Identification of a Bidirectional Splicing Enhancer: Differential Involvement of SR Proteins in 5′ or 3′ Splice Site Activation (original) (raw)
Abstract
The adenovirus E1A pre-mRNA undergoes alternative splicing whose modulation occurs during infection, through the use of three different 5′ splice sites and of one major or one minor 3′ splice site. Although this pre-mRNA has been extensively used as a model to compare the transactivation properties of SR proteins, no _cis_-acting element has been identified in the transcript sequence. Here we describe the identification and the characterization of a purine-rich splicing enhancer, located just upstream of the 12S 5′ splice site, which is formed from two contiguous 9-nucleotide (nt) purine motifs (Pu1 and Pu2). We demonstrate that this sequence is a bidirectional splicing enhancer (BSE) in vivo and in vitro, because it activates both the downstream 12S 5′ splice site through the Pu1 motif and the upstream 216-nt intervening sequence (IVS) 3′ splice site through both motifs. UV cross-linking and immunoprecipitation experiments indicate that the BSE interacts with several SR proteins specifically, among them 9G8 and ASF/SF2, which bind preferentially to the Pu1 and Pu2 motifs, respectively. Interestingly, we show by in vitro complementation assays that SR proteins have distinct transactivatory properties. In particular, 9G8, but not ASF/SF2 or SC35, is able to strongly activate the recognition of the 12S 5′ splice site in a BSE-dependent manner in wild-type E1A or in a heterologous context, whereas ASF/SF2 or SC35, but not 9G8, activates the upstream 216-nt IVS splicing. Thus, our results identify a novel exonic BSE and the SR proteins which are involved in its differential activity.
Alternative splicing is a widespread mechanism for controlling gene expression in higher eucaryotes that allows the formation of various messenger RNA isoforms from a single transcribed gene (1, 9). Control of alternative splicing is predicted to require both _cis_- and _trans_-acting factors, which regulate the choice of 5′ or 3′ splice sites (5′ss or 3′ss, respectively) either positively or negatively (4, 35). An increasing number of exonic or intronic _cis_-acting sequences in the pre-mRNA sequences of various genes have been described in recent years, including splicing enhancers (see below for references) and splicing silencers (10, 15, 37, 40, 64, 65), the former being the most characterized _cis_-acting sequences. Many splicing enhancers are purine rich and are present in exons downstream of weak 3′ss, whose utilization is then favored (16, 44, 63, 69, 78, 81). Non-purine-rich enhancers, also present downstream of weak 3′ss or in introns downstream of the 5′ss have also been identified (15, 23, 32, 45, 52, 54, 58). Finally, it has been shown recently that splicing enhancers may be present in exonic sequences of constitutively spliced pre-mRNA (50, 60). Up to recently however, no exonic enhancer located upstream of weak 5′ss and activating them had been well characterized.
Protein factors which have been proposed to be involved in alternative splicing include the SR proteins, a family of essential splicing factors (for reviews see references 21 and 49). The SR family includes a dozen members with designations between SRp20 and SRp75, which contain one or two RNP-type RNA binding domains in their amino-terminal parts and an arginine/serine-rich domain (RS domain) in their carboxy-terminal regions. In constitutive splicing, SR proteins are first involved in the formation of the early prespliceosomal complex, the E complex (66), where they recruit the U1 snRNP to the 5′ss. This has been well documented for ASF/SF2, which develops concomitant interactions with the U1 70,000 molecular weight subunit (via protein-protein contact between their respective RS domains) and with the pre-mRNA substrate around the 5′ss (through poorly specific RNA-protein interactions) (39, 42, 85). Then, SR proteins are involved in spliceosome formation by forming bridges across introns and exons, likely via protein-protein interactions with the U1 snRNP bound at the 5′ss on one hand and with the small subunit of U2AF factor bound at the 3′ss on the other hand (67, 77, 79).
SR protein involvement in alternative splicing regulation was postulated after the demonstration that some individual SR proteins influence the choice between multiple 5′ss when added to in vitro splicing assay mixtures (22, 27, 43, 82, 83) or when transiently overexpressed in cultured cells (5, 75). Furthermore, it has been shown that some SR proteins bind to natural purine-rich enhancers specifically and that they are able to activate the splicing of upstream introns under the control of these enhancers (28, 44, 48, 56, 63, 66, 69). Interestingly, evidence has been accumulated to show that individual SR proteins exhibit distinct properties, for instance, in committing specific pre-mRNA to splicing pathways (20), and that they have the ability to recognize distinct RNA sequences. By using a SELEX approach, high-affinity binding sites for ASF/SF2, SRp40, SC35, 9G8, and SRp20 and its Drosophila homolog RBP1 have been identified (7, 33, 70, 71). Importantly, it has been shown that single or multiple copies of most binding sites, except that of SC35, function as SR-dependent enhancers in complementation assays (7, 70, 71). An approach complementary to the classical SELEX approach, the so called “functional” SELEX, has also been used to identify splicing enhancers (14, 46, 59, 73). Taken together, all these studies have led to the characterization of distinct classes of SR-specific enhancers, varying from highly purine-rich motifs recognized by ASF/SF2 to pyrimidine-rich motifs recognized by SRp20, with more-balanced sequences recognized by SC35 or 9G8. Therefore, SR proteins, either individually or in the form of complexes with other SR or SR-like polypeptides, should have the ability to regulate multiple splicing events through the use of a large variety of RNA motifs present within exons or introns.
Although there is evidence for SR protein involvement in natural alternative pre-mRNA models, there are only a few examples in which the identity and the role of the factors directly involved in the regulating process have been demonstrated unambiguously. doublesex pre-mRNA splicing regulation by RBP1 and Tra/Tra2 regulators through their binding to doublesex repeat elements represents one of the best-characterized examples (48). In contrast, it has been shown for other systems that SR proteins are collectively involved or that one particular SR species (the others were not analyzed) is able to bind and to regulate splicing. Due to the complexity of the SR protein family (for example the SRp30 proteins are formed by at least four distinct species), however, these studies do not provide a complete view of the individual involvement of various SR proteins within splicing regulation, and further analyses are required to characterize more extensively natural alternative splicing systems and the _trans_-acting factors which are specifically involved.
