Identification of a Linear Heparin Binding Domain for Human Respiratory Syncytial Virus Attachment Glycoprotein G (original) (raw)
Abstract
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract disease in infants and young children worldwide. Infection is mediated, in part, by an initial interaction between attachment protein (G) and a highly sulfated heparin-like glycosaminoglycan (Gag) located on the cell surface. Synthetic overlapping peptides derived from consensus sequences of the G protein ectodomain from both RSV subgroups A and B were tested by heparin-agarose affinity chromatography for their abilities to bind heparin. This evaluation identified a single linear heparin binding domain (HBD) for RSV subgroup A (184A→T198) and B (183K→K197). The binding of these peptides to Vero cells was inhibited by heparin. Peptide binding to two CHO cell mutants (pgsD-677 and pgsA-745) deficient in heparan sulfate or total Gag synthesis was decreased 50% versus the parental cell line, CHO-K1, and decreased an average of 87% in the presence of heparin. The RSV-G HBD peptides were also able to inhibit homologous and heterologous virus infectivity of Vero cells. These results indicate that the sequence 184A/183K→198T/K197 for RSV subgroups A and B, respectively, defines an important determinant of RSV-G interactions with heparin.
Human respiratory syncytial virus (RSV), a member of the genus Pneumovirus within the family Paramyxoviridae, is the leading cause of lower respiratory tract infection in infants and young children worldwide (8). Currently, there are no effective licensed vaccines. During clinical trials in the 1960s, children inoculated with a formalin-inactivated RSV vaccine were left unprotected and developed exacerbated disease associated with eosinophilia upon subsequent exposure to wild-type virus (7, 23, 25). Since that time, many investigators have worked to gain a better understanding of the mechanisms involved in the development of severe bronchiolitis sometimes observed during the course of natural infection. One important aspect of this process is identifying the steps required for attachment and infection of target cells.
RSV-G is one of three glycoproteins found on the surface of the virion and is synthesized as a core protein of 298 amino acids. RSV-G then undergoes extensive N- and O-linked glycosylation prior to expression on the cell surface as a type II integral membrane protein (8, 9, 38, 50). RSV-G has been shown to function as an attachment protein (29). Many have speculated that the receptor-binding domain of RSV-G may be located between amino acids 164H→C176. The principal evidence supporting this speculation is based on the observation that this region is exactly conserved among all wild-type RSV isolates sequenced to date (21). While no specific receptor has been described that recognizes the G glycoprotein, it was recently shown that RSV could bind to immobilized heparin (27). In vivo, heparin is primarily located in the granules of mast cells and basophils. However, heparan sulfate, a related compound, is found on the surface of most mammalian cell types and in the extracellular matrix (17). Many viruses, including herpesviruses (16, 28, 31, 51), human immunodeficiency viruses (33, 36, 37), flaviviruses (6), picornaviruses (20), and alphaviruses (3, 26), utilize heparan sulfate to mediate attachment and infection of target cells. Heparin binding proteins are known to interact with heparin via electrostatic charge interactions generated between the negatively charged sulfate groups on heparin and the positively charged amino acids within the protein’s heparin binding domain (HBD) (5, 16, 45). Interestingly, the ectodomain of the RSV-G protein contains a cluster of positively charged amino acids (180P→K233) (27) which falls within an immunodominant region of RSV-G. It has been postulated that RSV-G–heparin binding interactions are mediated via this clustering of basic amino acids within the RSV-G ectodomain (27). However, there has been no experimental evidence to corroborate this assumption. Therefore, it is the purpose of this study to identify potential linear HBDs within the ectodomain of the RSV-G protein and to determine if the clustering of positively charged amino acids is involved in RSV-Gag interactions.
MATERIALS AND METHODS
Cells, virus, and purified viral proteins.
Vero cells were grown in Eagle’s medium containing Earle’s salts (EMEM) (Mediatech Inc., Herndon, Va.) and 10% fetal bovine serum (FBS) (Intergen, Purchase, N.Y.). The following Chinese hamster ovary (CHO) cells were grown in Ham’s F12 medium (Mediatech Inc.) containing 10% FBS: K1, the parental CHO cell line; pgsD-677, which contains a defect in GlcNAc and GlcA transferase and is heparan sulfate negative while producing three to four times the normal amount of chondroitin sulfate; and pgsA-745, which is xylosyl transferase deficient, producing approximately 1% of wild-type Gag (13, 14). Human RSV strains A2 and 18537 were prepared by inoculating Vero cells at a multiplicity of infection between 0.1 and 1 (32). Virus was concentrated as previously described (32) or pelleted directly from the tissue culture supernatant and resuspended in EMEM containing 1% FBS, 100 mM MgSO4, and 50 mM HEPES (Bio-Whittaker, Walkersville, Md.). Infectious titers were determined following inoculation of Vero cell monolayers and reported as 50% tissue culture infectious doses (TCID50) or PFU by methods previously described (19).
Purified attachment (G) protein (0.31 mg/ml) from the A2 strain of RSV grown in Vero cells and polyclonal rabbit anti-G antiserum was supplied by Lederle-Praxis Biologicals (West Henrietta, N.Y.) (29).
Synthesis of RSV-G overlapping peptides derived from G protein ectodomain sequence.
