Development and Characterization of Human Primary Cholangiocarcinoma Cell Lines (original) (raw)
Abstract
Cholangiocarcinoma (CCA) is the second most common primary liver tumor and is associated with late diagnosis, limited treatment options, and a 5-year survival rate of around 30%. CCA cell lines were first established in 1971, and since then, only 70 to 80 CCA cell lines have been established. These cell lines have been essential in basic and translational research to understand and identify novel mechanistic pathways, biomarkers, and disease-specific genes. Each CCA cell line has unique characteristics, reflecting a specific genotype, sex-related properties, and patient-related signatures, making them scientifically and commercially valuable. CCA cell lines are crucial in the use of novel technologies, such as three-dimensional organoid models, which help to model the tumor microenvironment and cell-to-cell crosstalk between tumor-neighboring cells. This review highlights crucial information on CCA cell lines, including: i) type of CCA (eg, intra- or extrahepatic), ii) isolation source (eg, primary tumor or xenograft), iii) chemical digestion method (eg, trypsin or collagenase), iv) cell-sorting method (colony isolation or removal of fibroblasts), v) maintenance-medium choice (eg, RPMI or Dulbecco's modified Eagle's medium), vi) cell morphology (eg, spindle or polygonal shape), and vii) doubling time of cells.
Cholangiocarcinoma (CCA) is a heterogeneous group of malignancies originating from the biliary tree.1 CCA accounts for approximately 3% of all malignancies of the gastrointestinal system and is the second most common primary hepatic tumor, after hepatocellular carcinoma.2,3 The prevalence of CCA varies broadly by region; while the prevalence is 1.6 per 100,000 people in the United States, it could be as high as 85 to 90 per 100,000 people in northeastern Thailand, where infection with Opisthorchis viverrini, a CCA risk factor, is endemic.4 Several additional risk factors for cholangiocarcinogenesis have been identified, such as Clonorchis sinensis infection, bile duct cyst, primary sclerosing cholangitis, hepatolithiasis, cholelithiasis, and inflammatory bowel disease.5 Despite the development of state-of-the-art therapies, the 5-year survival rate of CCA is still under 30%.6,7
CCAs are commonly classified, according to anatomic location along the biliary tree, as extrahepatic or intrahepatic (EHCCA or IHCCA).8 EHCCAs are subcategorized as perihilar or distal.8 CCAs can also be subclassified based on macroscopic growth pattern (mass forming, periductal infiltrating, intraductal, or mixed), cell of origin (cholangiocytes, goblet cells, hepatic stem cells, or biliary tree stem/progenitor cells), and/or microscopic features (adeno, squamous, adenosquamous, mucinous, undifferentiated, or sarcomatous).9,10
Although the importance of organ-specific or cancer-specific microenvironment has been recently highlighted due to cell-to-cell crosstalk, two-dimensional cancer cell lines are one of the best sources for investigating the respective cancer type.11,12 CCA cell lines have been used for almost 50 years to: i) better understand CCA properties, ii) investigate treatment options, iii) model the disease in vitro, and iv) generate in vivo xenograft models.13, 14, 15, 16 RPMI 7451 is the first known CCA cell line isolated by George Eugene Moore and his team.17 Dr. Moore, an oncologist, surgeon, accomplished scientist, and director of the RPMI, pioneered many cancer studies, established several additional cancer cell lines, and formulated RPMI 1640, a widely used cell-growth medium.18, 19, 20, 21, 22
There is currently a growing interest in CCA-related basic and translational research, due to poor outcomes with the currently available treatment options. For recently developed novel experimental models [eg, three-dimensional (3D) organoids] and treatments, CCA cell lines have been used for the identification of novel mechanistic pathways, biomarkers, and disease-specific genes. Therefore, previously isolated human primary CCA cell lines, their isolation methods, and known essential characteristics are summarized herein to help researchers in their future studies.
Human Primary CCA Cell Lines
The first CCA cell line was established about a half-century ago, in 1971 in the United States17; more than half of currently available CCA cell lines were generated in Japan between 1975 and 1990. To date, researchers from Italy, Germany, China, South Korea, and Thailand have reported on the isolation of CCA cell lines and related studies. In the past 2 decades, researchers have reported the isolation of multiple CCA cell lines in Thailand more than in any other country, possibly due to the high prevalence of the disease in that geographic region.4
Each CCA cell line is unique in reflecting a specific genotype, sex-related properties, and patient-related signatures, making them especially valuable for scientific and commercial applications. It would undoubtedly be helpful to have an international and collective cell bank that includes cell-identification analysis from each study to appropriately define the CCA properties and prevent any misidentification of the existing cell lines.23 For instance, the ETK-1 CCA cell line was retracted in 2004 after a short tandem repeat polymorphism analysis revealed that the ETK-1 and SSP-25 cell lines were identical.24,25 However, later, it was understood that SSP-25 should have been retracted instead of ETK-1 because the SSP-25 and RBE cell lines were isolated from the same patient, but their short tandem repeat profiling results did not match (https://cell.brc.riken.jp/en/rcb/rbessp-25_announce, last accessed May 5, 2022). As a result, the ETK-1 cells have been distributed under the name of SSP-25 since 2004, but it is unclear when that cell line was initially misidentified. It is also unclear what happened to the SSP-25 cell line. Similarly, the M156, M213, and M214 cell lines were believed to have been isolated from three separate patients; however, results from recent short tandem repeat profiling showed that they were isolated from the same patient.26
Cross-contamination of cell lines is a long-standing problem that has resulted in scientific errors27 and may initiate inaccurate data chains. Various levels of the cell-culturing process have been blamed for cross-contamination, including: i) accidental inoculation, ii) mislabeling, iii) confusion in freeze-thawing, iv) working with more than one cell line in a biosafety cabinet at the same time, and v) contamination of the stock bottle of media with cells.28,29 Additionally, cancer cells may carry a greater risk for cross-contamination due to their greater capacity for proliferation. Even a minimal amount of inoculation of cancer cell lines could suppress other cell lines in the culture and take their place over time. Therefore, the preservation and maintenance of the cell line is as important and challenging as is the establishment of one.
In a different case, CHGS was misidentified as a CCA cell line in 2015 by Zach et al30 and listed in the Cellosaurus database (accession number CVCL_M272; https://web.expasy.org/cellosaurus, last accessed May 5, 2022). However, the first mention of CHGS, in 1988 by Katoh et al,31 indicates that CHGS is a CCA tissue line passaged in mice, and that cancer cells have not been isolated from this tissue line. Similarly, the CC-CL-1 cell line was studied as a CCA cell line in a research study; however, none of the references cited in the study contain information regarding the CC-CL-1 cell line, generating doubts about its credibility.32
In addition to primary CCA cell lines, derivative CCA cell lines have been established for drug resistance studies. QBC939/ADM is doxorubicin resistant; HuCCT1-G100, YSCCC-G100, RTFK-1, KKU-M139/GEM, KKU-213B/GEM, MT-CHC01R1.5, and SNU-1196/GR are gemcitabine resistant; and KKU-M055/46 and KKU-213B/246 are 5-fluorouracil–resistant CCA cell lines.16,33, 34, 35, 36, 37 The KKU-213L5 cell line is a derivative of the KKU-213A cell line, with high metastatic activity.38
Tables 1 and 2 summarize the CCA cell lines available between 1971 and 2000, and between 2001 and 2021, respectively, from the English and non-English published literature. Tables 1 and 2 include: i) the type of CCA (eg, intra- or extrahepatic), ii) isolation source (eg, primary tumor or xenograft), iii) tissue-digestion method (eg, trypsin or collagenase), iv) cell-sorting and -purification methods (colony isolation or removal of fibroblasts), v) maintenance-medium choice (eg, RPMI or DMEM), vi) cell morphology (eg, spindle or polygonal shape), and vii) doubling time of cells.
Table 1.
