CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2 (original) (raw)
Abstract
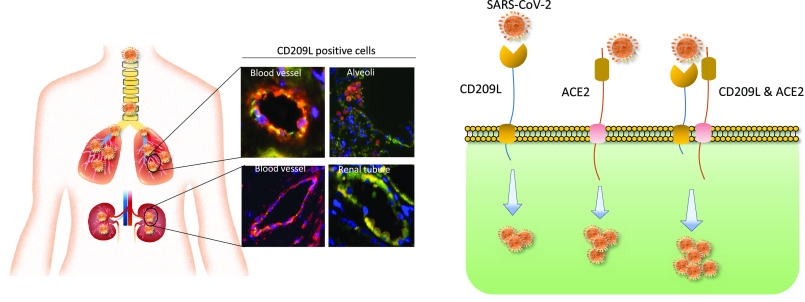
As the COVID-19 pandemic continues to spread, investigating the processes underlying the interactions between SARS-CoV-2 and its hosts is of high importance. Here, we report the identification of CD209L/L-SIGN and the related protein CD209/DC-SIGN as receptors capable of mediating SARS-CoV-2 entry into human cells. Immunofluorescence staining of human tissues revealed prominent expression of CD209L in the lung and kidney epithelia and endothelia. Multiple biochemical assays using a purified recombinant SARS-CoV-2 spike receptor-binding domain (S-RBD) or S1 encompassing both N termal domain and RBD and ectopically expressed CD209L and CD209 revealed that CD209L and CD209 interact with S-RBD. CD209L contains two _N_-glycosylation sequons, at sites N92 and N361, but we determined that only site N92 is occupied. Removal of the _N_-glycosylation at this site enhances the binding of S-RBD with CD209L. CD209L also interacts with ACE2, suggesting a role for heterodimerization of CD209L and ACE2 in SARS-CoV-2 entry and infection in cell types where both are present. Furthermore, we demonstrate that human endothelial cells are permissive to SARS-CoV-2 infection, and interference with CD209L activity by a knockdown strategy or with soluble CD209L inhibits virus entry. Our observations demonstrate that CD209L and CD209 serve as alternative receptors for SARS-CoV-2 in disease-relevant cell types, including the vascular system. This property is particularly important in tissues where ACE2 has low expression or is absent and may have implications for antiviral drug development.
Short abstract
In human endothelial cells, CD209L acts as a receptor for SARS-CoV-2; together with ACE2, it can function as a co-receptor. Blocking CD209L activity inhibited virus entry, indicating a novel target for development of antiviral drugs.
Introduction
The outbreak of coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), constitutes a serious ongoing threat to global public health and has generated a major worldwide socioeconomic impact.1−3 The morbidity and mortality of SARS-CoV-2 are associated with acute respiratory distress syndrome (ARDS) and other complications such as coagulopathy, thrombosis, and multiorgan failure in COVID-19 patients.3−6 Although the role of the vascular system, particularly endothelial cells, in the pathogenesis of COVID-19 remains largely unknown, emerging evidence suggests that SARS-CoV-2 directly attacks the vascular system.7−9 Severe endothelial injury, vascular thrombosis with microangiopathy, occlusion of alveolar capillaries, and angiogenesis were distinctively observed in lung autopsies of COVID-19 patients,8 underscoring the critical importance of the vasculature system in the pathogenesis of COVID-19.
Human angiotensin-converting enzyme 2 (ACE2) is known to interact with the surface spike (S) protein of SARS and also acts as an entry receptor for SARS-CoV-2.11−13 While it was previously reported that ACE2 is widely expressed in the lung, vascular system, and other organs,14 recent studies demonstrated that ACE2 is expressed at very low levels and only in a small subset of lung epithelial cells15 and low-to-undetectable levels in endothelial cells,16 suggesting that SARS-CoV-2 entry into and infection of certain human cells may be occurring via alternative receptors or a combination of multiple receptors and/or enhancers. Consistent with this idea, neuropilin receptors,17,18 CD147/basigin,19 and heparin sulfate20 are reported to facilitate SARS-CoV-2 entry. Neuropilin receptors are highly expressed in endothelial and neuronal cells and play major roles in vascular endothelial growth factor (VEGF)-dependent angiogenesis and semaphorin-dependent axon guidance.21 CD147 is expressed in erythrocytes22,23 and endothelial cells of the brain and acts as a receptor for plasmodium.22,24 In addition to ACE2, alternative receptors that function as points of entry have been reported for other coronaviruses, such as human NL-63 and SARS-CoV.25 These include CD209L (also known as L-SIGN) and CD209 (also known as DC-SIGN).26−29 CD209L and CD209 are members of the C-type lectin superfamily and are implicated as mediators of viral pathogenesis.25,30 While CD209L is highly expressed in human type II alveolar cells and the endothelial cells of the lung, liver, and lymph nodes,31,32 CD209 is primarily expressed in dendritic cells and tissue-resident macrophages, including alveolar macrophages,33 dermal macrophages,34 and peripheral blood mononuclear cells.35,36 Despite their differential expression profiles, CD209L and CD209 share 79.5% amino acid sequence homology. The most distinguishing region of CD209L and CD209 is the C-type lectin domain (CRD), which functions as a calcium-dependent glycan-recognition domain.37 It is thought that the highly conserved EPN motif (Glu-Pro-Asn) on the CRD domain is responsible for the recognition of mannose-, fucose- or galactose-containing structures.38 However, despite a high degree of homology of the amino acid residues in the CRD of CD209L and CD209, there is evidence for differential recognition of oligosaccharide structures by these receptors. For example, CD209L appears to prefer high-mannose oligosaccharides but not complex glycans, especially those containing antennary fucose epitopes, such as LewisX (LeX), whereas CD209 binds to fucose and LeX.39,40
In this manuscript, we demonstrate that the receptor-binding domain (RBD) of the SARS-CoV-2 S protein binds to CD209L and CD209, mediating SARS-CoV-2 entry. CD209L is expressed in human endothelial cells and mediates endothelial cell adhesion, capillary tube formation, and sprouting. CD209L contains two sequons (NXT/S, X ≠ P) that provide the potential for _N_-linked glycosylation at sites N92 and N361. We determined that only the N92 site is occupied and that high-mannose glycans are present at this site. Removal of _N_-glycans from the cell surface enhanced the binding of S-RBD with CD209L. We further show that CD209L interacts with ACE2, in tissues where both are present, and thereby propose a role for heterodimerization of CD209L and ACE2 in virus entry and infection. These findings suggest that CD209L and CD209 represent novel potential therapeutic targets against COVID-19 and have implications for antiviral drug design.
Results
CD209L Is Expressed in Human Lung Epithelial and Endothelial Cells and Regulates the Angiogenic Properties of Endothelial Cells
To investigate the potential involvement of CD209L and CD209 in COVID-19, we first examined expression of CD209L in human tissues from SARS-CoV-2 target organs, which include lung endothelial and epithelial cells, renal vessels, tubules, glomeruli, and the temporal artery, via immunofluorescence staining. Lung tissue was costained with antibodies against CD209L and MUC1, the latter serving as a marker for type II alveolar cells,41,42 and was examined using laser microscopy. Isotope-labeled antibody served as a control and showed negligible staining of the lung tissue (S. Figure 1A). The specificity of CD209L antibody was validated in cell culture in HUVEC-TERT cells. Knockdown of CD209L eliminated the immunoreactivity of CD209L antibody in Western blotting (S. Figure 1B). Our results showed prominent expression of CD209L in the MUC1 positive alveolar cells (Figure 1A). We also observed expression of CD209L in pulmonary capillaries (Figure 1B), the endothelium of the small and medium sized temporal artery (Figure 1C), and renal arterioles (Figure 1D). CD31 served as a marker of endothelial cells (Figure 1B–D. Moreover, we show that CD209L is expressed in renal proximal tubular epithelial cells, which were marked by aquaporin 1 (S. Figure 2A). However, CD209L was not observed in the glomerular capillaries, and minimal expression was noted in the mesangial area, probably in the infiltrating immune cells (S. Figure 2B). The results demonstrate the prominent expression of CD209L in type II alveolar cells and pulmonary endothelium as well as renal vessels and renal tubular cells, which are also potential target cells of SARS-CoV-2.
Figure 1.
