A Novel Genetic Screen for snRNP Assembly Factors in Yeast Identifies a Conserved Protein, Sad1p, Also Required for Pre-mRNA Splicing (original) (raw)
Abstract
The assembly pathway of spliceosomal snRNPs in yeast is poorly understood. We devised a screen to identify mutations blocking the assembly of newly synthesized U4 snRNA into a functional snRNP. Fifteen mutant strains failing either to accumulate the newly synthesized U4 snRNA or to assemble a U4/U6 particle were identified and categorized into 13 complementation groups. Thirteen previously identified splicing-defective prp mutants were also assayed for U4 snRNP assembly defects. Mutations in the U4/U6 snRNP components Prp3p, Prp4p, and Prp24p led to disassembly of the U4/U6 snRNP particle and degradation of the U6 snRNA, while prp17-1 and prp19-1 strains accumulated free U4 and U6 snRNA. A detailed analysis of a newly identified mutant, the sad1-1 mutant, is presented. In addition to having the snRNP assembly defect, the sad1-1 mutant is severely impaired in splicing at the restrictive temperature: the RP29 pre-mRNA strongly accumulates and splicing-dependent production of β-galactosidase from reporter constructs is abolished, while extracts prepared from sad1-1 strains fail to splice pre-mRNA substrates in vitro. The sad1-1 mutant is the only splicing-defective mutant analyzed whose mutation preferentially affects assembly of newly synthesized U4 snRNA into the U4/U6 particle. SAD1 encodes a novel protein of 52 kDa which is essential for cell viability. Sad1p localizes to the nucleus and is not stably associated with any of the U snRNAs. Sad1p contains a putative zinc finger and is phylogenetically highly conserved, with homologues identified in human, Caenorhabditis elegans, Arabidospis, and Drosophila.
Four small nuclear ribonucleoprotein particles (snRNPs) are required for splicing the majority of mRNA precursors in the eukaryotic nucleus (40). These are the U1, U2, U4/U6, and U5 snRNPs, which consist of one (or, in the case of U4/U6, two) U snRNAs complexed with a number of stably associated protein components. A group of eight proteins, collectively referred to as the core or Sm proteins, are associated with the U1, U2, U4, and U5 snRNAs (32). A 69-kDa protein from human cells (21), the vertebrate SMN1 and associated SIP1 proteins (13, 31), and the related yeast Brr1p (47) have also been shown to associate with several snRNAs, albeit more loosely than the Sm proteins. U6 is associated with a separate group of core proteins, which are structurally related to but distinct from the canonical Sm proteins (11, 55). In addition to the common proteins, individual U snRNPs contain particle-specific proteins, which exhibit a variety of binding affinities for their respective U snRNPs (reviewed in reference 71).
Studies with yeast and human cells have demonstrated that snRNPs form the core structure of the spliceosome, with U snRNAs directly participating in intron recognition and splice site alignment (reviewed in reference 37). In addition to U snRNAs, numerous proteins have been implicated in splicing in higher eukaryotes, while genetic screens and biochemical analyses with yeast allowed the identification and characterization of over 40 spliceosomal proteins (reviewed in reference 70).
While the function of spliceosomal snRNPs has been extensively investigated, the process of U snRNP assembly is less well understood (reviewed in reference 39). In vertebrate cells, the U1, U2, U4, and U5 snRNAs are transcribed by RNA polymerase II, acquire a monomethyl (m7G) cap structure cotranscriptionally, and are rapidly exported to the cytoplasm. U snRNA export is mediated by at least one specific factor, which is not shared with tRNA, 5S rRNA, and mRNA export pathways (25) and requires a nuclear cap binding complex (24) and the Ran/TC4 GTP exchange factor RCC1 (10), while an involvement of importin-α (17) and the importin-β homologue CRM1 (14) has been proposed. Once in the cytoplasm, U1, U2, U4, and U5 snRNAs associate with the common Sm proteins. The spinal muscular atrophy disease gene product SMN and its associated protein SIP1 (31) were recently shown to be involved in the assembly of the Sm core domain on the U snRNAs in the cytoplasm (13). Assembly of the Sm proteins on the U snRNAs triggers hypermethylation of the m7G cap structure of U snRNAs to a trimethyl 2,2,7mG (TMG) cap. The methylase involved is external to the snRNP and recognizes the Sm core (49). The Sm core domain and the TMG cap constitute a bipartite nuclear localization signal which targets the newly assembled snRNPs to the nucleus. The pathway of nuclear import followed by the snRNPs is distinct from, but shares common factors with, the protein import pathway (38, 48). Further modifications of the snRNAs involving 3′ trimming (45, 74) and base and sugar modifications, take place either before or after the import step. At least some of the U snRNP-specific proteins are transported to the nucleus independently of the U snRNAs (26, 27) and probably join the core snRNP in the nucleus to form the mature, functional particle. In contrast to the polymerase II-transcribed U snRNAs, U6 is a polymerase III transcript and does not appear to leave the nucleus. Methylation of the gamma phosphate at the 5′ end of the RNA (59), addition of UMP residues and formation of a 2′-3′ cyclic phosphate at the 3′ end (33), internal modifications, protein association, and assembly with the U4 snRNA and snRNP proteins appear to take place in the nucleus.
The ease of genetic manipulations in the yeast Saccharomyces cerevisiae makes it an ideal system for identifying factors involved in snRNP biogenesis. Even though little is known about the snRNP biosynthetic pathway in this organism, the available data point to an evolutionary conservation. The yeast U6, similarly to its mammalian counterpart, is transcribed by RNA polymerase III (7, 41). The U1, U2, U4, and U5 snRNAs have a TMG cap (72) and associate with common as well as particle-specific proteins, some of which have clear mammalian homologues (5, 51, 52, 55). 3′ end processing events have been reported for the U2 (47) and U5 (8) snRNAs. The U2 snRNA processing event is affected by mutation in the BRR1 gene, while the U5 snRNA maturation depends on RNase III. Mutation or depletion of a number of U snRNP proteins results in destabilization of U snRNAs (see, e.g., references 5, 51, and 52).
Extensive screens for splicing-defective mutants have not so far unravelled factors involved in snRNP biosynthesis, with the exception of snRNP protein subunits (47). This is probably due to the fact that U snRNP turnover is extremely slow (U snRNAs have a half-life of several hours in yeast [60, 47]), and therefore, a block in U snRNP biosynthesis is not expected to affect splicing until several hours later. We report here a novel screening procedure for mutants defective in the U snRNP biosynthetic pathway which monitors the fate of an inducible U4 snRNA. A number of mutants in which newly synthesized U4 snRNA either is destabilized or fails to assemble with the U6 snRNP and accumulates as free U4 snRNP were identified. A detailed analysis of one of these mutants showed that the mutated protein is essential and evolutionarily highly conserved and is required for splicing.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strains used in this study are shown in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| DJY36a | MATa ade ura3 prp2-1 |
| SpJ3.33a | MATa his leu2 lys2 ura3-52 prp3-1 |
| SpJ4.41a | MATa leu2 ura3-52 prp4-1 |
| SpJ6.66a | MATa ade his lys2 ura3-52 prp6-1 |
| SpJ8.31a | MATa his3 leu2 ura3-52 prp8-1 |
| PRP17ab | MATa ade2-101 his3-Δ200 leu lys ura3-52 prp17-1 |
| PRP18ab | MATa ade2-101 his3-Δ200 ura3-52 prp18-1 |
| PRP19ab | MATa ade2-101 his3-Δ200 ura3-52 prp19-1 |
| PRP20b | MATα ade2-101 his3-Δ200 ura3-52 prp20-1 |
| PRP21ab | MATa ade2-101 his3-Δ200 ura3-52 prp21-1 |
| PRP22ab | MATa ade2-101 his3-Δ200 ura3-52 prp22-1 |
| PRP24ab | MATa ade2-101 his3-Δ200 ura3-52 prp24-1 |
| VL682Cc | MATa his-11,15, leu2-3,-112 lys2-801 trp1 ura3-52 prp33-1 |
| S5d | MATa ade2 his leu2 lys1 nsp1-1::URA3 |
| Nop1-tsd | MATα his3 leu2 ura3 nop1-5::URA3 |
| D163e | MATα ade2-10 lys2 met4-10 trp1 ura3-52 rna12 |
| B-8302f | MATa _cyc1_-NLS cyc7-67 ura3-52 lys5-10 nip1-1 |
| 313d | MATα ade lys leu trp1 ura nsp49::TRP1 HIS3::nsp49-313 |
| MGD353-13Dg | MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 |
| BSY251h | MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 snr14::TRP1 pBS377[pCEN-_LEU2-SNR14_] |
| BSY253h | MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 snr14::TRP1 pBS379[pCEN-LEU2-SNR14*] |
| BSY295i | MATa ade2 arg4 leu2-3,112 ura3-52 TRP1::GAL-SNR14* |
| BSY307i | MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 snp1::TRP1 pBS410[pCEN-_TRP1_-ProtA::_SNP1_] |
| BSY360i | MATα ade2 ade5 his3-Δ1 leu2-3,112 ura3-52 TRP1::GAL-SNR14* |
| BSY387h | MATa ade2 arg4 leu2-3,112 ura3-52 TRP1::GAL-SNR14* sad1-1 |
| BSY466h | MATα ade2 arg4 leu2-3,112 trp1-289 ura3-52 sad1::TRP1 pBS772[pCEN-_URA3_-ProtA::_SAD1_] |
| BSY467h | MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 sad1::TRP1 pBS773[pCEN-URA3-SAD1::ProtA] |
U4* was constructed by mutating positions 76 to 85 of the wild-type U4 snRNA to CUCUUAAGCCAUAG. U4*, under the control of the U4 promoter on a CEN-ARS plasmid, was introduced into a SNR14::TRP1 strain by plasmid shuffling, creating strain BSY253. The doubling time of BSY253 on yeast extract-peptone-dextrose at 30°C is identical to that of strain BSY251, which bears the wild-type SNR14 gene, while there were no growth differences observed at 16 or 37°C or on minimal medium.