Human Primary Cholangiocarcinoma Cell Lines Established between 1971 and 2000
| Cell no. | Year | Country | Cell name | Diagnosis | Isolation source | Digestion method | Sorting method | Maintenance medium | Cell morphology | Doubling time | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1971 | USA | RPMI 7451 | CCA | — | — | — | Eagle's MEM + 10% FBS + nonessential amino acids + 60 pg/mL gentamicin | Tightly adherent, monolayer, polygonal shaped cells | — | 17,39,40 |
| 2 | 1975 | Japan | H-1 or H1 | CCA | — | — | — | — | — | — | 40,41 |
| 3 | 1981 | Japan | OZ | IHCCA | Ascites | No digestion | Colony isolation by trypsin and EDTA-soaked filter; other cells removed by enzymatic and mechanical treatments | Williams' E + 10% newborn calf serum + P/S | Large nucleus with 1 to 2 nucleoli; dark cytoplasm; high nucleus/cytoplasm ratio; pavement-like proliferation. Abundant production of gel-like substance | 48 hours | 42 |
| 4 | 1984 | Germany | EGI-1 | CCA | Primary tumor | — | — | MEM + 10% FBS + 2× MEM amino acids (essential and nonessential) + 4 mmol/L l-glutamine + 1 mmol/L sodium pyruvate | Monolayer, adherent, polymorphic cells | 45–50 hours | 43 |
| 5 | 1985 | Japan | HChol-Y1 | CCA | Primary tumor | No digestion | Fibroblasts spontaneously disappeared in 2 months | Ham's F12 | Uniform, monolayer cells; including abundant granules; prominent round nucleus with multiple nucleoli | 52 hours | 44 |
| 6 | 1985 | Germany | SK-ChA-1 or WITT | EHCCA | Ascites | No digestion | Light trypsin treatment | CMRL + 15% + P/S | Adherent, spindle- to polygonal shaped, polymorphic cells; proliferating as single adherent cells or small clusters | 48 hours | 45,46 |
| 7 | 1987 | Japan | KMCH-1 | IHCCA + HCC | Primary tumor | 0.5 U/mg type IV collagenase in PBS, 37°C, 30 minutes | Colony isolation by trypsin | DMEM + 20% FBS + 35 μmol/L sodium bicarbonate + P/S | Large, round nucleus with multiple nucleoli; abundant clear cytoplasm; pavement-like proliferation; microvilli on the luminal surfaces | 39 hours | 47 |
| 8 | 1988 | Japan | HuH-28 | IHCCA | Primary tumor (frozen-thawed) | 500 U/mL trypsin | — | RPMI 1640 + 20% FBS + %0.2 lactalbumin hydrolysate | Spindle- to polygonal shaped cells | 80 hours | 48 |
| 9 | 1989 | Japan | HuCC-T1 | IHCCA | Ascites | No digestion | IS-RPMI medium used to eliminate fibroblasts | RPMI 1640 + 0.2% lactalbumin hydrolysate | Polygonal to spindle-shaped, abundant and clear cytoplasm, proliferation in pavement arrangement | 74 hours | 49,50 |
| 10 | 1989 | Japan | MEC | CCA | Pleural effusion | No digestion | — | RPMI 1640 + 20% FBS | Polymorphic, epithelial-like cells; high nucleus/cytoplasm ratio | 40 hours | 51 |
| 11 | 1990 | USA | PCI:SG231 | IHCCA | Primary tumor | Type II collagenase, 37°C, 3 hours | — | α MEM (Earle's salts) with nucleosides + 10% FBS + 60 pg/mL gentamicin | Tightly adherent, monolayer, polygonal shaped cells; nonadherent cell clusters that formed three-dimensional tubular structures resembling bile ducts | — | 40 |
| 12 | 1991 | Thailand | HuCCA-1 | IHCCA | Primary tumor | No digestion | Fibroblasts gradually decreased with each passage and disappeared in a month | Ham's F12 + P/S | Monolayer, adherent, polygonal shaped epithelial cells with occasionally multiple, round to oval nuclei; granule-filled cytoplasms; piling up of cells and occasional gland-like appearances | 55 hours | 52 |
| 13 | 1991 | Japan | KMBC | EHCCA | Xenograft | Collagenase 150–250 U/mg, 37°C, 90–110 minutes | Fibroblasts scraped away and completely disappeared after several passage | DMEM + 5% FBS + 12 mmol/L sodium bicarbonate + P/S | Polymorphic epithelial-like cells; one or more large, irregular, round to oval nuclei with a few prominent nucleoli; relatively poor, round to polygonal cytoplasm; pavement-like proliferation; tubular formation | 30 hours | 53,54 |
| 14 | 1992 | USA | CC-LP-1 | IHCCA | Primary tumor | 0.05% (w/v) collagenase type IV, 0.002% (w/v) DNase type 1, 90 minutes | Centrifugation on double-layer (75%/100% v/v) Ficoll-Hypaque gradients; fibroblasts removed by differential trypsinization | DMEM + 15% FBS + 2 mmol/L l-glutamine + antibiotics | Cobblestone-like proliferation of monolayers; stratification of cells in some areas | 180 hours | 55 |
| 15 | 1992 | CC-SW-1 | 72 hours | ||||||||
| 16 | 1992 | Japan | KMC-1 | IHCCA | Xenograft | 0.5 mg/nL type IV collagenase in PBS, 37°C, 60–80 minutes | Tumor cells suppressed the fibroblasts by the time | DMEM + 10% or 5% FBS | Monolayer, pavement-like proliferation; clear cytoplasm, oval-shaped nuclei; tubular formation; some cells with mucin in the cytoplasm | 54 hours | 56 |
| 17 | 1993 | China | QBC939 | EHCCA | Primary tumor | Type I collagenase, DNase I, plasminase | Fibroblasts gradually decreased by passaging | RPMI 1640 + 10% FBS | Monolayer, polymorphic, adherent cells; round or oval nuclei; high nucleus/cytoplasm ratio | 24 hours | 57,58 |
| 18 | 1994 | Japan | TK | EHCCA | Ascites | No digestion | Colony isolation using 0.33% agar | RPMI 1640 + 15% FBS + 2 mmol/L glutamine + 1 mmol/L sodium pyruvate | Monolayer, adherent proliferation; forming gland-like structures; lobated or dark, large nuclei | 29 hours | 59 |
| 19 | 1995 | Japan | OCUCh-LM1 | EHCCA | Primary tumor | No digestion | Fibroblasts gradually decreased and disappeared in 1 month | DMEM + 10% FBS + 2 mmol/L l-glutamine + 0.5 mmol/L sodium pyruvate + P/S | Monolayer, pavement-like proliferation; clear cytoplasm and oval nuclei | 31 hours | 60 |
| 20 | 1995 | Japan | TFK-1 | EHCCA | Primary tumor | 1000 U/mL dispase, 37°C, 30 minutes | Fibroblasts removed by mechanical scraping and differential attachment selection with trypsin | RPMI 1640 + 10% FBS + P/S | Polygonal epithelial monolayers; pavement-like proliferation | 37 hours | 61 |
| 21 | 1996 | Japan | KMCH-2 | IHCCA + HCC | Primary tumor | 150–250 U/mg collagenase in PBS, 37°C, 90–110 minutes | Fibroblasts removed by mechanical scraping | DMEM + 5% FBS + 12 mmol/L sodium bicarbonate + P/S | Monomorphic polygonal cells; large round to oval nuclei with a few prominent nucleoli; pavement-like proliferation | 44 hours at 20th passage; 32 hours at 55th passage | 62 |
| 22 | 1997 | Japan | ETK-1 | IHCCA | Ascites | No digestion | Limiting dilution | RPMI 1640 + 10% FBS + 10 mmol/L HEPES + 2 mmol/L l-glutamine + 0.1 mmol/L nonessential amino acids + 1 mmol/L sodium pyruvate + 0.005 mmol/L β-mercaptoethanol | Small polygonal cells; round to oval nuclei and prominent nucleoli; uniform, monolayer with a pavement-like proliferation | 71 hours | 41 |
| 23 | 1997 | Japan | ICBD-1 | EHCCA | Primary tumor | No digestion | Fibroblasts gradually decreased and disappeared in 1 month | RPMI 1640 + 10% FBS + 0.5 mmol/L sodium bicarbonate + 2 mmol/L l-glutamine + P/S | Large round to oval nuclei with a few nucleoli; relatively poor, polygonal to oval cytoplasm; pavement-like proliferation | 31.5 hours | 63 |
| 24 | 1997 | Japan | SSP-25 | IHCCA | Primary tumor | No digestion | Fibroblasts gradually disappeared, limiting dilution | RPMI 1640 + 10% FBS | Spindle-shaped cells | 64 hours | 64 |
| 25 | RBE | Monolayer of polygonal cells, pavement-like proliferation | 45 hours | ||||||||
| 26 | 1998 | Japan | TBCN-1 | EHCCA | Primary tumor | — | — | RPMI 1640 + 10% FBS | — | 54 hours | 65 |
| 27 | 1999 | Thailand | KKU-213A | IHCCA | Primary tumor | 0.25% trypsin-EDTA, 37°C, 1 hour | — | DMEM + 10% FBS + A/A | Small, spindle-shaped cells | 23 hours | 26 |
| 28 | KKU-213B | Irregular polygonal cells; high nucleus to cytoplasmic ratio; patch-like structure | 24.5 hours | ||||||||
| 29 | KKU-213C | Irregular polygonal cells; high nucleus to cytoplasmic ratio | 25.6 hours | ||||||||
| 30 | 1999 | Japan | YSCCC | CCA | — | — | — | RPMI 1640 + 10% FBS | Lymphocyte-like | — | 66, _https://cellbank.brc.riken.jp/cell_bank/CellInfo/?cellNo_=_RCB1549&lang_=, last accessed May 5, 2022 |
| 31 | 2000 | Japan | HBDC | EHCCA | Ascites | No digestion | Centrifugation on triple-layer (75%/100%/25) Ficoll-Hypaque gradients; colony isolation using porcelain cloning rings | Williams' E + 10% FBS + 2 ng/mL HGF + 2 mmol/L l-glutamine + 6 mmol/L glucose + 0.5 mmol/L sodium bicarbonate + 100 ng/mL kanamycin + 10 ng/mL fungizone | Polygonal or spindle-shaped polymorphic cells; occasional large vacuoles in the cytoplasm; forming small clusters or clumps; one or more large, irregular, round or oval nuclei with a few prominent nucleoli; pavement-like proliferation | 32 hours | 67 |
Table 2.