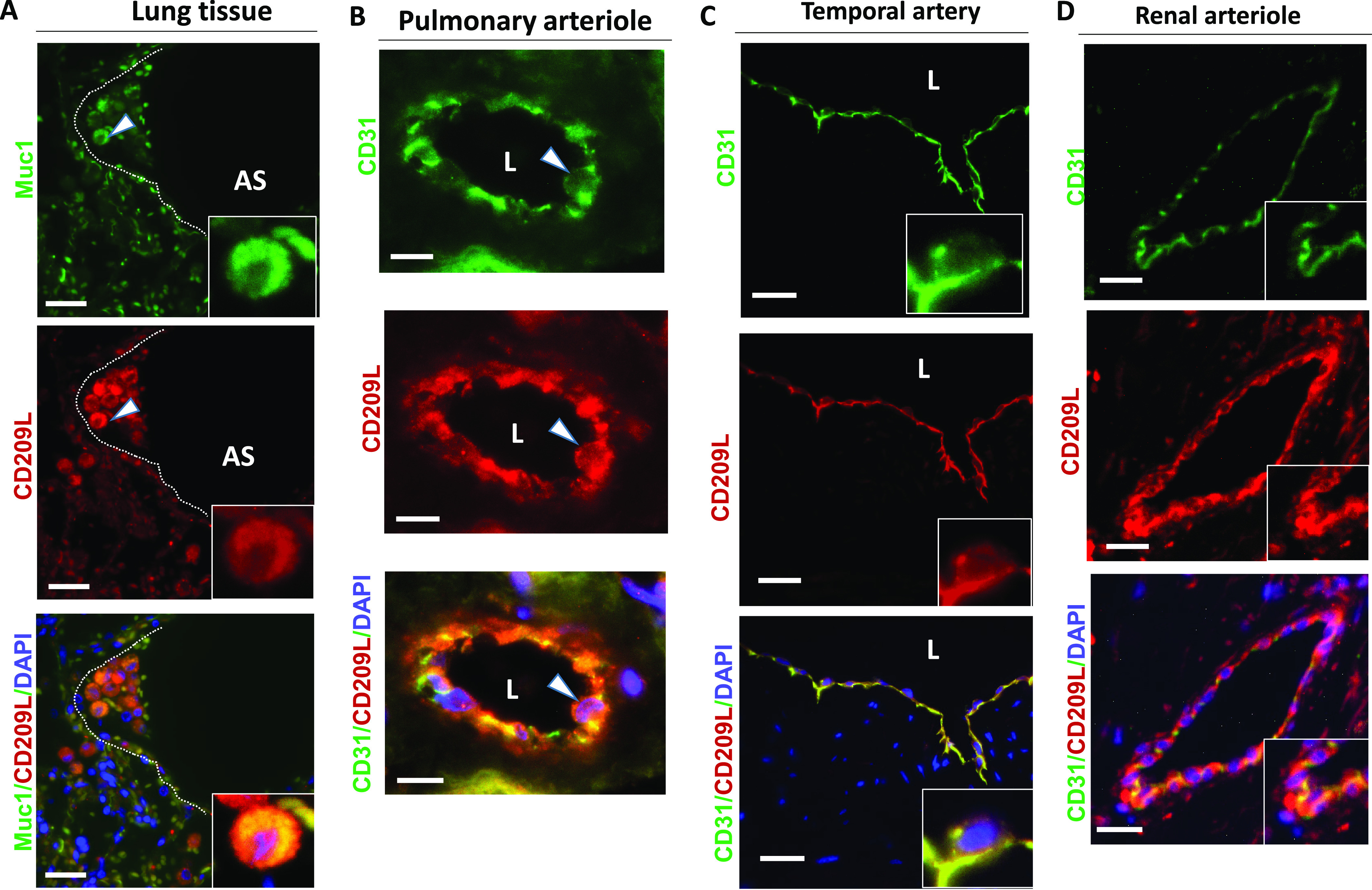
CD209L is expressed in lung as well as renal epithelial and endothelial cells: PFA fixed human lung, renal, and temporal arteriole tissues were subjected to immunofluorescence staining. Lung tissue stained with anti-MUC1, anti-CD31, and anti-CD209L antibodies. (A) Type II alveoli epithelial cells of alveoli were positive for CD209L (red) and MUC1 (green). (B) Pulmonary arteriole endothelial cells were positive for CD31 (green) and CD209L (red). (C) Endothelial cells of the temporal arteriole were stained with CD209L (green) and CD31 (red). (D) Renal endothelial cells were positive for CD209L (red) and CD31 (green). White arrowhead points to the alveolar cell, AS = alveolar space, and white dotted line corresponds to alveolar septa. Image magnification, white bars = 50 μm.
CD209 was expressed only in a subset of type MUC1 positive II alveolar cells (S. Figure 2A). Furthermore, unlike CD209L, which is highly expressed in endothelial cells (Figure 1C,D), we did not observe expression of CD209 in the pulmonary or renal arterioles (data not shown). A distinct expression of CD209 was observed in the limited proximal tubular epithelial cells in kidneys (S. Figure 3B), but glomerular capillaries were also mostly negative for CD209 (S. Figure 3C). Overall, these results suggest that CD209 has a limited expression profile compared to CD209L in the organs examined.
Considering that CD209L is highly expressed in endothelial cells, we examined the potential role of CD209L in regulation of the angiogenic responses of endothelial cells. First, we confirmed expression of CD209L in human umbilical endothelial cells immortalized with telomerase (HUVEC-TERT cells)43 by Western blot analysis (S. Figure 4A). However, expression of ACE2 in these cells was very low, and multiple weak protein bands potentially corresponding to ACE2 were only detected after long exposure times (S. Figure 4B). Additionally, we compared ACE2 expression in HUVEC-TERT cells to lung carcinoma cell lines, A549 and H1299 cells. ACE2 was detected in the cell lysates of A549 and HT1299 but not in the HUVEC-TERT cells (S. Figure 4B). To address the functional importance of CD209L in endothelial cells, we knocked down CD209L via an shRNA strategy (S. Figure 4C) and tested key angiogenic characteristics of endothelial cells, including cell adhesion, capillary tube formation/in vitro angiogenesis, and cell migration. To determine the role of CD209L in cell adhesion, we generated a Myc-tagged soluble CD209L encompassing the ectodomain of CD209L (sCD209L) (Figure 2). We coated 24-well plates with sCD209L and incubated HUVEC-TERT cells expressing control shRNA or CD209L-shRNA. After 30 min of incubation, unadhered cells were washed off the plates, cells were fixed, and the number of adherent cells was quantified. The result showed that depletion of CD209L in HUVEC-TERT cells significantly decreased cell adhesion (S. Figure 6A), suggesting that CD209L mediates endothelial cell–cell contact. Next, we subjected these cells to an in vitro angiogenesis assay. HUVEC-TERT cells expressing CD209L-shRNA displayed considerably reduced capillary tube formation compared to HUVEC-TERT cells expressing control shRNA (S. Figure 5B). Capillary tube formation is a complex dynamic process that involves cell–cell adhesion, cell proliferation, and cell migration. Therefore, we examined the effect of knockdown of CD209L in the migration of HUVEC-TERT cells. Knockdown of CD209L resulted in a robust increase in cell migration (S. Figure 5C). Given that remodeling of the actin cytoskeleton into filopodia, formation lamellipodia (i.e., cytoplasmic protrusions that contain a thick cortical network of actin filaments), and a stress fiber assembly play vital roles in endothelial cell migration,44 we investigated actin stress fiber formation by staining of the cells with phalloidin. Consistent with the observed effect of knockdown of CD209L in cell migration, silencing of CD209L also increased cytoplasmic protrusions at the leading edge of HUVEC-TERT cells (S. Figure 6). These data demonstrate that CD209L serves important roles in the regulation of angiogenic properties of endothelial cells.
Figure 2.