The GAL-U4* gene was constructed by fusing the GAL-U1 promoter upstream of the U4* coding sequence, as reported previously for the GAL-U2 construct (57). Strain BSY295 was created by integrating the GAL-U4* gene at the TRP1 locus of the wild-type strain BSY17. The 5′ end of U4* was determined by primer extension, while the lack of 3′-end-extended products was verified by Northern blotting following electrophoretic fractionation in a denaturing polyacrylamide gel.
Construction of the collection of temperature-sensitive mutants has been previously described (35).
U4/U6 assembly assay.
To test for U4/U6 assembly defects, strains were grown individually on yeast extract-peptone-lactate-glycerol (for the BSY295 derivatives) or on minimal lactate-glycerol medium (for strains bearing the GAL-U4* gene on a plasmid) at 23°C to mid-log phase. When strains were shifted to the nonpermissive temperature, an equal volume of medium prewarmed to 51°C was added and growth was continued for the time indicated at 37°C. Subsequently, galactose was added to a 2% final concentration, and growth was continued for another 2 h at 37°C to allow expression of the U4* snRNA. Cells (1 × 108 to 1.5 × 108) were then collected, resuspended in 250 μl of RNA extraction buffer (100 mM LiCl, 1 mM EDTA, 100 mM Tris-Cl [pH 7.5], 0.2% sodium dodecyl sulfate [SDS]), and broken with 250 μl of phenol-chloroform-isoamyl alcohol and 250 μl of acid-washed, silicon-coated glass beads, using a cooled shaker (Braun). The aqueous phase was mixed with one-third volume of loading dye (50% glycerol, 0.02% bromophenol blue) and loaded on a 5% nondenaturing polyacrylamide–Tris-borate-EDTA gel containing 5% glycerol. Electrophoresis took place overnight for 1,000 V · h at 4°C with 0.5× Tris-borate-EDTA as the running buffer. The gel was then soaked for 1 h in 20 mM NaPO4 (pH 6.5)–8.3 M urea–0.1% SDS at 37°C, followed by 1 h in 25 mM NaPO4 (pH 6.5), and electrotransferred to a nylon membrane (GeneScreen) according to the manufacturer’s instructions. Hybridization to 5′-end-labelled oligonucleotide probes was performed in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt solution–0.2% SDS overnight at 37°C, followed by two 5-min washes in 6× SSC–0.2% SDS at 37°C and one 15-min wash in 3× SSC–0.2% SDS at 37°C. Results were quantified by using a PhosphorImager. The oligonucleotide probes used were DT168, complementary to the U1 snRNA (56); ol23, complementary to U4* (34); ol14 (5′ CGGTCTGGTTTATAAT 3′), complementary to wild-type U4 in the region mutated in U4*; ol5 (5′ TCATCCTTATGCAGGG 3′), complementary to U6 snRNA; and ol25 (5′ GAATTCGACAGGTTATCAGC 3′), complementary to the GAL10 mRNA.
Assessment of in vivo splicing efficiency.
Extraction of RNA for Northern blotting (67) and gel electrophoresis and transfer (53) were performed as described previously. To detect splicing defects, a randomly primed probe derived from the intron-containing RP29 gene was used for hybridization, as described previously (9), while the GAL10 mRNA was detected with the 5′-end-labelled oligonucleotide probe ol25.
To measure the extent of in vivo splicing deficiency, a sad1-1 strain was transformed with the HZ18 and Acc0 reporter constructs bearing the β-galactosidase gene interrupted by an intron behind a galactose-inducible promoter (30, 64). An intronless vector (pLG-SD5) and a vector without a β-galactosidase-coding sequence were used as a controls. Cells grown to mid-exponential phase on yeast extract-peptone-lactate-glycerol medium were shifted to 37°C for 30 min, after which galactose was added and growth was continued for 2 h at 37°C. Measurement of β-galactosidase activity was performed as described previously (28).
Extracts, immunoprecipitation, in vitro splicing, and immunolocalization.
Yeast whole-cell extracts were prepared as described previously (57).
To detect association of Sad1p with U snRNAs, extracts were prepared from strains expressing ProtA-Sad1p, Sad1p-ProtA, or ProtA–U1-70K (55) or from wild-type strains. Immunoprecipitation and extraction of coprecipitating RNAs were performed as described previously (35) with the exception that the NaCl concentration in the immunoprecipitation and washing buffer was either 50, 150, 300, or 600 mM. The presence of U1, U2, U4, U5, and U6 snRNAs in the total extract, immunoprecipitate, and immune supernatant was analyzed by primer extension as described previously (34). While U1-70K associated with the U1 snRNA under all salt conditions tested, no association of Sad1p with any of the spliceosomal U snRNAs was detected in parallel reactions.
For in vitro splicing reactions, capped, internally labelled pre-mRNA substrates derived from the ACT1 gene or the RP51A gene were produced by in vitro transcription and purified on a polyacrylamide gel (57, 68). Four microliters of extract was incubated with 20,000 cpm of pre-mRNA substrate in a buffer containing 60 mM KPO4 (pH 7.0), 3 mM MgCl2, 2 mM cordycepin, 3% polyethylene glycol, and 1 mM spermidine for 30 min at 25°C. Subsequently, RNA was extracted by proteinase K digestion and phenol-chloroform-isoamylalcohol extraction, precipitated with ethanol, and analyzed on an 8% polyacrylamide gel (46).
To assess the subcellular localization of Sad1p, exponentially growing cells expressing ProtA-Sad1 or Sad1-ProtA and wild-type cells were fixed with 4% formaldehyde for 1 h at 25°C, spheroplasted, and processed for immunofluorescence as described previously (18). Rabbit anti-ProtA antibody (Sigma; 1:200 dilution) and mouse monoclonal antibody 66, which detects the nucleolar Nop1p (22) (a kind gift of J. P. Aris; 1:100 dilution) were used at a 1:100 dilution. Goat antirabbit antibody coupled to fluorescein isothiocyanate (Amersham) and goat antimouse antibody coupled to the fluorochrome Cy3 (Sigma) were used at 1:200 and 1:2,000 dilutions respectively.
Cloning, disruption, and tagging of SAD1.
To clone SAD1, a strain bearing the sad1-1 allele was transformed with a genomic S. cerevisiae library (3), and cells were directly selected for growth at 37°C. Six partially overlapping plasmids, all of which conferred temperature resistance to the _sad1-1_-bearing strain, were recovered. To localize further the complementing activity, four subclones were constructed from the smallest complementing insert and tested for complementation of the sad1-1 temperature sensitivity. A 2.2-kb _Cla_I-_Bam_HI fragment was the minimal complementing subclone. Both strands of this _Cla_I-_Bam_HI fragment were sequenced from the original as well as further subclones by using vector and internal oligonucleotide primers.