Human Primary Cholangiocarcinoma Cell Lines Established between 2001 and 2021
| Cell no. | Year | Country | Cell name | Diagnosis | Isolation source | Digestion method | Sorting method | Maintenance medium | Cell morphology | Doubling time | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2001 | Republic of Korea | Choi-CK | IHCCA | Primary tumor | No digestion | Differential trypsinization, scraping, or G418 treatment (100 μg/mL) | Opti-MEM + 10% FBS + 30 mmol/L sodium bicarbonate + antibiotics | Polygonal and compact cells with indistinct cell boundaries | — | 75 |
| 2 | Cho-CK | Polygonal and compact cells with indistinct cell boundaries | |||||||||
| 3 | JCK | Polygonal morphology and some spindle-shaped cells | |||||||||
| 4 | SCK | Monolayer, epithelioid or spindle-shaped cells; fewer polygonal shaped cells | |||||||||
| 5 | 2002 | Thailand | HubCCA-1 | IHCCA | Intrahepatic biliary fluid | — | — | Ham's F12 + 20% FBS + P/S | Adherent, monolayer, polygonal to spindle-shaped cells | — | 76,77 |
| 6 | 2002 | Thailand | KKU-100 | IHCCA | Primary tumor | 0.025% trypsin-EDTA, 37°C, 1 hour | Fibroblasts removed with a cell scraper and by differential trypsinization | Ham's F12 + 20% FBS + P/S | Compact, polygonal to spindle-shaped cells; floating or clumping in a confluent monolayer; large nucleus containing two to five nucleoli and a clear cytoplasm | 72 hours | 77,78 |
| 7 | 2002 | Republic of Korea | SNU-245 | EHCCA | Primary tumor | No digestion | Differential trypsinization used to remove fibroblasts | RPMI 1640 + 10% FBS | Monolayer, adherent cells; trabecular arrangement with acinar formation | 54 hours | 79 |
| 8 | SNU-1079 | IHCCA | Monolayer, adherent cells; pleomorphic appearance with multiple cytoplasmic processes; numerous cytoplasmic vacuoles in some cells; some multinucleated cells | 72 hours | |||||||
| 9 | SNU-1196 | EHCCA | Monolayer, adherent cells; trabecular pattern consisted of spindle to polygonal shaped cells having vesicular nuclei and multiple small nucleoli | 48 hours | |||||||
| 10 | 2004 | Japan | TKKK | IHCCA | — | — | — | DMEM (low glucose) + 10% FBS | Adherent, monolayer, epithelial-like cells; pavement-like proliferation | — | 80 |
| 11 | 2005 | Thailand | KKU-M055 | IHCCA | — | — | — | Ham's F-12 + 10% FBS + P/S | — | — | 81 |
| 12 | KKU-OCA17 | ||||||||||
| 13 | 2005 | Japan | TBCN6 | EHCCA | Xenograft | 0.25% trypsin in PBS, 37°C, 10 minutes | — | DMEM + 10% FBS | Adherent, monolayer, polygonal cells | 38 hours | 82 |
| 14 | TGBC-47 | ||||||||||
| 15 | 2006 | Thailand | KKU-M139 | CCA | Primary tumor | — | — | Ham's F12 or RPMI 1640 + 10% FBS + P/S | Adherent, monolayer, polygonal cells; pavement-like proliferation | 17 hours | 16,76 |
| 16 | 2006 | Thailand | RMCCA-1 | IHCCA | Primary tumor | No digestion | Differential trypsinization used to remove fibroblasts | Ham's F12 + 20% FBS + EGF + 250 μg/mL amphotericin + P/S | Circular to spindle shape with many processes and ornamental fringes; granulated nucleus and cytoplasm | 48 hours | 83 |
| 17 | 2007 | China | HKGZ-CC | IHCCA | Primary tumor | 1200–2000 U/mL collagenase and DNase in 10 mL/g DPBS | — | DMEM | Adherent, monolayer, epithelial-like cells | 48 hours | 84 |
| 18 | 2007 | Japan | IHGGK | IHCCA | — | — | — | RPMI 1640 + 10% FBS + P/S | Adherent, monolayer, epithelial-like cells | 85, http://www2.idac.tohoku.ac.jp/dep/ccr/TKGdate/TKGvo106/0623.html, last accessed May 5, 2022 | |
| 19 | 2009 | Thailand | CL-2 | CCA | — | — | — | DMEM + 15% FBS + P/S | — | — | 86,87 |
| 20 | CL-6 | ||||||||||
| 21 | CL-19 | ||||||||||
| 22 | 2010 | Japan | NCC-BD1 | EHCCA | Xenograft | No digestion | Fibroblasts removed by mechanical scraping | RPMI 1640 + 10% FBS + 2 mmol/L l-glutamine + P/S | Adherent, monolayer, epithelial-like cells; pavement-like proliferation | — | 88 |
| 23 | NCC-BD2 | EHCCA | |||||||||
| 24 | NCC-CC1 | IHCCA | |||||||||
| 25 | NCC-CC3-1 | IHCCA | |||||||||
| 26 | NCC-CC3-2 | IHCCA | |||||||||
| 27 | NCC-CC4-1 | IHCCA | |||||||||
| 28 | 2013 | China | HCCC-9810 | IHCCA | — | — | — | RPMI 1640 + 10% FBS + P/S | — | 20 hours | 89 |
| 29 | 2015 | Italy | MT-CHC01 | IHCCA | Xenograft | 200 U/mL collagenase, 3 hours, 37°C | — | Knockout/DMEM/F-12 + 10% FBS + P/S | Monolayer, adherent cells | 40 hours | 90 |
| 30 | 2016 | USA | ICC1 | IHCCA | — | Trypsin, 30 minutes, 37°C | — | RPMI or DMEM/F12 + 5% FBS + P/S | — | 91 | |
| 31 | ICC2 | ||||||||||
| 32 | ICC5 | ||||||||||
| 33 | 2016 | China | ZJU-0826 | EHCCA | Primary tumor | — | Fibroblasts eliminated by differential trypsinization and differential attachment | RPMI 1640 + 10% FBS + antibiotics | Monolayer, adherent, homogeneous cells with characteristic loose pleomorphic cells and rare multinucleated cells | 63 hours | 92 |
| 34 | ZJU-1125 | IHCCA | Monolayer, epithelial-like, adherent cells; polygonal, occasionally multinucleated | 44 hours | |||||||
| 35 | 2017 | Germany | CCC-5 | EHCCA | Pleural effusion | No digestion | — | DMEM + 20% FBS: KFSM - 2:1 | Monolayer, spindle- to polygonal shaped cells, pavement-like proliferation; anaplastic, multinucleated giant cells | 60 hours | 93 |
| 36 | 2017 | Thailand | KKU-023 | IHCCA | Primary tumor | 1 mg/mL collagenase, 37°C, 30–45 minutes | Fibroblasts aseptically removed with a cell scraper | Ham's F12 + 10% FBS + P/S + nonessential amino acids + 12.5 mmol/L HEPES + 50 μg/mL cefazolin + 10 μg/mL ciprofloxacin + 2.5 μg/mL amphotericin B + 5 μmol/L ROCK inhibitor | Ovoid to cuboid shape polygonal cells; seldom multinucleated; forming compact monolayer with occasional multinucleated cells | 34 hours | 94 |
| 37 | KKU-452 | EHCCA | Spindle-shaped cells; seldom multinucleated; segregated and spread surround | 17 hours | |||||||
| 38 | 2018 | China | ICC-1 | IHCCA | Primary tumor | No digestion | — | — | — | — | 73 |
| 39 | ICC-2 | ||||||||||
| 40 | 2019 | Thailand | KKK-D049 | IHCCA | Xenograft | 1000 U/mL collagenase + 0.1 mg/mL DNase I, 3 hours | Stromal cells sequentially removed by partial trypsinization and mechanical removal | DMEM + 10% FBS + P/S | Epithelial-like cells; high nuclear to cytoplasmic ratio; tight clustering | — | 95 |
| 41 | KKK-D068 | Epithelial-like, polygonal and spindle-shaped cells; high nuclear to cytoplasmic ratio | — | ||||||||