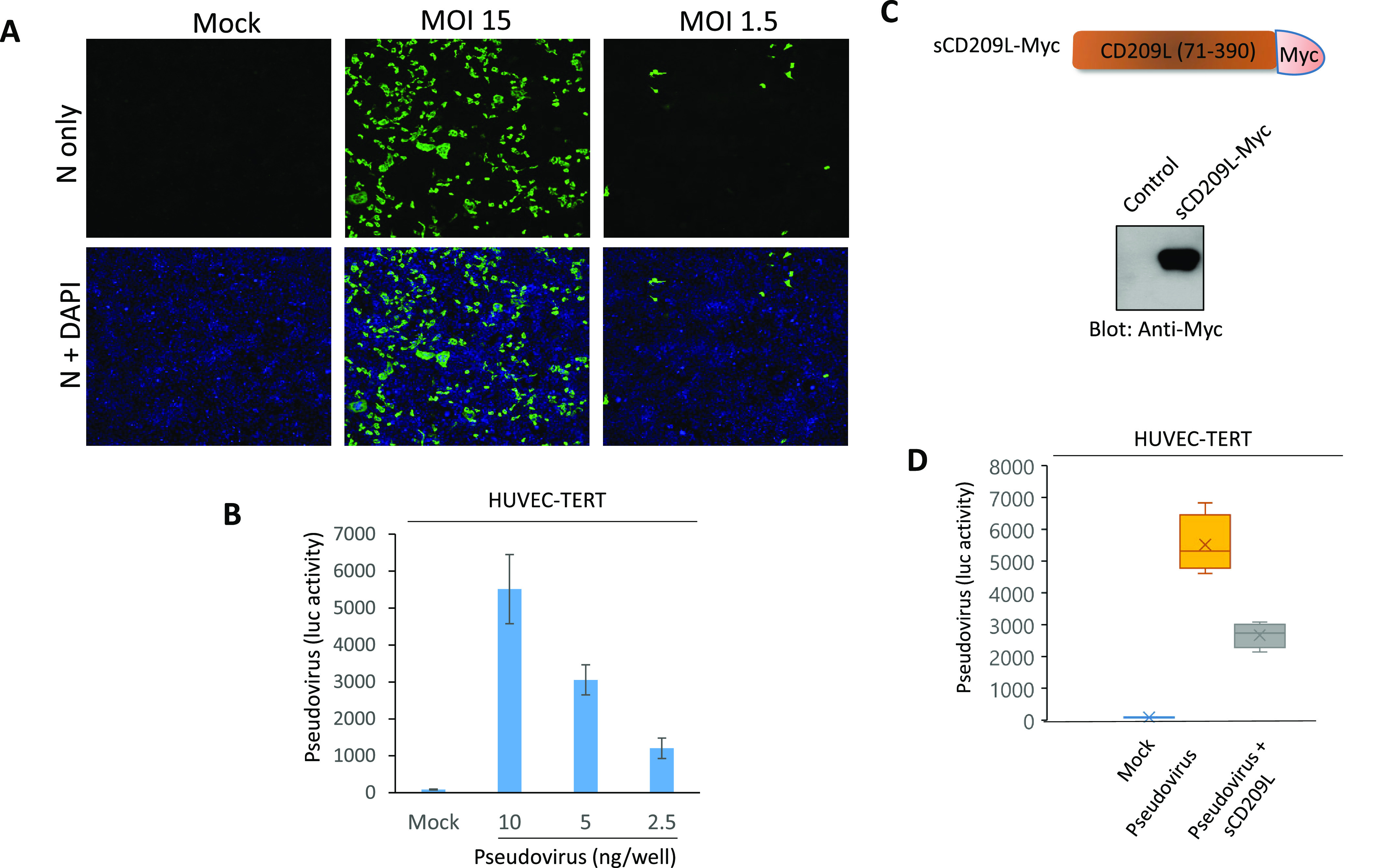
Endothelial cells are permissive to SARS-CoV-2 virus, and soluble CD209L neutralizes viral entry. (A) HUVEC-TERT cells seeded in chamber slides were mock-infected or infected with SARS-CoV-2 at the indicated MOIs. Cells were fixed at 1 day post infection and stained with an antibody directed against the viral nucleoprotein, N (green). Cell nuclei were stained with DAPI (blue). Image magnification 50 μm. (B) HUVEC-TERT cells (2 × 104/well, 96-well plate, and quadruple/group) were infected with pseudovirus with different concentrations. After 24 h, cells were processed and subjected to a luciferase activity assay, and representative data are shown. (C) Schematic of Myc-tagged soluble CD209 (sCD209L-Myc) and Western blot analysis of sCD209L-Myc. (D) HUVEC-TERT cells (2 × 104/well, 96-well plate, and triplicate/group) were infected with mock, pseudotyped virus (10 ng/mL) with control conditioned medium (CM) or CM containing sCD209L (approximate concentration 1.5 μg/mL). After 24 h, cells were analyzed for luciferase activity. P < 0.05.
CD209L Mediates SARS-CoV-2 Infection in Endothelial Cells
It has been proposed that the vascular system might be a direct target of SARS-CoV-2 infection.8 To explore if human endothelial cells are permissive to SARS-CoV-2, we infected HUVEC-TERT cells with SARS-CoV-2 at various multiplicities of infection (MOIs). A robust infection was observed at 1 day post infection, when the cells were infected with SARS-CoV-2 at a high MOI (MOI = 15). Even a lower MOI of 1.5 led to detectable infection levels at 1 day post infection, indicating that HUVEC-TERT cells are permissive to SARS-CoV-2 infection (Figure 2A). Moreover, we generated SARS-CoV-2 S-pseudotyped lentiviral particles and measured viral entry into HUVEC-TERT cells. SARS-CoV-2 S-pseudotyped lentiviral particles infected HUVEC-TERT cells in a concentration-dependent manner (Figure 2B). To investigate if CD209L expression in HUVEC-TERT cells promotes SARS-CoV-2 S-mediated entry, we carried out neutralization assays using a soluble form of CD209L (sCD209L) (Figure 2C). sCD209L reduced viral entry by 48% (Figure 2D). Our data demonstrate that HUVEC-TERT endothelial cells are permissive to SARS-CoV-2, and sCD209L reduces SARS-CoV-2 S-pseudotyped viral entry.
To further examine the role of CD209L in viral entry, we infected HUVEC-TERT cells, in which CD209L had been knocked down by shRNA, and parental control cells with SARS-CoV-2 S-pseudotyped lentiviral particles. The result showed that loss of CD209L in HUVEC-TERT cells markedly reduced viral entry (Figure 3A). These data were confirmed by SARS-CoV-2 infection studies. Immunofluorescence analysis using an antibody directed against the viral nucleocapsid (N) protein revealed that silencing of CD209L led to a substantial decrease of SARS-CoV-2 infection in HUVEC-TERT cells (Figure 3B). Next, we asked whether expression of ACE2 in HUVEC-TERT cells contributes to viral entry by CD209L; albeit, its expression in HUVEC-TERT cells is very low (S. Figure 4A). To address the relative contribution of ACE2 in SARS-CoV-2 entry in HUVEC-TERT cells, we knocked down ACE2 alone (S. Figure 7A) or CD209L alone by shRNA (S. Figure 1B) and subjected the cells to a viral entry assay. The result showed that, while depletion of CD209L significantly reduced viral entry, knockdown of ACE2 had only a minor effect on the viral entry (S. Figure 7B), indicating that viral entry in HUVEC-TERT cells is mainly mediated by CD209L.
Figure 3.
