Microcompartments in Prokaryotes: Carboxysomes and Related Polyhedra (original) (raw)
All cyanobacteria and many chemoautotrophs contain polyhedral inclusion bodies that are bound by a unilamellar protein shell (15, 63). Isolation and enzymatic analysis of the bodies from Halothiobacillus neapolitanus (previously Thiobacillus neapolitanus) revealed that they are filled with ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO); therefore they were given the name “carboxysomes” (59). Subsequent studies of both cyanobacteria and chemoautotrophic bacteria have led to the well-accepted conclusion that the “organelles” or microcompartments function to enhance the catalytic properties of the RuBisCO they contain, although the mechanism of this catalytic enhancement is unclear (51, 65). Localization and characterization of the genes encoding carboxysome components has underscored the apparent common function of these bodies in carboxysome-containing autotrophic bacteria. More surprising is the finding that a number of heterotrophic prokaryotes harbor genes homologous to those for carboxysome shell proteins (9, 32, 62). Under proper growth conditions, these bacteria produce polyhedral inclusion bodies that are morphologically similar to carboxysomes, although the cells expressing these bodies contain no RuBisCO and do not fix CO2 via the Calvin-Benson-Bassham cycle as a major part of their carbon metabolism. This review evaluates the evidence that relates carboxysome structure to function in the carbon metabolism of autotrophic prokaryotes and examines similarities to newly discovered particles found in heterotrophs. The possibility is explored that microcompartmentalization of key metabolic enzymes by carboxysomes and their relatives is a more widely utilized regulatory mechanism in prokaryotes than was previously envisioned.
CARBOXYSOMES OF CHEMOAUTOTROPHS
The first carboxysomes that were purified and studied biochemically are those of the obligate chemoautotrophic bacterium H. neapolitanus (59, 60). In thin sections, the bodies appear to be approximately 120 nm in diameter and are most often observed as regular hexagons with a solid, granular interior that is surrounded by a characteristic 3- to 4-nm-thick protein shell. Analysis of carboxysomes by electron microscopy, gel electrophoresis, and immunochemical methods revealed that the particles are filled with type I RuBisCO, which is composed of eight large and eight small subunits. Individual holoenzyme molecules are clearly discernible in negatively stained intact carboxysomes and in the area surrounding broken particles (Fig. 1) (25, 27, 60). Under some conditions, the enzyme appears to be in a paracrystalline array, but it is not clear whether such a well-defined molecular assembly of enzyme molecules is of physiological significance or merely a preparation artifact. The apparent arrangement of enzyme molecules in a layer adjoining the inside of the shell led Holthuijzen et al. (27) to suggest that the carboxysome interior is hollow. While this intriguing idea cannot be completely ruled out, densitometry measurements of polypeptides in denaturing polyacrylamide gels showed that RuBisCO accounts for approximately 60% of the carboxysomal protein (13). In addition, calculations that take into account the dimensions of carboxysomes and of type I RuBisCO are consistent with the notion that approximately 300 molecules of the enzyme can be encapsulated in each polyhedral body, if one assumes that the particle is completely filled (63).
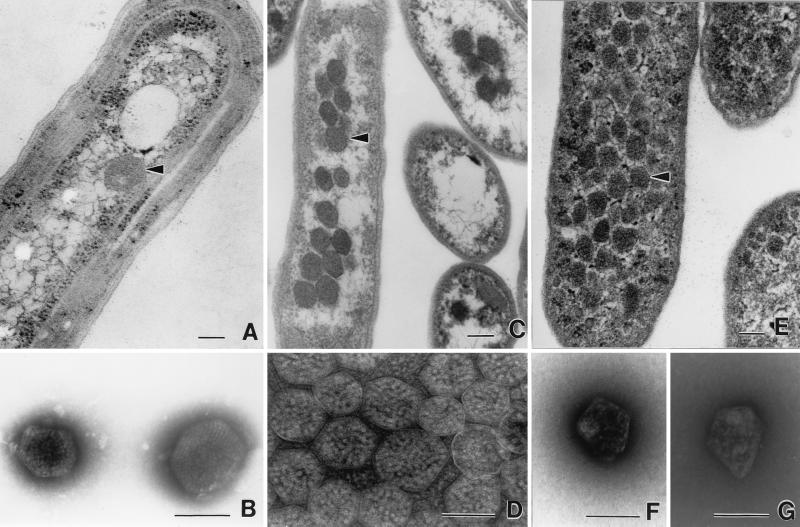
FIG. 1.
Transmission electron micrographs of carboxysomes and enterosomes. (A) Thin section of a cell of Synechococcus strain PCC7942 (fixed cells kindly supplied by George Espie), showing a typical carboxysome (arrowhead). (B) Negatively stained carboxysomes from lysed cells of A. nidulans (now Synechococcus). Molecules of RuBisCO are visible inside. Micrograph kindly supplied by Elisabeth Gantt. (C) Thin sections of H. neapolitanus grown in air, showing aggregation of carboxysomes (arrowhead) in the nucleoid region of the cell. (D) Negative stain of carboxysomes isolated from H. neapolitanus. RuBisCO assemblies are visible inside. (E) Thin section of S. enterica serovar Typhimurium LT2 grown on propanediol under aerobic conditions. Many polyhedral bodies (enterosomes [arrowhead]) are visible throughout the cytoplasm. They are less regular than carboxysomes and slightly smaller. (F and G) Negatively stained, isolated enterosomes from S. enterica serovar Typhimurium LT2. Note the irregular shape. Contents appear to be of variable sizes. Photographed from preparation kindly supplied by Greg Havemann. Panels A, C, and E are all printed at the same magnification, as are panels B, D, F, and G. Bars, 100 nm.
The 3- to 4-nm-thick proteinaceous shell maintains its polyhedral shape during isolation from disrupted carboxysomes by density gradient ultracentrifugation. As revealed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), this structure is composed of at least six polypeptides (two of 5 kDa and one each of 15, 60, 85, and 130 kDa) that comprise 17% of the total carboxysomal protein (5, 12, 13). No lipid component has ever been shown to be associated with the shell.
To date, highly purified carboxysomes have been isolated only from H. neapolitanus and from two Nitrobacter strains (18, 61). Using several independent methods, the carboxysome preparations from H. neapolitanus were shown to be homogeneous and to contain 9 to 15 polypeptides, of which 9 can be visualized under all gel-staining conditions (13, 28). Besides the shell polypeptides, RuBisCO accounts for two polypeptides with a large and a small subunit present in 1:1 stoichiometry as expected for form I of the enzyme (11–13). Earlier findings indicating the presence of all Calvin-Benson-Bassham cycle enzymes within carboxysomes (8) have subsequently been attributed to cellular contamination, and to date, RuBisCO is the only enzymatic activity unequivocally associated with carboxysomes (28). A previous report that carboxysomes of Nitrobacter agilis contain DNA (76), taken together with the presumed icosahedral shape of the particles, made it tempting to suggest that carboxysomes are evolutionarily related to bacteriophages. However, subsequent studies have led to the conclusion that the DNA associated with the particles was a contaminating section of the bacterial chromosome (26), and it is currently thought that carboxysomes do not contain a nucleic acid component. Both of these examples point to the importance of using pure preparations of the particles for structural and functional studies. Furthermore, cryoelectron microscopic studies demonstrated that H. neapolitanus carboxysomes are not icosahedrons but are D6 polyhedrons that display a sixfold axis of symmetry along the top view and two twofold axes of symmetry perpendicular to the sixfold axis. No known virus features this type of geometry, thereby further discounting a viral origin for the particles (A. M. Paredes, F. Soyer, H. C. Aldrich, S. Ludtke, H. Tsuruta, W. Chiu, and J. M. Shively, Abstr. Annu. Meet. Am. Soc. Microbiol. 2001, abstr. 135J-4, 2001) The morphological characteristics of carboxysomes seem to be quite similar for particles from different bacterial species.