| 42 | KKK-D131 | Epithelial-like, polygonal and spindle-shaped cells; high nuclear to cytoplasmic ratio | — | ||||||||
| 43 | KKK-D138 | Epithelial-like, polygonal and spindle-shaped cells; high nucleus-to-cytoplasm ratio | — | ||||||||
| 44 | 2021 | Italy | 82.3 | IHCCA | Xenograft | Collagenase 200 U/mL, 3 hours, 37°C | 10% FBS + P/S + gemcitabine | Monolayer, adherent, epithelial-like cells | 53 hours | 74 |
Overview of Isolation Methods
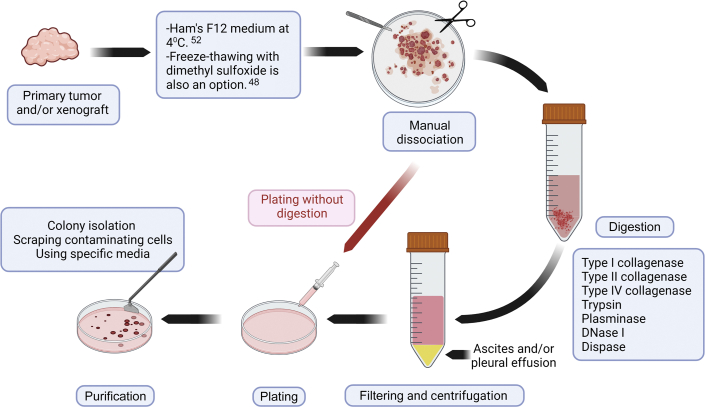
The general principles of the CCA cell–isolation protocol are summarized in Figure 1. CCA cell isolation requires either a solid sample (eg, primary tumor or xenograft) or a liquid sample (eg, ascites or pleural effusion). Samples can come from autopsy, surgery, paracentesis, or animal harvesting. Samples can be transferred into Ham's F12 culture medium at 4°C.52 It is better to process the samples as soon as possible to avoid ischemia-related problems in cells. If the CCA sample needs to be preserved, it can be freeze-thawed in 10% dimethyl sulfoxide in culture medium48 or can be preserved in histidine-tryptophan-ketoglutarate organ-preservation solution at 4°C until tissue processing.68
Figure 1.
CCA cell isolation method. CCA cell lines can be isolated from either fresh samples (eg, HuCCA-1 cell line) or frozen-thawed samples (eg, huh-28 cell line) from primary tumors or from xenografts, kept in different mediums, after manual dissociation, plated with or without digestion.
CCA cell lines can be isolated using several methods.44,47 Herein, the general perspective of isolation protocols is briefly summarized. All tissue processing should be performed under aseptic conditions in a biological safety cabinet with sterile or disposable surgical instruments. The first and the most essential step of CCA cell isolation from solid samples is cutting the tissue into small pieces (>1 mm in diameter).44, 45, 46, 47 After that, the protocol can be continued either with or without enzymatic digestion. For the protocols without enzymatic digestion, a stainless-steel mesh is helpful for releasing CCA cells.44 Chemical digestion usually proceeds at 37°C, and the digestion time is dependent on the concentration and type of the enzyme used (Table 1). Several types of collagenases (types I, II, and IV) and trypsin have been successfully used for chemical digestion (Table 1). Plasminase to digest fibrin clumps and DNase I to lyse DNA leaked into cell suspension can be included in the dissociation protocol (Figure 1).57
After chemical dissociation, the cell suspension should be filtered with 70 μm of mesh and centrifuged47; the pellet can then be resuspended in a variety of media conditions [RPMI, Dulbecco's modified Eagle medium, minimum essential medium, William's, and Ham's F12].42, 43, 44,47,49 Stromal fibroblasts are the primary contaminating cells in CCA cell cultures.69 After cell attachment, CCA cells can be purified using scraping of the contaminating cells, colony isolation, and/or passaging cells until all contaminating cells have been removed (Figure 1).42,44,49
Need for More CCA Cell Lines
Cell lines provide indispensable in vitro model systems for the assessment of features of cancer cells, since they are direct descendants of the primary tumors.12 Under the right conditions, most of the phenotypic and genotypic properties of the primary tumor are preserved in cancer cell lines.11 After >50 years of research, these features have allowed for the establishment of thousands of cell lines from various malignancies, of which only 70 to 80 belong to the CCA family (Tables 1 and 2). While each new cell line is not necessarily superior to previous lines, collectively, they have been important for the understanding of common and differing features of CCA, including: i) antigenic variations, ii) new genotypes, and iii) enzymes that sustain tumorigenic growth to drug sensitivities or resistances.
One reason that CCA remains difficult to cure is the lack of a precise understanding of oncogenesis.70 The established CCA cell lines can be utilized to identify the properties of cells that can help in predicting positive or negative responses to antigen- or pathway-targeted therapies. CCA cell lines could be powerful in determining many aspects of CCA phenotypes, especially when used in a 3D microenvironment in the presence of other liver cells to study the impact of adjacent cells in the tumor milieu.71 The 3D tumor organoid system can be used for the identification of candidate novel therapies for future clinical trials.72
In a recent study, an established IHCCA cell line was used for understanding the impact of RA190, a proteasome subunit ADRM1 inhibitor that induces cell apoptosis, in a pathway-targeted therapy model.73 Although the outcomes of that in vitro study were encouraging, the study did not evaluate the roles of the tumor microenvironment and other CCA cell lines to determine whether a genotypic difference within the CCA lines may have modulated (increased or decreased) the response and sensitivity to RA190.73 A counterexample is evidenced by the recent discovery in Italy of a novel IHCCA cell line characterized by the presence of genes encoding resistance to fluorouracil, carboplatin, and oxaliplatin—drugs actively and frequently used in CCA treatment.74
Future Perspectives
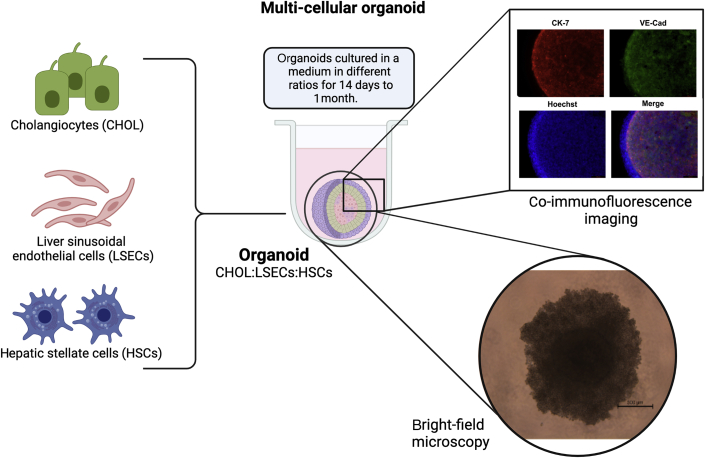
The establishment of new, well-defined, and well-characterized CCA cell lines is crucial in enhancing the understanding of this aggressive disease. The establishment of oncogenesis through the cancer cells provides information on their drug-sensitivity and drug-resistance mechanism, and helps to identify possible novel drug targets. The inclusion of tumor stromal cells, such as hepatic stellate cells, liver sinusoidal endothelial cells, and tumor-associated macrophages, in the CCA cell lines makes these culture models more representative of the CCA tumor microenvironment (ie, 3D tumor organoids)71 (Figure 2). Therefore, the inclusion of multiple other cell types will help to determine the ways in which CCA cells interact with other neighboring cells and alter their genotypes, phenotypes, and transcriptomic profiles.