One pre-mRNA model which has been frequently used to analyze and compare SR protein activities is the adenoviral E1A pre-mRNA. During early and late periods of adenovirus infection, a splicing modulation which gives rise to various mRNAs obtained from the alternative usage of three competing 5′ss (3, 13), as well as a major downstream and a minor upstream 3′ss, occurs (26, 68, 74). We have previously shown that the 13S-to-9S transition is triggered through a titration of SR proteins (mainly SRp30, 9G8, SC35, and ASF/SF2) by major-late transcripts which accumulate in nuclei late in infection (25, 38). Numerous studies have analyzed the ability of individual SR proteins to promote various alternative splicing patterns of E1A pre-mRNA in vitro (19, 31, 82) or in vivo (5, 62, 75, 84). However, almost no study had been dedicated to the identification of putative _cis_-acting elements on the E1A pre-mRNA (55). Here we report the identification of a purine-rich sequence located immediately upstream of the 12S 5′ss. We demonstrate that this element positively regulates both the downstream 12S 5′ss and the minor upstream 3′ss in vitro and in vivo. Alternative activation of one or the other splicing reaction occurs through alternative binding of different SR proteins (in particular 9G8, ASF/SF2, and SC35 proteins) on two motifs of the enhancer. This is the first characterization of a bidirectional splicing enhancer (BSE) and of its specific _trans_-acting factors. These results provide strong evidence for the contrasting roles of various SR proteins in 5′ss or 3′ss regulation.
MATERIALS AND METHODS
Constructs.
Constructs for in vitro splicing assays were derived from the Sp4 plasmid, which contains almost the complete E1A unit (61). Mutants were obtained by insertion of synthetic oligonucleotides containing the modified sequence between _Bst_XI and _Dra_III sites (mBstX-Dra) or between a _Dra_III and a _Bgl_II site created 17 bp downstream the 12S 5′ss (BSE mutants). Constructs with a mutated 13S 5′ss were obtained by replacing _Acc_I fragments of the different mutants with the same fragment from the Sp4/13S- plasmid (55). Constructs with duplicated 12S splice sites were derived from the Sp1 plasmid (61). The 13S 5′ss was removed by an _Acc_I digestion followed by mild S1 nuclease digestion (the chosen deletion extends from −13 to +4 relative to this site) and then by insertion of a _Bam_HI-_Xba_I-_Sal_I linker. In the D/D construct, DNA fragments from positions −4 to +36 nucleotides (nt) relative to the 12S 5′ss were inserted in both _Bam_HI and _Xba_I sites. Derived plasmids containing the wild-type or mutant BSE were constructed by replacing the _Xba_I fragment of the D/D vector with synthetic oligonucleotides recreating the E1A sequence from positions −20 to +8 nt relative to the 12S 5′ss.
Constructs for UV cross-linking experiments were obtained by insertion of synthetic oligonucleotides corresponding to a wild-type or mutant BSE between _Sac_I and _Pst_I sites of the pGEM-3Zf(+) vector (Promega). For transfection experiments, _Eco_RI-_Hin_dIII fragments from Sp4 and derivatives, containing the inserted E1A unit, were transferred into the pXJ41 vector (a polylinker-modified version of the pXJ40 plasmid described in reference 80).
In vitro splicing assays.
In vitro splicing assays using Sp4 transcripts and derivatives were done as previously described (55, 61). In order to obtain efficient 216-nt intervening sequence (IVS) splicing, 10 μl of nuclear extracts (NE) efficient for this reaction, supplemented with 2 μl of cytoplasmic fraction S100, was used, and the salt concentrations were adjusted to 48 mM KCl and 2.6 mM MgCl2. Standard NE were checked and gave qualitatively similar results (data not shown). Complementation assays were done with a mixture of S100 fraction (8 μl), 15 to 45% ammonium sulfate nuclear fraction (3 μl) and NE (1 μl), and KCl at a concentration of 60 mM. Baculovirus-purified recombinant proteins (300 to 500 ng) (7, 8, 24) were preincubated with the reaction mixture for 5 to 10 min at room temperature before the addition of the transcript. In vitro splicing with Sp1-derived transcripts was performed for 90 min, and complementations were done in limiting conditions with 7 μl of S100 and 5 μl of NE supplemented with 300 to 500 ng of recombinant proteins.
UV cross-linking.
UV cross-linking and immunoprecipitation experiments were performed as described previously (7). RNA probes were radiolabeled with either [α-32P]ATP or [α-32P]CTP, as indicated in text and figure legends.
Transfections and RT-PCR analysis.
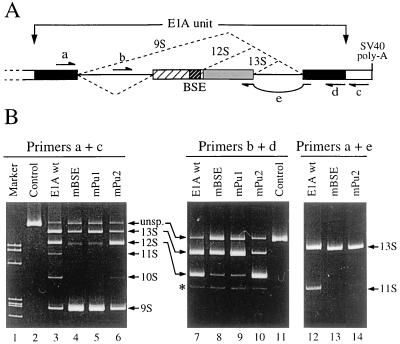
HeLa cells were grown on Dulbecco’s modified Eagle’s medium supplemented with 2.5% fetal calf serum. Cells (∼106/60-mm-diameter plate) were transfected by the CaCl2 procedure with 4 μg of E1A reporter construct and 4 μg of carrier pBlueScript SK plasmid DNA (Stratagene). Approximately 10% of cells were transfected, as checked by cotransfection of a _lacZ_-expressing vector. At 48 h after transfection, cells were harvested and total RNA was extracted with Trizol reagent (Gibco-BRL) and treated with DNase. Reverse transcription (RT) was carried out on 1.5 μg of total RNA with Moloney murine leukemia virus reverse transcriptase (Gibco-BRL) for 1 h at 42°C. Different sets of primers were used for specific analysis of the different splicing reactions (as indicated in the legend for Fig. 5), according to the primer used for the RT. Primer sequences were as follows: a, 5′-TTTGGACCAGCTGATCGAAG-3′; b, 5′-GAGTCTGTAATGTTGGCGGT-3′; c, 5′-TAACCATTATAAGCTGCAAT-3′; d, 5′-AAGCTTGGGCTGCAGGTCGA-3′; e, 5′-TTCAGAACACAGGACTGTAG-3′. The cycle numbers were kept to a minimum (15 to 19 cycles) to avoid strong amplification bias for the different PCR fragments. Products were resolved on 6% (30/1) polyacrylamide gels and visualized by SYBR Green I (Molecular Probes) staining.
FIG. 5.