Consensus sequences were generated for the G protein amino acid sequence deduced from G gene nucleotide sequence data for RSV subgroup A and B viruses (Fig. 1). The subgroup A strains used to generate the consensus sequence were A2, Long, 1734, 5857, 6190, 6256, 642, and 6614. The sequences used to generate the subgroup B consensus sequence were 18537, 8/60, 9320, nm1355, wv10010, wv15291, and wv4843. The peptide sets consisted of amino acids 58 to 298 of the RSV Ga or amino acids 58 to 292 of the Gb glycoprotein, respectively. All peptides included biotin-SGSG at their amino termini and 15 amino acids of RSV G ectodomain sequence with a 5-amino-acid overlap and offset. Peptides were synthesized by 9-fluorenylmethoxycarbonyl solid-phase chemistry (Chiron Technologies, San Diego, Calif.) with an average purity of 80 to 90%. The peptide pools were designated A or B, and individual peptides from each pool were designated a or b, corresponding to the RSV subgroup from which they were derived. The peptide pools for subgroup A were set up as follows, starting from the carboxy terminus: pool A1 (a1 to a9; 240T→Q298), pool A2 (a10 to a18; 189I→L249), pool A3 (a19 to a27; 144S→T198), pool A4 (a28 to a36; 98G→N153), and pool A5 (a37 to a44; 58A→T106). The subgroup B pools were set up as follows: pool B1 (b1 to b9; 238K→T292), pool B2 (b10 to b18; 192K→S247), pool B3 (b19 to b27; 148T→P202), pool B4 (b28 to b36; 102S→K157), and pool B5 (b37 to b45; 58A→H112). Peptide 3vnm (YFARGPGIHIRKRKN), a reverse-oriented human immunodeficiency virus type 1 V3 loop peptide (307N→Y321), was used as a negative control. Peptide Vn (346A→G358), representing the mammalian consensus HBD motif, XBBXBX (bold letters represent basic residues), derived from the vitronectin HBD, was used as a positive control. The sequence of the RSV-Ga cysteine noose peptide was 164HFEVFNFVPCSICSNNPTCWAICKRI189.
FIG. 1.
RSV G glycoprotein ectodomain consensus sequences. Fifteen-amino-acid long overlapping peptides were generated for both RSV subgroups. Consensus peptides (RSVCONSA and -B) are aligned with subgroup A (strain A2) and subgroup B (strain 18537) viruses to demonstrate homology of the consensus peptides with wild-type virus. Peptides are numbered 1 through 44 (subgroup A) or 1 through 45 (subgroup B) starting from the carboxy terminus. The highlighted sequence represents the exactly conserved region (amino acids 164 to 176), and conserved cysteine residues are marked by black dots. The putative heparin binding region of the RSV-G ectodomain (amino acids 187 to 217) (27), characterized by a cluster of basic amino acids, is defined by the black box. Peptides for subgroup A (solid lines) and subgroup B (dashed lines) indicate the positions of the peptides that bound heparin.
Heparin-agarose affinity chromatography (HAAC).
Assays involving the use of 1 ml of pooled (100 μg/ml) or individual (10 μg/ml) peptides were run as described previously (27) with some modifications. Heparin-agarose or unlabeled Sepharose CL4B (Sigma, St. Louis, Mo.) columns were equilibrated with carbonate-bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3 [pH 9.6]) prior to the addition of peptide. The columns were then washed with 20 column volumes of carbonate-bicarbonate buffer (pH 9.6) containing 0.1% Triton X-100 and 100 mM NaCl, followed by 10 column volumes of carbonate-bicarbonate buffer (pH 9.6) without detergent or salt to avoid interference with plate coating (see below). Peptides were eluted with carbonate-bicarbonate buffer (pH 9.6) containing 2 mg of heparin (porcine intestinal mucosa; Mr = 6,000; Sigma). Optical density (OD) values from a no-peptide control were subtracted from all peptide-containing wells. Positive OD values were equal to or greater than twice the OD value of the negative-control peptide.
Peptide enzyme-linked immunosorbent assay (ELISA).
Eluted peptide fractions were subjected to serial twofold dilutions in carbonate-bicarbonate buffer (pH 9.6), starting with the undiluted material. Immunolon I plates (Dynatech, Chantilly, Va.) were coated with 50 μl of each peptide dilution overnight at 4°C, washed with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-T), and then blocked with PBS containing 5% nonfat dry milk (BLOTTO) for 1 h at 37°C or overnight at 4°C. Bound biotinylated peptide was detected following the addition of 50 μl of avidin-horseradish peroxidase (HRP) conjugate (1:500) (Kirkegaard and Perry Laboratories, Gaithersburg, Md.) in BLOTTO plus 0.05% Tween 20 for 1 h at 37°C. After being washed with PBS-T, the plates were developed with 100 μl of ABTS [2,2′-azinobis (3-ethylbenzthiazolinesulfonic acid)] peroxidase substrate (Kirkegaard and Perry Laboratories) and read at 405 nm on a Vmax kinetic plate reader (Molecular Devices, Menlo Park, Calif.). Positive OD values were determined as described for HAAC.
Cell binding ELISA.
Peptides were tested for their ability to bind to Vero or various CHO cell lines. Briefly, 96-well tissue culture plates were seeded with 2 × 104 cells per well and incubated overnight. For Vero cells, the medium was removed and the monolayers were fixed overnight by drying them at 37°C or by adding 100 μl of 80% methanol (MeOH) per well for 30 min at 4°C prior to blocking with 5% BLOTTO. Peptide binding was assayed by ELISA as described above. Briefly, pooled or individual peptides were diluted to 100 and 10 μg/ml, respectively, prior to incubation on fixed cell monolayers overnight at 4°C or for 1 h at 37°C. After being washed with PBS-T, their specific attachment was detected following the addition of avidin-HRP (1:500). Heparin inhibition of peptide binding was carried out by diluting peptides in diluent containing the indicated concentrations of heparin just prior to assaying them by ELISA for cell binding. Due to high background binding levels of peptides to MeOH-fixed CHO cells, these cells were fixed subsequent to the peptide binding reaction. Peptides were diluted in EMEM–2% bovine serum albumin and then reacted with CHO cells for 1 h at 4°C and the cells were washed five times with PBS and then MeOH fixed. Heparin inhibition of peptide binding and peptide detection were carried out as described previously.
Detection of purified RSV-G binding to Vero cells was done with a monospecific polyclonal rabbit anti-G antiserum (1:1,000) followed by a goat anti-rabbit HRP conjugate (1:1,000) (Kirkegaard and Perry Laboratories) and 100 μL of ABTS as a substrate. Positive OD values were determined as described for HAAC.
Infectivity inhibition assay.
Peptides were assayed for their abilities to inhibit RSV A2 or 18537 infectivity. The peptides were diluted to 50 μM in EMEM–1% FBS, and 50 μl was added in quadruplicate to Vero cells in a 96-well plate. After a 45-min incubation at 37°C, 50 μl of RSV A2 or 18537 virus (100 TCID50) was added to peptide-treated and untreated control wells. The virus-peptide mixture was allowed to adsorb for 2 h at 37°C, after which the cells were washed and overlaid with EMEM containing 1% FBS and 1% methylcellulose. Three days postinoculation, the cells were fixed and stained with 1% crystal violet. Plaques were counted, and percent inhibition of virus infectivity of treated wells was determined versus untreated control wells.