Figure 2.
CCA organoids with multiple cell lines. Three-dimensional (3D) CCA organoids include multiple cell lines, such as CCA cell lines (cholangiocytes; CHOL), liver sinusoidal endothelial cells (LSECs), and hepatic stellate cells (HSCs) (representative). Other liver cells, such as hepatocytes and Kupffer cells, can also be added, if needed. These organoids can be made scaffold-free without any biomaterial (eg, Matrigel) in a low-binding plate using culture mediums and can be kept for 14 to 28 days. Evidence suggests that CCA cells express cytokeratin (CK)-7, a cholangiocyte marker, and organoids can be stained with other cellular markers (eg, vascular endothelial cadherin; VE-Cad) that are present in organoids. CCA organoids can be exposed to immune cells, as seen at the bottom right. Bright-field microscopy shows a 3D CCA organoid exposed to mast cells. Scale bar = 500 μm. Original magnification, ×10.
The establishment of well-characterized CCA cell lines with a close resemblance to primary tumors, especially in a 3D microenvironment, will provide an infinite capacity for replicability, limiting the use of small animal in vivo studies and making them prime materials for CCA research.
Author Contributions
A.I. drafted the manuscript. A.Y., D.S., and E.A. wrote and reviewed the manuscript; B.E. developed the concept and wrote and critically reviewed the manuscript. All of the authors approved the final version.
Footnotes
Partially supported by an ASTS Faculty Development grant (B.E.); Indiana University Health Values Fund for Research award VFR-457-Ekser (B.E.); an Indiana University Health Foundation Jerome A. Josephs Fund for Transplant Innovation grant (B.E.); a Hickam Endowed Chair, Gastroenterology, Medicine, Indiana University grant (G.A.); an Indiana University Health–Indiana University School of Medicine Strategic Research Initiative grant (G.A.); Senior Career Scientist award IK6 BX004601 (G.A.); VA merit award 5I01BX000574 (G.A.); US Department of Veteran's AffairsCareer Scientist AwardIK6BX005226 (H.F.); VA merit award 1I01BX003031 (H.F.); Biomedical Laboratory Research and Development Service NIH grants DK108959 (H.F.), DK119421 (H.F.), DK054811 (G.A. and S.G.), DK115184 (G.A. and S.G.), DK076898 (G.A. and S.G.), DK107310 (G.A. and S.G.), DK110035 (G.A. and S.G.), DK062975 (G.A. and S.G.), and AA028711 (G.A. and S.G.); a Partners Seeking a Cure grant (G.A.); CPRIT grant RP210213 (S.C.); and the Richard L. Roudebush VA Medical Center (Indianapolis, IN).
Disclosures: None declared.
The views expressed in this article are those of the authors and do not necessarily represent the views of the US Department of Veterans Affairs.
References
- 1.Sato K., Glaser S., Alvaro D., Meng F., Francis H., Alpini G. Cholangiocarcinoma: novel therapeutic targets. Expert Opin Ther Targets. 2020;24:345–357. doi: 10.1080/14728222.2020.1733528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan S.A., Tavolari S., Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. 2019;39(Suppl 1):19–31. doi: 10.1111/liv.14095. [DOI] [PubMed] [Google Scholar]
- 3.Fabris L., Sato K., Alpini G., Strazzabosco M. The tumor microenvironment in cholangiocarcinoma progression. Hepatology. 2021;73(Suppl 1):75–85. doi: 10.1002/hep.31410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirstein M.M., Vogel A. Epidemiology and risk factors of cholangiocarcinoma. Visc Med. 2016;32:395–400. doi: 10.1159/000453013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyson G.L., El-Serag H.B. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173–184. doi: 10.1002/hep.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loosen S.H., Vucur M., Trautwein C., Roderburg C., Luedde T. Circulating biomarkers for cholangiocarcinoma. Dig Dis. 2018;36:281–288. doi: 10.1159/000488342. [DOI] [PubMed] [Google Scholar]
- 7.Ahn D.H., Bekaii-Saab T. Biliary cancer: intrahepatic cholangiocarcinoma vs. extrahepatic cholangiocarcinoma vs. gallbladder cancers: classification and therapeutic implications. J Gastrointest Oncol. 2017;8:293–301. doi: 10.21037/jgo.2016.10.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kendall T., Verheij J., Gaudio E., Evert M., Guido M., Goeppert B., Carpino G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019;39(Suppl 1):7–18. doi: 10.1111/liv.14093. [DOI] [PubMed] [Google Scholar]
- 9.Krasinskas A.M. Cholangiocarcinoma. Surg Pathol Clin. 2018;11:403–429. doi: 10.1016/j.path.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Cardinale V., Carpino G., Reid L., Gaudio E., Alvaro D. Multiple cells of origin in cholangiocarcinoma underlie biological, epidemiological and clinical heterogeneity. World J Gastrointest Oncol. 2012;4:94–102. doi: 10.4251/wjgo.v4.i5.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masters J.R. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- 12.Mirabelli P., Coppola L., Salvatore M. Cancer cell lines are useful model systems for medical research. Cancers (Basel) 2019;11:1098. doi: 10.3390/cancers11081098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buranrat B., Prawan A., Kukongviriyapan U., Kongpetch S., Kukongviriyapan V. Dicoumarol enhances gemcitabine-induced cytotoxicity in high NQO1-expressing cholangiocarcinoma cells. World J Gastroenterol. 2010;16:2362–2370. doi: 10.3748/wjg.v16.i19.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panrit L., Plengsuriyakarn T., Martviset P., Na-Bangchang K. Inhibitory activities of plumbagin on cell migration and invasion and inducing activity on cholangiocarcinoma cell apoptosis. Asian Pac J Trop Dis. 2018;11:430–435. [Google Scholar]
- 15.Kotawong K., Chaijaroenkul W., Muhamad P., Na-Bangchang K. Cytotoxic activities and effects of atractylodin and beta-eudesmol on the cell cycle arrest and apoptosis on cholangiocarcinoma cell line. J Pharmacol Sci. 2018;136:51–56. doi: 10.1016/j.jphs.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Wattanawongdon W., Hahnvajanawong C., Namwat N., Kanchanawat S., Boonmars T., Jearanaikoon P., Leelayuwat C., Techasen A., Seubwai W. Establishment and characterization of gemcitabine-resistant human cholangiocarcinoma cell lines with multidrug resistance and enhanced invasiveness. Int J Oncol. 2015;47:398–410. doi: 10.3892/ijo.2015.3019. [DOI] [PubMed] [Google Scholar]
- 17.Fogh J., Fogh J.M., Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59:221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 18.Moore G.E., Merrick S.B., Woods L.K., Arabasz N.M. A human squamous cell carcinoma cell line. Cancer Res. 1975;35:2684–2688. [PubMed] [Google Scholar]
- 19.Morgan R.T., Woods L.K., Moore G.E., McGavran L., Quinn L.A., Semple T.U. A human gallbladder adenocarcinoma cell line. In Vitro. 1981;17:503–510. doi: 10.1007/BF02633511. [DOI] [PubMed] [Google Scholar]
- 20.Moore G.E. Tumors. J Am Coll Surg. 1998;186:219–221. doi: 10.1016/s1072-7515(98)00073-8. [DOI] [PubMed] [Google Scholar]
- 21.Moore G.E., Minowada J. Historical progress and the future of human cell culture research. Hum Cell. 1992;5:313–333. [PubMed] [Google Scholar]