The BSE exhibits the same effect in vivo as in vitro. (A) Representation of the minigene used in transfection experiments. The E1A unit (see also Fig. 1 legend) was subcloned in the pXJ41 vector (80) under the control of the human cytomegalovirus promoter, followed by simian virus 40 (SV-40) polyadenylation signals. Primers used for RT and PCR are represented by arrows. The position of the BSE is indicated. (B) Results of RT-PCR. Wild-type (wt) E1A or mutants mBSE, mPu1, and mPu2 (see Fig. 1B for sequences) were transfected in HeLa cells (see Materials and Methods for details). Total RNA was extracted and analyzed by RT-PCR followed by nondenaturing polyacrylamide gel electrophoresis. Lanes 2 and 11, PCR control with pXJ41-E1A plasmid as a matrix; lane 1, pSP65/MspI marker (501, 489, 404, 242, 228, 223 and 190 bp). Different sets of primers were used according to the various splicing reactions analyzed. In each panel, the downstream primer was used for both RT and PCR. Primers a and c, general E1A splicing pattern; primers b and d, 13S and 12S mRNA analysis; primers a and e, 13S and 11S mRNA analysis.
RESULTS
Identification of a BSE.
We have shown previously that the splicing of the upstream intron of the E1A pre-mRNA, delimited by the 9S 5′ss and a weak 3′ss located 216 nt downstream (Fig. 1A) absolutely requires the presence of the 13S 5′ss located 260 nt downstream but not that of the 12S 5′ss, which is located at a more appropriate distance (122 nt) (55). Thus, this suggested that the activation of 216-nt IVS splicing does not result from a simple process of exon definition and that other elements must be involved. To search for other _cis_-acting elements, a preliminary set of deletions of variable length, extending between the 216-nt IVS 3′ss and the 13S 5′ss, was tested. We were able to define a region surrounding the 12S 5′ss, from the _Bst_XI site to the _Xma_I site (positions 930 to 1006 on the viral genome), but excluding the splice site, which appears to be involved in the activation of the upstream splicing reaction (data not shown). Because of the presence of a purine-rich sequence upstream of the 12S 5′ss, we focused our attention on the region between the _Bst_XI site and the 12S 5′ss (Fig. 1B). This region was subjected to mutations, and the splicing of the resulting constructs was analyzed in vitro (Fig. 1C). The splicing of the wild-type E1A gives rise to the classical pattern of alternative products (schematized in Fig. 1A) resulting from the major 13S reaction, the minor 12S reaction, and the 216-nt IVS reaction, which occurs predominantly on the 13S mRNA and generates the well-represented 11S mRNA (Fig. 1C, lane 1). As shown in Fig. 1C, mutation of the region upstream of the purine-rich element (mBstX-Dra; lane 2) did not affect the 216-nt IVS splicing and significantly decreased the amount of 12S mRNA which migrates just below 13S exon 1. In contrast, destruction of the purine-rich element by two sets of mutations (m*BSE and mBSE) resulted in a dramatic decrease of both the 12S and 216-nt IVS reactions (lanes 3 and 4, respectively). Because the purine-rich sequence could be separated into two contiguous 9-nt elements (Pu1 and Pu2 motifs; Fig. 1B), we mutated each motif independently. Mutation of the Pu1 motif (GACGACGAG) led to a severe inhibition of the 12S splicing but affected the upstream 216-nt IVS reaction more weakly (Fig. 1C, lane 5), indicating that activation of both reactions may be uncoupled. In contrast, mutation of Pu2 element (GAUGAAGAG) did not interfere with the 12S splicing and even improved it slightly, whereas it inhibited the 216-nt IVS reaction significantly (compare lane 6 to lane 1). This suggested that the single Pu1 motif is involved in the 12S splicing activation, whereas both Pu1 and Pu2 are required for an optimal activation of the 216-nt IVS reaction, most likely through the activation of its weak 3′ss. Finally, analysis of the purine-rich sequence mutation in the context of an inactivated 12S 5′ss (lanes 9 and 10) shows that the regulation of the 216-nt IVS splicing is independent of the presence or absence of a functional 12S 5′ss. Because of the independent upstream and downstream enhancer activities of this exonic sequence, we called it a BSE.
FIG. 1.
A purine-rich sequence separately activates two distinct splicing reactions. (A) Structure of the Sp4 transcript and mRNA for alternative splicing reactions of the adenovirus type 2 E1A unit. Boxes represent the exons, and lines represent the introns. Note that sequences represented by hatched boxes and grey boxes act as intronic sequences when 9S or 12S splicing reactions occur. The hairpin structure of the 216-nt intron is indicated (12). Restriction sites: B, _Bst_XI; D, _Dra_III; X, _Xma_I. Different mRNA isoforms obtained by alternative splicing of the Sp4 transcript are schematized below the pre-mRNA structure. (B) Sequence of the 12S 5′ss region of the E1A unit. Nucleotides 936 to 978 (in the adenovirus type 2 viral genome) of the wild-type (wt) sequence are represented. Exonic nucleotides are in uppercase, and intronic nucleotides are in lowercase. The BSE is underlined and divided into two halves, namely, Pu1 and Pu2. Only the mutated nucleotides of the different constructs are indicated. (C) In vitro splicing assays. Splicing reactions were carried out with various transcripts, as indicated above the lanes. Lanes 1 to 8, transcripts with wild-type splice sites; lanes 9 and 10, transcripts with mutant 12S 5′ss; lanes 11 to 16, transcripts with mutant 13S 5′ss. Products of the different splicing reactions are indicated along the sides of the gels. Asterisks, mRNA obtained by using two cryptic 5′ss located between the 12S and the 13S 5′ss (55); arrowhead, a premature transcription termination.