CYANOBACTERIAL CARBOXYSOMES
Polyhedral inclusion bodies were first found in Nostoc pruniforme by Jensen and Bowen in 1961 (31) and have since become recognized as a characteristic feature of all cyanobacteria (15). Gantt and Conti observed an ordered paracrystalline array of “doughnut-shaped” particles within the polyhedral structures of Anacystis nidulans (now Synechococcus) (23). In 1976, Codd and Stewart demonstrated that RuBisCO activity copurifies with the polyhedral bodies of Anabaena cylindrica and established that they are carboxysomes (16). Ultrastructurally, the carboxysomes of cyanobacteria resemble those of the chemoautotrophs, although there seems to be a wider range of carboxysome sizes, particularly among the filamentous species. Purification of carboxysomes from cyanobacteria seems to be complicated by the high content of internal membranes, and as a result compositional studies of homogeneous preparations have been greatly hampered. SDS-PAGE analysis of enriched fractions of carboxysomes from Chlorogloeopsis fritschii (37) and Synechococcus strain PCC7942 (49) revealed RuBisCO as the major protein component. The C. fritschii carboxysomes contain an additional 12 to 15 polypeptide bands, none of which were identified at the time the study was published. The preparation of Synechococcus carboxysomes contains more than 30 polypeptide bands, of which 10 were reported to have molecular masses similar to those observed in H. neapolitanus carboxysomes (49). The identity of the remaining bands is unknown, but unless Synechococcus carboxysomes are considerably more complex in structure than those of Chlorogloeopsis and of the thiobacilli, it is likely that many of the bands arise from contaminating cell membrane fragments reported to be present in the enriched fraction. No other enzyme activity of the Calvin-Benson-Bassham cycle has been reported to be associated with cyanobacterial carboxysomes. However, low levels of carbonic anhydrase (CA) appear to copurify with the particles from Synechococcus (49) and Synechocystis (66). The presence of CA is consistent with several lines of experimentation and is the central premise of a model for CO2 concentration discussed below. In contrast, the majority of CA activity measured in C. fritschii was found to be particulate and to have sedimentation properties on sucrose gradients that differ from those of the carboxysome fraction, leading Lanaras et al. to suggest that the bulk of CA activity is associated with membrane vesicles that were probably created during disruption of the cells (38). Despite several attempts aimed at detecting CA in homogeneous preparations of carboxysomes from H. neapolitanus and N. agilis (11, 13, 18), this enzyme has yet to be detected in the carboxysomes of chemoautotrophs. While genetic studies of cyanobacterial carboxysomes have been most revealing, a comparative examination of a membrane-free, homogeneous preparation of cyanobacterial carboxysomes is needed to directly address questions about their composition with some finality.
CARBOXYSOME FUNCTION
Since the original observation that carboxysomes contain RuBisCO, the overriding question has been that of the metabolic purpose that is served by packaging the main carbon assimilation enzyme into a microcompartment. Two possibilities are immediately obvious: (i) the carboxysome serves as a storage form of excess RuBisCO, or (ii) by being placed in the carboxysome, the kinetic abilities of the enzyme are somehow enhanced. Initial evidence indicating carboxysome functionality included experiments which demonstrated that isolated H. neapolitanus particles are active in CO2 fixation despite the apparent stability of their shell (59) and that total cellular RuBisCO activity levels and carboxysome copies per cell increase on CO2 limitation (7, 11). Similar results were obtained with the facultative chemoautotroph, Thiobacillus intermedius (now Thiomonas intermedia). When grown heterotrophically, both RuBisCO protein and carboxysomes disappeared from the cells; transfer back to autotrophic growth conditions was accompanied by rapid reappearance of particles and enzyme activity (53). Moreover, the formation of stable complexes between RuBisCO, CO2, Mg2+, and the transition state analog 2-_C_-carboxyarabinitol-1,5-bisphosphate (CABP) in intact carboxysomes demonstrates that all of the enzyme packaged within the carboxysome is in its activated state (12). When carboxysomes are treated with CABP in situ after chloroform permeabilization of H. neapolitanus cells, both carboxysomal and free enzyme bind a full complement of activating 14CO2, providing further proof that the carboxysomal enzyme is active in vivo (12). It has been suggested that in cyanobacteria, most RuBisCO is packaged in carboxysomes (2, 34), while it has been demonstrated that in H. neapolitanus, the fraction of cellular RuBisCO that is carboxysome bound varies with the availability of inorganic carbon (7, 12).
Since cyanobacteria and chemoautotrophic bacteria utilize RuBisCO as their central carbon-assimilating enzyme through the Calvin-Benson-Bassham cycle, it is reasonable to assume that carboxysomes perform similar or identical functions in both types of prokaryotes. This notion is supported for Synechococcus strain PCC7942 mutants that have aberrant or no carboxysomes and require elevated CO2 levels for growth (50, 67). Subsequently, a number of other mutations in carboxysome genes of cyanobacteria and chemoautotrophs were studied (51), and the results unequivocally demonstrate the active role of carboxysomes in CO2 fixation, although the mechanism that drives the particle's role is less clear.
Several facts about bacterial RuBisCO have served as the basis for hypotheses that address the mechanism of carboxysome function.
(i) Type I RuBisCO from different prokaryotes demonstrates a rather wide range of Km values for CO2 and often displays low affinity for this substrate (Km > 150 μM) (68). It is possible that the carboxysomal enzyme represents a high-affinity form of RuBisCO. However, comparison of the basic kinetic parameters of RuBisCO from purified H. neapolitanus carboxysomes and of free enzyme indicates that once corrections in specific activity are made for the carboxysome, little or no difference is observed between the catalytic potential of particle-bound and free enzyme (13, 25). A more detailed and rigorous kinetic comparison of the two enzyme forms from additional prokaryotic sources is needed to resolve the question whether significant differences exist between the kinetic constants of particle-bound and free RuBisCO.
(ii) RuBisCO must bind a nonsubstrate CO2 at an activation site for optimal activity. This activation brings about the formation of a ternary complex of enzyme, CO2, and Mg2+ that optimizes catalysis at the active site (58). It is possible that carboxysome-bound RuBisCO is maintained in a conformation that mimics the activated form and results in a microcompartment filled with active enzyme regardless of the external CO2 concentration. The fact that isolated carboxysomes display similar activation kinetics to the free enzyme argues against this mechanism, although it is possible that during purification the conformation-modifying factor is altered, requiring carboxysome-bound enzyme in vitro to be activated just like the free enzyme.
(iii) RuBisCO is a bifunctional enzyme that also catalyzes the competing, apparently nonproductive fixation of O2 (1, 24). Perhaps the carboxysomal shell differentially blocks oxygen from reaching the enzyme located inside, thereby enhancing its carboxylase activity through exclusion of the competing substrate. The oxygenase activity of isolated carboxysomes from H. neapolitanus was assessed with an oxygen electrode and compared to that of free RuBisCO. In vitro, the two enzyme forms have approximately the same level of ribulose 1,5-bisphosphate-dependent oxygenase activity (11, 13). However, since the potentiometric determination of oxygenase activity is technically difficult and not particularly sensitive, only a very narrow range of assay conditions have been tested. The use of the more accurate isotope-based simultaneous assay has suggested that the specificity factor for carboxysomes may be significantly higher than that of RuBisCO that is released from the particles. In addition, Marcus et al. reported that the RuBisCO activity of a carboxysome-enriched fraction from Synechocystis strain PCC 6803 is considerably less sensitive to competitive inhibition by oxygen than is the same fraction after rupture of the carboxysomes (41). This report is consistent with the idea that the proteinaceous shell is capable of differentiating between O2 and CO2, although it is difficult to imagine a molecular mechanism that accomplishes such discrimination. This problem is somewhat compounded by the fact that ribulose 1,5-bisphosphate and 3-phosphoglycerate, both charged and relatively large molecules, are capable of crossing the shell with only limited resistance to diffusion (57).