- 22.Pincock S. George Eugene Moore. Lancet. 2008;372:442. [Google Scholar]
- 23.Dirks W.G., Faehnrich S., Estella I.A., Drexler H.G. Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines. ALTEX. 2005;22:103–109. [PubMed] [Google Scholar]
- 24.Yoshino K., Iimura E., Saijo K., Iwase S., Fukami K., Ohno T., Obata Y., Nakamura Y. Essential role for gene profiling analysis in the authentication of human cell lines. Hum Cell. 2006;19:43–48. doi: 10.1111/j.1749-0774.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- 25.Huang C.K., Iwagami Y., Aihara A., Chung W., de la Monte S., Thomas J.M., Olsen M., Carlson R., Yu T., Dong X., Wands J. Anti-tumor effects of second generation β-hydroxylase inhibitors on cholangiocarcinoma development and progression. PLoS One. 2016;11:e0150336. doi: 10.1371/journal.pone.0150336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sripa B., Seubwai W., Vaeteewoottacharn K., Sawanyawisuth K., Silsirivanit A., Kaewkong W., Muisuk K., Dana P., Phoomak C., Lert-Itthiporn W., Luvira V., Pairojkul C., Teh B.T., Wongkham S., Okada S., Chamgramol Y. Functional and genetic characterization of three cell lines derived from a single tumor of an Opisthorchis viverrini-associated cholangiocarcinoma patient. Hum Cell. 2020;33:695–708. doi: 10.1007/s13577-020-00334-w. [DOI] [PubMed] [Google Scholar]
- 27.Markovic O., Markovic N. Cell cross-contamination in cell cultures: the silent and neglected danger. In Vitro Cell Dev Biol Anim. 1998;34:1–8. doi: 10.1007/s11626-998-0040-y. [DOI] [PubMed] [Google Scholar]
- 28.Rojas A., Gonzalez I. Cell line cross-contamination: a detrimental issue in current biomedical research. Cell Biol Int. 2018;42:272. doi: 10.1002/cbin.10904. [DOI] [PubMed] [Google Scholar]
- 29.Nelson-Rees W.A., Daniels D.W., Flandermeyer R.R. Cross-contamination of cells in culture. Science. 1981;212:446–452. doi: 10.1126/science.6451928. [DOI] [PubMed] [Google Scholar]
- 30.Zach S., Birgin E., Rückert F. Primary cholangiocellular carcinoma cell lines. J Stem Cell Res Transplant. 2015;2:1013. [Google Scholar]
- 31.Katoh H., Shinbo T., Otagiri H., Saitoh M., Saitoh T., Ishizawa S., Shimizu T., Satoh A., Tazawa K., Fujimaki M. Character of a human cholangiocarcinoma, CHGS, serially transplanted to nude mice. Hum Cell. 1988;1:101–105. [PubMed] [Google Scholar]
- 32.Caca K., Feisthammel J., Klee K., Tannapfel A., Witzigmann H., Wittekind C., Mössner J., Berr F. Inactivation of the INK4a/ARF locus and p53 in sporadic extrahepatic bile duct cancers and bile tract cancer cell lines. Int J Cancer. 2002;97:481–488. doi: 10.1002/ijc.1639. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z.H., He Y.P., Zhou Y., Zhang P., Qin H. Establishment and identification of the human multi-drug-resistant cholangiocarcinoma cell line QBC939/ADM. Mol Biol Rep. 2011;38:3075–3082. doi: 10.1007/s11033-010-9975-7. [DOI] [PubMed] [Google Scholar]
- 34.Sato J., Kimura T., Saito T., Anazawa T., Kenjo A., Sato Y., Tsuchiya T., Gotoh M. Gene expression analysis for predicting gemcitabine resistance in human cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2011;18:700–711. doi: 10.1007/s00534-011-0376-7. [DOI] [PubMed] [Google Scholar]
- 35.Saiki Y., Yoshino Y., Fujimura H., Manabe T., Kudo Y., Shimada M., Mano N., Nakano T., Lee Y., Shimizu S., Oba S., Fujiwara S., Shimizu H., Chen N., Nezhad Z.K., Jin G., Fukushige S., Sunamura M., Ishida M., Motoi F., Egawa S., Unno M., Horii A. DCK is frequently inactivated in acquired gemcitabine-resistant human cancer cells. Biochem Biophys Res Commun. 2012;421:98–104. doi: 10.1016/j.bbrc.2012.03.122. [DOI] [PubMed] [Google Scholar]
- 36.Varamo C., Peraldo-Neia C., Ostano P., Basiricò M., Raggi C., Bernabei P., Venesio T., Berrino E., Aglietta M., Leone F., Cavalloni G. Establishment and characterization of a new intrahepatic cholangiocarcinoma cell line resistant to gemcitabine. Cancers (Basel) 2019;11:519. doi: 10.3390/cancers11040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namwat N., Amimanan P., Loilome W., Jearanaikoon P., Sripa B., Bhudhisawasdi V., Tassaneeyakul W. Characterization of 5-fluorouracil-resistant cholangiocarcinoma cell lines. Chemotherapy. 2008;54:343–351. doi: 10.1159/000151541. [DOI] [PubMed] [Google Scholar]
- 38.Uthaisar K., Vaeteewoottacharn K., Seubwai W., Talabnin C., Sawanyawisuth K., Obchoei S., Kraiklang R., Okada S., Wongkham S. Establishment and characterization of a novel human cholangiocarcinoma cell line with high metastatic activity. Oncol Rep. 2016;36:1435–1446. doi: 10.3892/or.2016.4974. [DOI] [PubMed] [Google Scholar]
- 39.Fogh J., Wright W.C., Loveless J.D. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 40.Storto P.D., Saidman S.L., Demetris A.J., Letessier E., Whiteside T.L., Gollin S.M. Chromosomal breakpoints in cholangiocarcinoma cell lines. Genes Chromosomes Cancer. 1990;2:300–310. doi: 10.1002/gcc.2870020408. [DOI] [PubMed] [Google Scholar]
- 41.Enjoji M., Nakashima M., Honda M., Sakai H., Nawata H. Hepatocytic phenotypes induced in sarcomatous cholangiocarcinoma cells treated with 5-azacytidine. Hepatology. 1997;26:288–294. doi: 10.1002/hep.510260206. [DOI] [PubMed] [Google Scholar]
- 42.Homma S., Nagamori S., Fujise K., Yamazaki K., Hasumura S., Sujino H., Matsuura T., Shimizu K., Kameda H., Takaki K. Human bile duct carcinoma cell line producing abundant mucin in vitro. Gastroenterol Jpn. 1987;22:474–479. doi: 10.1007/BF02773816. [DOI] [PubMed] [Google Scholar]
- 43.Okaro A.C., Deery A.R., Hutchins R.R., Davidson B.R. The expression of antiapoptotic proteins Bcl-2, Bcl-X(L), and Mcl-1 in benign, dysplastic, and malignant biliary epithelium. J Clin Pathol. 2001;54:927–932. doi: 10.1136/jcp.54.12.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi N., Morioka H., Ohkura H., Hirohashi S., Kawai K. Establishment and characterization of the human cholangiocarcinoma cell line HChol-Y1 in a serum-free, chemically defined medium. J Natl Cancer Inst. 1985;75:29–35. [PubMed] [Google Scholar]
- 45.Knuth A., Gabbert H., Dippold W., Klein O., Sachsse W., Bitter-Suermann D., Prellwitz W., Meyer zum Büschenfelde K.H. Biliary adenocarcinoma. Characterisation of three new human tumor cell lines. J Hepatol. 1985;1:579–596. doi: 10.1016/s0168-8278(85)80002-7. [DOI] [PubMed] [Google Scholar]
- 46.Vickers S.M., Jhala N.C., Ahn E.Y., McDonald J.M., Pan G., Bland K.I. Tamoxifen (TMX)/Fas induced growth inhibition of human cholangiocarcinoma (HCC) by gamma interferon (IFN-gamma) Ann Surg. 2002;235:872–878. doi: 10.1097/00000658-200206000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakami T., Yano H., Maruiwa M., Sugihara S., Kojiro M. Establishment and characterization of a human combined hepatocholangiocarcinoma cell line and its heterologous transplantation in nude mice. Hepatology. 1987;7:551–556. doi: 10.1002/hep.1840070322. [DOI] [PubMed] [Google Scholar]
- 48.Kusaka Y., Tokiwa T., Sato J. Establishment and characterization of a cell line from a human cholangiocellular carcinoma. Res Exp Med (Berl) 1988;188:367–375. doi: 10.1007/BF01851205. [DOI] [PubMed] [Google Scholar]