To delete SAD1, two constructs were made in which an _Hpa_I-_Pfl_MI or an _Hpa_I-_Bgl_II fragment was deleted from the SAD1 open reading frame and replaced by the TRP1 selectable marker. Linear fragments bearing the sad1::TRP1 alleles were used to transform a wild-type yeast strain to Trp prototrophy. Trp+ transformants were tested for correct integration events by Southern analysis. To construct the ProtA-Sad1 fusion proteins, a 395-bp fragment encoding two immunoglobulin G binding domains of Staphylococcus aureus ProtA was fused in frame to the SAD1 coding sequence either immediately before the first AUG (ProtA-Sad1) or immediately before the stop codon (Sad1-ProtA). The original SAD1 promoter and 3′ flanking sequences were retained in both constructs. Constructs were sequenced to confirm that no mutations were introduced during the PCR amplification steps. The ProtA-Sad1 and Sad1-ProtA fusion genes in a centromeric-ARS plasmid were used to transform a heterozygous sad1::TRP1/SAD1 diploid. After sporulation and dissection, haploids bearing the sad1::TRP1 allele complemented by either the amino-terminal or carboxy-terminal fusion genes were viable and exhibited no pronounced growth defect.
Sequence analysis.
Sequence analysis was performed with the Genetics Computer Group package.
Nucleotide sequence accession numbers.
Accession numbers for SAD1 from different species are as follows: S. cerevisiae protein, P43589; S. cerevisiae gene, D50617; S. cerevisiae expressed sequence tag (EST), T38687; human ESTs, AA176284, AA206156, AA243275, T09081, AA181173, F12552, T74457, T05174, and H57235; mouse ESTs, W65854 and AA107884; Drosophila ESTs, AA541001 and AA264758; Arabidopsis thaliana gene, AL021712; Caenorhabditis elegans ESTs, C66020 and C40457; and C. elegans cosmid, AF040640.
RESULTS
Assay for snRNP assembly.
In order to identify S. cerevisiae mutants conditionally defective in the assembly of U snRNPs (snRNP assembly-defective [SAD] mutants), we devised an assay for formation of the U4/U6 particle.
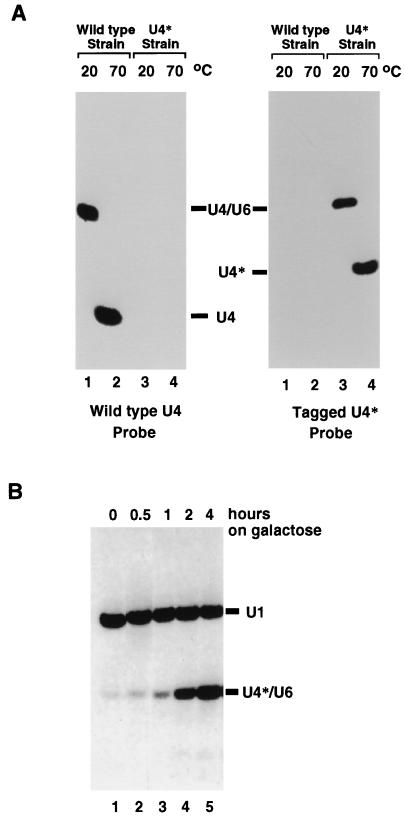
Nearly all of the U4 snRNA present in a wild-type yeast cell is associated with the U6 snRNA in the U4/U6 particle, where the two RNAs are held together through extensive base pairing (6) (see below). We reasoned that in a Sad− mutant, newly synthesized U4 snRNA would be unable to associate with U6 snRNA and would therefore accumulate as a free species or be degraded. To assay for the status of the U4 snRNA in a given strain, we made use of a nondenaturing electrophoretic system which enables the separation of the U4/U6 RNA complex from free U4 snRNA (see Materials and Methods) (6). This is depicted in Fig. 1A (left panel), where total cellular RNA was fractionated on such a native gel, transferred to nitrocellulose, and hybridized with a U4-specific probe. The migrations are different for U4 RNA kept at below 20°C or heated to 70°C prior to loading (Fig. 1A, left panel, compare lanes 1 and 2). In the sample kept at the lower temperature, the retarded U4 snRNA comigrates with a fraction of the U6 snRNA to which it is associated (data not shown). This association, which results from base pairing, is disrupted by heating, with a Tm of approximately 55°C (data not shown) (6). Nearly all of the U4 snRNA is associated with the U6 snRNA in a cell, as very low levels of free U4 snRNA are detectable in the sample kept at the lower temperature (Fig. 1A, left panel, lane 1 [free U4 RNA is detectable only after longer exposure]).
FIG. 1.
Assay for U4/U6 assembly. (A) Total cell RNA was extracted from a wild-type yeast strain (lanes 1 and 2) or a strain in which the U4 snRNA gene was replaced by U4* (lanes 3 and 4). After RNA extraction at 4°C, RNA was incubated for 10 min at either 20°C (lanes 1 and 3) or 70°C (lanes 2 and 4) prior to being loaded on a native polyacrylamide gel. After transfer to nylon membranes, identical filters were hybridized with an oligonucleotide probe specific for the wild-type U4 snRNA (left panel) or the U4* snRNA (right panel). (B) Total cell RNA was extracted from strain BSY295 grown for 0, 0.5, 1, 2, and 4 h in the presence of galactose. After native gel electrophoresis and transfer to a nylon membrane, the expression of the U4* snRNA was detected with a radioactively labelled oligonucleotide complementary to U4*. A labelled U1-specific oligonucleotide probe was included, as U1 snRNA served as a loading control.
To monitor the fate of newly synthesized U4 snRNA, we constructed an inducible tagged U4 snRNA species. We first selected regions of the U4 snRNA that were not phylogenetically conserved, not known to be involved in interaction with other splicing factors, and not sensitive to mutation (20, 23, 66, 69). These regions were mutated to give four different U4 variants harboring mutations allowing the specific detection of the tagged U4 snRNA with an antisense probe (see below) as well as to create a restriction site that enabled discrimination from the wild-type gene. After extensive testing (see below), a single tagged U4 snRNA, hereafter referred to as U4*, was retained for further study. It carries six substitutions and three nucleotide insertions in the middle part of the U4 snRNA (see Materials and Methods) (35). U4* is fully functional, as it can replace the wild-type U4 snRNA and does not confer any growth phenotype under the conditions tested (see Materials and Methods). U4* assembles with the U6 snRNA, and the RNA complex has the same Tm as wild-type U4/U6 (data not shown). A set of oligonucleotide probes which allowed specific detection of either the wild-type (Fig. 1A, left panel) or the tagged (Fig. 1A, right panel) U4 snRNA was designed. U4* was placed under the control of the GAL regulatory sequences. A single copy of the GAL-U4* construct was integrated at the TRP1 locus of the wild-type strain BSY17, creating strain BSY295. This strain retains a functional copy of the wild-type U4 snRNA encoded at the original U4 locus. When strain BSY295 is grown in the absence of galactose, only trace amounts of U4* are detected (Fig. 1B, lane 1). U4* is strongly induced upon addition of galactose and assembles with the U6 snRNA (Fig. 1B, lanes 2 to 5). Similar kinetics of induction of the U4* snRNA are observed when the GAL-U4* gene is introduced into the wild-type BSY17 strain on a CEN-ARS plasmid (data not shown). The U4* snRNA produced under the control of the GAL regulatory sequences has a single 5′ end, identical to that of wild-type U4, and no 3′-end heterogeneity (data not shown). Two hours of galactose induction was chosen for further experiments. At this time point, U4* is easily detected (Fig. 1B, lane 4), and its amount corresponds to less than 10% of the wild-type U4 snRNA levels (data not shown).
Testing of splicing-defective mutants for U4/U6 assembly defects.