Because the Pu1 and Pu2 motifs of the BSE differ by 2 nt only (Fig. 1B), we replaced the wild-type element by a duplication of either the Pu1 or Pu2 motif (lanes 7 and 8) to assess the level of specificity of these sequences. Interestingly, the activation of the 12S splicing appears to be more sensitive to the primary sequence of the elements than the activation of 216-nt IVS splicing, which is only weakly affected (compare the 12S and 11S mRNA variations in lanes 7 and 8 to those in lane 1). In particular, even the duplication of the Pu1 motif resulted in a 12S splicing inhibition, suggesting that the inclusion of a second Pu1 motif too close to the 12S 5′ss could be deleterious (lane 7). Whereas the existence of a splicing enhancer for the 216-nt IVS reaction was not unexpected because of the weakness of its 3′ss (26), the identification of a 12S splicing enhancer seemed rather surprising. As the 12S 5′ss is strongly competed by the predominant 13S 5′ss in vitro when the wild-type Sp4 transcript is used, it was important to determine the effects of the BSE in the absence of a functional 13S 5′ss (Fig. 1C, lanes 11 to 16). Note that with the 13S mutated transcript, the 216-nt IVS reaction became almost undetectable, as shown previously (55). In its wild-type context, the 12S splicing became predominant in the mutant 13S 5′ss transcript (lane 11). However, mBSE or mPu1 mutations induced an almost complete (lane 12) or a dramatic (lane 13) 12S splicing inhibition, as observed previously (lanes 3 to 5), whereas the Pu2 motif mutation was without effect (lane 14). This confirms that only the Pu1 motif is highly required for the occurrence of the 12S reaction. As expected, Pu1 or Pu2 duplication led to effects that were less strong than those in the presence of the 13S 5′ss, but replacement of the Pu1 motif by a second Pu2 motif still resulted in a significant 12S splicing inhibition (lane 16), whereas Pu1 duplication led only to a weaker 12S inhibition (lane 15). Thus we have identified a BSE, and our data show that both Pu1 and Pu2 motifs are involved differentially in upstream and downstream activations.
SR proteins interact with the BSE.
To analyze whether cellular factors, in particular SR proteins, interact with the BSE, we performed UV cross-linking experiments with proteins of NE or cytoplasmic fraction S100 and a 55-nt probe containing labeled wild-type or mutant BSE. We observed that the BSE cross-linked with a high-molecular-weight protein from S100 (Fig. 2A, lane 1). In contrast, the BSE interacted efficiently with proteins of about 35 and 40 kDa present in NE (lane 3), or in S100 supplemented with purified total SR proteins (lane 2), suggesting that the two bands could represent SRp30 and SRp40 proteins, respectively. This binding was sequence specific because signals were almost totally abolished with a mutated BSE (lane 4) or by the addition of an excess of a cold homologous competitor, but not by the addition of a mutated competitor (data not shown). We further confirmed the identity of these nuclear adducted proteins by immunoprecipitating them with an SR-specific antibody, the monoclonal antibody 10H3 (7) (data not shown). The facts that Tra2 protein was not detected in purified SR protein preparations (72) and that a recombinant SRp40 added to S100 extract cross-linked to the BSE (data not shown) indicate that the 40-kDa protein detected in NE mainly represents SRp40 rather than Tra2. Interestingly, mutation of the Pu1 motif alone resulted in a decrease in SRp30 band intensity while the SRp40 band was less affected (lane 5); the converse happened with the mutated Pu2 sequence (lane 6). This suggested first that Pu1 and Pu2 motifs correspond to real entities of the BSE and second that SRp40 is not primarily involved in BSE-dependent 12S activation because it binds to the Pu2 motif preferentially. As expected, duplications of Pu1 or Pu2 motifs resulted in an increase of the signals observed relative to those observed with a single Pu1 or Pu2 motif (compare lanes 7 to 6 and 8 to 5).
FIG. 2.
The BSE binds several SR proteins specifically. (A) UV cross-linking experiments were done with [α-32P]ATP-labeled wild-type (wt) or various mutated BSE RNAs (see Fig. 1B for sequences) in the presence of the S100 fraction (lane 1), S100 supplemented by a total-SR preparation (lane 2), or NE (lanes 3 to 8). (B) Immunoprecipitation assay with [α-32P]CTP-labeled wild-type BSE. Standard UV cross-linking reaction mixtures with NE were immunoprecipitated with antibodies specific for individual SR proteins as indicated at the top of each lane (9G8, ASF/SF2, or SC35). The amounts of immunoprecipitated loaded samples correspond to about four times the amounts of control samples. (C) Immunoprecipitation assays with [α-32P]ATP-labeled probes. Experimental conditions were as described for panel B, with BSE and derivatives used as probes, as indicated above each gel. Only 9G8 and ASF/SF2 antibodies were used in this experiment. The time exposure for lanes 5 and 6 corresponds to three times that for lane 4.
It was of interest to analyze the ability of the three major SRp30 proteins to interact with BSE and derivatives. Thus, we immunoprecipitated the cross-linked SR proteins with specific antibodies directed against each of these factors. With [α-32P]CTP labeling of BSE, which leads to a labeling of the Pu1 motif only, we observed an efficient binding with 9G8 (Fig. 2B, lane 2) and weaker binding with ASF/SF2 or SC35 (lanes 3 and 4), whereas uniform [α-32P]ATP labeling resulted in more-comparable interactions with 9G8 and ASF/SF2 (Fig. 2C, lanes 2 and 3). To determine what the preferential 9G8 and ASF/SF2 targets are, we used other BSE-derived sequences. We show that 9G8 binds the Pu1 probe more efficiently than ASF/SF2 by using the mPu2 and Pu1 x2 probes (Fig. 2C, lanes 5 and 6 and 8 and 9, respectively), in agreement with the fact that the Pu1 motif GACGACGAG is very similar to a 9G8 consensus motif AGAC(G/U)ACGAPy identified by SELEX (7). With the duplicated Pu2 motif, we observed the converse situation (lanes 11 and 12). Thus, our data indicate that endogenous 9G8 and ASF/SF2 present in NE bind preferentially to the Pu1 and Pu2 motifs, respectively, but that they also bind more moderately to the respective BSE counterpart, whereas SRp40 interacts with the Pu2 motif preferentially (Fig. 2C; compare lanes 1 and 10 to lanes 4 and 7). Our results also show that other SR proteins do not bind significantly to the BSE, although a weak band perhaps corresponding to the cross-linking of SRp55 was detected in certain conditions (Fig. 2A, lanes 2 and 5).
Differential splicing activation by SRp30 proteins.