Sequence analysis of heparin binding peptides.
Peptides representing the putative HBDs of the RSV-G glycoprotein were compared to known RSV subgroup A and B G protein sequences by using the University of Wisconsin Genetics Computer Group program.
RESULTS
RSV synthetic-peptide HAAC.
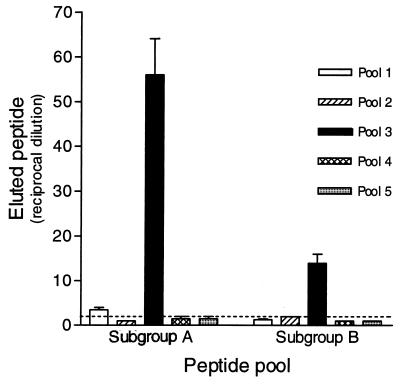
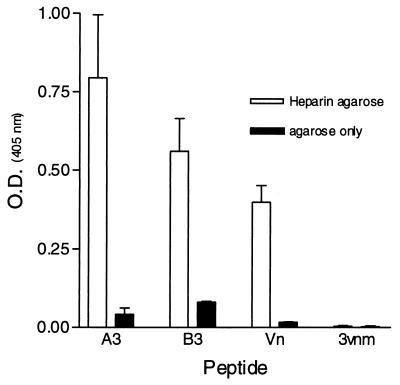
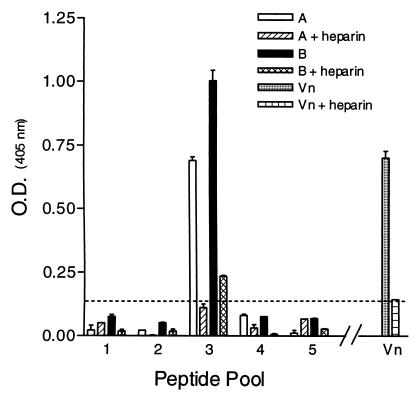
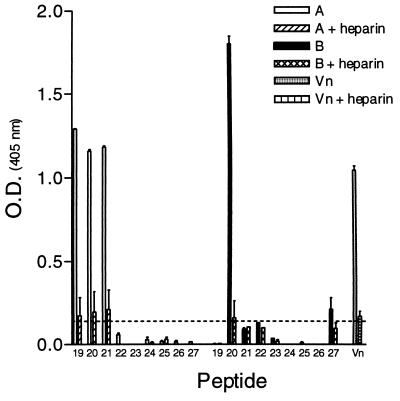
In an attempt to determine regions on the RSV-G protein important for heparin binding, a series of overlapping peptides representing the consensus sequence for Ga and Gb were synthesized (Fig. 1). Using HAAC, the peptide pools were tested for their ability to bind immobilized heparin. Figure 2 demonstrates that the heparin binding activity for subgroup A and B peptide pools resides primarily within pool 3 (A3 and B3), spanning amino acids 144S→T198 for subgroup A and 148T→P202 for subgroup B. The interactions of pools A3 and B3 are considered heparin specific, as the peptides were eluted with heparin and did not bind to unlinked Sepharose CL-4B (Fig. 3). Individual peptides comprising pool 3 for both subgroups were then examined for their abilities to bind immobilized heparin (Fig. 4). The results show that pool A3 peptides a19, a20, and a21 bound to heparin corresponding to amino acids 175SICSNPTCWAICKRIPNKKPGKKT198 (bold letters represent basic residues), of the Ga glycoprotein. Examination of the individual pool B3 peptides shows that peptide b20 (183KSICKTIPSNKPKKK197) and b27 (148TKPRSKNPPKKPKDD162) were the only peptides with significant heparin binding activity (Fig. 4).
FIG. 2.
HAAC of pooled biotinylated peptides. One milliliter of peptide (100 μg/ml) in carbonate-bicarbonate buffer (pH 9.6) was run over heparin agarose columns. The columns were washed with 20 column volumes of carbonate-bicarbonate buffer (pH 9.6) containing 100 mM NaCl and 0.1% Triton X-100 followed by 10 column volumes of carbonate-bicarbonate buffer only. Bound peptides were eluted in carbonate-bicarbonate buffer containing 2 mg of heparin/ml. Eluted biotinylated peptides were adsorbed to microtiter plates, and endpoint titers were determined by ELISA. Values above the dashed line (twice background) are considered positive. The data are from a representative experiment of at least three experiments, with the error bars indicating the standard error of the mean of quadruplicate wells.
FIG. 3.
Demonstration of the specificity of the HAAC for RSV and other heparin binding peptides. Peptide pools A3 and B3 (100 μg/ml) and control peptides Vn (positive control) and 3vnm (negative control) (10 μg/ml) were reacted with heparin agarose or unlinked CL4B agarose as described in the legend to Fig. 2. Data are from one of two separate experiments, with error bars indicating the standard error of the mean.
FIG. 4.
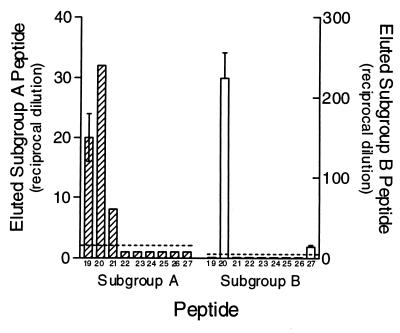
HAAC of individual biotinylated peptides comprising pool 3. The individual peptides (10 μg/ml) were tested for their abilities to bind heparin agarose. Peptides that specifically bound heparin were eluted and detected as described in the legend to Fig. 2. Values equal to or above the dashed line (twice background) are considered positive. The data are from a representative experiment of at least three experiments, with the error bars indicating the standard error of the mean of quadruplicate wells.
Binding of RSV G heparin binding peptides to cells.