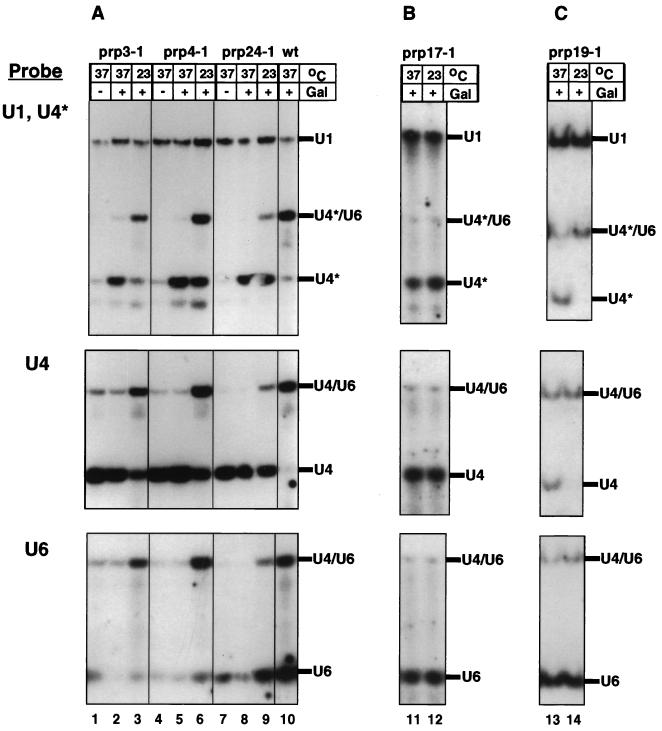
To assess our strategy, we first tested whether mutations in known splicing factors affect the assembly of newly synthesized U4 snRNA into a U4/U6 particle. A number of proteins involved in splicing are found stably associated with the mature U4/U6 snRNP and are likely to contribute to the assembly and/or stability of the particle. Other splicing factors could contribute to the disassembly-reassembly of the U4/U6 hybrid which accompanies each round of splicing (50, 73). We used the U4/U6 assembly assay described above to analyze 13 previously characterized splicing-defective mutants and five nonrelated thermosensitive mutants (see Materials and Methods) for U4/U6 assembly defects. Each mutant, transformed with the GAL-U4* reporter gene on a plasmid, was grown on selective medium to mid-log phase at the permissive temperature and shifted for 30 min to the nonpermissive temperature (37°C). U4* transcription was subsequently turned on by addition of galactose, and growth was continued for 2 h at 37°C. Total RNA was extracted, fractionated on a native gel, and analyzed by Northern blotting with probes specific for the U4*, U4, and U6 snRNAs. While the majority of the splicing-defective mutants analyzed (namely, the prp2, prp6, prp8, prp18, prp20, prp21, prp22, prp33 mutants) showed no defects in U4/U6 assembly, five mutant strains exhibited an abnormal phenotype. None of the control thermosensitive mutations unrelated to splicing produced a U4/U6 assembly phenotype, demonstrating the specificity of the assay.
In Fig. 2A, the U4/U6 assembly phenotype conferred by the prp3-1, prp4-1, and prp24-1 mutations, which are in genes encoding components of the U4/U6 or free U6 snRNP (1, 2, 58), is shown. In all three mutants grown at the nonpermissive temperature, U4* fails to associate with U6 and accumulates as free U4* (Fig. 2A, upper panel, lanes 2, 5, and 8). A partial block in U4*/U6 formation is already detectable at the permissive temperature (lanes 3, 6, and 9). A similar behavior is seen for the endogenous U4 (Fig. 2A, middle panel), most of which has been synthesized before the temperature shift. Hybridization with a U6-specific probe demonstrates that the levels of U6 in these cells are drastically reduced (Fig. 2A, lower panel) (1, 4). Accumulation of free U4 in these strains probably results from the instability of the U6 snRNA.
FIG. 2.
U4/U6 assembly defects of splicing-defective mutants. The prp3-1, prp4-1, and prp24-1 strains and a wild-type (wt) strain (A), the prp17-1 strain (B), and the prp19-1 strain (C), all transformed with the galactose-inducible U4* gene, were grown for 30 min at 37°C (lanes 1, 2, 4, 5, 7, 8, 10, 11, and 13) or left at 23°C (lanes 3, 6, 9, 12, and 14). Production of U4* was subsequently induced by addition of galactose (Gal) (except in lanes 1, 4, and 7), and growth was continued at the same temperature for another 2 h. Total cell RNA was extracted, separated on a native polyacrylamide gel, transferred to a nylon membrane, and hybridized with radioactively labelled oligonucleotides complementary to U4* and U1 (loading control) snRNAs (upper panels). The same filters (after removal of the U1 and U4* probes) were hybridized with an oligonucleotide probe specific to the wild-type U4 snRNA (middle panels) and subsequently with a U6-specific probe (lower panels).
A different mutant phenotype is observed for prp17-1 and prp19-1 (Fig. 2B and C). While free U6 snRNA levels are not significantly affected (lower panels), a fraction of the U4* synthesized after the shift to the nonpermissive temperature appears as free U4* (upper panels, lanes 11 and 13). The prp17-1 mutant already exhibits the snRNP assembly defect at the permissive temperature (lane 12), as has been previously reported for its splicing defect, while the prp19-1 mutant is defective only at the nonpermissive temperature (lane 13). Probing for the endogenous U4 snRNA demonstrates accumulation of free wild-type U4 in the prp17-1 and prp19-1 mutants, as is observed for the prp3-1, prp4-1, and prp24-1 mutants (middle panels). These factors are therefore required for efficient assembly or stability of the U4/U6 particle.
This analysis demonstrated that the reporter U4* construct can be used to identify _trans_-acting mutations preventing the formation of wild-type levels of the U4*/U6 snRNP associated with accumulation of free U4* snRNA and/or instability of the U6 snRNA. However, because the endogenous and reporter U4 snRNAs were affected in the same way, we conclude that none of the mutant alleles tested affected specifically the assembly of the newly synthesized U4/U6 particle.
Screening for novel SAD mutants.
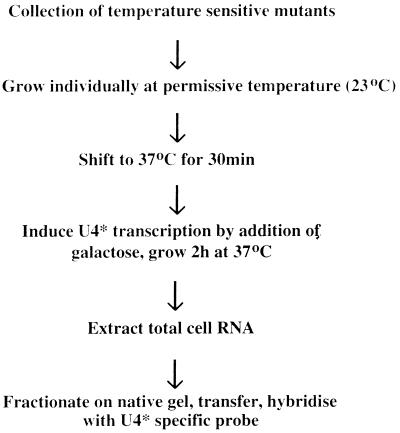
In order to identify genes required for the U4/U6 assembly pathway, a collection of 246 temperature-sensitive mutants was generated by mutagenizing strain BSY295. These mutants were screened by the procedure depicted in Fig. 3. Table 2 summarizes the results of the screen. Twenty mutant strains, which fell into 18 complementation groups, were isolated. Two major mutant phenotypes were observed.
FIG. 3.
Procedure used to identify strains defective in U4/U6 particle assembly.
TABLE 2.
Strains identified in the U4/U6 assembly screena
| Strain(s) | U4* snRNA | Gal10 mRNA | RP29 mRNA | Segregation | Gene |
|---|---|---|---|---|---|
| At5 | None | None | wt (low levels) | ND | ND |
| At104 | None | None | wt (low levels) | ND | ND |
| At142 | None | None | wt (low levels) | ND | ND |
| Pt84 | None | None | wt (low levels) | ND | ND |
| Ac94 | None | None | wt | ND | ND |
| At20 | None | wt | wt | ts linked | BDFL |
| At134 | None | wt | wt | ND | ND |
| At265 | None | wt | wt | ts linked | ND |
| Pt6 | None | wt | wt | ND | ND |
| Pt39, At205 | None | wt | wt | ts linked | ND |
| At81 | None | wt | None | ts linked | ND |
| Ac106 | None | wt | None | ND | ND |
| Pt51 | Free | wt | wt | ts linked | On chromosome II |
| At80 | Free | wt | wt | ts linked | ND |
| At173 | Free | wt | wt | ND | ND |
| At201 | Free | wt | wt | ND | ND |
| At164, Pt4 | Free | wt | None | ts linked | SEC53 |
| At216 | Free | wt | Pre-mRNA accumulation | ts linked | SAD1 |
(i) In 12 complementation groups, no U4* snRNA was detected. The presence of the U4* gene in these strains was verified by PCR (data not shown). The inability to accumulate the U4* snRNA could be due to either a transcriptional defect or an assembly defect which would lead to degradation of the nonassembled U4*. The GAL10 gene, which is under the control of the same regulatory sequences as U4*, was correctly transcribed for seven of these complementation groups (data not shown), excluding a global transcriptional defect as well as a defect in the pathway of induction by galactose. The characterization of one such mutant, carrying bdf1-1, has been reported elsewhere (34).
(ii) In six complementation groups, a fraction (20 to 50%) of the newly synthesized U4* snRNA was unable to assemble with U6 and accumulated as free U4* snRNA. We describe here the characterization of one such mutant, which we refer to as the sad1-1 mutant.
sad1-1 prevents the assembly of newly synthesized U4 snRNA in U4/U6 complexes.