Results of Fig. 1 show that the activation of 12S and 216-nt IVS reactions through the BSE can be uncoupled, suggesting that the splicing activations could be due to different _trans_-acting factors. To analyze the effect of different SRp30 proteins, we added individual SR proteins to assay mixtures containing limiting amounts of SR proteins (see Materials and Methods) and we analyzed the splicing of the wild-type E1A transcript. We observed that each SRp30 species induced the major 13S splicing at the same level (Fig. 3A; compare lanes 2, 3, and 4 to control lane 1). However, 9G8 reactivated significantly the 12S splicing and not the 216-nt IVS reaction (lane 2), whereas ASF/SF2 or SC35 (lanes 3 and 4) or a mixture of both proteins (data not shown) reactivated the 216-nt IVS splicing (11S mRNA) but not the 12S splicing. This is in agreement with the good match between the Pu1 motif and 9G8-specific target sequences and with the fact that the single Pu1 motif is required for optimal 12S splicing (see above). The 12S activation by 9G8 is sequence dependent, because changing the Pu1 motif in Pu2 in the Pu2 x2 transcript resulted in a strong 12S splicing decrease in the presence of 9G8 (lane 6). As expected from the results shown in Fig. 1, the activation of the 216-nt IVS reaction by ASF/SF2 and SC35 was not strongly affected by such a purine substitution (compare lanes 7 and 8 to 3 and 4). As shown in Fig. 3B, similar results were obtained for the 12S reaction when the 13S 5′ss was mutated. In particular, only 9G8 (lane 2) activated strongly the 12S splicing in a Pu1-dependent manner, since the 12S activation is weaker with the Pu2 x2 transcript than with the wild-type transcript (lanes 6 and 2). In contrast, the 12S splicing observed with SC35 or ASF/SF2 was weaker and BSE independent (compare lanes 3 and 4 to lanes 7 and 8). Finally, we observed that all splicing assays performed in the presence of 9G8 (Fig. 3, lanes 2 and 6) allowed a significant amount of 9S IVS in the upper part of the gel to be detected, indicating that the common 9S or 216-nt IVS 5′ss is well recognized. Therefore the absence of activation of the 216-nt IVS reaction by 9G8 observed in Fig. 3A is not due to a silencing of the upstream 5′ss but rather to an absence of activation of the 3′ part of the 216-nt intron.
FIG. 3.
SR proteins activate 12S and 216-nt IVS splicing reactions differentially. (A) In vitro splicing of transcripts with wild-type (wt) splice sites. Complementation of the splicing of two different transcripts (wt Sp4 [lanes 1 to 4] and Pu2 x2 transcript [lanes 5 to 8]) was performed in limiting conditions (lanes 1 and 5, assays without SR protein), as described in Materials and Methods, or with fixed amounts of individual purified recombinant SR proteins 9G8, ASF/SF2, and SC35, as indicated at the top of each lane. (B) Same experiment as that shown in panel A, but with mutant 13S 5′ss transcripts. Lanes 1 and 5, assays without supplemented SR protein.
9G8 activates the BSE-dependent 5′ss in a heterologous context.
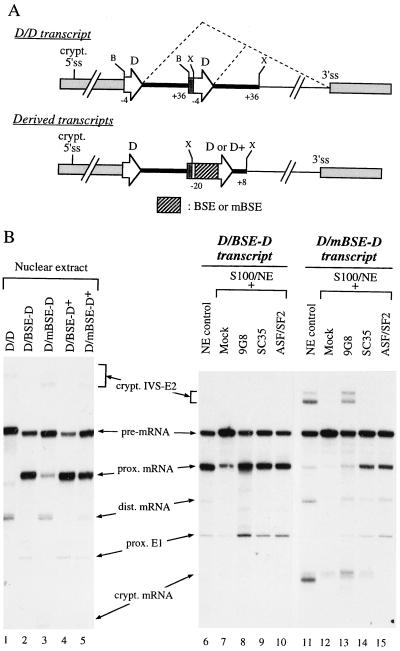
To analyze further the role of the BSE and SRp30 proteins in the activation of the downstream 12S splicing reaction, we inserted the BSE and the contiguous 12S 5′ss in a heterologous context. We used a simplified E1A transcript (Fig. 4A) in which the 13S 5′ss was replaced by two BSE-lacking 12S 5′ss in the control transcript (D/D). With this transcript, the distal site was preferentially recognized by the splicing machinery from NE (Fig. 4B, lane 1). Strikingly, the insertion of the BSE in its natural position upstream of the proximal 5′ss resulted in a strong splicing activation and in a complete shift toward the proximal site (lane 2), whereas the mutated BSE had only a poor effect (lane 3). As expected, the presence of the wild-type enhancer became more dispensable for proximal 5′ss selection when the strength of this site improved (compare lanes 4 and 5 to lanes 2 and 3).
FIG. 4.
9G8 factor activates 12S splicing specifically in a BSE-dependent manner in a heterologous context. (A) Schematic representation of the transcript with duplicated 12S 5′ss. Two copies of the BSE-lacking 12S 5′ss (D) each surrounded by 4 exonic nucleotides (open region at base of arrowhead) and by 36 intronic nucleotides (thick line) were inserted between _Bam_HI (B) and _Xba_I (X) sites of a linker at the place of the natural E1A 13S 5′ss, to give rise to the D/D construct. Light grey boxes and thin line: natural exonic and intronic regions of the E1A unit, respectively. The proximal site was next replaced by a wild-type (D) or an improved (D+) 12S 5′ss, preceded by the wild-type or mutant BSE (mBSE), giving rise to four new constructs: D/BSE-D, D/mBSE-D, D/BSE-D+, and D/mBSE-D+. (B) In vitro splicing assays. Standard experiments using NE (lanes 1 to 6 and 11) or complementation experiments with a fixed amount of SR proteins (lanes 7 to 10 and 12 to 15) were performed as described in Materials and Methods. Splicing products resulting from the utilization of the proximal (prox.), distal (dist.), or cryptic (crypt.) 5′ss (located 88 nt upstream of the distal site) are indicated alongside the gel. Lanes 7 and 12, assays without SR protein.
