crawford R.K., Zhou J., Miller B.L., Greicius M.D. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62(1):42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Fox P.T., Miller K.L., Glahn D.C., Fox P.M., Mackay C.E. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Vidaurre D., Beckmann C.F., Glasser M.F., Jenkinson M., Miller K.L. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 2013;17(12):666–682. doi: 10.1016/j.tics.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S.S., Williamson P.D., Spencer D.D., Mattson R.H. Human hippocampal seizure spread studied by depth and subdural recording: the hippocampal commissure. Epilepsia. 1987;28:479–489. doi: 10.1111/j.1528-1157.1987.tb03676.x. [DOI] [PubMed] [Google Scholar]
- Spreng R.N., Turner G.R. Structural covariance of the default network in healthy and pathological aging. J. Neurosci. 2013;33(38):15226–15234. doi: 10.1523/JNEUROSCI.2261-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranahan A.M. Models and mechanisms for hippocampal dysfunction in obesity and diabetes. Neuroscience. 2015;309:125–139. doi: 10.1016/j.neuroscience.2015.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Huster R., Yu Q., Segall J.M., Calhoun V.D. Function–structure associations of the brain: evidence from multimodal connectivity and covariance studies. NeuroImage. 2014;102:11–23. doi: 10.1016/j.neuroimage.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregellas J.R., Smucny J., Harris J.G., Olincy A., Maharajh K., Kronberg E. Intrinsic hippocampal activity as a biomarker for cognition and symptoms in schizophrenia. Am. J. Psychiatr. 2014;171(5):549–556. doi: 10.1176/appi.ajp.2013.13070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub P.E., Eickhoff S.B., Laird A.R., Fox M., Wiener M., Fox P. Minimizing within-experiment and within-group effects in activation likelihood estimation meta-analyses. Hum. Brain Mapp. 2011;33(1):1–13. doi: 10.1002/hbm.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes R.P., Hoover W.B., Szigeti-Buck K., Leranth C. Nucleus reuniens of the midline thalamus: link between the medial prefrontal cortex and the hippocampus. Brain Res. Bull. 2007;71(6):601–609. doi: 10.1016/j.brainresbull.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Fu K., Liu H., Xing F., Zhang S. Brain structural changes and their correlation with vascular disease in type 2 diabetes mellitus patients: a voxel-based morphometric study. Neural Regen. Res. 2014;9(16):1548–1549. doi: 10.4103/1673-5374.139482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Zhang Z., Xu Q., Yang F., Sun K., Lu G. More severe extratemporal damages in mesial temporal lobe epilepsy with hippocampal sclerosis than that with other lesions. Medicine. 2016;95(10):e3020–9. doi: 10.1097/MD.0000000000003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Brickman A.M., Luchsinger J., Ferrazzano P., Pichiule P., Yoshita M. The brain in the age of old: the hippocampal formation is targeted differentially by diseases of late life. Ann. Neurol. 2008;64(6):698–706. doi: 10.1002/ana.21557. (York, N.Y.: 1991) 19, 1462–1473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gennatas E.D., Kramer J.H., Miller B.L., Seeley W.W. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hippocampal Network Model: Structural MACM BrainMap Workspace.
Temporal Lobe Epilepsy Hippocampal Network Structural MACM BrainMap Workspace.
Schizophrenia Hippocampal Network Structural MACM BrainMap Workspace.
Alzheimer's Disease Hippocampal Network Structural MACM BrainMap Workspace.
Mild Cognitive Impairment Hippocampal Network Structural MACM BrainMap Workspace.
Hippocampal Network Model Masks.
Supplementary Table 1
BrainMap's VBM database arranged by ICD-10 codes, total number of papers per codable disease, total number of experiments per codable disease, and total number of foci reported per codable disease.