A potential alternative mechanism might exist that can be viewed as a variant of hypothetical mechanism 2. The enzyme packed within the carboxysome could be forced into a conformation that favors the binding of CO2 over that of O2. This would explain the observed kinetics without the need to postulate a transport process for the carboxysomal shell that discriminates between the two gases. More detailed analyses of the shell structure and its chemical and physical properties are required to determine whether differential permeability or binding of CO2 plays a role in carboxysome function.
(iv) The form of inorganic carbon used as a substrate by RuBisCO is CO2, rather than bicarbonate (68, 69). The rate of carboxylation is therefore limited by the available intracellular CO2 concentration, which, in turn, is determined by the HCO3−/CO2 equilibrium and overall intracellular concentration of HCO3−. In the very early days of proposing a molecular function for carboxysomes, it was suggested that perhaps CA is sequestered with RuBisCO inside the carboxysome. Such an arrangement would ensure a rapid conversion of HCO3− to CO2 locally at the site of the carboxylation reaction, thereby creating a ready source of CO2 for RuBisCO (11, 13, 15, 20, 33, 38, 56). Despite the attractiveness of the hypothesis, CA does not appear to colocalize with carboxysomes from several chemoautotrophs and cyanobacteria (6, 13, 18, 22, 38). Nevertheless, several groups have reported CA activity in cyanobacterial carboxysome preparations and have taken this as evidence for the role of the particles in an inorganic carbon-concentrating mechanism (CCM) in those organisms (2, 10, 34, 49, 66). Carboxysomes are thought to represent the second stage of the CCM that rapidly converts HCO3− to CO2 to feed the RuBisCO-catalyzed carboxylation reaction. Kaplan and coworkers have elaborated a detailed quantitative model for the CCM and carboxysome function, which is consistent with many of the data so far collected (20, 33, 34, 56). However, to quote Raven, “This model of the functioning of the carboxysome still has a few untidy ends…” (55). Since the nature of the CCM and the role proposed for carboxysomes have been recently reviewed (2, 34), only the salient details that pertain to carboxysomes will be covered here in hopes of stimulating further research that will clear up the “untidy ends.”
It is the generally accepted view that cyanobacteria and perhaps chemoautotrophs (29, 34) are able to concentrate the relatively low levels of inorganic carbon, typically less than 15 μM, that are found in most natural environments to levels of intracellular bicarbonate that exceed 1mM. This concentration process involves the action of active transporters thought to be associated with the cytoplasmic membrane. The predicted energy-dependent intracellular accumulation of HCO3− has been experimentally demonstrated in a number of cyanobacteria and in H. neapolitanus (10, 29, 35, 42). Moreover, ectopic expression of human CA in the cytoplasm of Synechococcus strain PCC7942 yields transformants that have the high-CO2-requiring (HCR) phenotype and leak CO2 into the surrounding medium (47). It is thought that the high intracellular concentration of bicarbonate generated by the CCM is in disequilibrium with CO2, leading to conversion of HCO3− back to CO2 by the overexpressed enzyme and to the observed leakage of the gas from the cells. Low concentrations of the CA inhibitor ethoxyzolamide reverse the effects of induced expression of CA. If in normal cells CA is located exclusively inside the carboxysome, leakage of CO2 after its conversion from HCO3− might be prevented either by the carboxysome shell or by the limited diffusion of CO2 through the tightly packed proteins comprising the carboxysome core. The proposed requirement for either hypothetical diffusion barrier is a bit troublesome in that the membrane would have to be directionally permeable to an uncharged small molecule (in the first scenario) or the CA would have to be located exclusively in the center region of the carboxysome (in the second case). To date, no direct evidence has been reported to support either one of these hypotheses.
Mutational studies have provided additional experimental evidence for the view that the carboxysome is an integral component of the CCM in cyanobacteria and chemoautotrophs (43, 51). In a number of induced mutants that display an HCR phenotype, HCO3− is accumulated to high intracellular concentration but cannot be used efficiently. Many of these mutants contain morphologically altered carboxysomes or lack these structures altogether. In Synechococcus, the mutations map upstream of the genes for RuBisCO large and small subunit (50). Several putative carboxysome genes are clustered in this region (see below and Fig. 2), but a direct correlation between mutant phenotype and nonfunctional bona fide carboxysome protein genes has not yet been established. This leaves in question the exact role of the mutant gene products as carboxysome components or as assembly factors. To date, no genes that encode CA have been mapped to the genome region near the cluster of carboxysome genes.
FIG. 2.
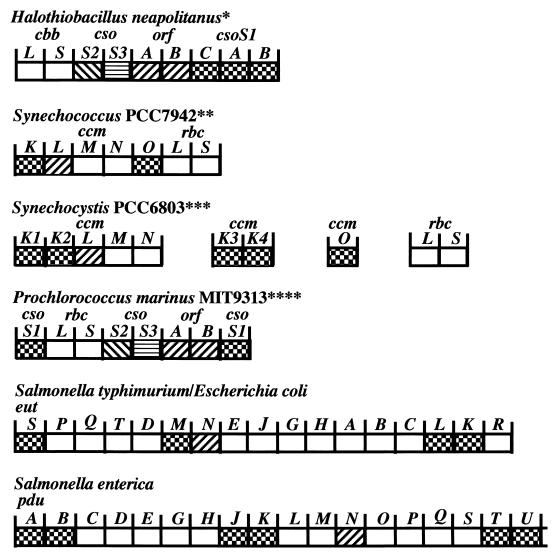
Organization of carboxysome genes and the genes of related polyhedra. Gene similarity is indicated by the different fill patterns. Gene homologies were assigned based on multiple sequence alignments that considered residue and positional identities. The deduced ORFA and ORFB peptides are 22 to 30% identical to CcmL gene products; CsoS1-like peptides are 50 to 99% identical (62). All of the genes are read from left to right. We have taken the liberty of calling the Prochlorococcus carboxysome genes csoS1, ORFAB, csoS2, and csoS3 instead of ccmK, ccmL, ccmM, and ccmN, respectively, since they are more closely related to the former. Abbreviations: cbb, Calvin-Benson-Bassham cycle in eubacteria and archaea cbbL and cbbS, large and small subunits of RuBisCO; cso, carboxysome; ORF, unidentified open reading frame; ccm, carbon-concentrating mechanism; rbcL and rbcS, large and small subunits of RuBisCO in cyanobacteria; pdu, propanediol utilization; eut, ethanolamine utilization. ∗, Two different sulfur bacteria exhibit the same organization, and a third exhibits the same except that the cso genes are not located downstream of cbbLS. ∗∗, Synechococcus strain PCC 7002 has the same organization as Synechococcus strain PCC7942, except that ccmO is missing from the cluster and rbcX resides between rbcL and rbcS. The ccmO gene may reside elsewhere on the genome of PCC7002 (see Synechocystis strain PCC6803). Additional ccmK genes could potentially reside elsewhere on the genome in both PCC7942 and PCC7002. Synechococcus strains PCC6301 possesses two ORFs immediately upstream of rbcL and rbcS, which appear to be equivalent to ccmO. Additional upstream sequencing has not been reported. ∗∗∗, Synechocystis strain PCC6803 possesses two additional ccmK genes and a ccmO gene completely separate from the ccmKLMN cluster. Nostoc punctiforme exhibits the same organization as Synechocystis strain PCC6803, except that ccmO is found immediately downstream of ccmN and not elsewhere on the genome. Furthermore, ccmO possess only a single ccmK homolog rather than two, as is the case with the other ccmO. ∗∗∗∗, Prochlorococcus strain MED4 shows the same organization as MIT9313, except the downstream csoS1 is missing. Synechococcus WH8102 shows the same organization as Prochlorococcus MIT9313. It seems likely that Synechococcus strain WH7803 will also show the same organization, but the downstream region has not been sequenced.