Each of the peptide pools was then examined for the ability to bind Vero cells. Peptide pools A3, B3, and the positive control peptide, Vn, bound to Vero cells, as determined by ELISA (Fig. 5), whereas all the other peptide pools for both subgroups as well as the negative-control peptide, 3vnm, were not able to bind. Furthermore, the reactivity of pools A3, B3, and Vn with cell surface molecules was inhibited by the addition of soluble heparin (2 mg/ml), suggesting that this interaction was likely mediated via cellular, heparin-like Gags (Fig. 5). Reactivity with Vero cells was inhibited by 84, 76, and 84% for pool A3, pool B3, and Vn, respectively.
FIG. 5.
Reactivities of pooled biotinylated peptides with Vero cells. Subgroup A and B peptide pools (100 μg/ml) were diluted in BLOTTO–0.05% Tween 20 with or without the addition of heparin (2 mg/ml) and tested for their abilities to bind to Vero cells. Bound peptides were detected with avidin conjugated with HRP (1:500) and ABTS as a substrate. Values at or above the dashed line (twice background) are considered positive. The data are from a representative experiment of at least three experiments, with the error bars indicating the standard error of the mean of quadruplicate wells.
Individual subgroup A (a19, a20, and a21) and B (b20) peptides were also able to bind to Vero cells. All the other individual pool A3 peptides were negative. Likewise, all the remaining subgroup B3 peptides were considered negative, with the exception of peptide b27, which bound only weakly to Vero cells (Fig. 6). The binding of peptides a19, a20, and a21 was inhibited by 87, 85, and 83%, respectively, by the addition of 2 mg of heparin/ml (Fig. 6). Heparin decreased the reactivity of peptide b20 and b27 by 90 and 48%, respectively. However, even though peptide b27 binding was decreased 48% in the presence of heparin, a direct comparison of the overall reactivity of untreated b27 peptide with Vero cells was approximately 90% less than that of b20. Interestingly, the degree of binding inhibition of purified RSV G glycoprotein in a similar assay as well as heparin inhibition of whole-virus binding to unfixed Vero cells was approximately 50% (unpublished data). However, considering the extensive glycosylation and secondary structure of the purified protein as well as the multimeric nature of G on the native virion, we cannot rule out non-heparin-mediated attachment that might account for the incomplete inhibition. Of note, peptides spanning the conserved region (164H→C176) or the entire cysteine noose region (164H→I189 [data not shown]) did not appear to react with Vero cells.
FIG. 6.
Reactivities of individual biotinylated heparin binding peptides with Vero cells. The individual peptides from pool A3 and B3 shown previously to interact with heparin were tested for their abilities to bind Vero cells in the presence (diagonally hatched bars, subgroup A; crosshatched bars, subgroup B) and absence (open bars, subgroup A; solid bars, subgroup B) of 2 mg of heparin/ml. Individual peptides were tested at a concentration of 10 μg/ml and detected as described in the legend to Fig. 5. Values at or above the dashed line (twice background) are considered positive. The data are from a representative experiment of at least three experiments, with the error bars indicating the standard error of the mean of quadruplicate wells.
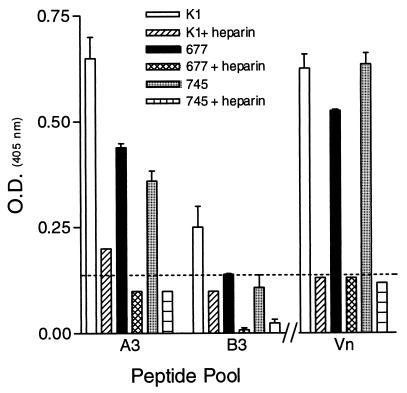
In an attempt to determine the specific Gag requirements for the RSV-G peptides, we measured the reactivity of the peptide pools with three CHO cell lines, two of which contained defects in their abilities to express particular Gags. The results shown in Fig. 7 demonstrate that the peptide pools A3 and B3 reacted with each of the cell lines examined while all the other pools did not bind (data not shown). The reactivity of pools A3 and B3 with both the heparan sulfate-deficient (pgsD-677) and Gag-deficient (pgsA-745) cell lines was approximately 60 and 47% that of the parental (K1) cell lines for pools A3 and B3, respectively. In addition, heparin reduced the reactivity of pool A3 by 70% for each of the three CHO cell lines and reduced pool B3 reactivity with CHO-K1 cells by a total of 60%, reduced that with pgsD-677 cells by 92%, and reduced that with pgsA-745 cells by 84% (Fig. 7). Interestingly, the positive-control peptide, Vn, also reacted strongly with all three CHO cell lines and did not exhibit a significant decrease in binding to either of the Gag-deficient CHO cell lines (Fig. 7). The binding of the Vn peptide was decreased by 84, 74, and 82% for the K1, pgsD-677, and pgsA-745 CHO cell lines, respectively, when heparin was added.
FIG. 7.
Reactivities of pooled biotinylated peptides with various CHO cell lines. Pooled peptides were reacted with various CHO cells, and after fixation, bound peptides were detected as described in the legend to Fig. 5. Values at or above the dashed line (twice background) are considered positive. The data are from a representative experiment of at least three experiments, with the error bars indicating the standard error of the mean of quadruplicate wells.
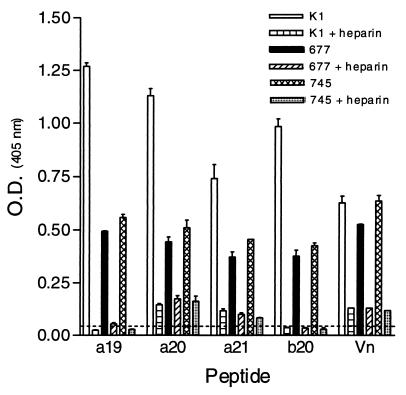
Examination of the individual peptides from pools A3 and B3 with CHO-K1, pgsD-677, and pgsA-745 cell lines revealed that only peptides a19 to a21 and b20 bound (Fig. 8). All the other individual peptides within pools A3 and B3 were unable to bind (data not shown). On average, the reactivities of a19 to a21 and b20 were reduced by 60 and 50% for pgsD-677 and pgsA-745 cells, respectively, compared to the parental CHO K1 cell line. Furthermore, the reactivity of a19 to a21 and b20 decreased an average of 92, 80, and 88% in the presence of heparin for CHO K1, pgsD-677, and pgsA-745 cells, respectively (Fig. 8). In contrast to the Vero cell data (Fig. 6), peptide b27 did not react with any of the CHO cell lines tested.