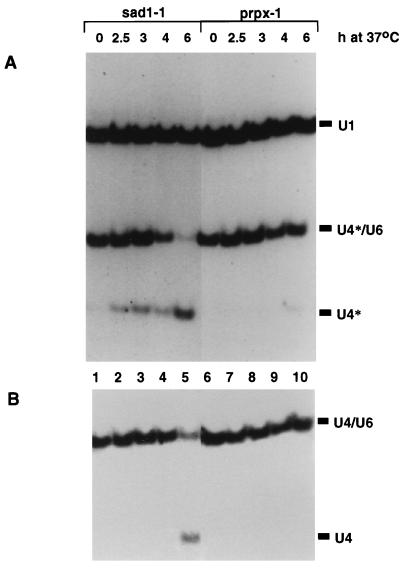
A strain carrying the sad1-1 mutation was grown either at the permissive temperature (Fig. 4, lanes 1) or for 0.5, 1, 2, and 4 h at 37°C (lanes 2 to 5), after which U4* transcription was induced and allowed to proceed for 2 h at the same temperature. All of the U4* produced in a wild-type strain at 37°C (data not shown), in an unrelated thermosensitive strain (lanes 6 to 10; prpx-1 is a splicing-defective strain isolated from the same collection) or in the _sad1-1_-containing strain grown at the permissive temperature (lanes 1) correctly assembles with the U6 snRNA. In contrast, at the nonpermissive temperature, the sad1-1 allele induces accumulation of free U4*, which, at later time points, corresponds to the majority of the U4* produced (lanes 5). Wild-type U4 snRNA, which at the earlier time points represents mostly U4 synthesized before the temperature shift (because of the long half-life of U snRNAs [60]), is much less affected than U4* (compare the ratio of U4/U6 to free U4 in Fig. 4A and B). This indicates that sad1-1 affects the assembly of the newly synthesized U4/U6 particle rather than the stability of the existing U4/U6 particles or the reassembly of U4/U6 which accompanies each round of splicing.
FIG. 4.
The sad1-1 mutant is defective in the assembly of newly synthesized U4* snRNA. The sad1-1 strain and the unrelated temperature-sensitive and splicing-defective prpx-1 strain were grown for 0.5, 1, 2, and 4 h at 37°C. Subsequently, galactose was added and growth was continued for a further 2 h at 37°C. Following RNA extraction, separation on a native acrylamide gel, and transfer, labelled oligonucleotide probes were used to detect the U4* snRNA and U1 snRNA, which serves as a loading control (A). The same filter (after removal of the U1 and U4* probes) was hybridized with an oligonucleotide specific for the U4 snRNA (B).
sad1-1 confers a splicing defect.
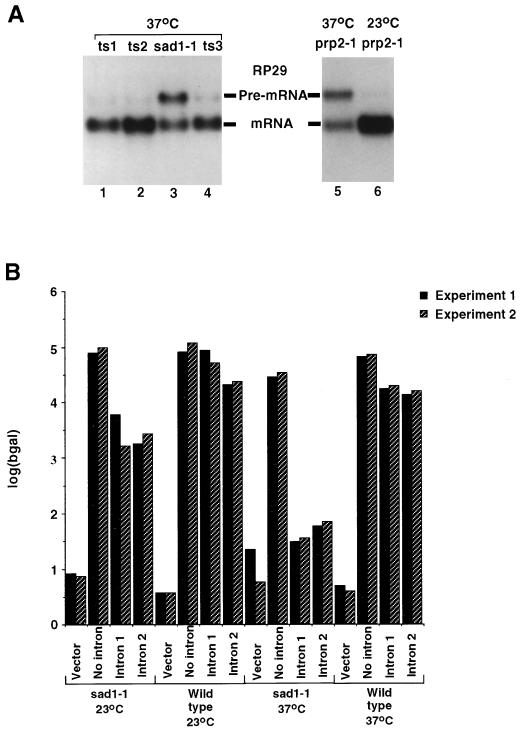
The U4/U6 assembly defect of the sad1-1 mutant would be expected to affect splicing due to depletion of the U4/U6 particle only after several hours at the nonpermissive temperature. To test whether sad1-1 confers a splicing defect independently of the U4/U6 assembly defect, pre-mRNA accumulation was assessed in sad1-1 cells 2.5 h after the temperature shift, when the majority of the U4/U6 particle present in the cell is still unaffected (Fig. 4B, lane 2). Strains carrying sad1-1 (Fig. 5A, lane 3), three unrelated temperature-sensitive alleles (lanes 1, 2, and 4), and the well-characterized splicing defective prp2-1 mutation (lane 5) were grown for 2.5 h at the nonpermissive temperature. Total RNA was extracted, fractionated on a denaturing agarose gel, transferred to nitrocellulose, and hybridized with a radioactively labelled fragment of the intron-containing RP29 gene. Similar to prp2-1 (lane 5), sad1-1 provoked a strong accumulation of the unspliced RP29 pre-mRNA (lane 3) and therefore confers a splicing defect. None of the other putative sad mutants identified showed a splicing defect (data no shown).
FIG. 5.
The sad1-1 mutant is defective in splicing. (A) Total cell RNAs extracted from the sad1-1 strain (lane 3), three unrelated temperature-sensitive strains (lanes 1, 2, and 4) and the prp2-1 strain (lane 5) grown for 2.5 h at the nonpermissive temperature were resolved on an agarose-formaldehyde gel, transferred to nitrocellulose, and hybridized with a radioactively labelled fragment of the RP29 gene. As a control, RNAs isolated from the prp2-1 strain grown at the permissive temperature (23°C) are presented (lane 6). The positions of migration of the RP29 pre-mRNA and mRNA are indicated. (B) The sad1-1 strain or an isogenic wild-type strain was transformed with either vector alone, a plasmid containing the coding region of the β-galactosidase gene under the control of the GAL10 promoter, the same plasmid in which an intron deriving from the RP51A gene was introduced in the lacZ coding region (intron 1) (62), or the same plasmid in which a synthetic intron was introduced in the lacZ coding region (intron 2) (28). The strains were either grown continually at 23°C or grown for 30 min at 37°C. In either case, galactose was added subsequently and growth was continued for 2 h at the same temperature. Levels of β-galactosidase (bgal) activity produced in these cells are shown (in duplicate).
The sad1-1 mutant strain was backcrossed to the parental wild-type strain BSY295. After sporulation and dissection, four complete tetrads were analyzed for the segregation of the snRNP assembly defect, the splicing defect, and the temperature sensitivity. Cosegregation of all three mutant phenotypes was observed (data not shown), indicating that they are due to mutation in a single gene.
To measure more precisely the level of splicing inhibition in sad1-1 cells and to establish whether this effect was intron specific, the sad1-1 mutant and the isogenic wild-type strain (BSY295) were transformed with three reporter plasmids. Two of the reporter plasmids contain a β-galactosidase open reading frame interrupted by an intron derived from the RP51A gene (64), while the control reporter expresses a continuous β-galactosidase open reading frame. Expression of the reporter genes was induced for 2 h either at the permissive temperature or after incubation of the cells for 30 min at the restrictive temperature, and β-galactosidase activity was determined (Fig. 5B). While expression of the intronless reporter gene did not vary significantly, splicing of the intron-containing reporter genes was more than 10-fold reduced in the sad1-1 background at the permissive temperature compared to that in the wild-type cells. When _sad1-1_-harboring cells were grown at the nonpermissive temperature, the β-galactosidase activity of the intron-containing reporter genes dropped almost to background levels, thus establishing that sad1-1 confers a strong temperature-sensitive splicing defect. Primer extension analysis with RNA extracted from sad1-1 cells expressing the reporter gene at the nonpermissive temperature showed accumulation of pre-mRNA, rather than splicing intermediates, demonstrating that sad1-1 is defective in the first step of splicing (33a).