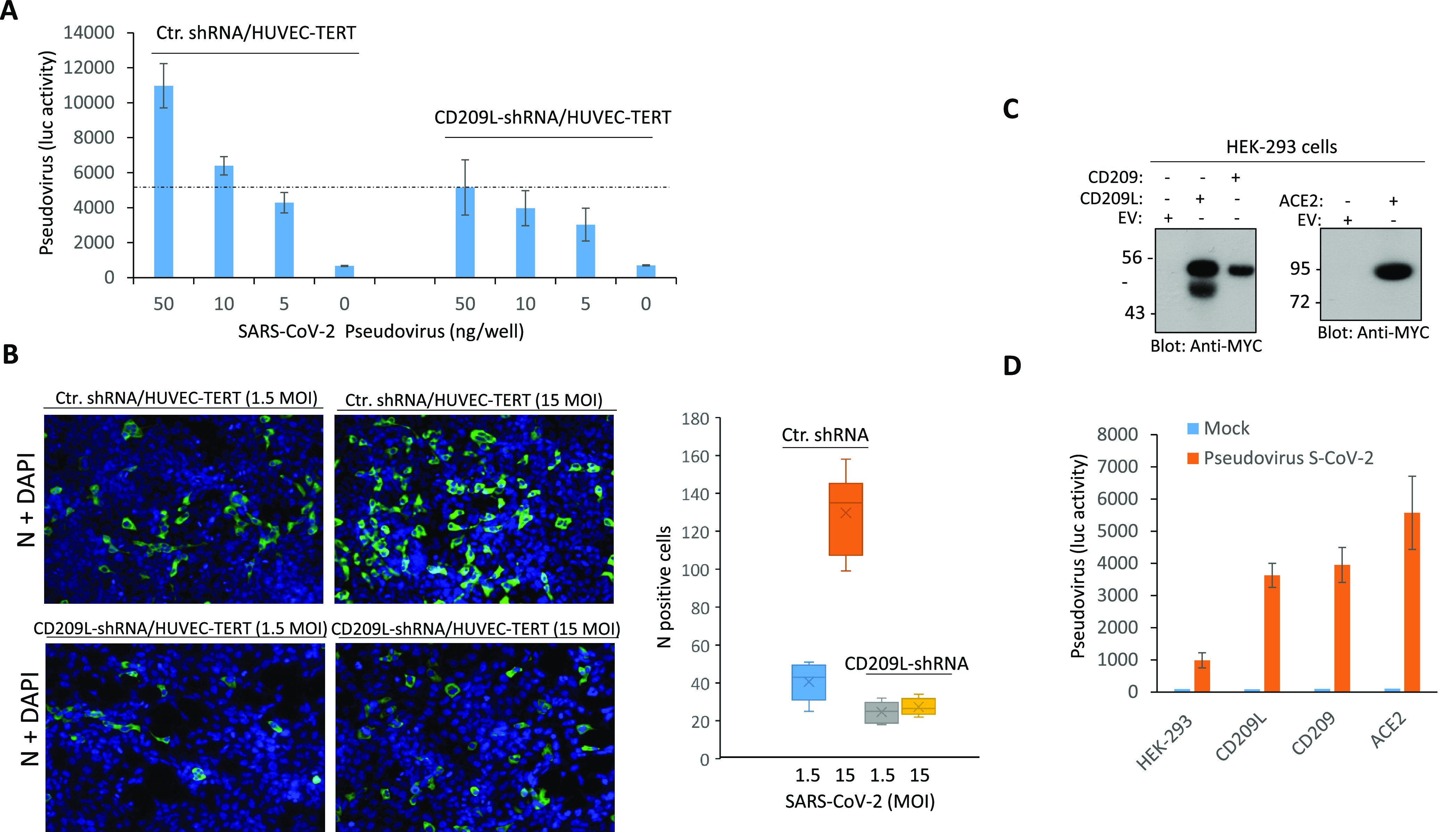
CD209L mediates SARS-CoV-2 entry and infection in endothelial cells. (A) HUVEC-TERT cells expressing control shRNA or CD209L-shRNA (2 × 104/well, 96-well plate, and triplicate/group) were infected with different amounts of SARS-CoV-2 pseudotyped lentivirus. After 24 h, cells were processed and subjected to luciferase activity, and representative data are shown. (B) HUVEC-TERT cells expressing control shRNA or CD209L-shRNA seeded in chamber slides (triplicate/group) were infected with SARS-CoV-2 at the indicated MOIs. Cells were fixed at 1 day post infection and stained with an antibody directed against the viral nucleoprotein, N (green). Cell nuclei were stained with DAPI (blue). Image magnification 50 μm. Quantification of N protein positive cells is shown. P < 0.05. (C) Western blot analysis showing expression of CD209L, CD209, and ACE2. (D) HEK-293 cells expressing CD209L, CD209, or ACE2 (2 × 104/well, 96-well plate, and quadruple wells/group) were infected with pseudotyped virus (10 ng/mL) or mock virus. After 24 h, cells were analyzed for luciferase activity. P < 0.05.
To further illustrate the role of CD209L and CD209 in viral entry compared to ACE2, we overexpressed CD209L, CD209, or ACE2 in HEK-293 cells (Figure 3C) and compared the individual roles of these receptors in SARS-CoV-2 S-mediated entry, using SARS-CoV-2 S-pseudotyped lentiviral particles. The results showed that both CD209L and CD209 were able to facilitate SARS-CoV-2 S-pseudotyped virus entry (Figure 3D). However, we consistently observed a higher S-pseudotyped virus entry in HEK-293 cells expressing ACE2 (Figure 3D). Altogether, our data demonstrate that CD2209L and CD209 can mediate SARS-CoV-2 entry, and thus, even tissues lacking ACE2 can serve as infection sites.
CD209L and CD209 Act as Receptors for SARS-CoV-2 Spike
Given that CD209L facilitated SARS-CoV-2 entry, we investigated whether SARS-CoV-2 can physically interact with CD209L. Specifically, we asked whether the RBD domain of SARS-CoV-2-S binds to CD209L. We used multiple binding assays to determine if SARS-CoV-2 S interacts with CD209L. First, we generated a chimeric soluble S-RBD-Fc-Myc (Figure 4A) and tested its binding with CD209L in an immunoprecipitation assay using whole cell lysate derived from HEK-293 cells ectopically expressing CD209L (Figure 4A). The result showed that S-RBD-Fc domain binds to CD209L (Figure 4B). An unrelated Fc-chimeric protein, Fc-TMIGD1, did not bind to CD209L (S. Figure 8A), indicating that the binding of S-RBD-Fc-Myc with CD209L is mediated by the S-RBD and not by the Fc protein. To exclude that the observed binding between CD209L and S-RBD is a potential artifact of the Fc chimera system, we created a purified recombinant HIS tag S-RBD protein (Figure 4C) and analyzed CD209L binding via far-Western blot analysis. The result showed that S-RBD-HIS strongly interacted with CD209L (Figure 4D). To further demonstrate the interaction of CD209L with S protein, we generated an S1-Myc construct, which is composed of an N-terminal domain and RBD (amino acids 19–685), and analyzed its binding with CD209L expressed in HEK-293 cells. The result showed that, similar to Fc-S-RBD, S1 binds to CD209L, indicating that RBD is the main domain involved in the binding of S protein with CD209L (S. Figure 9B). Additionally, S-RBD interacted with CD209L endogenously expressed in HUVEC-TERT cells (S. Figure 9C), indicating that the binding of S-RBD with CD209L expressed in HEK-293 cells is not an artifact of overexpression of CD209L in HEK-293 cells. Next, we compared the binding of S-RBD-HIS with CD209L, CD209, and ACE2 in a dot blot assay. S-RBD-HIS interacted with CD209L, CD209, and ACE2 in a concentration-dependent manner (Figure 4E). The strongest signals were observed with Fc-ACE2-Myc (Figure 4E), suggesting a higher affinity between S-RBD and ACE2 compared to CD209L and CD209.
Figure 4.
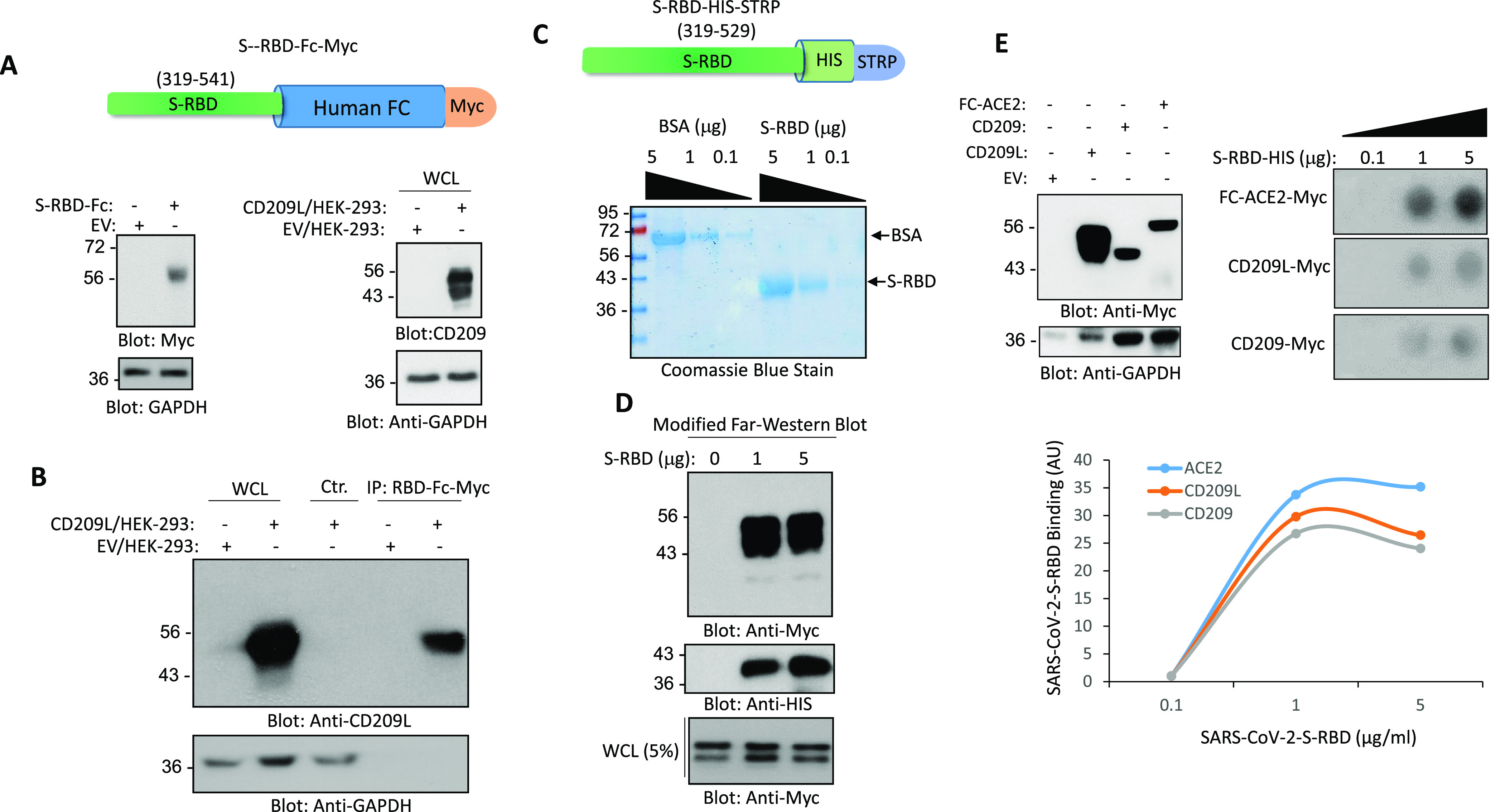
CD209L and CD209 bind to SARS-CoV-2-S-RBD. (A) Schematic of Fc-CoV-2-S-RBD; expression of Fc-CoV-2-S-RBD and CD209L in HEK-293 is shown. (B) Immunoprecipitation assay demonstrates the binding of CD209L with Fc-CoV-2-S-RBD. (C) Schematic and Coomassie blue stain of CoV-2-S-RBD-HIS are shown. (D) Far-Western blot analysis shows the binding of HIS-STRP-tagged CoV-2-S-RBD with CD209L. (E) Western blot analysis of CD209L, CD209, and Fc-ACE2 ectopically expressed in HEK-293 cells. (F) CoV-2-S-RBD-HIS applied onto a PFVD membrane with varying concentrations as indicated in the figure legend via a dot blot apparatus. The membranes, after blocked with 5% BSA, were incubated with cell lysates derived from HEK-293 cells expressing Fc-ACE-2-Myc, CD209L-Myc, or CD209-Myc, and the binding of ACE2, CD209L, and CD209 to CoV-2-S-RBD was detected with anti-Myc antibody. Quantification of the dot blots is shown. AU, arbitrary unit. ImageJ software was used to quantify the dot blots.