Next we analyzed the action of the three BSE-interacting SRp30 proteins on the D/BSE-D transcript in suboptimal splicing conditions (Fig. 4B). The addition of each of the individual SR proteins, 9G8, SC35, or ASF/SF2, resulted in an efficient recognition of the proximal 5′ss for the three factors (lanes 8 to 10 compared to lane 7). However, the activation by 9G8 protein is strongly BSE dependent because mutation of the BSE resulted in a dramatic decrease of splicing at the proximal site and a shift toward the distal site and a cryptic site located at the beginning of the transcript (compare lane 13 to lane 8), as in NE (lane 11). In contrast, splicing obtained with SC35 and ASF/SF2 was less affected by the BSE mutation (compare lanes 14 and 15 to lanes 9 and 10), suggesting that the splicing observed with these two SR proteins was due, primarily at least, to their general property of favoring the use of the proximal site in competing 5′ss transcripts. Thus, we conclude that, among the SRp30 proteins which bind to the BSE, only 9G8 exhibits a strong BSE-dependent downstream transactivatory property whether the BSE and the contiguous 12S 5′ss are positioned in their wild-type context (Fig. 3) or in a heterologous context (Fig. 4).
The BSE presents the same activity in vivo as in vitro.
In order to find out if the BSE identified in vitro is also active in vivo, we inserted the E1A unit and derivatives into an expression vector and performed transfection experiments with HeLa cells (Fig. 5A; see Materials and Methods for details). Total extracted RNA was analyzed by RT-PCR with a limited number (15 to 19) of cycles, and the results are shown in Fig. 5B. A first general analysis with external primers a and c revealed all expected mRNA products, i.e., major 13S, 12S, and 9S mRNA, as well as minor 11S and 10S mRNA resulting from the 216-nt IVS reaction (lane 3). With these primers however, synthesis of the DNA corresponding to the 9S mRNA was favored during the RT and PCR processes due to the small size of the DNA, so that a limited imbalance in 13S and 9S mRNA production with mutated BSE constructs was amplified (lanes 4 to 6 and data not shown). Use of primers b and d (lanes 7 to 11) and a and e (lanes 12 to 14) allowed a more quantitative comparison of 12S and 216-nt IVS reactions, respectively. Whereas the 12S splicing appeared to be efficient in vivo on the wild-type E1A construct (Fig. 5B, lanes 3 and 7), mutation of the BSE led to a severe 12S inhibition (more than 85%; compare lanes 4 to 3 and 8 to 7). Similar results were obtained when the BSE was mutated in m*BSE (data not shown) or mPu1 (lanes 5 and 9) constructs. In contrast, as observed previously in vitro (Fig. 1), the Pu2 motif mutation significantly improved the 12S splicing efficiency relative to that of 13S (compare lanes 10 to 7 and 6 to 3). This confirms that the Pu2 motif may have a slight deleterious effect on the 12S 5′ss activation, either by binding one SR protein too close to the 12S 5′ss or by decreasing the 9G8 binding on the Pu1 motif. Effects of BSE mutations on the 216-nt IVS splicing were analyzed with primers a and e, and the analysis revealed 13S and 11S products (lanes 12 to 14). The data show a strong inhibition (more than 90%) of the 216-nt IVS splicing occurring on the 13S mRNA when the BSE or Pu2 motif is mutated (compare lanes 12 to 14). Although the 10S mRNA is still weakly detected with the mPu2 construct by using external primers a and c (lane 6), the 216-nt IVS splicing appeared to be more sensitive to either a Pu1 or Pu2 mutation in vivo than in vitro (Fig. 1). Finally, the 9S splicing reaction was also stimulated, especially with the mutated BSE or Pu1 constructs (lanes 4 and 5), and this stimulation is likely the consequence of both 12S and 216-nt IVS splicing inhibition. Note, however, that the use of primers specific for 9S splicing indicated that this activation is weaker than that observed with primers a and c (data not shown). Taken together, our results demonstrate that the BSE is fully functional in vitro and in vivo and that it is absolutely required to promote 12S 5′ss activation and efficient 12S splicing, as well as 216-nt IVS 3′ss activation.
DISCUSSION
By searching for predicted cis elements involved in the splicing activation of the E1A pre-mRNA 216-nt intron, we have identified a novel enhancer element located in an exonic sequence close to the 12S 5′ss, which exhibits upstream and downstream activity (Fig. 6A). Until now, only one bidirectional element has been clearly identified, i.e., that in the rat FGFR-2 gene (6), which mediates upstream exon IIIb activation and downstream exon IIIc repression independently. The factors which bind to this intronic element have not been identified. It should also be noted that an exonic splicing element within the alternative exon 6 of β-tropomyosin has been shown very recently to be important for the use of both weak upstream 3′ss and downstream 5′ss, but in this example, the analysis of the positive effect on 5′ss recognition was not uncoupled from that on 3′ss recognition (63).
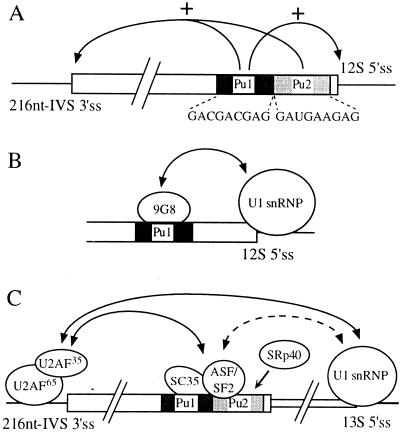
FIG. 6.
Model for alternative effect of SR proteins on E1A 12S and 216-nt IVS splicing reactions through the BSE. (A) General activation mechanism of both splicing reactions by the BSE. Only the region from the 216-nt IVS 3′ss to the 12S 5′ss is schematized, with lines representing the introns and boxes representing the exon. Sequences of Pu1 and Pu2 motifs of the BSE are indicated under the exon. Both halves of the BSE are necessary to activate efficiently the upstream 216-nt IVS splicing reaction, whereas only the Pu1 motif is required for 12S reaction activation. (B) Activation hypothesis for the downstream 12S splicing reaction. 9G8 binds to the first part of the BSE (Pu1 motif) and stabilizes the binding of the U1 snRNP at the 5′ splice site, either directly by interaction with one component of the U1 snRNP or indirectly. (C) Hypotheses for the coordinated activation of the upstream splicing reaction. ASF/SF2, SC35, or possibly SRp40 proteins bind to the BSE and enhance the recognition of the weak 216-nt IVS 3′ss, likely by protein-protein interactions with U2AF35. We do not know if the 13S 5′ss-mediated activation involves direct interactions with the splicing machinery at the 216-nt IVS 3′ss, as a simple exon definition process, or if interactions with the BSE activation complex occur first.