Clearly, most currently available evidence is consistent, albeit indirectly, with carboxysomes acting as an integral part of the CCM in autotrophic prokaryotes. However, until a physical link between CA and carboxysomes can be clearly and reproducibly demonstrated by immunological, biochemical, and/or genetic means, it is just as likely that the kinetic enhancement of RuBisCO that leads to efficient use of the available intracellular inorganic carbon pool is satisfied by one of the alternative mechanisms discussed above.
GENES FOR CARBOXYSOME COMPONENTS
Numerous carboxysome genes have been identified, sequenced, and mapped. Mutational analysis of Synechococcus strain PCC7942 revealed that a cluster of genes consisting of ccmK, ccmL, ccmM, ccmN, and ccmO is located immediately upstream from the RuBisCO large and small-subunit genes (rbcL and rbcS, respectively, in cyanobacteria) (50). Their proximity to the RuBisCO genes was taken to mean that these genes encode products needed for carboxysome structure and function, although specific roles could not be assigned to their products. In the chemoautotrophs, micropeptide sequencing of purified carboxysome proteins provided the means of identifying one of the carboxysome shell protein genes, csoS1, which is located downstream from the RuBisCO large- and small-subunit genes (cbbL and cbbS, respectively, in chemoautotrophs) (19). Sequence analysis revealed significant homology between the deduced products of the csoS1 gene and the ccmK and ccmO genes of Synechococcus, lending further indirect support to the view that the proteins encoded by these cyanobacterial genes are components of the carboxysomal shell (64).
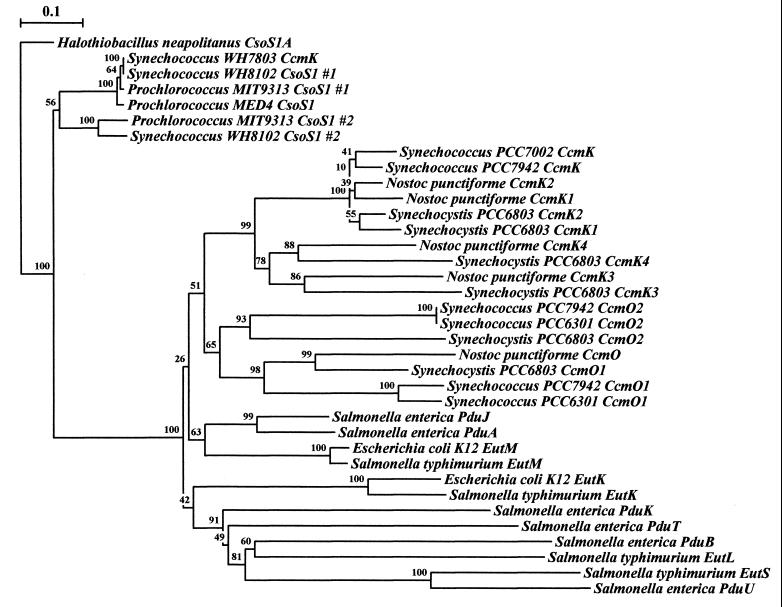
Detailed analysis of the genome region surrounding the csoS1 locus in H. neapolitanus has shown that this gene (now called csoS1A) has adjacent homologs, named csoS1B and csoS1C, which encode practically identical protein products and appear to be the result of gene duplications. The primary sequences of CsoS1A and CsoS1C differ by only one residue, and CsoS1B is 90% homologous to and 12 residues longer than CsoS1A and CsoS1C (64). The CsoS1 family of shell proteins seems to be a defining structural characteristic for carboxysomes and related microcompartments, since homologous genes have been found in all organisms that harbor carboxysomes and carboxysome-like bodies (62).
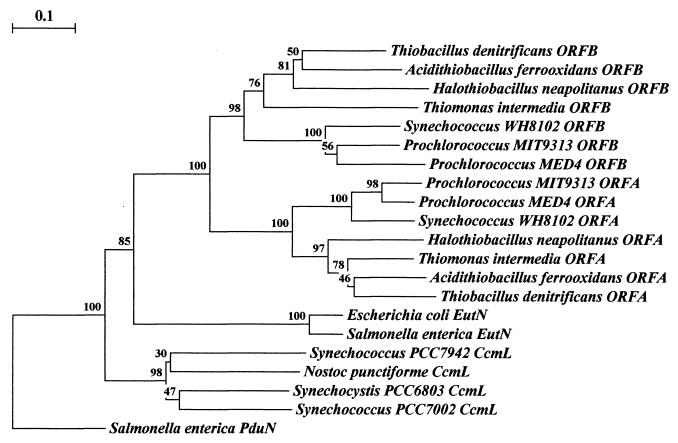
Two additional shell genes, csoS2 and csoS3, have been identified in H. neapolitanus through reverse genetics, expression of recombinant protein, and immunoelectron microscopy (4, 5). The csoS2 gene encodes the 85- and 130-kDa polypeptides evident in SDS-PAGE gels of H. neapolitanus carboxysomes. The observed difference between the molecular masses of the two polypeptides seems to be due to differential levels of posttranslational glycosylation (4). A 60-kDa protein that is a minor component of the carboxysomal shell is encoded by the csoS3 gene. Two open reading frames (ORFs), termed ORFA and ORFB, are located between csoS3 and csoS1C (Fig. 2) and apparently are duplicated genes that code for small polypeptides. The proteins have not yet been identified in H. neapolitanus carboxysomes. The deduced products of ORFA and ORFB have approximately 47% homology at the amino acid level and have homologs in many other carboxysome-containing prokaryotes (Fig. 2 and 3). These seven genes and the two ORFs are located in a contiguous genomic stretch of 8 kbp that features a putative promoter upstream from the cbbL gene and a termination sequence downstream from csoS1B (64). This arrangement of carboxysome genes into an operon (the cso operon) is also found in three other members of the former genus Thiobacillus, namely, Thiomonas intermedia, Acidothiobacillus ferrooxidans, and Thiobacillus denitrificans. T. denitrificans has a cso operon that lacks the cbbL and cbbS genes and, somewhat surprisingly, is the only known bacterium containing this operon that does not form carboxysomes (62). However, to our knowledge, a rigorous examination of this organism under various growth conditions has not been undertaken, so that the complete absence of carboxysomes is not certain.
FIG. 3.
Phylogenetic tree of the conserved region of CsoS1-like polypeptides. Rooting was done arbitrarily using the conserved region of H. neapolitanus csoS1A, which codes for a polypeptide component of the carboxysome shell, which has greater than 90% similarity/identity to each of the known CsoS1 polypeptide regions. Several Synechococcus and Prochlorococcus species seem to group out near H. neapolitanus as well. Respective cyanobacterial and enteric CsoS1 homologs each seem to group together into respective major branches, with conserved protein clusters implying similarity of function and/or origin. The data for each bootstrap value were resampled 1,000 times, with the evolutionary distance scale (in the upper left-hand corner) horizontally representing the number of fixed substitutions per site. Sequence alignment was performed with ClustalW 1.8 (70), and phylogenetic trees were constructed with Treecon (72, 73).