ACE2 is described as a main entry receptor for SARS-CoV-2;13 we asked whether CD209L can associate with ACE2. To answer this question, we carried out a coimmunoprecipitation assay and demonstrated that CD209L can form a heterodimer with ACE2 (S. Figure 10), suggesting both ACE2-independent and ACE2-dependent roles for CD209L in SARS-CoV-2 entry and infection. However, whether CD209L directly interacts with ACE2 or its interaction with ACE is aided by additional proteins requires further investigation. To investigate the mechanism of CD209L interaction with ACE2, we asked whether the CRD domain on CD209L is involved in the binding of ACE2. We treated HEK-293 cells expressing CD209L-Myc with EGTA that specifically removes calcium and, as a result, disables the mannose recognition capability of the CRD domain. The result showed that the removal of calcium had no significant impact on the binding of ACE2 with CD209L, indicating that the CRD is not involved in the CD209L interaction with ACE2 (S. Figure 11).
CD209L Is _N_-Glycosylated, and _N_-Glycosylation Interferes with Interaction of CD209L with SARS-CoV2 Spike Protein
CD209L interacts with glycoproteins via its C-type lectin domain, and it is itself subject to _N_-glycosylation.37 We determined the occupancy of the potential _N_-glycosylation sites on our CD209L construct and investigated whether _N_-glycosylation plays a role in the interaction of CD209L with spike protein. After treatment of the immunoprecipitated protein with PNGase F in the presence of H217O (which removes _N_-linked glycans and converts N → D, incorporating 18O at the formerly glycosylated site, resulting in a mass shift of 3 u), we detected only the formerly glycosylated version of the peptide spanning the N92 _N_-glycosylation sequon (S. Figure 12). The results indicated that the CD209L protein is fully _N_-glycosylated at site N92. Furthermore, nUPLC-MS/MS analyses of CD209L digests enabled detection of a glycopeptide bearing high-mannose-type _N_-linked glycosylation at site N92 (Figure 5A). The dominant glycoform was Man8. In contrast, although the peptide that includes the N361 site was observed, no deamidation/18O incorporation was detected after the PNGase F treatment (S. Figure 12). This result indicated that the N361 site is unoccupied.
Figure 5.
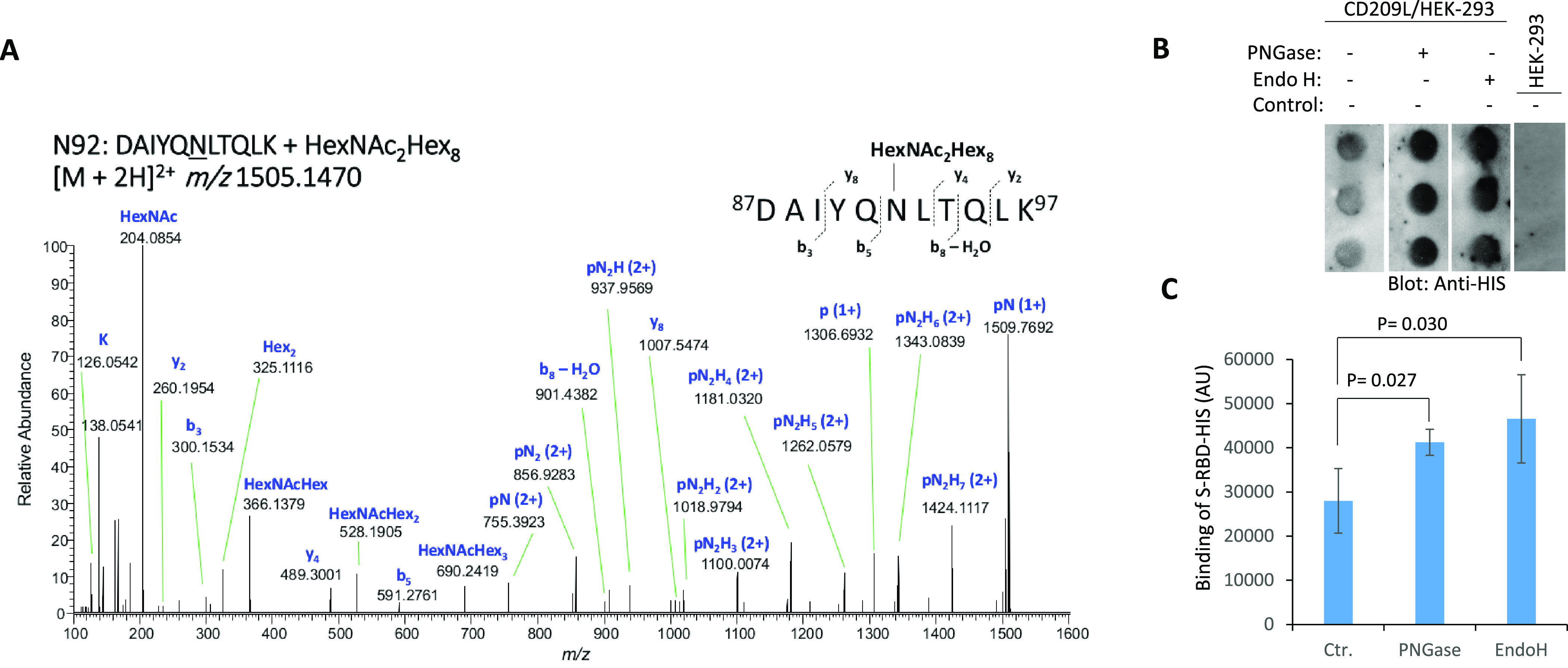
CD209L is _N_-glycosylated, and _N_-glycosylation modulates its interaction with SARS-CoV2 spike protein. (A) HCD MS/MS spectrum of m/z 1505.1470, assigned as the [M + 2H]2+ of the glycopeptide 87DAIYQNLTQLK97 + HexNAc2Hex8, corresponding to CD209L site N92 bearing a Man8 glycan. Stepped-collision energy (15/25/35% NCE) was used. The fragment ions do not retain the glycan moiety. p = peptide, N = N-acetylhexosamine (HexNAc), H = hexose (Hex). (B) Dot blot of CD209L/HEK-293 cell lysate under control conditions (no treatment) or treated with PNGase F or Endo H. Dot blot of HEK-293 cell lysate was used as a negative control. Following treatment of lysate, the spike protein receptor-binding domain (S-RBD-HIS) was incubated with the immobilized lysate, followed by detection with anti-HIS antibody. (C) Quantification of blots is shown. AU, arbitrary unit. ImageJ software was used to quantify the dot blots.