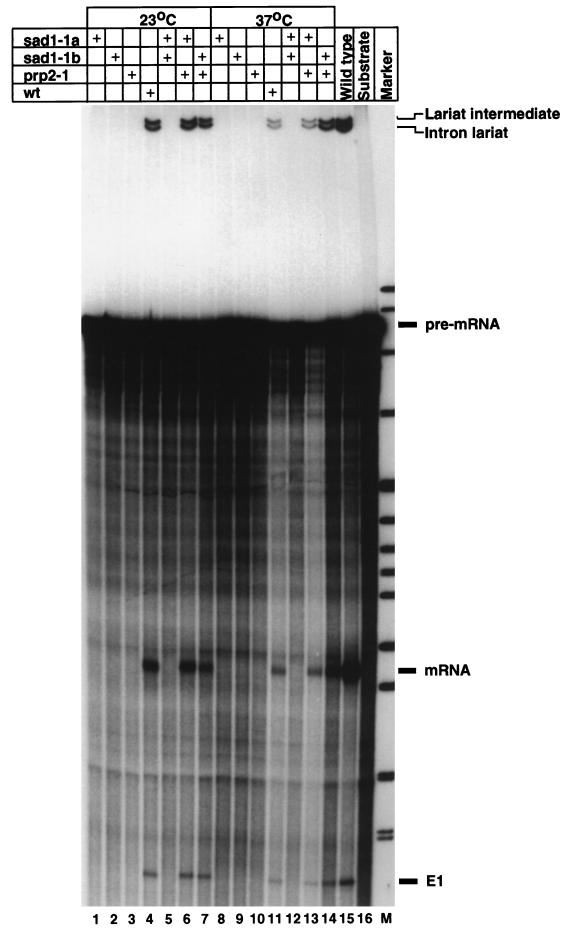
The in vivo splicing defect of sad1-1 mutant cells was corroborated by in vitro experiments. Extracts were prepared from two different strains harboring the sad1-1 allele as well as from prp2-1 and wild-type cells grown either at the permissive temperature or for 2.5 h at the nonpermissive temperature. An internally labelled fragment of the actin pre-mRNA (68) was incubated with the different extracts for 30 min at 25°C. The products of the reaction were extracted and separated on a denaturing polyacrylamide gel (Fig. 6). While extracts made from wild-type cells splice the actin pre-mRNA in vitro (lanes 4 and 11), extracts from sad1-1 or prp2-1 cells at either the permissive or restrictive temperature show no in vitro splicing activity (lanes 1 to 3 and 8 to 10). As with the in vivo experiments, no splicing intermediates are detected, pointing to a defect in the first step of splicing. Mixing experiments show that prp2-1 extracts complement the sad1-1 extracts (Fig. 6, lanes 6, 7, 13, and 14), while sad1-1 extracts do not complement each other (lanes 5 and 12). Similar results were obtained when an RP51-based pre-mRNA was used as the in vitro splicing substrate (data not shown). With this substrate, which was in general more efficiently spliced, low levels of splicing were observed with extracts from sad1-1 and prp2-1 cells grown at the permissive temperature, in accordance with the low levels of in vivo splicing measured with the β-galactosidase reporter genes under similar conditions (Fig. 5B).
FIG. 6.
Extracts made from sad1-1 cells do not splice a pre-mRNA substrate in vitro. Extracts were prepared from two sad1-1 isolates, from a prp2-1 strain, and from a wild-type (wt) strain grown either at the permissive temperature (lanes 1 to 7) or for 2.5 h at the nonpermissive temperature. Four microliters of extract was incubated with a radioactive actin pre-mRNA for 30 min at 25°C. In lanes 5, 6, 7, 12, 13, and 14, 2 μl each of the indicated extracts was used. Subsequently, RNA was deproteinized and analyzed on a denaturing polyacrylamide gel. The positions of migration of the pre-mRNA substrate, the exon 1 and lariat intermediates, and the mRNA and intron lariat products of the reaction are indicated.
We therefore conclude that, in addition to an snRNP assembly defect, sad1-1 confers a pronounced splicing defect both in vivo and in vitro.
SAD1 encodes an essential, phylogenetically conserved protein.
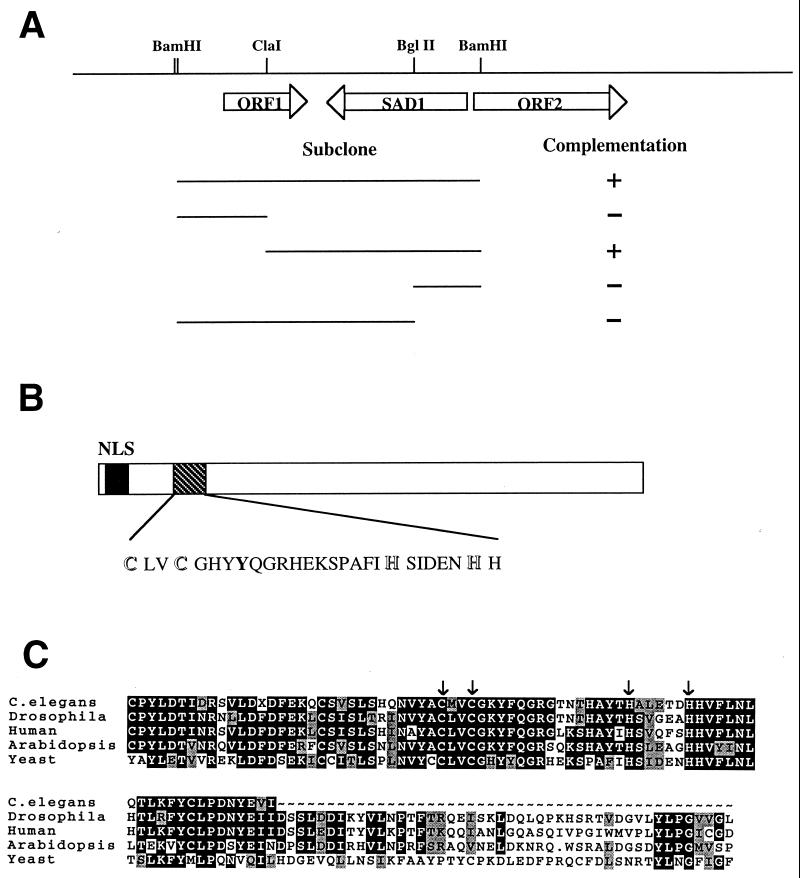
The SAD1 gene was cloned by complementation of the temperature sensitivity of sad1-1 cells, using a genomic library. Six partially overlapping genomic clones were recovered, all of which conferred temperature resistance to sad1-1 cells. Different subclones were constructed and tested for their ability to complement sad1-1 (Fig. 7A). The complementing activity was localized to a 2.2-kb fragment, which was completely sequenced on both strands, revealing an open reading frame of 1,354 bp. Hybridization to an ordered phage library localized the complementing fragment to chromosome VI, which was confirmed by comparison to the complete yeast chromosome VI sequence (42).
FIG. 7.
Cloning and sequence analysis of SAD1. (A) Restriction map of the chromosomal locus containing the SAD1 gene. The extents of the subclones constructed are shown below the map. Complementation of the temperature sensitivity of the sad1-1 strain by the subclones is indicated on the right. (B) Schematic representation of the SAD1 protein. The position of the putative nuclear localization sequence (NLS) and the position and sequence of the putative zinc finger motif are indicated. (C) Alignment of the region surrounding the putative zinc finger in Sad1p and its homologues from C. elegans, Drosophila, Arabidopsis, and human. The positions of the conserved cysteine and histidine residues are marked by arrows. The two mouse ESTs identified encode a peptide identical to the first 40 amino acids deduced from the human ESTs and are not shown. Regions outside this N-terminal block are less well conserved. Identical residues are presented in white letters in a black background, while conservative substitutions are denoted by gray shading.
To demonstrate that the complementing fragment contains the SAD1 gene, rather than an extragenic suppressor, integration of the LEU2 marker was targeted to the chromosomal locus of the complementing fragment in a wild-type haploid strain. Correct integration was verified by Southern analysis. The resulting strain was crossed to a sad1-1 mutant strain, and tetrad analysis was performed. In seven complete tetrads tested, LEU2 always cosegregated with the wild-type SAD1 allele, demonstrating linkage of the cloned fragment to SAD1.
The SAD1 gene codes for a protein of 52 kDa named Sad1p. The N terminus of the protein contains a putative nuclear localization signal and a putative zinc finger-like domain of the C2H2 type (Fig. 7B). Database searches revealed a number of EST sequences from human, mouse, Drosophila, C. elegans, and an A. thaliana cosmid whose deduced products exhibit significant similarity to the N terminus of the yeast Sad1p (see Materials and Methods). All ESTs from a given species appear to be derived from the same gene and probably represent orthologues. In Fig. 7C, the amino acid sequences deduced from the database entries have been aligned to the N terminus of Sad1p. The C2H2 zinc finger appears to be conserved.
A null allele of SAD1 was constructed by replacing the majority of the SAD1 open reading frame with the TRP1 marker. A diploid heterozygous for the deletion was constructed. Tetrad analysis showed that in 22 complete tetrads dissected, only two of four spores grew into colonies, and none of the growing spores carried the TRP1 marker (data not shown). The spores carrying the sad1 deletion germinated but arrested after a few cell divisions. SAD1 is therefore essential for cell viability.
Sad1p localizes to the nucleus and is not an snRNP protein.
In order to study the association of Sad1p with other cellular components and its subcellular localization, Sad1p was tagged by fusing its N or C terminus to two immunoglobulin G binding domains derived from S. aureus ProtA. Both fusion proteins (ProtA-Sad1p and Sad1p-ProtA, respectively), driven by the SAD1 promoter, are fully functional, as they complement a sad1 disruption. A haploid strain carrying the sad1 null allele complemented by ProtA-Sad1p or Sad1p-ProtA on a plasmid was used for further experiments. Western blot analysis showed that both fusion proteins appear as single bands of the expected molecular weight (data not shown).