FIG. 8.
Reactivities of individual biotinylated peptides with various CHO cell lines. Individual peptides from pools A3 and B3 shown previously to interact with heparin were tested for their abilities to bind various CHO cell lines. Individual peptides were tested at a concentration of 10 μg/ml and, after fixation, were detected as described in the legend to Fig. 5. Values at or above the dashed line (twice background) are considered positive. The data are from a representative experiment of at least three experiments, with the error bars indicating the standard error of the mean of quadruplicate wells.
Effects of RSV-G heparin binding peptides on virus infectivity.
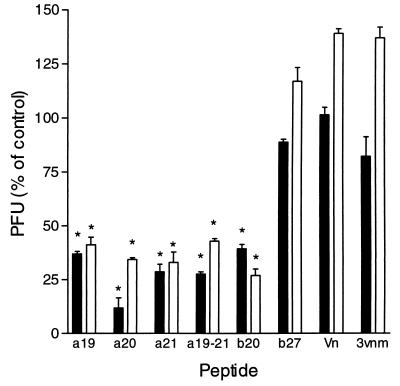
To determine if the RSV-G HBD peptides could inhibit virus infectivity by blocking the interaction between infectious virus and cellular Gags, an infectivity inhibition assay was carried out. Three RSV-Ga peptides (a19, a20, and a21) inhibited the homologous subgroup A virus (strain A2) infectivity by 60 to 90% (Fig. 9). Peptide b20 was able to inhibit the infectivity of heterologous A2 virus by 60%. In the reciprocal experiment, peptide b20 reduced homologous subgroup B virus (strain 18537) infectivity by 81%, and a19, a20, and a21 were able to inhibit 18537 infectivity by 70, 75, and 76%, respectively (Fig. 7). Peptide b27 or the Vn peptide, both of which contain the mammalian consensus HBD motif (XBBXBX), did not inhibit A2 or 18537 infectivity. Furthermore, the peptide pools 1, 2, 4, and 5 for each subgroup did not inhibit A2 or 18537 infectivity, nor did the remainder of the peptides in pool A3 or B3 (data not shown). It should be noted that pools A3 and B3 (data not shown) or a mixture of peptides a19 to a21 did not further decrease A2 or 18537 virus infectivity over that of the individual peptides (a19, a20, a21, or b20). The results of this assay were then tested for significance based on the analysis of variance of the test peptides versus the inhibitory effect of control peptide 3vnm followed by the post hoc Tukey test to measure the differences among groups. The results for peptides a19, a20, a21, and b20 were considered significant, with P values of <0.05.
FIG. 9.
Inhibition of homologous and heterologous RSV infectivity by heparin binding peptides. Vero cells were preincubated with 50 μM peptide per well in 50 μl. Peptide was allowed to adsorb for 45 min at 37°C, followed by the addition of 100 TCID50 of virus (solid bars, strain A2; open bars, strain 18537) for 2 h. The cells were washed and overlaid with EMEM–1% FBS–1% methylcellulose for 3 days before being fixed and stained with 1% crystal violet. 3vnm was used as a negative control for peptide inhibition of virus infectivity. Infectivity was determined as a percentage versus an untreated control. The error bars represent the standard error of the mean of quadruplicate measurements from three separate experiments. The test peptide results were considered significant, with P values of <0.05 versus control peptides (*).
Sequence analysis.
The linear amino acid sequences 175SICSNPTCWAICKRIPNKKPGKKT198 for subgroup A viruses and 183KSICKTIPSNKPKKK197 for subgroup B viruses contained significant heparin binding activity. A second potential HBD represented by peptide b27 (148T→D162) was found just upstream of the conserved region within the subgroup B RSV-G ectodomain. However, this peptide bound only weakly in the Vero cell ELISA (Fig. 6), did not bind to any of the CHO cell lines (Fig. 8), and did not inhibit homologous or heterologous virus infectivity (Fig. 9). Alignment of the subgroup A and B consensus HBD sequences (184A/183K→T198/K197) yielded the following consensus sequence for the linear RSV HBD: XXICBXIPXXBPXBB, where B is a basic amino acid and X is usually an uncharged residue (Table 1). The density of basic residues was high, with 40% of the putative HBDs composed of basic residues. An important consideration is that the negative-control peptide, 3vnm, contained 33% basic amino acids, not including any histidine residues. This peptide failed to bind to immobilized heparin, Vero cells, or CHO cells, suggesting that the presence of basic residues alone is insufficient for heparin binding. In contrast, Vn bound to immobilized heparin and Vero cells, as well as parental and Gag-deficient CHO cells, and the binding of Vn was inhibited by heparin.
TABLE 1.
Linear sequence alignment of various viral and mammalian HBDs
| HBDa | Locationb | RSV peptide | Sequencec | Consensusd motif | Reference(s)e |
|---|---|---|---|---|---|
| Viral | RSV-A 184–198 | a19–21 | ---AICKRIPNKK-PGKKT---- | XBBX | |
| RSV-B 183–197 | b20 | --KSICKTIPSNK-PKKK----- | XBX, XBBBX | ||
| RSV-B 148–162 | b27 | --TKPRSKNPPKKPKDD------ | XBBXBX | ||
| PRV 263–271 | -----PRRSVRLR-W-------- | XBBBXXBX | 30, 52 | ||
| HSV1 142–152 | ------IR-CRFRNSTRM----- | XBXBXBX | 46 | ||
| Mammalian | Vn 346–358 | ------AKKQRFRHRNRKG---- | XBBXBX | 43 |
Comparison of the subgroup A peptide HBD sequences against homologous G ectodomain sequences indicated that the cysteine, prolines, and basic residues found within the RSV-Ga HBD, 184A→T198, were 100% conserved. Likewise, the RSV-Ga HBD sequence was nearly 100% conserved among all sequences examined, with the exception of two subgroup A viruses, each of which contained a single conservative change (192N→S or 198K→R). Examination showed that the RSV-Gb HBD, 183K→K197, was 100% conserved for all but one strain B virus, wv10010, which contained the mutation 190P→S. The homology between the subgroup A and B HBDs was approximately 60%.