The carboxysomal genes of the three cyanobacterial species, Synechocystis strain PCC6803, Synechococcus strain PCC7942, and Synechococcus strain PCC7002, do not share the same arrangement as those of the thiobacilli. Instead, the ccmKLMN cluster described above appears upstream from the rbcL and rbcS genes in the two Synechococcus strains (Fig. 2), while in Synechocystis strain PCC6803 the same cluster is located at a distance from the rbcL and rbcS genes (51). There is no evidence at present that links the clustered genes in a single transcriptional unit. The ccmK gene, which probably encodes a carboxysomal shell protein, is represented four times in the Synechocystis strain PCC6803 genome but occurs as a single copy in Synechococcus strain PCC7942 cluster, although it may appear elsewhere in the genome (50). The function of the ccmL gene product (homolog of the ORFA and ORFB products) is unknown. Inactivation of this gene in Synechococcus strain PCC7942 results in an HCR phenotype with elongated carboxysomes variants (46, 48, 50).
The ccmM gene is part of the ccm cluster in both Synechococcus species and in Synechocystis strain PCC6803 (39). The deduced amino acid sequences of CcmM proteins from these species are 41 to 47% homologous, with the one from Synechococcus strain PCC7942 being somewhat smaller than those from the other two cyanobacteria. The N terminus of CcmM from Synechococcus strain PCC7002 has significant sequence homology to the CA of Methanosarcina thermophilia, while its C-terminal end contains four repeated stretches of 85 residues which each displays up to 30% homology to RbcS. The ccmM gene product is likely to be a component of carboxysomes, as demonstrated by the cross-reactivity of a carboxysome-enriched fraction from Synechococcus strain PCC7942 with antibodies that had been raised against recombinant 58-kDa CcmM polypeptide (39). The similarities of the repeated C-terminal domains to RbcS suggest that CcmM may play a structural role in the carboxysome through protein-protein interactions with the RuBisCO large subunit. The homology to CA raises the tempting possibility that the CcmM protein acts as the carboxysomal CA; however, more traditional CA genes and their protein products have also been proposed for Synechococcus strain PCC 7942 and Synechocystis strain PC6803. To date, homologs to ccmM have not be found in other carboxysome-containing bacteria (39); however, the sequence structure of its product suggests a very important role for the protein in the carboxysome, and future functional analyses will certainly prove to be enlightening.
The ccmN gene, whose inactivation in Synechococcus strain PCC7942 leads to the HCR phenotype (21, 51), encodes a protein of 23.5 kDa. Like the other members of the ccm cluster, this gene has homologs in Synechococcus strain PCC7002 and Synechocystis strain PCC6803.
In the Synechococcus strain PCC7942 genome, ccmO is found at the end of the ccmKLMN cluster just upstream from rbcL, whereas this gene is absent from the ccm clusters of Synechococcus strain PCC7002 and Synechocystis strain PCC6803. It is thought to encode a protein that is necessary for carboxysome assembly (40) and has considerable homology to CsoS1 and CcmK.
Based on these data, the conjecture that cyanobacterial carboxysome gene organization follows the ccmKLMN cluster pattern, while the chemoautotrophic bacteria feature cso operons, was generally accepted until very recently. Advances in total genome sequencing of cyanobacteria have provided four additional examples of carboxysome gene clusters for in scripto studies. Work by teams from the DOE Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html/index.html) has resulted in the elucidation of the complete genome sequence for two strains of the marine cyanobacterial species Prochlorococcus marinus and of another Synechococcus strain. Prochlorococcus is probably the most abundant cyanobacterial genus in the world's oceans and has a photosystem that uses chlorophylls a and b but lacks the phycobilins characteristic of most other cyanobacteria (14). Synechococcus strain WH8102 is a member of an open-ocean motile group that is phylogenetically distinct from other Synechococcus strains (75). In contrast to the ccm cluster described for Synechococcus strains PCC7942 and PCC7002 and Synechocystis strain PCC6803, the carboxysome genes in P. marinus MIT9313 and Synechococcus strain WH8102 are grouped into clusters that are nearly identical to the carboxysome operons of the thiobacilli, except for the placement of the obvious gene duplications (Fig. 2). The genome of an additional marine Synechococcus, Synechococcus strain WH7803, has been partially sequenced and seems likely to contain a carboxysome gene cluster similar to that of thiobacilli, although complete analysis will have to await sequencing of the downstream region. In addition to the conserved gene order within the carboxysome operon, a high degree of sequence conservation exists between these genes and their apparent homologs in the _cso_-type cluster. Phylogenetic trees constructed for the csoS1 gene of H. neapolitanus and for ORFA- and ORFB-like genes clearly show that carboxysome genes arranged in the _cso_-type operon of the thiobacilli are more closely related, regardless of the organisms in which they are found, than are their apparent counterparts in organisms that feature the _ccm_-type cluster (Fig. 3 and 4). In addition, homologs of the ccmM gene of Synechococcus strain PCC7942 and Synechocystis strain PCC6803 are not detectable by BLAST homology searches of the chemoautotrophs and of cyanobacteria that possess the thiobacillus-type operon. These facts make it seem likely that lateral gene transfer of the cso operon as a unit has been a rather common and perhaps recent occurrence among autotrophic prokaryotes while the arrangement of carboxysome genes in cyanobacteria that contain the _ccm_-type cluster suggests a different evolutionary path. Although it is tempting to hypothesize that the carboxysomes encoded by these two different gene clusters may be compositionally and functionally distinct, such speculation must await a careful biochemical analysis.
FIG. 4.
Phylogenetic tree of the ORFA and ORFB polypeptides and their relatives. Rooting was done arbitrarily through Salmonella enterica serovar Typhimurium PduN, which groups individually and away from the other families of polypeptides. Clustering the polypeptides into groupings respective of species physiology may be seen among the cyanobacteria, the thiobacilli, and the enteric polypeptides. Uniform clustering of protein homologs by relative physiology occurs with a high degree of confidence. Of the major branches of protein groupings, all are represented by bootstrap values greater than 75%. The data for each bootstrap value were resampled 1,000 times, with the evolutionary distance scale in the upper left-hand corner representing the number of fixed substitutions per site, as a measure of horizontal distance. Sequence alignment was performed with ClustalW 1.8 (70) and phylogenetic trees were constructed with Treecon (72, 73).
REGULATION OF CARBOXYSOME GENE EXPRESSION AND CARBOXYSOME FORMATION
Two observations led to the conclusion that the regulation of carboxysome gene expression must be actively and tightly controlled: the relative amounts of cso operon gene products in the carboxysome are not equal (13, 28, 46, 61), and RuBisCO activity and carboxysome numbers per cell increase in response to inorganic carbon limitation in most prokaryotes that contain these bodies (7, 45, 54). Several possible regulatory schemes can be envisioned to explain these observations: (i) differential strength of ribosomal binding sites in the individual genes of the polycistronic master transcript, (ii) premature transcription termination mediated by secondary structures located on the 3′ ends of individual coding sequences, (iii) processing of the master mRNA into smaller mRNA species with differential stability or translatability, and (iv) cryptic promoters located within the operon that differentially drive transcription of internal coding regions. To date, no rigorous analysis of carboxysome gene expression has been conducted, but available data collected from work with H. neapolitanus suggest that differential ribosome binding and mRNA processing might occur (3–5, 19). In cyanobacteria that contain the ccmKLMN gene cluster, coordinated expression of carboxysome genes would seem to be somewhat more complicated, given the dispersed nature of the gene locations. Clearly, much work remains to be done on this very important aspect of carboxysome molecular biology.