To investigate the role of CD209L _N_-glycosylation in its interaction with the SARS-CoV-2-S-RBD, a dot blot analysis was performed, in which CD209L/HEK-293 or control HEK-293 cell lysates were immobilized on a PVDF membrane, followed by incubation with the spike protein receptor-binding domain (S-RBD-HIS). Treatment with PNGase F or Endo H, which remove both complex and high-mannose _N_-linked glycans or only high-mannose _N_-linked glycans, respectively, led to an increase in the binding of SARS-CoV-2 S-RBD in a statistically significant manner compared to control conditions (Figure 5B,C). Together, the data suggest that high-mannose _N_-linked glycans on CD209L and/or the SARS-CoV2 S-protein hinder the interaction of CD209L and the S-protein, while the absence of high-mannose glycans favors the CD209L-S protein interaction.
Discussion
In this study, we demonstrate that CD209L is broadly expressed in human pulmonary and kidney tissues, in both endothelial cells and epithelial cells. CD209L and CD209 can serve as receptors for SARS-CoV-2, as evidenced by their interactions with the purified RBD domain of the SARS-CoV-2 S. Recognition of CD209L and CD209 by the RBD domain of SARS-CoV-2 S mediates virus entry. Endothelial cells that lack ACE2 are permissive to SARS-CoV-2 infection. Interfering with CD209L expression or function with shRNA or soluble CD209L in endothelial cells inhibited SARS-CoV-2 virus entry. Similarly, ectopic expression of CD209L and CD209 in HEK-293 cells increased SARS-CoV-2 entry, which underscores the potential roles of these receptors in SARS-CoV-2 in infection of human cells. This idea is consistent with the previous studies which demonstrated that CD209L interacts with Ebola virus surface glycoprotein and mediates endothelial cell infection.45,46 The actions of host viral entry receptors, through multiple mechanisms such as endocytosis and fusion, can result in viral replication in host cells, whereas cell surface structures (e.g., glycans and lectins) can serve as adhesion receptors, binding to virus and significantly enhancing virus entry into the target cells via their interactions with features on the viral surface (e.g., spike protein) or with the entry receptors (e.g., ACE2). CD209L and CD209 have been described as attachment factors for multiple viruses, including Marburg and SARS.25,26 Subsequent studies demonstrated CD209L and CD209 themselves can act as entry receptors for SARS independent of ACE2.32,48 Our results indicate that CD209L and CD209, by acting as entry receptors, mediate transinfection of SARS-CoV-2 in endothelial cells. However, CD209L in other cell types could function as an attachment factor, but the present study has not investigated this mechanism of CD209L in viral entry. Interestingly, a recent study demonstrates that CD209L can also mediate cisinfection of SARS-CoV-2 in monocytes and T-lymphocytes.49 Another important aspect of CD209L involvement in SARS-CoV-2 infection is the demonstration of the interaction of CD209L with ACE2. This suggests that CD209L is capable of facilitating SARS-CoV-2 entry in both ACE2-dependent and -independent manners. However, further detailed studies are needed to fully elaborate the ACE2-independent function of the CD209L and CD209 in SARS-CoV-2 viral entry and infection.
When compared to CD209L or CD209, ACE2 appeared to have a stronger binding to SARS-CoV-2 and also mediated SARS-CoV-2 entry in HEK-293 cells more efficiently. Nevertheless, in agreement with the proposed role of CD209L and CD209 in SARS-CoV-2 infection, multiple biochemical assays demonstrated the direct physical interaction of CD209L and CD209 with SARS-COV-2 S-RBD protein. CD209L and CD209 showed similar interactions with SARS-CoV-2 S-RBD. Thus, CD209L and CD209 can serve as alternative entry receptors for SARS-CoV-2 spike protein. In agreement with our findings, a recent report by van Kooyk and colleagues also reported an ACE2-independent binding between CD209L and SARS-CoV-2 S protein.50 Furthermore, while the preprint of this manuscript had been shared in the bioRxiv (https://www.biorxiv.org/content/10.1101/2020.06.22.165803v1), another group also reported the binding of SARS-COV-2 S protein with CD209 in a glycosylation-dependent manner, although they did not determine the site occupancies or the glycoform distributions.51 Another important feature of CD209L interaction with SARS-CoV-2 S protein is that N -glycosylation of CD209L appears to hinder its interaction with the SARS-CoV-2 S protein, suggesting that differential _N_-glycosylation of CD209L (and/or the S-protein) due to differences in cell types, pathology, individuals, or species could influence the overall interaction of CD209L with SARS-CoV-2. One possible mechanism for the function of _N_-glycosylation of CD209L is that _N_-glycosylation generates a hindrance for the CRD domain and hence reduces its ability to recognize the mannose-rich SARS-CoV-2 S protein. However, further structure–function studies are needed to elucidate the possible mechanism by which _N_-glycosylation of CD209L impacts its recognition of SARS-CoV-2-S protein.
ACE2 was originally reported to be expressed in various cell types, including endothelial cells of the heart, kidneys, and testis52 and in lung alveolar epithelial cells.14 However, more recent studies showed that ACE2 is expressed at relatively low levels or is undetectable in endothelial cells of lung, liver, skin, and intestine16 and lung tissue.15 We observed a strong staining of CD209L in alveolar type II epithelial lung cells, renal proximal epithelial cells, and blood vessels, which might provide potential routes of entry for SARS-CoV-2. However, we did not observe CD209 expression in blood vessels or lung epithelial cells. Previous studies have shown that CD209 is mostly expressed in dendritic cells, tissue-resident macrophages, and B cells,53,54 suggesting that these cell types could be targeted by SARS-CoV-2 via recognition of CD209.
Recent studies suggest that the vascular system is a major site of attack by SARS-CoV-2.7,8 COVID-19 patients suffer from distinct endothelial cell injury (i.e., endothelitis) and altered angiogenesis8 with widespread microvascular thrombosis.3,4 These observations, coupled with the fact that vascular endothelial dysfunction also plays crucial roles in the pathogenesis of COVID-19,55 underscore the role of endothelial cells in the pathobiology of SARS-CoV-2 infection and also suggest a therapeutic opportunity.
Conclusion
Our present study reveals that CD209L not only can act as a SARS-COV-2 entry receptor but also performs critical functions in the angiogenic responses of endothelial cells. This suggests that SARS-COV-2, by exploiting CD209L, could subvert CD209L function in endothelial cells, leading to endothelial cell injury and altered angiogenesis. However, further studies such as cellular context and physical interaction of CD209L and CD209 with spike protein and ACE2 are required to fully understand the role of CD209 and CD209L in SARS-CoV-2, as we so far have used only a cell line to link CD209L to viral entry in endothelial cells. Investigations of the molecular and cellular effects of SARS-CoV-2 on the vascular system are thus critical to defining the selective roles of CD209L and CD209, in both the presence and absence of ACE2 in this and other systems, in order to explore the therapeutic potential of CD209L and CD209 as targets for combatting COVID-19.
Materials and Methods
Full details of the plasmids, antibodies, and procedures used are provided in the supplemental data. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE56 partner repository with the data set identifier PXD021309.
Statistical Analyses
Experimental data were subjected to Student’s t test or one-way analysis of variance analysis, where appropriate, with representation from at least three independent experiments. p < 0.05 was considered significant.
Safety Statement
No unexpected or unusually high safety hazards were encountered.
Acknowledgments
This work was supported in part through grants from CTSI grant 1 UL1 TR001430 and Malory Fund, Department of Pathology, Boston University (to N.R.): a grant from The Evans Center for Interdisciplinary Biomedical Research ARC on COVID-19, and NIH grants from the National Institute for Allergy and Infectious Diseases (NIAID) R01 AI064099 and National Institute for Aging R01 AG060890 (to S.G.); NIH grants from NIAID R01 AI133486 and National Center for (NCATS) CTSI UL1 TR001430, and awards from Evergrande MassCPR subaward 280870.5116795.0025 and Fast Grants (to E.M.); NIH Heart, Lung and Blood Institute grant R01 HL132325 (to V.C.), NIH grants from the Institute of General Medical Sciences R24 GM134210, the Office of the Director S10 OD021728 and BUSM CTSI 1 UL1 TR001430 COVID-19 Related Research Award (to C.E.C.); NIH/NIAID grant R01 AI146779 and a Massachusetts Consortium on Pathogenesis Readiness (MassCPR) grant (to A.G.S.) and NIH training grants: T32 GM007753 for B.M.H. and T.M.C; T32 AI007245 for J.F.; and T32 HL007035 for E.L.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01537.