Immunoprecipitation experiments were carried out at different salt concentrations to assess whether Sad1p is found associated with snRNAs. No detectable levels of U1, U2, U4, U5, or U6 snRNAs were coprecipitated with either ProtA-Sad1p or Sad1p-ProtA, while a tagged U1-70K protein processed in parallel precipitated the U1 snRNA under the four different salt conditions tested (data not shown). It is therefore very unlikely that Sad1p is a stably associated snRNP protein.
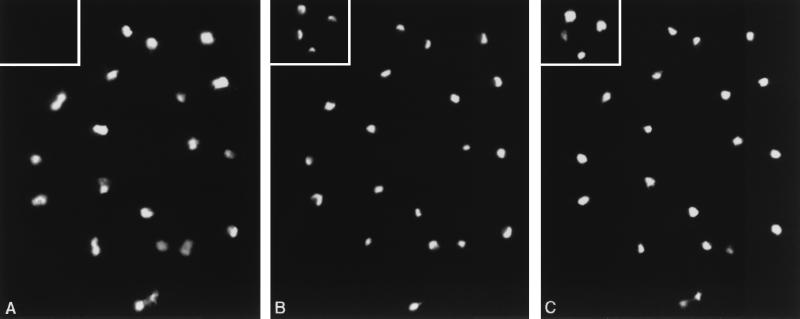
The ProtA-tagged Sad1 proteins were used to assess the localization of Sad1p. ProtA-Sad1p (Fig. 8A) is found in the nucleus (as judged by DAPI [4′,6-diamidino-2-phenylidole] staining [Fig. 8C]). Colocalization experiments with Nop1 (fibrillarin) (Fig. 8B) showed that Sad1p is not excluded from the nucleolus. No background staining is detectable in a wild-type strain (inset in Fig. 8A). Similar results were obtained for Sad1p-ProtA (data not shown). Sad1p is therefore a non-snRNP-associated nuclear protein.
FIG. 8.
Subcellular localization of SAD1p. A strain expressing ProtA-Sad1 and a wild-type strain (inset) were fixed for immunolocalization. Staining of the same cells with an anti-ProtA rabbit antibody (A), an anti-Nop1 monoclonal antibody (B), and DAPI (C) is shown.
DISCUSSION
We describe here a novel assay which allows the identification of factors required for the biosynthesis of spliceosomal snRNPs in yeast. Screening a collection of temperature-sensitive mutants resulted in the isolation of the sad1-1 mutant, a strain impaired in U4/U6 particle assembly.
While transcription and accumulation of the U4 snRNA appear to be unaffected, the U4 snRNA synthesized in sad1-1 cells at the nonpermissive temperature fails to assemble properly with the U6 snRNA and accumulates as free U4. A U4 particle is normally not detectable in wild-type cells, possibly due to the rapid kinetics of association of the newly synthesized U4 with the U6 snRNA, which is in excess (58). In a sad1-1 background, the U4/U6 particle assembled before the temperature shift appears to be stable at the nonpermissive temperature, pointing to a requirement of Sad1p for U4/U6 biosynthesis, rather than stability of the particle or reassembly after each round of splicing. Deletion of BRR1, which encodes a protein associated with all spliceosomal U snRNAs, results in degradation of newly synthesized U snRNAs (47). Similarly, depletion of the core U snRNP protein Smd1p (52), Smd3p (51), or SmEp (5) leads to destabilization of U snRNAs. In these mutants, failure to assemble the core U snRNP particle possible leads to degradation of U snRNAs. We have identified seven temperature-sensitive mutants which exhibit a similar phenotype: the levels of newly synthesized U4 are severely reduced at the nonpermissive temperature. In contrast, the accumulation of U4 snRNA in the sad1-1 mutant could be explained if Sad1p acted later in the biosynthetic pathway, when assembly of a core particle on U4 snRNA had already taken place, protecting the RNA moiety from degradation.
Sad1p appears to have a dual role in the cell. In addition to its involvement in the assembly of the U4/U6 particle, Sad1p is also required for splicing: sad1-1 cells accumulate endogenous pre-mRNAs at the nonpermissive temperature; the splicing of an intron-containing reporter gene is severely diminished in sad1-1 cells already at the permissive temperature, while it is abolished at the restrictive temperature; and additionally, extracts of sad1-1 cells fail to splice a pre-mRNA substrate in vitro. The accumulation of pre-mRNA, rather than splicing intermediates, indicates that Sad1p is required for the first step of splicing. We have so far been unable to immunodeplete Sad1p from extracts to a level that would inhibit splicing in vitro (data not shown). It is therefore formally possible that the splicing defect conferred by sad1-1 is indirect. It is, however, unlikely that the sad1-1 splicing defect is a consequence of the U4/U6 assembly defect (e.g., because of reduced levels of the U4/U6 particle), as it is evident early after the shift to the nonpermissive temperature, when there is still an abundant supply of wild-type U4/U6 and only a low level of free newly synthesized U4* (compare Fig. 4, lanes 2, to Fig. 5B). On the other hand, the U4/U6 assembly defect does not appear to be a nonspecific secondary effect of the defective splicing of sad1-1 mutants (e.g., due to the depletion of an intron-containing assembly factor), as the majority of the splicing mutants tested have no U4/U6 assembly defect. A dual requirement for U snRNA biosynthesis and splicing is expected for U snRNP protein components. For instance, the U snRNA-associated Brr1p, Smd1p, Smd3p, and SmE (5, 47, 51, 52) are required for splicing as well as snRNP biosynthesis. However, we could not detect an association of Sad1p with any of the spliceosomal U snRNAs under different salt conditions, making it unlikely that Sad1p is a stably associated snRNP protein.
Sad1p is essential for cell viability, and it localizes to the nucleus at steady state, consistent with an involvement in splicing and in a late step in snRNP assembly. Sequence analysis reveals, in addition to a nuclear localization signal, the presence of a putative zinc finger motif of the C2H2 type at the N terminus of the protein, similar to the one present in the splicing factors Prp6p, Prp9p, Prp11p, and the U1C protein (29, 44, 61). The role of this motif in spliceosomal proteins is unknown, but it is speculated to mediate interactions with U snRNAs or the mRNA substrate or to be involved in protein-protein interactions. Homologues of Sad1p were identified in Arabidopsis, C. elegans, Drosophila, and human, all of which contain the C2H2 motif.
Thirteen splicing-defective prp strains were tested for their ability to assemble the U4/U6 particle at the nonpermissive temperature. While the majority of the splicing mutants tested had no U snRNP assembly defects, two groups of mutants exhibited a SAD phenotype.
Prp3p, Prp4p, and Prp24p are associated with the U6 and U4/U6 particles and have been proposed to be involved in promoting dissociation-reassociation of U4 with U6 snRNAs during the spliceosomal cycle (1, 2, 16, 50, 58). Consistent with their proposed role, we show that the vast majority of U4 snRNA present in prp3-1, prp4-1, or prp24-1 cells at the nonpermissive temperature was not associated with U6. While at the permissive temperature, assembly of the U4/U6 particle took place (albeit inefficiently, especially for prp24-1 cells), soon after a shift to the nonpermissive temperature, both the U4 snRNA present from before the temperature shift and newly synthesized U4 snRNA accumulated as free U4. The levels of U6 snRNA were severely reduced at the nonpermissive temperature (4), while free U4 snRNA was stable.
Strains harboring the prp17-1 or prp19-1 allele (67) are also defective in the assembly of the U4/U6 snRNP, but, in contrast to prp3-1, prp4-1, and prp24-1 strains, they accumulate both free U4 and U6 snRNAs. Prp17p is required for the second step of splicing and genetically interacts with the U5 snRNP (15). Prp19p, which is not tightly associated with any U snRNA (63), becomes associated with the spliceosome concomitantly with or just after dissociation of the U4 snRNA (63) and is present in extracts in a complex with a number of unidentified proteins (62). The prp17-1 or prp19-1 mutation could be blocking the reassociation of U4/U6 following splicing. While mutations in the U4/U6 snRNP components Prp3p, Prp4p, and Prp24p probably destabilize the particle and expose U6 snRNA to degradation, mutations in Prp17p and Prp19p might block the spliceosome at a stage where U4 has dissociated from U6.
sad1-1 appears to be unique among the splicing-defective mutations tested in affecting the assembly of newly synthesized U4 into the U4/U6 particle rather than the stability of the assembled U4/U6 snRNP or the reassociation of U4 and U6 following each round of splicing. The involvement of Sad1p in U4/U6 assembly could be independent from its function in splicing. Alternatively, the block imposed in splicing in the sad1-1 mutant could result in titrating a factor that is rate limiting for U4/U6 assembly. Elucidation of the role of Sad1 has to await a more detailed analysis of its biochemical function and would be aided by the characterization of its higher eukaryotic homologues and their role in snRNP assembly in vertebrates. Additionally, it is hoped that characterization of the remaining mutants identified by the screen will shed more light on the U snRNP assembly pathway in yeast.