The regulatory gene cbbR, which encodes a transcriptional regulator of the LysR type, plays a role in controlling expression of Calvin-Benson-Bassham cycle genes in several autotrophic bacteria (17, 30, 71, 74). Homologs of cbbR have been noted in some carboxysome-containing cyanobacteria and in the thiobacilli (3). With the exception of A. ferrooxidans and T. denitrificans, the cbbR gene is not typically present in the region directly upstream of the cso operon and the ccmKLMN cluster, as would be expected if its product were actively involved in controlling carboxysome gene expression. Further analysis is needed to determine if the cbbR homologue observed upstream of the H. ferrooxidans and T. denitrificans cso operon is involved in regulating carboxysome gene expression.
Little is known about the mechanism or sequence of events during carboxysome assembly. A model for carboxysome assembly in Anabaena based on electron microscopic examination suggests that the shell assembles first, followed by the insertion of RuBisCO molecules to fill the polyhedral body (46). Similar electron microscopic observations of Synechococcus cells led Orus et al. to suggest that RuBisCO molecules first assemble into a polyhedral aggregate that is subsequently coated with the shell (44, 45). With the data presently available, it is difficult to favor either assembly model. Many carboxysome genes have now been identified and characterized and can be modified in a defined fashion. In addition, a number of carboxysome proteins have been overexpressed as recombinant proteins. In particular, recent experiments in which the entire H. neapolitanus cso operon was heterologously expressed in Escherichia coli resulted in the formation of carboxysome-like structures with associated RuBisCO activity (H. C. Aldrich, S. Elvington, H. E. MacInnes, R. Szabady, K. Feder, L. McDowell, and J. M. Shively, Abstr. 59th Annu. Meet. Microsc. Soc. Am., abstr., 2001). These powerful tools are the prerequisites for a combined biochemical and genetic approach to reproduction of carboxysome assembly in vitro and dissection of the in vivo assembly process of these prokaryotic inclusion bodies.
ENTEROSOMES: CARBOXYSOMES ARE NOT JUST FOR AUTOTROPHS ANY MORE
Of the many findings that have resulted from sequence analysis of carboxysome genes, probably the most striking and unexpected one is the identification of shell gene homologs in a number of obligately heterotrophic enteric bacteria (62). The pduA gene of Salmonella enterica serovar Typhimurium is a member of the propanediol utilization (pdu) operon and encodes a protein that is highly homologous to the carboxysomal shell proteins CsoS1 and CcmK. Subsequent analysis of the pdu operon in S. enterica serovar Typhimurium has shown that pduB, pduJ, pduK, pduN, pduT, and pduU have significant homology to carboxysome genes (9, 32). Likewise, genes that are components of the ethanolamine metabolism pathway in Salmonella (36) and E. coli (62) are related to carboxysome genes. The operon comparison in Fig. 2 and the phylogenetic tree constructs in Fig. 3 and 4 show that ORFA, ORFB, and the csoS1 genes of the _cso_-type operons are related to genes from a wide variety of autotrophic and heterotrophic prokaryotes. In addition, polyhedral bodies that bear a strong resemblance to carboxysomes have been observed in Salmonella during growth on propanediol or ethanolamine and in E. coli and Klebsiella during growth on ethanolamine (9, 36, 62). Since in these three genera no RuBisCO activity has been reported and no genes that could encode a functional RuBisCO enzyme appear to be present, the obvious questions are those of a function for carboxysome gene products in these enteric bacteria and the identity of the protein(s) harbored in the carboxysome-like structures, which we propose to call enterosomes. A partial answer is provided by recent work conducted by Bobik and coworkers, who have correlated the induction of the pdu operon with the occurrence of enterosomes on a metabolic shift of S. enterica serovar Typhimurium to 1,2-propanediol (32). Immunogold labeling indicated that the S. enterica enterosome contains diol dehydratase, an enzyme central to the catabolism of propanediol (9). With the help of antibodies raised against recombinant PduA, this protein was localized to the periphery of the enterosome, thereby confirming its role as a shell protein (G. D. Havemann, E. M. Sampson, and T. A. Bobik, Abstr. Annu. Meet. Am. Soc. Microbiol. 2001, abstr. 35/K4, 2001). Although it has been suggested that enterosomes formed during the growth of Salmonella on ethanolamine contain the key catabolic enzyme ethanolamine ammonia-lyase, at present no experimental evidence exists to directly support this view.
The precise advantage that enterosomes present for these organisms by presumably sequestering specific metabolic enzyme activities is not clear. Possibilities include concentration of CO2 within the microcompartments to supply an as yet unknown CO2-requiring reaction, sequestration of toxic by-products of propanediol or ethanolamine degradation reactions as a protective mechanism, and protection from inhibiting oxygen of diol dehydratase or ethanolamine ammonia-lyase (9, 32, 36, 52, 62). The first possibility seems unlikely, since CO2 is not a substrate for any of the known reactions involved, and while containment of toxic by-products is plausible, the protection of compartmentalized enzymes from oxygen would draw a direct analogy to a possible function of carboxysomes in cyanobacteria and chemoautotrophs. Alternatively, other enzymes of ethanolamine and propanediol catabolism might be present in their respective enterosomes. Increased concentration of substrates at the active sites of these enzymes might enhance their catalytic efficiency through a process somewhat similar to channeling. This mechanism would be analogous to the proposed role of carboxysomes in the CCM of cyanobacteria. Clearly, direct experimental evidence is needed to establish the exact mechanism(s) by which microcompartmentalization of enzymes into carboxysomes and enterosomes provides catalytic advantages to the cell.
SUMMARY
Nearly 30 years ago, the polyhedral bodies observed in cyanobacteria and some chemoautotrophs were shown to contain the primary carbon-assimilating enzyme of the organisms. Since that time, our knowledge of prokaryotic autotrophic metabolism and of carboxysome function has greatly increased. It has become increasingly clear that these organisms play an important part in maintaining the global carbon balance, but a more complete understanding of the molecular mechanisms by which they assimilate CO2 is needed to fully fathom the profound effect these autotrophic prokaryotes have on Earth's environment. Many questions remain unanswered about the details of carboxysome function, such as the biochemical mechanism of CO2 fixation enhancement and whether carboxysomes perform the same function in all autotrophic prokaryotes. The hypothesis most often presented, that carboxysomes contain carbonic anhydrase in addition to RuBisCO, needs to be tested rigorously in the absence of contaminating cellular membranes. Experimentation needs to be initiated that will examine the molecular interactions driving carboxysome biogenesis and regulatory strategies that ensure a rapid response to changes in cellular needs for CO2 fixation.
The fact that the carboxysomal shell has functional homologs in heterotrophic prokaryotes with completely unrelated metabolisms strongly supports the view of its unique property, whose nature remains to be elucidated. Its potential ability to selectively exclude oxygen would certainly be consistent with the needs of the enzymes from three different metabolic pathways that are microcompartmentalized. However, it is difficult to conceive a proteinaceous membrane that can selectively exclude oxygen while allowing CO2 and ribulose 1,5-bisphosphate to enter the particle. Given the recent interest in materials capable of performing complex selective chemistry at the nanoscale level, it is possible that a better understanding of bacterial microcompartmentalization will lead to the development of new materials with a wide range of applications in medicine, agriculture, and engineering.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support for part of the work reported herein by the U.S. Department of Agriculture (grant 92-37306-7663 to J.M.S.), the National Science Foundation (grant MCB-9513481 to J.M.S.), and the T. W. Bennett Distinguished Professor Fund (to G.C.C.).