- Twelve additional figures including material and methods (PDF)
Author Contributions
N.R., R.A., E.M., S.G., V.C., K.B.C., and C.E.C. were involved in the experimental design, data interpretation, and writing and editing of the manuscript. N.R., R.A., M.A.N., W.Y., J.B., E.L.S., Q.Z., J.O., K.B.C., and C.X. performed the experiments. J.F., B.M.H., T.M.C., and A.G.S. provided protein and reagents.
The authors declare no competing financial interest.
Notes
Data availability: Data and reagents are available from the corresponding author upon request. Mass spectrometry data are available via ProteomeXchange with identifier PXD021309.
Supplementary Material
References
- Bhatraju P. K.; Ghassemieh B. J.; Nichols M.; Kim R.; Jerome K. R.; Nalla A. K.; Greninger A. L.; Pipavath S.; Wurfel M. M.; Evans L.; Kritek P. A.; West T. E.; Luks A.; Gerbino A.; Dale C. R.; Goldman J. D.; O’Mahony S.; Mikacenic C. Covid-19 in Critically Ill Patients in the Seattle Region-Case Series. N. Engl. J. Med. 2020, 382, 2012. 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang-Hua Y.; Le-Min W.; Ai-Bin L.; Zhu G.; Riquan L.; Xu-You Z.; Wei-Wei R.; Ye-Nan W. Severe acute respiratory syndrome and venous thromboembolism in multiple organs. Am. J. Respir. Crit. Care Med. 2010, 182 (3), 436–7. 10.1164/ajrccm.182.3.436. [DOI] [PubMed] [Google Scholar]
- Ng K. H.; Wu A. K.; Cheng V. C.; Tang B. S.; Chan C. Y.; Yung C. Y.; Luk S. H.; Lee T. W.; Chow L.; Yuen K. Y. Pulmonary artery thrombosis in a patient with severe acute respiratory syndrome. Postgrad. Med. J. 2005, 81 (956), e3. 10.1136/pgmj.2004.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N.; Li D.; Wang X.; Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemostasis 2020, 18 (4), 844–847. 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Wang X.; Yang P.; Zhang S. COVID-19 Complicated by Acute Pulmonary Embolism. Radiology: Cardiothoracic Imaging 2020, 2 (2), e200067. 10.1148/ryct.2020200067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z.; Flammer A. J.; Steiger P.; Haberecker M.; Andermatt R.; Zinkernagel A. S.; Mehra M. R.; Schuepbach R. A.; Ruschitzka F.; Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395 (10234), 1417–1418. 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M.; Verleden S. E.; Kuehnel M.; Haverich A.; Welte T.; Laenger F.; Vanstapel A.; Werlein C.; Stark H.; Tzankov A.; Li W. W.; Li V. W.; Mentzer S. J.; Jonigk D. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120. 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V.; Kwon H.; Prado P.; Hagelkruys A.; Wimmer R. A.; Stahl M.; Leopoldi A.; Garreta E.; Hurtado Del Pozo C.; Prosper F.; Romero J. P.; Wirnsberger G.; Zhang H.; Slutsky A. S.; Conder R.; Montserrat N.; Mirazimi A.; Penninger J. M. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181 (4), 905–913.e7. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K.; Imai Y.; Rao S.; Gao H.; Guo F.; Guan B.; Huan Y.; Yang P.; Zhang Y.; Deng W.; Bao L.; Zhang B.; Liu G.; Wang Z.; Chappell M.; Liu Y.; Zheng D.; Leibbrandt A.; Wada T.; Slutsky A. S.; Liu D.; Qin C.; Jiang C.; Penninger J. M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11 (8), 875–9. 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Moore M. J.; Vasilieva N.; Sui J.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; Choe H.; Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426 (6965), 450–4. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H. R.; Zhu Y.; Li B.; Huang C. L.; Chen H. D.; Chen J.; Luo Y.; Guo H.; Jiang R. D.; Liu M. Q.; Chen Y.; Shen X. R.; Wang X.; Zheng X. S.; Zhao K.; Chen Q. J.; Deng F.; Liu L. L.; Yan B.; Zhan F. X.; Wang Y. Y.; Xiao G. F.; Shi Z. L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579 (7798), 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I.; Timens W.; Bulthuis M. L.; Lely A. T.; Navis G.; van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203 (2), 631–7. 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F.; Méar L.; Edvinsson Å.; Micke P.; Uhlén M.; Lindskog C.. The protein expression profile of ACE2 in human tissues. bioRxiv 2020, 2020.03.31.016048v2. [DOI] [PMC free article] [PubMed]
- Ganier C.; Du-Harpur X.; Harun N.; Wan B.; Arthurs C.; Luscombe N.; Watt F.; Lynch M.. CD147 (BSG) but not ACE2 expression is detectable in vascular endothelial cells within single cell RNA sequencing datasets derived from multiple tissues in healthy individuals. bioRxiv 2020, 2020.05.29.123513.
- Daly J. L.; Simonetti B.; Klein K.; Chen K.-E.; Williamson M. K.; Anton-Plagaro C.; Shoemark D. K.; Simon-Gracia L.; Bauer M.; Hollandi R.; Greber U. F.; Horvath P.; Sessions R. B.; Helenius A.; Hiscox J. A.; Teesalu T.; Matthews D. A.; Davidson A. D.; Collins B. M.; Cullen P. J.; Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (Washington, DC, U. S.) 2020, 370, 861. 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L.; Ojha R.; Pedro L. D.; Djannatian M.; Franz J.; Kuivanen S.; van der Meer F.; Kallio K.; Kaya T.; Anastasina M.; Smura T.; Levanov L.; Szirovicza L.; Tobi A.; Kallio-Kokko H.; Osterlund P.; Joensuu M.; Meunier F. A.; Butcher S. J.; Winkler M. S.; Mollenhauer B.; Helenius A.; Gokce O.; Teesalu T.; Hepojoki J.; Vapalahti O.; Stadelmann C.; Balistreri G.; Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (Washington, DC, U. S.) 2020, 370, 856. 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Chen W.; Zhou Y.-S.; Lian J.-Q.; Zhang Z.; Du P.; Gong L.; Zhang Y.; Cui H.-Y.; Geng J.-J.; Wang B.; Sun X.-X.; Wang C.-F.; Yang X.; Lin P.; Deng Y.-Q.; Wei D.; Yang X.-M.; Zhu Y.-M.; Zhang K.; Zheng Z.-H.; Miao J.-L.; Guo T.; Shi Y.; Zhang J.; Fu L.; Wang Q.-Y.; Bian H.; Zhu P.; Chen Z.-N.. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020, 2020.03.14.988345.