DISCUSSION
In order to map regions of RSV-G important for heparin binding, we used two sets of synthetic overlapping peptides representing the ectodomains of both RSV subgroups. We identified two linear sequences, 184AICKRIPNKKPGKKT198 for subgroup A viruses and 183KSICKTIPSNKPKKK197 for subgroup B viruses, as being important for RSV heparin binding. When subgroup A and B HBD peptide sequences were compared with G glycoprotein sequences from their respective subgroups, the HBD region was conserved among the majority of sequences examined. Although RSV-G A and B HBD sequences have only 60 to 70% sequence homology, the HBDs spatially mapped to nearly identical locations on their respective proteins (42). Interestingly, the HBDs are proximal to the conserved region (164H→C176) including the cysteine noose, which suggests that this site might play a critical role in the function of RSV-G HBDs. In our assays a peptide representing the conserved region (164H→C176) or the entire cysteine noose region (164H→I189) of RSV-G did not appear to bind to Vero cells. Assuming this peptide folded correctly, these experiments did not support the idea that this region interacts with cellular receptors (18). However, an important limitation of peptides is that they may not represent native proteins in terms of conformation or glycosylation. Thus, further binding studies will be necessary to determine if the proximity of the linear HBD to the cysteine noose region is important for heparin binding and whether there are conformational determinants involved in RSV-heparin interactions.
Analysis of several mammalian heparin binding proteins has yielded two consensus sequences, XBBXBX and XBBBXXBX, where B is almost always a basic residue and X is usually an uncharged hydrophobic residue (for a review, see reference 5). Several viral HBDs have been mapped and experimentally shown to bind heparin (15, 16, 30, 35, 39, 46, 52). Many of these viral HBDs do not conform to either of the mammalian consensus HBD linear sequence motifs (Table 1). Interestingly, for RSV-G, peptide b27 is a proline-rich peptide that contains an XBBXBX mammalian HBD motif, yet this peptide bound 16-fold less than peptide b20 in the HAAC assay. Furthermore, peptide b27 bound only weakly to Vero cells and did not bind to any of the CHO cells assayed, whereas the Vn HBD peptide, also containing an XBBXBX mammalian HBD motif, reacted strongly with both Vero and CHO cells and was inhibited by heparin. Thus, these data suggest that heparin binding is not limited to linear XBBXBX or XBBBXXBX motifs (6, 26, 47). Furthermore, factors such as conformation, accessibility of the basic residues to heparin, and possibly proline content may also influence viral protein interactions with cellular Gags. Likewise, our data suggest that our heparin binding peptides preferentially bind heparin; however, the RSV HBD peptide requirements for heparin may not be absolute. Peptide binding to pgsD-677 cells was reduced but not completely abrogated. This finding was not unexpected, as chondroitin sulfate, which is overexpressed on the pgsD-677 cells, was able to partially elute the RSV HBD peptides from heparin agarose (data not shown). In addition, these peptides may interact with an as-yet-unknown cellular protein. Unexpectedly, the RSV HBD peptides also bound to the pgsA-745 CHO cells. Work is currently under way to explain this paradox. Interestingly, preliminary data from gel electrophoresis of cell lysates stained with toluidine blue revealed the presence of a single high-molecular-mass proteoglycan (>250 kDa) that stains in pgsA-745 cells that was also one of multiple bands present in the K1 cell line, suggesting that these cells are Gag deficient but may not be Gag negative. These data could explain, in part, the binding of RSV and Vn HBD peptides to these cells. Little work has been done to model viral HBDs; thus, the minimal sequence requirements for RSV-G heparin binding are as yet unclear (6, 16). Further studies will be required to better characterize RSV peptide interactions with CHO cells in hopes of better understanding virus interactions with cellular glycosaminoglycans.
Initial attempts to understand how RSV interacts with heparin were focused on the mechanism by which heparin inhibits virus infectivity. Recent reports have created some controversy over this mechanism. One report suggests that the heparin inhibition of RSV is the result of heparin binding to RSV-G, thereby blocking attachment and/or infectivity of the virus (27). Our data lend further support to this mechanism of RSV heparin inhibition. In contrast, a second report argues that virus grown in Hep2 cells results in the production of viral heparin-like molecules associated with RSV-G. In this scenario, exogenous heparin binds to the cell surface, blocking interactions with viral proteoglycans (2). Our data could also suggest that cellular Gags might be complexed with RSV-G via interactions with intrinsic HBDs. Many viruses acquire cellular membrane components while budding from the plasma membrane of the cell (1, 4), and it is possible that RSV acquires cellular heparan sulfate while budding from the plasma membrane. Acquisition of cellular heparan sulfate by RSV could benefit the virus in at least two ways: (i) heparan sulfate binding could act to stabilize glycoprotein conformation (48) and (ii) heparan sulfate may mask an important functional site from the host response (48). Interestingly, evidence was recently presented from studies with RSV B1/cp52, a subgroup B G-SH deletion mutant, suggesting that the G glycoprotein is not absolutely required for virus infectivity (24). This finding raises some interesting questions, the first being what the functional role of RSV-G is and the second being how this function relates to RSV-G-heparin interactions.
The most obvious functional role for RSV-G interactions with heparan sulfate involves receptor binding, tissue tropism, and determination of the extent of viral infection within the respiratory tract. Heparan sulfate is the membrane-associated cellular homologue of heparin, and it consists of a heterogeneous population of molecules that differ in chain length, hexuronic acid composition, and degree of sulfation (12). As reported previously for dengue virus (6), RSV seems to bind best to highly sulfated forms of heparan sulfate, as inhibition of RSV infectivity is more sensitive to heparinase treatment than to heparitinase (27). This distinction in Gag lyase sensitivity implies that the degree of sulfation may be one of the primary factors influencing binding avidity (3). In fact, given the molecular heterogeneity of heparan sulfate and its varied expression on different cell types (47), it seems plausible that RSV could differentially increase the probability of virus attachment to cells based on the form of cellular heparan sulfate expressed. Thus, binding interactions between RSV HBDs and cellular Gags might influence not only viral tropism but virulence as well. It would seem that even though RSV-G is not required for infectivity, it does confer a selective advantage, allowing the virus to spread and easily attach to neighboring cells. It should also be noted that non-heparin-dependent virus-cell interactions might influence attachment and infectivity. Examination of non-heparin-dependent interactions between RSV-G and cell surface molecules is currently under way.