- 49.Miyagiwa M., Ichida T., Tokiwa T., Sato J., Sasaki H. A new human cholangiocellular carcinoma cell line (HuCC-T1) producing carbohydrate antigen 19/9 in serum-free medium. In Vitro Cell Dev Biol. 1989;25:503–510. doi: 10.1007/BF02623562. [DOI] [PubMed] [Google Scholar]
- 50.Nakabayashi H., Taketa K., Yamane T., Miyazaki M., Miyano K., Sato J. Phenotypical stability of a human hepatoma cell line, HuH-7, in long-term culture with chemically defined medium. Gan. 1984;75:151–158. [PubMed] [Google Scholar]
- 51.Yoshida K., Tomizawa H., Ota T., Nagashima T., Kikuchi H., Watanabe H., Hashizaki K., Yonaha A. [Establishment and characterization of human cholaginocarcinoma, MEC, producing carbohydrate antigen 19-9] Hum Cell. 1990;3:346–351. [PubMed] [Google Scholar]
- 52.Sirisinha S., Tengchaisri T., Boonpucknavig S., Prempracha N., Ratanarapee S., Pausawasdi A. Establishment and characterization of a cholangiocarcinoma cell line from a Thai patient with intrahepatic bile duct cancer. Asian Pac J Allergy Immunol. 1991;9:153–157. [PubMed] [Google Scholar]
- 53.Yano H., Maruiwa M., Murakami T., Fukuda K., Ito Y., Sugihara S., Kojiro M. A new human pleomorphic hepatocellular carcinoma cell line, KYN-2. Acta Pathol Jpn. 1988;38:953–966. doi: 10.1111/j.1440-1827.1988.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 54.Yano H., Maruiwa M., Iemura A., Mizoguchi A., Kojiro M. Establishment and characterization of a new human extrahepatic bile duct carcinoma cell line (KMBC) Cancer. 1992;69:1664–1673. doi: 10.1002/1097-0142(19920401)69:7<1664::aid-cncr2820690705>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 55.Shimizu Y., Demetris A.J., Gollin S.M., Storto P.D., Bedford H.M., Altarac S., Iwatsuki S., Herberman R.B., Whiteside T.L. Two new human cholangiocarcinoma cell lines and their cytogenetics and responses to growth factors, hormones, cytokines or immunologic effector cells. Int J Cancer. 1992;52:252–260. doi: 10.1002/ijc.2910520217. [DOI] [PubMed] [Google Scholar]
- 56.Iemura A., Maruiwa M., Yano H., Kojiro M. A new human cholangiocellular carcinoma cell line (KMC-1) J Hepatol. 1992;15:288–298. doi: 10.1016/0168-8278(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 57.Wang S. [Establishment of extrahepatic cholangiocarcinoma cell line] Chin J Exp Surg. 1997;14:67–68. [Google Scholar]
- 58.Wang B., Yang R., Wu Y., Li H., Hu Z., Chen Y., Zou S. Sodium valproate inhibits the growth of human cholangiocarcinoma in vitro and in vivo. Gastroenterol Res Pract. 2013;2013:374593. doi: 10.1155/2013/374593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe M., Chigusa M., Takahashi H., Nakamura J., Tanaka H., Ohno T. High level of CA19-9, CA50, and CEA-producible human cholangiocarcinoma cell line changes in the secretion ratios in vitro or in vivo. In Vitro Cell Dev Biol Anim. 2000;36:104–109. doi: 10.1290/1071-2690(2000)036<0104:HLOCCA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 60.Yamada N., Chung Y.S., Arimoto Y., Sawada T., Seki S., Sowa M. Establishment of a new human extrahepatic bile duct carcinoma cell line (OCUCh-LM1) and experimental liver metastatic model. Br J Cancer. 1995;71:543–548. doi: 10.1038/bjc.1995.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saijyo S., Kudo T., Suzuki M., Katayose Y., Shinoda M., Muto T., Fukuhara K., Suzuki T., Matsuno S. Establishment of a new extrahepatic bile duct carcinoma cell line, TFK-1. Tohoku J Exp Med. 1995;177:61–71. doi: 10.1620/tjem.177.61. [DOI] [PubMed] [Google Scholar]
- 62.Yano H., Iemura A., Haramaki M., Momosaki S., Ogasawara S., Higaki K., Kojiro M. A human combined hepatocellular and cholangiocarcinoma cell line (KMCH-2) that shows the features of hepatocellular carcinoma or cholangiocarcinoma under different growth conditions. J Hepatol. 1996;24:413–422. doi: 10.1016/s0168-8278(96)80161-9. [DOI] [PubMed] [Google Scholar]
- 63.Takiyama I., Terashima M., Ikeda K., Kawamura H., Kashiwaba M., Tamura G., Suto T., Nakashima F., Sasaki R., Saito K. Establishment and characterization of a new human extrahepatic bile duct carcinoma cell line (ICBD-1) Oncol Rep. 1998;5:463–467. doi: 10.3892/or.5.2.463. [DOI] [PubMed] [Google Scholar]
- 64.Enjoji M., Sakai H., Nawata H., Kajiyama K., Tsuneyoshi M. Sarcomatous and adenocarcinoma cell lines from the same nodule of cholangiocarcinoma. In Vitro Cell Dev Biol Anim. 1997;33:681–683. doi: 10.1007/s11626-997-0125-z. [DOI] [PubMed] [Google Scholar]
- 65.Koike N., Todoroki T., Kawamoto T., Yoshida S., Kashiwagi H., Fukao K., Ohno T., Watanabe T. The invasion potentials of human biliary tract carcinoma cell lines: correlation between invasiveness and morphologic characteristics. Int J Oncol. 1998;13:1269–1274. doi: 10.3892/ijo.13.6.1269. [DOI] [PubMed] [Google Scholar]
- 66.Sugiyama H., Onuki K., Ishige K., Baba N., Ueda T., Matsuda S., Takeuchi K., Onodera M., Nakanuma Y., Yamato M., Yamamoto M., Hyodo I., Shoda J. Potent in vitro and in vivo antitumor activity of sorafenib against human intrahepatic cholangiocarcinoma cells. J Gastroenterol. 2011;46:779–789. doi: 10.1007/s00535-011-0380-3. [DOI] [PubMed] [Google Scholar]
- 67.Jiao W., Yakushiji H., Kitajima Y., Ogawa A., Miyazaki K. Establishment and characterization of human hilar bile duct carcinoma cell line and cell strain. J Hepatobiliary Pancreat Surg. 2000;7:417–425. doi: 10.1007/s005340070038. [DOI] [PubMed] [Google Scholar]
- 68.Mangus R.S., Tector A.J., Agarwal A., Vianna R., Murdock P., Fridell J.A. Comparison of histidine-tryptophan-ketoglutarate solution (HTK) and University of Wisconsin solution (UW) in adult liver transplantation. Liver Transpl. 2006;12:226–230. doi: 10.1002/lt.20552. [DOI] [PubMed] [Google Scholar]
- 69.Vaquero J., Aoudjehane L., Fouassier L. Cancer-associated fibroblasts in cholangiocarcinoma. Curr Opin Gastroenterol. 2020;36:63–69. doi: 10.1097/MOG.0000000000000609. [DOI] [PubMed] [Google Scholar]
- 70.Zabron A., Edwards R.J., Khan S.A. The challenge of cholangiocarcinoma: dissecting the molecular mechanisms of an insidious cancer. Dis Model Mech. 2013;6:281–292. doi: 10.1242/dmm.010561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sato K., Zhang W., Safarikia S., Isidan A., Chen A.M., Li P., Francis H., Kennedy L., Baiocchi L., Alvaro D., Glaser S., Ekser B., Alpini G. Organoids and spheroids as models for studying cholestatic liver injury and cholangiocarcinoma. Hepatology. 2021;74:491–502. doi: 10.1002/hep.31653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., Speed T., Spellman P.T., DeVries S., Lapuk A., Wang N.J., Kuo W.L., Stilwell J.L., Pinkel D., Albertson D.G., Waldman F.M., McCormick F., Dickson R.B., Johnson M.D., Lippman M., Ethier S., Gazdar A., Gray J.W. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu G.Y., Wang X., Zheng S.S., Gao X.M., Jia Q.A., Zhu W.W., Lu L., Jia H.L., Chen J.H., Dong Q.Z., Lu M., Qin L.X. RA190, a proteasome subunit ADRM1 inhibitor, suppresses intrahepatic cholangiocarcinoma by inducing NF-KB-mediated cell apoptosis. Cell Physiol Biochem. 2018;47:1152–1166. doi: 10.1159/000490210. [DOI] [PubMed] [Google Scholar]