- Clausen T. M.; Sandoval D. R.; Spliid C. B.; Pihl J.; Painter C. D.; Thacker B. E.; Glass C. A.; Narayanan A.; Majowicz S. A.; Zhang Y.; Torres J. L.; Golden G. J.; Porell R.; Garretson A. F.; Laubach L.; Feldman J.; Yin X.; Pu Y.; Hauser B.; Caradonna T. M.; Kellman B. P.; Martino C.; Gordts P. L. S. M.; Leibel S. L.; Chanda S. K.; Schmidt A. G.; Godula K.; Jose J.; Corbett K. D.; Ward A. B.; Carlin A. F.; Esko J. D.. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. bioRxiv 2020, 2020.07.14.201616. [DOI] [PMC free article] [PubMed]
- Karpanen T.; Heckman C. A.; Keskitalo S.; Jeltsch M.; Ollila H.; Neufeld G.; Tamagnone L.; Alitalo K. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J. 2006, 20 (9), 1462–72. 10.1096/fj.05-5646com. [DOI] [PubMed] [Google Scholar]
- Wanaguru M.; Liu W.; Hahn B. H.; Rayner J. C.; Wright G. J. RH5-Basigin interaction plays a major role in the host tropism of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (51), 20735–40. 10.1073/pnas.1320771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniweh Y.; Gao X.; Hao P.; Meng W.; Lai S. K.; Gunalan K.; Chu T. T.; Sinha A.; Lescar J.; Chandramohanadas R.; Li H. Y.; Sze S. K.; Preiser P. R. P. falciparum RH5-Basigin interaction induces changes in the cytoskeleton of the host RBC. Cell. Microbiol. 2017, 19 (9), e12747. 10.1111/cmi.12747. [DOI] [PubMed] [Google Scholar]
- Zenonos Z. A.; Dummler S. K.; Muller-Sienerth N.; Chen J.; Preiser P. R.; Rayner J. C.; Wright G. J. Basigin is a druggable target for host-oriented antimalarial interventions. J. Exp. Med. 2015, 212 (8), 1145–51. 10.1084/jem.20150032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amraei R.; Rahimi N. COVID-19, Renin-Angiotensin System and Endothelial Dysfunction. Cells 2020, 9 (7), 1652. 10.3390/cells9071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A.; Gramberg T.; Simmons G.; Möller P.; Rennekamp A. J.; Krumbiegel M.; Geier M.; Eisemann J.; Turza N.; Saunier B.; Steinkasserer A.; Becker S.; Bates P.; Hofmann H.; Pohlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78 (21), 12090. 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Y.; Huang Y.; Ganesh L.; Leung K.; Kong W. P.; Schwartz O.; Subbarao K.; Nabel G. J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004, 78 (11), 5642–50. 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S. A.; Tusell S. M.; Gillim-Ross L.; Hemmila E. M.; Achenbach J. E.; Babcock G. J.; Thomas W. D. Jr.; Thackray L. B.; Young M. D.; Mason R. J.; Ambrosino D. M.; Wentworth D. E.; Demartini J. C.; Holmes K. V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (44), 15748–53. 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S. A.; Hemmila E. M.; Holmes K. V. Human coronavirus 229E can use CD209L (L-SIGN) to enter cells. Advances in experimental medicine and biology 2006, 581, 265–9. 10.1007/978-0-387-33012-9_44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M.; Generous A.; Sinn P. L.; Cattaneo R. Connections matter--how viruses use cell-cell adhesion components. J. Cell Sci. 2015, 128 (3), 431–9. 10.1242/jcs.159400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann S.; Soilleux E. J.; Baribaud F.; Leslie G. J.; Morris L. S.; Trowsdale J.; Lee B.; Coleman N.; Doms R. W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (5), 2670–5. 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S. A.; Tusell S. M.; Gillim-Ross L.; Hemmila E. M.; Achenbach J. E.; Babcock G. J.; Thomas W. D. Jr.; Thackray L. B.; Young M. D.; Mason R. J.; Ambrosino D. M.; Wentworth D. E.; Demartini J. C.; Holmes K. V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (44), 15748–53. 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailleux L.; Pham-Thi N.; Bergeron-Lafaurie A.; Herrmann J.-L.; Charles P.; Schwartz O.; Scheinmann P.; Lagrange P. H.; de Blic J.; Tazi A.; Gicquel B.; Neyrolles O. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med. 2005, 2 (12), e381. 10.1371/journal.pmed.0020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soilleux E. J.; Morris L. S.; Lee B.; Pohlmann S.; Trowsdale J.; Doms R. W.; Coleman N. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. Journal of pathology 2001, 195 (5), 586–92. 10.1002/path.1026. [DOI] [PubMed] [Google Scholar]
- Bashirova A. A.; Geijtenbeek T. B.; van Duijnhoven G. C.; van Vliet S. J.; Eilering J. B.; Martin M. P.; Wu L.; Martin T. D.; Viebig N.; Knolle P. A.; KewalRamani V. N.; van Kooyk Y.; Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 2001, 193 (6), 671–8. 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummidi S.; Catano G.; Lam L.; Hoefle A.; Telles V.; Begum K.; Jimenez F.; Ahuja S. S.; Ahuja S. K. Extensive repertoire of membrane-bound and soluble dendritic cell-specific ICAM-3-grabbing nonintegrin 1 (DC-SIGN1) and DC-SIGN2 isoforms. Inter-individual variation in expression of DC-SIGN transcripts. J. Biol. Chem. 2001, 276 (35), 33196–212. 10.1074/jbc.M009807200. [DOI] [PubMed] [Google Scholar]
- Zelensky A. N.; Gready J. E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272 (24), 6179–217. 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Drickamer K. C-type lectin-like domains. Curr. Opin. Struct. Biol. 1999, 9 (5), 585–90. 10.1016/S0959-440X(99)00009-3. [DOI] [PubMed] [Google Scholar]
- Van Liempt E.; Imberty A.; Bank C. M.; Van Vliet S. J.; Van Kooyk Y.; Geijtenbeek T. B.; Van Die I. Molecular basis of the differences in binding properties of the highly related C-type lectins DC-SIGN and L-SIGN to Lewis X trisaccharide and Schistosoma mansoni egg antigens. J. Biol. Chem. 2004, 279 (32), 33161–7. 10.1074/jbc.M404988200. [DOI] [PubMed] [Google Scholar]
- Rahimi N. C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors. Biology (Basel, Switz.) 2021, 10 (1), 1. 10.3390/biology10010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. H.; Jung K. C.; Kim J. H.; Sung S. W.; Chung J. H.; Shim Y. S.; Lee S. D.; Chung D. H. Expression patterns of markers for type II pneumocytes in pulmonary sclerosing hemangiomas and fetal lung tissues. Arch. Pathol. Lab. Med. 2005, 129 (7), 915–9. 10.5858/2005-129-915-EPOMFT. [DOI] [PubMed] [Google Scholar]
- Jarrard J. A.; Linnoila R. I.; Lee H.; Steinberg S. M.; Witschi H.; Szabo E. MUC1 is a novel marker for the type II pneumocyte lineage during lung carcinogenesis. Cancer Res. 1998, 58 (23), 5582. [PubMed] [Google Scholar]
- Chandler K. B.; Leon D. R.; Meyer R. D.; Rahimi N.; Costello C. E. Site-Specific N-Glycosylation of Endothelial Cell Receptor Tyrosine Kinase VEGFR-2. J. Proteome Res. 2017, 16 (2), 677–688. 10.1021/acs.jproteome.6b00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamalice L.; Le Boeuf F.; Huot J. Endothelial cell migration during angiogenesis. Circ. Res. 2007, 100 (6), 782–94. 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- Simmons G.; Reeves J. D.; Grogan C. C.; Vandenberghe L. H.; Baribaud F.; Whitbeck J. C.; Burke E.; Buchmeier M. J.; Soilleux E. J.; Riley J. L.; Doms R. W.; Bates P.; Pohlmann S. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 2003, 305 (1), 115–23. 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- Alvarez C. P.; Lasala F.; Carrillo J.; Muniz O.; Corbi A. L.; Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002, 76 (13), 6841–4. 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. P.; Lohani M.; Cho M. W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007, 81 (21), 12029–39. 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépaut M.; Luczkowiak J.; Vivès C.; Labiod N.; Bally I.; Lasala F.; Grimoire Y.; Fenel D.; Sattin S.; Thielens N.; Schoehn G.; Bernardi A.; Delgado R.; Fieschi F.. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. bioRxiv 2020, 2020.08.09.242917. [DOI] [PMC free article] [PubMed]
- Chiodo F.; Bruijns S. C. M.; Rodriguez E.; Li R. J. E.; Molinaro A.; Silipo A.; Di Lorenzo F.; Garcia-Rivera D.; Valdes-Balbin Y.; Verez-Bencomo V.; van Kooyk Y.. Novel ACE2-Independent Carbohydrate-Binding of SARS-CoV-2 Spike Protein to Host Lectins and Lung Microbiota. bioRxiv 2020, 2020.05.13.092478.
- Gao C.; Zeng J.; Jia N.; Stavenhagen K.; Matsumoto Y.; Zhang H.; Li J.; Hume A. J.; Mühlberger E.; van Die I.; Kwan J.; Tantisira K.; Emili A.; Cummings R. D.. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv 2020, 2020.07.29.227462.
- Danilczyk U.; Eriksson U.; Crackower M. A.; Penninger J. M. A story of two ACEs. J. Mol. Med. (Heidelberg, Ger.) 2003, 81 (4), 227–34. 10.1007/s00109-003-0419-x. [DOI] [PubMed] [Google Scholar]
- Rappocciolo G.; Piazza P.; Fuller C. L.; Reinhart T. A.; Watkins S. C.; Rowe D. T.; Jais M.; Gupta P.; Rinaldo C. R. DC-SIGN on B lymphocytes is required for transmission of HIV-1 to T lymphocytes. PLoS Pathog. 2006, 2 (7), e70. 10.1371/journal.ppat.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel E. A.; van Gisbergen K. P.; Geijtenbeek T. B.; van Kooyk Y. Distinct functions of DC-SIGN and its homologues L-SIGN (DC-SIGNR) and mSIGNR1 in pathogen recognition and immune regulation. Cell. Microbiol. 2005, 7 (2), 157–65. 10.1111/j.1462-5822.2004.00480.x. [DOI] [PubMed] [Google Scholar]
- Zhou F.; Yu T.; Du R.; Fan G.; Liu Y.; Liu Z.; Xiang J.; Wang Y.; Song B.; Gu X.; Guan L.; Wei Y.; Li H.; Wu X.; Xu J.; Tu S.; Zhang Y.; Chen H.; Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020, 395 (10229), 1054–1062. 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Perez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaino J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47 (D1), D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.