While the primary role for RSV-G HBDs appears to involve direct interactions with cell surface molecules, it may also play a secondary role in immunomodulation. Several recent studies clearly demonstrate that G glycoprotein activation of CD4+ Th2 lymphocytes was largely responsible for the enhanced pulmonary eosinophilia seen after RSV challenge in a murine model of RSV infection (10, 11, 22, 34, 49). This adverse response was abrogated by the vaccination of mice with a vaccinia-G construct with amino acids 193 to 203 of the G protein deleted (41). Furthermore, a subgroup A peptide identical to the linear HBD sequence described here, 184A→T198, was able to prime mice for a CD4+-Th2 response, induce pulmonary eosinophilia, and stimulate a proliferative response in peripheral blood mononuclear cells in some human donors (44). Interestingly, several immune mediators, including cytokines and chemokines, perform their effector functions via an initial interaction with heparan sulfate (40). Thus, it is possible that RSV-G interactions with heparan sulfate on the surface of CD4+ T cells mediate subsequent cytokine or chemokine responses. Obviously, RSV-G interactions with cellular Gags need to be investigated further in order to better understand the mechanism by which the RSV-G HBDs (184A/183K→198T/K197) may prime for disease characterized by pulmonary eosinophilia.
While RSV-G may act to enhance attachment of RSV to cells, it does not appear to be absolutely required for infectivity. Interestingly, no wild-type virus has been isolated that does not express the G glycoprotein. Thus, it would seem that this protein must provide RSV with some selective advantage in vivo. To understand the biology of RSV, it will be important to understand not only the involvement of the G glycoprotein HBDs in virus attachment and infectivity but the role these domains play in the development of host immunity as well. By developing this understanding, important steps can then be taken to develop future therapeutic and preventive strategies against RSV-associated disease.
ACKNOWLEDGMENTS
We thank Michael Norcross and Hana Golding for critical reviews of the manuscript. We also thank Dan Speelman and Gerald E. Hancock from Wyeth-Lederle Pharmaceuticals for the purified G glycoprotein and the corresponding rabbit anti-G antiserum, Jeffrey Esko for providing the CHO cell lines, and Susette Audet for technical assistance.
REFERENCES
- 1.Azocar J, Essex M. Incorporation of HLA antigens into the envelope of RNA tumor viruses grown in human cells. Cancer Res. 1979;39:3388–3391. [PubMed] [Google Scholar]
- 2.Bourgeois C, Bour J B, Lidholt K, Gauthray C, Pothier P. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J Virol. 1998;72:7221–7227. doi: 10.1128/jvi.72.9.7221-7227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrnes A P, Griffin D E. Binding of Sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantin R, Fortin J-F, Lamontagne G, Tremblay M. The presence of host-derived HLA-DR1 on human immunodeficiency virus type 1 increases viral infectivity. J Virol. 1997;71:1922–1930. doi: 10.1128/jvi.71.3.1922-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardin A D, Weintraub H J R. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 7.Chin J, Magoffin R L, Shearer L A, Schieble J H, Lennette E H. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am J Epidemiol. 1969;89:449–463. doi: 10.1093/oxfordjournals.aje.a120957. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields Virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1313–1351. [Google Scholar]
- 9.Collins P L, Mottet G. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J Gen Virol. 1992;73:849–863. doi: 10.1099/0022-1317-73-4-849. [DOI] [PubMed] [Google Scholar]
- 10.Connors M, Kulkarni A B, Firestone C-Y, Holmes K L, Morse III H C, Sotnikov A V, Murphy B R. Pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of CD4+ T cells. J Virol. 1992;66:7444–7451. doi: 10.1128/jvi.66.12.7444-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connors M, Natalia A G, Kulkarni A B, Firestone C-Y, Morse III H C, Murphy B R. Enhanced pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of interleukin-4 (IL-4) and IL-10. J Virol. 1994;68:5321–5325. doi: 10.1128/jvi.68.8.5321-5325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad H E. Heparin binding proteins. San Diego, Calif: Academic Press; 1998. [Google Scholar]
- 13.Esko J D, Elgavish A, Prasthofer T, Taylor W H, Weinke J L. Sulfate transport-deficient mutants of Chinese hamster ovary cells. J Biol Chem. 1986;261:15725–15733. [PubMed] [Google Scholar]
- 14.Esko J D, Stewart T E, Taylor W H. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 1985;82:3197. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn S J, Burgett B L, Stein D S, Wilkinson K S, Ryan P. The amino-terminal one-third of pseudorabies virus glycoprotein gIII contains a functional attachment domain, but this domain is not required for the efficient penetration of Vero cells. J Virol. 1993;67:2646–2654. doi: 10.1128/jvi.67.5.2646-2654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn S J, Ryan P. A heterologous heparin-binding domain can promote functional attachment of a pseudorabies virus gC mutant to cell surfaces. J Virol. 1995;69:834–839. doi: 10.1128/jvi.69.2.834-839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher J T, Lyon M, Steward W P. Structure and function of heparan sulfate proteoglycans. Biochem J. 1986;236:313–325. doi: 10.1042/bj2360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman J J, Ferguson B L, Speelman D, Mills J. Determination of the disulfide bond arrangement of human respiratory syncytial virus attachment (G) protein by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Sci. 1997;6:1308–1315. doi: 10.1002/pro.5560060619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiung G D. Hsiung’s diagnostic virology. 4th ed. New Haven, Conn: Yale University Press; 1994. Virus assay, neutralization test, and antiviral assay; p. 46. [Google Scholar]