REFERENCES
- 1.Amichay D, Levitz R, Gurevitz M. Construction of a Synechocystis PCC6803 mutant suitable for the study of variant hexadecameric ribulose bisphosphate carboxylase/oxygenase enzymes. Plant Mol Biol. 1993;23:465–476. doi: 10.1007/BF00019295. [DOI] [PubMed] [Google Scholar]
- 2.Badger M R, Price G D. The CO2 concentrating mechanism in cyanobacteria and microalgae. Physiol Plant. 1992;90:529–536. [Google Scholar]
- 3.Baker S H, Jin S, Aldrich H C, Howard G T, Shively J M. Insertion mutation of the form I cbbL gene encoding ribulose bisphosphate carboxylase/oxygenase (RuBisCO) in Thiobacillus neapolitanus results in expression of form II RuBisCO, loss of carboxysomes, and an increased CO2 2requirement for growth. J Bacteriol. 1998;180:4133–4139. doi: 10.1128/jb.180.16.4133-4139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker S H, Lorbach S C, Rodriguez-Buey M, Williams D S, Aldrich H C, Shively J M. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch Microbiol. 1999;172:233–239. doi: 10.1007/s002030050765. [DOI] [PubMed] [Google Scholar]
- 5.Baker S H, Williams D S, Aldrich H C, Gambrell A C, Shively J M. Identification and localization of the carboxysome peptide Csos3 and its corresponding gene in Thiobacillus neapolitanus. Arch Microbiol. 2000;173:278–283. doi: 10.1007/s002030000141. [DOI] [PubMed] [Google Scholar]
- 6.Bedu S, Laurent B, Joset F, editors. Membranous and soluble carbonic anhydrase in a cyanobacterium, Synechocystis PCC 6803. III. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. [Google Scholar]
- 7.Beudeker R F, Cannon G C, Kuenen J G, Shively J M. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes, and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch Microbiol. 1980;124:185–189. [Google Scholar]
- 8.Beudeker R F, Kuenen J G. Carboxysomes: “Calvinosomes”? FEBS Lett. 1981;131:269–274. [Google Scholar]
- 9.Bobik T A, Havemann G D, Busch R J, Williams D S, Aldrich H C. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonfil D J, Ronen-Tarazi M, Sultemeyer D, Lieman-Hurwitz J, Schatz D, Kaplan A. A putative HCO3 transporter in the cyanobacterium Synechococcus sp. strain PCC 7942. FEBS Lett. 1998;430:236–240. doi: 10.1016/s0014-5793(98)00662-0. [DOI] [PubMed] [Google Scholar]
- 11.Cannon G C. Carboxysomes and CO2 fixation in Thiobacillus neapolitanus. Doctoral dissertation. Clemson, S.C: Clemson University; 1982. [Google Scholar]
- 12.Cannon G C, English R S, Shively J M. In situ assay of ribulose-1,5-bisphosphate carboxylase/oxygenase in Thiobacillus neapolitanus. J Bacteriol. 1991;173:1565–1568. doi: 10.1128/jb.173.4.1565-1568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannon G C, Shively J M. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol. 1983;134:52–59. [Google Scholar]
- 14.Chisholm S W, Rocap G, Moore L. Research abstracts from the DOE human genome program contractor-grantee workshop VIII. [Online.] 2001. Prochlorococcus: The smallest and most abundant photosynthetic microbe in the oceans. [Google Scholar]
- 15.Codd G A, Marsden W J N. The carboxysomes (polyhedral bodies) of autotrophic prokaryotes. Biol Rev. 1984;59:389–422. [Google Scholar]
- 16.Codd G A, Stewart W D P. Polyhedral bodies and ribulose 1,5-diphosphate carboxylase of the blue-green alga Anabaena cylindrica. Planta. 1976;130:323–326. doi: 10.1007/BF00387840. [DOI] [PubMed] [Google Scholar]
- 17.Dubbs J M, Bird T H, Bauer C E, Tabita F R. Interaction of CbbR and RegA∗ transcription regulators with the Rhodobacter sphaeroides cbbI promoter-operator region. J Biol Chem. 2000;275:19224–19230. doi: 10.1074/jbc.M002125200. [DOI] [PubMed] [Google Scholar]
- 18.Ebert A. Ribulose-1,5-bisphosphate carboxylase in Nitrobacter. Ph.D. dissertation. Hamburg, Germany: University of Hamburg; 1982. [Google Scholar]
- 19.English R S, Lorbach S C, Qin X, Shively J M. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 20.Fridlyand L, Kaplan A, Reinhold L. Quantitative evaluation of the role of a putative CO2-scavenging entity in the cyanobacterial CO2-concentrating mechanism. Biosystems. 1996;37:229–238. doi: 10.1016/0303-2647(95)01561-2. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg D, Kaplan A, Ariel R, Kessel M, Seijffers J. The 5′-flanking region of the gene encoding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is crucial for growth of the cyanobacterium Synechococcus sp. strain PCC 7942 at the level of CO2 in air. J Bacteriol. 1989;171:6069–6076. doi: 10.1128/jb.171.11.6069-6076.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuzawa H, Suzuki E, Komukai Y, Miyachi S. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc Natl Acad Sci USA. 1992;89:4437–4441. doi: 10.1073/pnas.89.10.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gantt E, Conti S F. Ultrastructure of blue-green algae. J Bacteriol. 1967;97:1486–1493. doi: 10.1128/jb.97.3.1486-1493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartman F C, Harpel M R. Structure, function, regulation, and assembly of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu Rev Biochem. 1994;63:197–234. doi: 10.1146/annurev.bi.63.070194.001213. [DOI] [PubMed] [Google Scholar]
- 25.Holthuijzen Y A, Kuenen J G, Konings W N. Activity of ribulose-1,5-bisphosphate carboxylase in intact and disrupted carboxysomes of Thiobacillus neapolitanus. FEMS Microbiol Lett. 1987;42:121–124. [Google Scholar]
- 26.Holthuijzen Y A, Maathuis F J M, Kuenen J G, Konings R N H, Konings W N. Carboxysomes of Thiobacillus neapolitanus do not contain extra chromosomal DNA. FEMS Microbiol Lett. 1986;35:193–198. [Google Scholar]
- 27.Holthuijzen Y A, VanBreeman J F L, Konings W N, VanBruggen E F J. Electron microscopic studies of carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:258–262. [Google Scholar]
- 28.Holthuijzen Y A, VanBreemen J F L, Kuenen J G, Konings W N. Protein composition of the carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:398–404. [Google Scholar]
- 29.Holthuijzen Y A, VanDissel-Emiliani E F M, Kuenen J G, Konings W N. Energetic aspects of CO2 uptake in Thiobacillus neapolitanus. Arch Microbiol. 1987;147:285–290. [Google Scholar]
- 30.Jeffke T, Gropp N H, Kaiser C, Grzeszik C, Kusian B, Bowien B. Mutational analysis of the cbb operon (CO2 assimilation) promoter of Ralstonia eutropha. J Bacteriol. 1999;181:4374–4380. doi: 10.1128/jb.181.14.4374-4380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen T E, Bowen C C. Organization of the centroplasm in Nostoc pruniforme. Proc Iowa Acad Sci. 1961;68:89–96. [Google Scholar]
- 32.Johnson C L, Pechonick E, Park S D, Havemann G D, Leal N A, Bobik T A. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol. 2001;183:1577–1584. doi: 10.1128/JB.183.5.1577-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan A, Friedberg D, Schwarz R, Ariel R, Seijffers J, Reinhold L. The “CO2 concentrating mechanism” of cyanobacteria: physiological, molecular and theoretical studies. Photosynth Res. 1989;17:243–255. [Google Scholar]
- 34.Kaplan A, Reinhold L. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:539–570. doi: 10.1146/annurev.arplant.50.1.539. [DOI] [PubMed] [Google Scholar]
- 35.Klughammer B, Sultemeyer D, Badger M R, Price G D. The involvement of NAD(P)H dehydrogenase subunits, NdhD3 and NdhF3, in high-affinity CO2 uptake in Synechococcus sp. PCC7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol Microbiol. 1999;32:1305–1315. doi: 10.1046/j.1365-2958.1999.01457.x. [DOI] [PubMed] [Google Scholar]