We have demonstrated that the downstream activation of the 12S mRNA reaction essentially depends on the Pu1 element of BSE, which is highly similar to the high-affinity 9G8 binding sequences (7, 59). In agreement with this result and cross-linking experiments (Fig. 2), we show that 9G8 strongly activates 12S mRNA splicing whereas ASF/SF2 or SC35 cannot activate the 12S reaction in its natural context (Fig. 3). However, this is not due to an inefficiency of these last two SR species to use the 12S 5′ss per se because this site was used efficiently by ASF/SF2 and SC35 either when it was moved to a heterologous context (Fig. 4) or in the presence of a large excess of these SR proteins (31, 75) (see below).
Up to now, only one clear natural example of downstream activation of a 5′ss has been demonstrated. In the Drosophila fruitless gene, the female-dependent 5′ss is activated by means of Tra/Tra2 repeat elements located between 38 and 238 nt upstream (34). A downstream activation has also been observed in chimeric models in which duplicated copies of an enhancer from α-tropomyosin exon 2 were inserted in an artificial adenovirus substrate (17) or in which the multipartite cardiac troponin T enhancer contained in exon 5 replaced the natural caldesmon enhancer (18). In this last model, the specific sequence, located 132 nt upstream of the activated 5′ss, is a short GACGACG motif, which is identical to the first 7 nt of our BSE Pu1 motif. Interestingly, such a downstream 5′ss activation could be the basis of more complicated systems. For instance, in the alternative polyadenylation of the calcitonin/calcitonin gene-related peptide pre-mRNA, it has been proposed that SRp20 binding to a polypyrimidine tract might stabilize the interaction of U1 snRNP with the enhancer 5′ss located just downstream (47). This could also be the case for the negative regulator of splicing of Rous sarcoma virus (37, 51).
What is the exact mechanism of the downstream activation by SR proteins or SR-related polypeptides? The general role of SR proteins in the constitutive formation of the earliest prespliceosomal complex E is well enough understood (39, 42, 66, 85). In contrast, the mechanism of action of SR proteins for selecting between several 5′ss is more obscure, specifically, when strong SR protein binding sites do not exist on the pre-mRNA or have not been defined. For instance, the presence of increasing amounts of ASF/SF2 promotes the splicing of proximal 5′ss among equivalent alternative sites (27, 43), and it has been proposed that this results in an increasing binding of U1 snRNP to all 5′ss indiscriminately (19). However, this mechanism could not be applied fully to the E1A pre-mRNA because in its natural context, the 12S 5′ss is not used efficiently in the presence of ASF/SF2, even when the 13S 5′ss is inactivated (Fig. 3). The weak ability of ASF/SF2 to activate 12S splicing is in apparent disagreement with previous observations performed either in vitro (31) or in vivo after the overexpression of ASF/SF2 (75). However, in contrast to both these previous studies, our experiments were performed with more-limiting amounts of active ASF/SF2 or SC35, so that the general property of these SR proteins to enhance the use of the most downstream 5′ss might be limited.
Conversely, when a strong binding site for one competent SR protein is present upstream of an alternative 5′ss (9G8 for the E1A 12S 5′ss), we can propose an activation mechanism in which the first event is an efficient binding of the competent SR protein. This binding could subsequently promote recognition of the close 5′ss by U1 snRNP and/or complex formation on the 5′ss (Fig. 6B). Whether the activating SR protein is involved as a single SR molecule in the U1 snRNP–5′ss complex formation or whether other molecules of the same SR species or SR-related polypeptides are also involved is not known.
We demonstrate that, in addition to its downstream activity, BSE exhibits an upstream activity towards the 216-nt intron through the binding of ASF/SF2 or SC35 on the BSE Pu1 and Pu2 motifs (Fig. 2 and 3). However, the whole activation mechanism is more complex than expected because we have shown previously that the 13S 5′ss also plays an important role in the activation of the 216-nt IVS 3′ss (55). Due to the occurrence of the exon definition process, which promotes very early cross talk between 3′ss and 5′ss through exonic sequences (57; reviewed in reference 2), the coordinated mechanism that we reveal for 216-nt IVS 3′ss activation is not unexpected (Fig. 6C). For instance, it can also occur for regulated alternative splicing of cassette exons or mutually exclusive exons containing exonic enhancers. However, in contrast to what was found for the E1A model, in which the 13S 5′ss is already a quite strong site, the downstream 5′ss of these models are frequently of weak strength and their improvement leads often to an almost constitutive splicing (53, 63).
Our results, as well as those obtained previously (55) strongly suggest that both the BSE and the 13S 5′ss function synergistically rather than additively to activate the 216-nt IVS 3′ss because a mutation of one of them severely affects 216-nt IVS splicing. Thus, it appears that the E1A pre-mRNA model differs from that of pre-mRNA substrates containing similar activatory elements: either doublesex repeats or SR-specific elements. Within these systems, it has been shown that activatory elements of a similar nature function additively rather than synergistically, implying that several elements work independently rather than coordinately (36). For the E1A model, in contrast, we can speculate that the enhancer complexes formed on the 13S 5′ss and BSE are different so that each of them brings specific activatory factors which could then act synergistically (Fig. 6C). The exact activation mechanism and the step at which the synergistic process takes place are not known. One possibility was that the BSE acts simply by strengthening the cis activity of 13S 5′ss, which is located at the upper limit (about 300 nt) for efficient involvement (2). This cannot be the only role of the BSE, however, because we have shown that large deletions in the E1A pre-mRNA which remove the BSE and 12S 5′ss but bring the 13S 5′ss closer to the 216-nt IVS do not allow the 216-nt IVS splicing to activate significantly (unpublished results).
A final particularity of the E1A model is that the 12S 5′ss, although ideally located, does not function for the cis activation of 216-nt IVS splicing (55). Interestingly, our results bring a plausible explanation for this feature. Indeed, although only 9G8 among the tested SRp30 proteins is able to activate the 12S 5′ss by interacting with the BSE, this SR is unable to promote the upstream activity of the BSE; the converse is true for ASF/SF2. This suggests that a coordinated process involving both the BSE and the 12S 5′ss for 216-nt IVS splicing activation, comparable to that involving the BSE and the 13S 5′ss, cannot be favored within the wild-type E1A RNA.