ACKNOWLEDGMENTS
We thank B. M. G. Luukkonen for analyzing the splicing defect of the sad1-1 mutant by primer extension, J. Venema for suggestions on immunofluorescence, and P. Lopez, M. Luukkonen, I. Mattaj, O. Puig, B. Rutz, and J. Salgado-Garrido for discussions and comments on the manuscript. We are grateful to J. Abelson and P. Legrain for prp mutant strains and to the EMBL services for their support.
This work was supported by EMBL.
REFERENCES
- 1.Anthony J G, Weidenhammer E M, Woolford J J. The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA. 1997;3:1143–1152. [PMC free article] [PubMed] [Google Scholar]
- 2.Banroques J, Abelson J N. PRP4: a protein of the yeast U4/U6 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1989;9:3710–3719. doi: 10.1128/mcb.9.9.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berges T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanton S, Srinivasan A, Rymond B C. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol Cell Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordonne R, Tarassov I. The yeast SME1 gene encodes the homologue of the human E core protein. Gene. 1996;176:111–117. doi: 10.1016/0378-1119(96)00230-2. [DOI] [PubMed] [Google Scholar]
- 6.Brow D A, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 7.Brow D A, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- 8.Chanfreau G, Elela S A, Ares M J, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng S C, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y, Dahlberg J, Lund E. Diverse effects of the guanine nucleotide exchange factor RCC1 on RNA transport. Science. 1995;267:1807–1810. doi: 10.1126/science.7534442. [DOI] [PubMed] [Google Scholar]
- 11.Cooper M, Johnston L H, Beggs J D. Identification and characterisation of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doye V, Wepf R, Hurt E C. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1051–1060. [DOI] [PubMed]
- 15.Frank D, Patterson B, Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol Cell Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghetti A, Company M, Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- 17.Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 18.Grandi P, Doye V, Hurt E C. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Z, Moerschell R P, Sherman F, Goldfarb D S. NIP1, a gene required for nuclear transport in yeast. Proc Natl Acad Sci USA. 1992;89:10355–10359. doi: 10.1073/pnas.89.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 21.Hackl W, Fischer U, Lührmann R. A 69-kD protein that associates reversibly with the Sm core domain of several spliceosomal snRNP species. J Cell Biol. 1994;124:261–272. doi: 10.1083/jcb.124.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henriquez R, Blobel G, Aris J P. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J Biol Chem. 1990;265:2209–2215. [PubMed] [Google Scholar]
- 23.Hu J, Xu D, Schappert K, Xu Y, Friesen J D. Mutational analysis of Saccharomyces cerevisiae U4 small nuclear RNA identifies functionally important domains. Mol Cell Biol. 1995;15:1274–1285. doi: 10.1128/mcb.15.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I W. A cap binding protein complexes mediates U snRNA nuclear export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 25.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kambach C, Mattaj I W. Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J Cell Biol. 1992;118:11–21. doi: 10.1083/jcb.118.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kambach C, Mattaj I W. Nuclear transport of the U2 snRNP-specific U2B" protein is mediated by both direct and indirect signalling mechanisms. J Cell Sci. 1994;107:1807–1816. doi: 10.1242/jcs.107.7.1807. [DOI] [PubMed] [Google Scholar]
- 28.Kandels-Lewis S, Séraphin B. Role of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 29.Legrain P, Choulika A. The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J. 1990;9:2775–2781. doi: 10.1002/j.1460-2075.1990.tb07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 32.Lührmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochem Biophys Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 33.Lund E, Dahlberg J E. 2′-, 3′-cyclic phosphates and non-templated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNAs. Science. 1992;255:327. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- 33a.Luukkonen, B. G. M. Personal communication.
- 34.Lygerou Z, Conesa C, Lesage P, Swanson R, Ruet A, Carlon M, Sentenac A, Séraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 36.Maddock J R, Roy J, Woolford J J. Six novel genes necessary for pre-mRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:1037–1044. doi: 10.1093/nar/24.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 38.Marshallsay C, Lührmann R. In vitro nuclear import of snRNPs: cytosolic factors mediate m3G-cap dependence of U1 and U2 snRNP transport. EMBO J. 1994;13:222–231. doi: 10.1002/j.1460-2075.1994.tb06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattaj I, Boelens W, Izaurralde E, Jarmolowski A, Kambach C. Nucleocytoplasmic transport and snRNP assembly. Mol Biol Rep. 1993;18:79–83. doi: 10.1007/BF00986760. [DOI] [PubMed] [Google Scholar]
- 40.Mattaj I W, Tollervey D, Séraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993;7:47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- 41.Moenne A, Camier S, Anderson G, Margottin F, Beggs J, Sentenac A. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 1990;9:271–277. doi: 10.1002/j.1460-2075.1990.tb08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami Y, Naitou M, Hagiwara H, Shibata T, Ozawa M, Sasanuma S, Sasanuma M, Tsuchiya Y, Soeda E, Yokoyama K E A. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat Genet. 1995;10:261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- 43.Nehrbass U, Fabre E, Dihlmann S, Herth W, Hurt E C. Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of nuclear pore protein NSP1. Eur J Cell Biol. 1993;62:1–12. [PubMed] [Google Scholar]
- 44.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Luhrmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuman D V H, Dahlberg J E. Nucleocytoplasmic transport and processing of small nuclear RNA. Mol Cell Biol. 1990;10:3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman A. Analysis of pre-mRNA splicing in yeast. In: Higgins S J, Hames B D, editors. RNA processing: a practical approach. I. Oxford, United Kingdom: IRL Press; 1994. pp. 179–195. [Google Scholar]
- 47.Noble S M, Guthrie C. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- 48.Palacios I, Weis K, Klebe C, Mattaj I W, Dingwall C. RAN/TC4 mutants identify a common requirement for snRNP and protein import. J Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plessel G, Fischer U, Lührmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raghunathan P L, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 51.Roy J, Zheng B, Rymond B, Woolford J. Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rymond B C. Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1993;90:848–852. doi: 10.1073/pnas.90.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Schwer B, Shuman S. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA. 1996;2:574–583. [PMC free article] [PubMed] [Google Scholar]
- 55.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA: pre mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosomal assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 58.Shannon K W, Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 59.Shimba S, Reddy R. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J Biol Chem. 1994;269:12419–12423. [PubMed] [Google Scholar]
- 60.Stutz F, Liao X C, Rosbash M. U1 small nuclear ribonucleoprotein particle-protein interactions are revealed in Saccharomyces cerevisiae by in vivo competition assays. Mol Cell Biol. 1993;13:2126–2133. doi: 10.1128/mcb.13.4.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang J, Abovich N, Fleming M L, Seraphin B, Rosbash M. Identification and characterization of a yeast homolog of U1 snRNP-specific protein C. EMBO J. 1997;16:4082–4091. doi: 10.1093/emboj/16.13.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarn W Y, Hsu C H, Huang K T, Chen H R, Kao H Y, Lee K R, Cheng S C. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarn W Y, Lee K R, Cheng S C. The yeast PRP19 protein is not tightly associated with small nuclear RNAs but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol. 1993;13:1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teem J L, Rosbash M. Expression of a β-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci USA. 1983;80:4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 66.Vankan P, McGuigan C, Mattaj I W. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 1990;9:3397–3404. doi: 10.1002/j.1460-2075.1990.tb07541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 68.Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1295. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wersig C, Bindereif A. Reconstitution of functional mammalian U4 small nuclear ribonucleoprotein: Sm protein binding is not essential for splicing in vitro. Mol Cell Biol. 1992;12:1460–1468. doi: 10.1128/mcb.12.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Will C, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 71.Will C L, Behrens S-E, Lührmann R. Protein composition of mammalian spliceosomal snRNPs. Mol Biol Rep. 1993;18:121–126. doi: 10.1007/BF00986766. [DOI] [PubMed] [Google Scholar]
- 72.Wise J A, Tollervey D, Maloney D, Swerdlow H, Dunn E J, Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983;35:743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- 73.Xie J, Beickman K, Otte E, Rymond B C. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 1998;17:2938–2946. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang H, Moss M L, Lund E, Dahlberg J E. Nuclear processing of the 3′-terminal nucleotides of pre-U1 RNA in Xenopus. Mol Cell Biol. 1992;12:1553–1560. doi: 10.1128/mcb.12.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]