- 74.Peraldo-Neia C., Massa A., Vita F., Basiricò M., Raggi C., Bernabei P., Ostano P., Casorzo L., Panero M., Leone F., Cavalloni G., Aglietta M. A novel multidrug-resistant cell line from an Italian intrahepatic cholangiocarcinoma patient. Cancers. 2021;13:2051. doi: 10.3390/cancers13092051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim D.G., Park S.Y., You K.R., Lee G.B., Kim H., Moon W.S., Chun Y.H., Park S.H. Establishment and characterization of chromosomal aberrations in human cholangiocarcinoma cell lines by cross-species color banding. Genes Chromosomes Cancer. 2001;30:48–56. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1053>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 76.Panichakul T., Intachote P., Wongkajorsilp A., Sripa B., Sirisinha S. Triptolide sensitizes resistant cholangiocarcinoma cells to TRAIL-induced apoptosis. Anticancer Res. 2006;26:259–265. [PubMed] [Google Scholar]
- 77.Panichakul T., Wanun T., Reutrakul V., Sirisinha S. Synergistic cytotoxicity and apoptosis induced in human cholangiocarcinoma cell lines by a combined treatment with tumor necrosis factor-alpha (TNF-alpha) and triptolide. Asian Pac J Allergy Immunol. 2002;20:167–173. [PubMed] [Google Scholar]
- 78.Sripa B., Leungwattanawanit S., Nitta T., Wongkham C., Bhudhisawasdi V., Puapairoj A., Sripa C., Miwa M. Establishment and characterization of an opisthorchiasis-associated cholangiocarcinoma cell line (KKU-100) World J Gastroenterol. 2005;11:3392–3397. doi: 10.3748/wjg.v11.i22.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ku J.L., Yoon K.A., Kim I.J., Kim W.H., Jang J.Y., Suh K.S., Kim S.W., Park Y.H., Hwang J.H., Yoon Y.B., Park J.G. Establishment and characterisation of six human biliary tract cancer cell lines. Br J Cancer. 2002;87:187–193. doi: 10.1038/sj.bjc.6600440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oka T., Yamamoto H., Sasaki S., Ii M., Hizaki K., Taniguchi H., Adachi Y., Imai K., Shinomura Y. Overexpression of beta3/gamma2 chains of laminin-5 and MMP7 in biliary cancer. World J Gastroenterol. 2009;15:3865–3873. doi: 10.3748/wjg.15.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tepsiri N., Chaturat L., Sripa B., Namwat W., Wongkham S., Bhudhisawasdi V., Tassaneeyakul W. Drug sensitivity and drug resistance profiles of human intrahepatic cholangiocarcinoma cell lines. World J Gastroenterol. 2005;11:2748–2753. doi: 10.3748/wjg.v11.i18.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ghosh M., Koike N., Tsunoda S., Hirano T., Kaul S., Kashiwagi H., Kawamoto T., Ohkohchi N., Saijo K., Ohno T., Ohno T., Miwa M., Todoroki T. Characterization and genetic analysis in the newly established human bile duct cancer cell lines. Int J Oncol. 2005;26:449–456. [PubMed] [Google Scholar]
- 83.Rattanasinganchan P., Leelawat K., Treepongkaruna S.A., Tocharoentanaphol C., Subwongcharoen S., Suthiphongchai T., Tohtong R. Establishment and characterization of a cholangiocarcinoma cell line (RMCCA-1) from a Thai patient. World J Gastroenterol. 2006;12:6500–6506. doi: 10.3748/wjg.v12.i40.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma S., Hu L., Huang X.H., Cao L.Q., Chan K.W., Wang Q., Guan X.Y. Establishment and characterization of a human cholangiocarcinoma cell line. Oncol Rep. 2007;18:1195–1200. [PubMed] [Google Scholar]
- 85.Kokuryo T., Senga T., Yokoyama Y., Nagino M., Nimura Y., Hamaguchi M. Nek2 as an effective target for inhibition of tumorigenic growth and peritoneal dissemination of cholangiocarcinoma. Cancer Res. 2007;67:9637–9642. doi: 10.1158/0008-5472.CAN-07-1489. [DOI] [PubMed] [Google Scholar]
- 86.Akarasereenont P., Aiamsa-ard T., Chotewuttakorn S., Thaworn A. Cholangiocarcinoma cell induced platelet aggregation via activation of thrombin and cyclooxygenase. Siriraj Med J. 2009;61:8–12. [Google Scholar]
- 87.Chaijaroenkul W., Viyanant V., Mahavorasirikul W., Na-Bangchang K. Cytotoxic activity of artemisinin derivatives against cholangiocarcinoma (CL-6) and hepatocarcinoma (Hep-G2) cell lines. Asian Pac J Cancer Prev. 2011;12:55–59. [PubMed] [Google Scholar]
- 88.Ojima H., Yoshikawa D., Ino Y., Shimizu H., Miyamoto M., Kokubu A., Hiraoka N., Morofuji N., Kondo T., Onaya H., Okusaka T., Shimada K., Sakamoto Y., Esaki M., Nara S., Kosuge T., Hirohashi S., Kanai Y., Shibata T. Establishment of six new human biliary tract carcinoma cell lines and identification of MAGEH1 as a candidate biomarker for predicting the efficacy of gemcitabine treatment. Cancer Sci. 2010;101:882–888. doi: 10.1111/j.1349-7006.2009.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu J., Han G., Liu H., Qin C. Suppression of cholangiocarcinoma cell growth by human umbilical cord mesenchymal stem cells: a possible role of Wnt and Akt signaling. PLoS One. 2013;8:e62844. doi: 10.1371/journal.pone.0062844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cavalloni G., Peraldo-Neia C., Varamo C., Casorzo L., Dell'Aglio C., Bernabei P., Chiorino G., Aglietta M., Leone F. Establishment and characterization of a human intrahepatic cholangiocarcinoma cell line derived from an Italian patient. Tumour Biol. 2016;37:4041–4052. doi: 10.1007/s13277-015-4215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saha S.K., Gordan J.D., Kleinstiver B.P., Vu P., Najem M.S., Yeo J.C., Shi L., Kato Y., Levin R.S., Webber J.T., Damon L.J., Egan R.K., Greninger P., McDermott U., Garnett M.J., Jenkins R.L., Rieger-Christ K.M., Sullivan T.B., Hezel A.F., Liss A.S., Mizukami Y., Goyal L., Ferrone C.R., Zhu A.X., Joung J.K., Shokat K.M., Benes C.H., Bardeesy N. Isocitrate dehydrogenase mutations confer dasatinib hypersensitivity and SRC dependence in intrahepatic cholangiocarcinoma. Cancer Discov. 2016;6:727–739. doi: 10.1158/2159-8290.CD-15-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y., Luo J., Dong X., Yang F., Zhang M., Zhao J., Wang Q., Zhou F., Sun J., Yang X. Establishment and characterization of two novel cholangiocarcinoma cell lines. Ann Surg Oncol. 2019;26:4134–4147. doi: 10.1245/s10434-019-07649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zach S., Grun J., Bauer A.T., Pilarsky C., Grutzmann R., Weng H., Dooley S., Wilhelm T.J., Gaiser T., Ruckert F. CCC-5, a new primary cholangiocellular cell line. Int J Clin Exp Pathol. 2017;10:2451–2460. [Google Scholar]
- 94.Saensa-Ard S., Leuangwattanawanit S., Senggunprai L., Namwat N., Kongpetch S., Chamgramol Y., Loilome W., Khansaard W., Jusakul A., Prawan A., Pairojkul C., Khantikeo N., Yongvanit P., Kukongviriyapan V. Establishment of cholangiocarcinoma cell lines from patients in the endemic area of liver fluke infection in Thailand. Tumour Biol. 2017;39 doi: 10.1177/1010428317725925. 1010428317725925. [DOI] [PubMed] [Google Scholar]
- 95.Vaeteewoottacharn K., Pairojkul C., Kariya R., Muisuk K., Imtawil K., Chamgramol Y., Bhudhisawasdi V., Khuntikeo N., Pugkhem A., Saeseow O.T., Silsirivanit A., Wongkham C., Wongkham S., Okada S. Establishment of highly transplantable cholangiocarcinoma cell lines from a patient-derived xenograft mouse model. Cells. 2019;8:496. doi: 10.3390/cells8050496. [DOI] [PMC free article] [PubMed] [Google Scholar]