- 36.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanaras T, Codd G A. Ribulose 1,5-bisphosphate carboxylase and polyhedral bodies of Chlorogloeopsis fritschii. Planta. 1981;153:279–285. doi: 10.1007/BF00383900. [DOI] [PubMed] [Google Scholar]
- 38.Lanaras T, Hawthornthwaite A M, Codd G A. Localization of carbonic anhydrase in the cyanobacterium Chlorogloeopsis fritschii. FEMS Microbiol Lett. 1985;26:285–288. [Google Scholar]
- 39.Ludwig M, Sultemeyer D, Price G D. Isolation of ccmKLMN genes from the marine cyanobacterium Synechococcus sp. PCC7002 and evidence that CcmM is essential for carboxysome assembly. J Phycol. 2000;36:1109–1118. [Google Scholar]
- 40.Marco E, Martinez I, Ronen-Tarazi M, Orus I, Kaplan A. Inactivation of ccmO in Synechococcus sp. strain PCC 7942 results in a mutant requiring high levels of CO2. Appl Environ Microbiol. 1994;60:1018–1020. doi: 10.1128/aem.60.3.1018-1020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcus Y, Berry J A, Pierce J. Photosynthesis and photorespiration in a mutant of the cyanobacterium Synechocystis PCC 6803 lacking carboxysomes. Planta. 1992;187:511–516. doi: 10.1007/BF00199970. [DOI] [PubMed] [Google Scholar]
- 42.Miller A G, Colman B. Active transport and accumulation of bicarbonate by a unicellular cyanobacterium. J Bacteriol. 1980;143:1253–1259. doi: 10.1128/jb.143.3.1253-1259.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohkawa H, Sonoda M, Katoh A, Ogawa T. The use of mutants in the analysis of the CO2-concentrating mechanism in cyanobacteria. Can J Bot. 1998;76:1035–1042. [Google Scholar]
- 44.Orus I, Rodriguez M L, Martinez F, Marco E. Biogenesis and ultrastructure of carboxysomes from wild type and mutants of Synechococcus sp. strain PCC7942. Plant Physiol. 1995;107:1159–1166. doi: 10.1104/pp.107.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orus M I, Rodriguez-Buey M L, Marco E, Fernandez-Valiente E. Changes in carboxysome structure and grouping and in photosynthetic affinity for inorganic carbon in Anabaena strain PCC 7119 (Cyanophyta) in response to modification of CO2 and Na+ supply. Plant Cell Physiol. 2001;42:46–53. doi: 10.1093/pcp/pce005. [DOI] [PubMed] [Google Scholar]
- 46.Price G D, Badger M R. Evidence for the role of carboxysomes in the cyanobacterial CO2-concentrating mechanism. Can J Bot. 1991;69:963–973. [Google Scholar]
- 47.Price G D, Badger M R. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype. Plant Physiol. 1989;91:505–513. doi: 10.1104/pp.91.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price G D, Badger M R. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC 7942: two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol. 1989;91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price G D, Coleman J R, Badger M R. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price G D, Howitt S M, Harrison K, Badger M R. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol. 1993;175:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price G D, Sultemeyer D, Klughammer B, Ludwig M, Badger M R. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- 52.Price-Carter M, Tingey J, Bobik T A, Roth J R. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purohit K, McFadden B A, Cohen A L. Purification, quaternary structure, composition, and properties of d-ribulose-1,5-bisphosphate carboxylase from Thiobacillus intermedius. J Bacteriol. 1976;127:505–515. doi: 10.1128/jb.127.1.505-515.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purohit K, McFadden B A, Shaykh M M. d-Ribulose-1,5-bisphosphate carboxylase and polyhedral inclusion bodies in Thiobacillus intermedius. J Bacteriol. 1976;127:516–522. doi: 10.1128/jb.127.1.516-522.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raven J A. Inorganic carbon acquisition by marine autotrophs. Adv Bot Res. 1997;27:85–208. [Google Scholar]
- 56.Reinhold L, Kosloff R, Kaplan A. A model for inorganic carbon fluxes and photosynthesis in cyanobacterial carboxysomes. Can J Bot. 1991;69:984–988. [Google Scholar]
- 57.Satoh R, Himeno M, Wadano A. Carboxysomal diffusion resistance to ribulose 1,5-bisphosphate and 3-phosphoglycerate in the cyanobacterium Synechococcus PCC7942. Plant Cell Physiol. 1997;38:769–775. [Google Scholar]
- 58.Schneider G, Lindqvist Y, Brändén C I. RUBISCO: structure and mechanism. Annu Rev Biophys Biomol Struct. 1992;21:119–143. doi: 10.1146/annurev.bb.21.060192.001003. [DOI] [PubMed] [Google Scholar]
- 59.Shively J M, Ball F, Brown D H, Saunders R E. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) in Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 60.Shively J M, Ball F L, Kline B W. Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J Bacteriol. 1973;116:1405–1411. doi: 10.1128/jb.116.3.1405-1411.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shively J M, Bock E, Westphal K, Cannon G C. Icosahedral inclusions (carboxysomes) of Nitrobacter agilis. J Bacteriol. 1977;132:673–675. doi: 10.1128/jb.132.2.673-675.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shively J M, Bradburne C E, Aldrich H C, Bobik T A, Mehlman J L, Jin S, Baker S H. Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can J Bot. 1998;76:906–916. [Google Scholar]
- 63.Shively J M, English R S. The carboxysome, a prokaryotic organelle: a mini review. Can J Bot. 1991;69:957–962. [Google Scholar]
- 64.Shively J M, Lorbach S C, Jin S, Baker S H. Carboxysomes: the genes of Thiobacillus neapolitanus. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer; 1996. pp. 56–63. [Google Scholar]
- 65.Shively J M, van Keulen G, Meijer W G. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu Rev Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- 66.So A K, Espie G S. Cloning, characterization and expression of carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Plant Mol Biol. 1998;37:205–215. doi: 10.1023/a:1005959200390. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki E, Fukuzawa H, Miyachi S. Identification of a genomic region that complements a temperature- sensitive, high CO2-requiring mutant of the cyanobacterium, Synechococcus sp. PCC7942. Mol Gen Genet. 1991;226:401–408. doi: 10.1007/BF00260652. [DOI] [PubMed] [Google Scholar]
- 68.Tabita F R. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: A different perspective. Photosynth Res. 1999;60:1–28. [Google Scholar]
- 69.Tabita F R, Gibson J L, Falcone D L, Lee B G, Chen J H. Recent studies on the molecular biology and biochemistry of CO2 fixation in phototrophic bacteria. FEMS Microbiol Rev. 1990;7:437–443. doi: 10.1111/j.1574-6968.1990.tb04950.x. [DOI] [PubMed] [Google Scholar]
- 70.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van den Bergh E R, Dijkhuizen L, Meijer W G. CbbR, a LysR-type transcriptional activator, is required for expression of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J Bacteriol. 1993;175:6097–6104. doi: 10.1128/jb.175.19.6097-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 73.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 74.van Keulen G, Girbal L, van den Bergh E R, Dijkhuizen L, Meijer W G. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J Bacteriol. 1998;180:1411–1417. doi: 10.1128/jb.180.6.1411-1417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waterbury J B, Watson S W, Valois F W, Franks D G. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Bull Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 76.Westphal K, Bock E, Cannon G C, Shively J M. Deoxyribonucleic acid in Nitrobacter carboxysomes. J Bacteriol. 1979;140:285–288. doi: 10.1128/jb.140.1.285-288.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]