- 20.Jackson T, Ellard F M, Abu G R, Brookes S M, Blakemore W E, Corteyn A H, Stuart D I, Newman J W I, King A M Q. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson T R, Johnson J E, Roberts S R, Wertz G W, Parker R A, Graham B S. Priming with secreted glycoprotein G of respiratory syncytial virus (RSV) augments interleukin-5 production and tissue eosinophilia after RSV challenge. J Virol. 1998;72:2871–2880. doi: 10.1128/jvi.72.4.2871-2880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapikian A Z, Mitchell R H, Chanock R M, Shvedoff R A, Stewart C E. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol. 1969;89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 24.Karron R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H W, Canchola J G, Brandt C D E A. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 26.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krusat T, Streckert H-J. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142:1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- 28.Langeland N, Moore L J. Reduction of HSV-1 binding to BHK cells after treatment with phosphatidylinositol-specific phospholipase C. FEBS Lett. 1990;277:253–256. doi: 10.1016/0014-5793(90)80859-h. [DOI] [PubMed] [Google Scholar]
- 29.Levine S, Klaiber-Franco R, Paradiso P R. Demonstration that the glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 30.Liang X, Babiuk L A, Zamb T J. Mapping of heparin-binding structures on bovine herpesvirus 1 and pseudorabies virus gIII glycoproteins. Virology. 1993;194:233–243. doi: 10.1006/viro.1993.1254. [DOI] [PubMed] [Google Scholar]
- 31.Lycke E, Johansson M, Svennerholm B, Lindahl U. Binding of herpes simplex virus to cellular heparan sulphate, an initial step in the adsorption process. J Gen Virol. 1991;72:1131–1137. doi: 10.1099/0022-1317-72-5-1131. [DOI] [PubMed] [Google Scholar]
- 32.Mbiguino A, Menezes J. Purification of human respiratory syncytial virus: superiority of sucrose gradient over percoll, renografin and metrizamide gradients. J Virol Methods. 1991;31:161–170. doi: 10.1016/0166-0934(91)90154-r. [DOI] [PubMed] [Google Scholar]
- 33.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy B R, Sotnikov A, Lawrence L, Banks S, Alling D, Prince G. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3–6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 35.Okazaki K, Honda E, Kono Y. Heparin-binding domain of bovoid herpesvirus 1 glycoprotein gIII. Arch Virol. 1994;134:413–419. doi: 10.1007/BF01310578. [DOI] [PubMed] [Google Scholar]
- 36.Patel M, Yanagishita M, Roderiquez G, Bou-Habib D C, Oravecz T, Hascall V C, Norcross M A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T cell lines. AIDS Res Hum Retroviruses. 1993;9:167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- 37.Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib D C, Mostowski H, Norcross M A. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol. 1995;69:2233–2239. doi: 10.1128/jvi.69.4.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satake M, Coligan J E, Elango N, Norrby E, Venkatesan S. Respiratory syncytial virus envelope glycoprotein (G) has a novel structure. Nucleic Acids Res. 1985;13:7795–7812. doi: 10.1093/nar/13.21.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawitzky D, Voigt A, Habermehl K-O. A peptide-model for the heparin-binding property of pseudorabies virus glycoprotein gIII. Immunology. 1993;182:285–292. doi: 10.1007/BF00191944. [DOI] [PubMed] [Google Scholar]
- 40.Selvan R S, Ihrcke N S, Platt J L. Heparan sulfate in immune responses. Ann N Y Acad Sci. 1996;797:127–139. doi: 10.1111/j.1749-6632.1996.tb52955.x. [DOI] [PubMed] [Google Scholar]
- 41.Sparer T E, Matthews S, Hussell T, Rae A J, Garcia-Barreno B, Melero J A, Openshaw P J M. Eliminating a region of respiratory syncytial virus attachment protein allows induction of protective immunity without vaccine-enhanced lung eosinophilia. J Exp Med. 1998;187:1921–1926. doi: 10.1084/jem.187.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullender W M, Mufson M A, Anderson L J, Wertz G W. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J Virol. 1991;65:5425–5434. doi: 10.1128/jvi.65.10.5425-5434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki S, Oldberg A, Hayman E G, Pierschbacher M D, Ruoslahti E. Complete amino acid sequence of human vitronectin deduced from cDNA. Similarity of cell attachment sites in vitronectin and fibronectin. Eur Mol Biol J. 1985;4:2519–2524. doi: 10.1002/j.1460-2075.1985.tb03965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebbey P W, Hagen M, Hancock G E. Atypical pulmonary eosinophilia is mediated by a specific amino acid sequence of the attachment (G) protein of respiratory syncytial virus. J Exp Med. 1998;188:1967–1972. doi: 10.1084/jem.188.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trybala E, Bergstrom T, Spillmann D, Svennerholm B, Olofsson S, Flynn S J, Ryan P. Mode of interaction between pseudorabies virus and heparan sulfate/heparin. Virology. 1996;218:35–42. doi: 10.1006/viro.1996.0163. [DOI] [PubMed] [Google Scholar]
- 46.Trybala E, Bergstrom T, Svennerholm B, Jeansson S, Glorioso J C, Olofsson S. Localization of a functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulfate. J Gen Virol. 1994;75:743–752. doi: 10.1099/0022-1317-75-4-743. [DOI] [PubMed] [Google Scholar]
- 47.van Kuppevelt T H, Dennissen M A B A, van Venrooij W J, Hoet R M A, Veerkamp J H. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. J Biol Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- 48.Van Regenmortel M H V, Neurath A R, editors. Immunochemistry of viruses II. Amsterdam, The Netherlands: Elsevier; 1990. [Google Scholar]
- 49.Waris M E, Tsou C, Erdman D D, Zaki S R, Anderson L J. Respiratory syncytial virus infection in BALB/c mice previously immunized with formalin-inactivated virus induces enhanced pulmonary inflammatory response with a predominant Th2-like cytokine pattern. J Virol. 1996;70:2852–2860. doi: 10.1128/jvi.70.5.2852-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wertz G W, Collins P L, Huang Y, Gruber C, Levine S, Ball L A. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci USA. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zsak L, Sugg N, Ben-Porat T. The different interactions of a gIII mutant of pseudorabies virus with several different cell types. J Gen Virol. 1992;73:821–827. doi: 10.1099/0022-1317-73-4-821. [DOI] [PubMed] [Google Scholar]