Taken together, our data strongly suggest that the BSE’s bidirectional activity is dependent on which SR proteins bind to the BSE, with 9G8 on one hand and ASF/SF2 and SC35 on the other displaying distinct downstream or upstream activities in the E1A pre-mRNA model that we analyzed. This does not mean that these activities correspond to absolute intrinsic properties of these SR proteins, however, because we and others have shown that 9G8 is also able to promote upstream 3′ss activation in the native doublesex pre-mRNA (48) or in chimeric substrates containing 9G8-specific enhancers (7, 59). Therefore, it is expected that specific characteristics of both SR proteins and E1A pre-mRNA are involved in the distinct properties of 9G8, ASF/SF2, and SC35. In addition to the establishment of different RNA-protein interactions with the BSE and 12S 5′ss, directly dependent of their RNA binding domains, it is likely that specific features related to the RS domains of these SR proteins are also important. Comparisons of RS domain activities have led to variable conclusions according to the specificity levels of the experiments. Thus, domain swaps between SC35 and ASF/SF2 by using a functional commitment assay (11) and studying the abilities of various RS domains to replace in vivo that of ASF/SF2 after genetic inactivation of endogenous ASF/SF2 (76) have shown a good interchangeability for the tested functions. In contrast, analyses of the abilities of various RS domains to activate in vitro enhancer-dependent splicing show that RS domains display distinct splicing activities (29). Therefore, it is possible that, according to the abilities of their RS domains to interact directly or indirectly with the U1 snRNP positioned downstream, 9G8 and ASF/SF2 may or may not activate U1 snRNP–12S 5′ss complex formation.
Finally, because our work is the first to establish a link between an in vitro effect of a given SR protein and a _cis_-acting element present on the E1A pre-mRNA, it was interesting to compare our results to the numerous studies which have used E1A pre-mRNA as a model to analyze alternative splicing activities of individual SR proteins (5, 19, 31, 62, 75, 82, 84). The strong 13S 5′ss activation versus the activation of distal 12S and 9S sites in the presence of an excess of ASF/SF2 or SC35 generally observed both in vitro (19, 31, 82) and in vivo (5, 62, 75, 84) has been ascribed to a strong position-dependent process that promotes the use of downstream 5′ss. In contrast to their role in 13S splicing regulation, the role of ASF/SF2 and SC35 in 216-nt IVS splicing has been poorly documented. Indeed, 216-nt IVS splicing activation after the progressive addition of ASF/SF2 to an S100 extract was observed only in one in vitro study (31), whereas larger amounts of ASF/SF2 in vitro (31) or overexpression in vivo (5, 62, 75, 84) did not favor this reaction. In these conditions, however, some alterations of particular splicing reactions have been observed (75). In contrast, the overexpression of SRp40 in vivo led to 216-nt IVS splicing activation (62, 84). Thus, all these results fit reasonably with the fact that ASF/SF2, SC35, and SRp40, play important roles in upstream 216-nt IVS splicing activation by interacting with one or both motifs of BSE.
Other SR proteins tested in vitro or in vivo, including SRp20, SRp55, and SRp75 (62, 82), as well as 9G8 (this study), have been shown to improve 12S splicing relative to 13S splicing, whereas SR protein p54 activates upstream 9S splicing (84). Improved 12S splicing by SRp20, -55, and -75 may result in an absence of systematic preference for the proximal 13S 5′ss, as well as in a better recognition of the 12S 5′ss relative to that by ASF/SF2 and SC35. However, because only SRp30 and SRp40 proteins bind efficiently to the BSE in conditions of competition between endogenous SR proteins present in a NE (Fig. 2), this indicates that SRp20, SRp55, and SRp75 should bind the BSE only moderately, if at all. In agreement with this, we have shown in complementation assays that SRp20 activates 12S splicing moderately, but in a manner which is not strictly dependent on the BSE (data not shown). In fact, it is possible that cis elements other than BSE are also present in the E1A pre-mRNA. For instance, we have some evidence that RNA sequences downstream of the 12S 5′ss are not neutral for 12S splicing, but no clear interactions of these sequences with nuclear proteins have been detected (unpublished results).
Finally, the identification of the BSE should also allow a better understanding of the modulation of E1A pre-mRNA alternative splicing which occurs during the early and late periods of infection. Due to the complexity of the SR protein family and to the variety of individual SR protein effects on E1A splicing, it was not possible to provide more-detailed data about the mechanism of SR protein titration (specially that of SRp30) that we proposed previously (38). More recently, it has been shown that dephosphorylation of SR proteins occurs later in infection and might also be responsible for adenoviral pre-mRNA splicing modulation (41). However, no significant SR protein dephosphorylation was detected during the infection cycle by us or others (30, 38).
In conclusion, results presented here allow us to identify a bidirectional enhancer in the E1A pre-mRNA which is absolutely required for the 12S mRNA splicing in vivo. Interestingly, the enhancer activity is regulated differentially by distinct SR proteins. Further experiments are required to determine the molecular bases of the specific properties of 9G8, ASF/SF2, and SC35 in this bidirectional enhancer system.
ACKNOWLEDGMENTS
This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpitaux Universitaires de Strasbourg, and the Association pour la Recherche contre le Cancer. C.F.B. was supported by fellowships from the French Ministère de l’Education Nationale et de la Recherche, the Association pour la Recherche contre le Cancer, and the Ligue Nationale Contre le Cancer.
We thank P. Blader, R. Gattoni, and F. Lejeune for critical reading of the manuscript. We are grateful to L. Kister for excellent technical assistance, to G. Mengus and I. Davidson for the gift of the pXJ41 vector, and to the staffs for oligonucleotide synthesis, sequencing, cell culture, and artwork for providing materials and technical help.
REFERENCES
- 1.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 2.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 3.Berk A J, Sharp P A. Structure of the adenovirus 2 early mRNAs. Cell. 1978;14:695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- 4.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 5.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 6.Carstens R P, McKeehan W L, Garcia-Blanco M A. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavaloc Y, Bourgeois C F, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 10.Chan R C, Black D L. Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chebli K, Gattoni R, Schmitt P, Hildwein G, Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989;9:4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow L T, Broker T R, Lewis J B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979;134:265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- 14.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirksen W P, Hampson R K, Sun Q, Rottman F M. A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J Biol Chem. 1994;269:6431–6436. [PubMed] [Google Scholar]
- 17.Dye D T, Buvoli M, Mayer S A, Lin C H, Patton J G. Enhancer elements activate the weak 3′ splice site of alpha-tropomyosin exon 2. RNA. 1998;4:1523–1536. doi: 10.1017/s1355838298980360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elrick L L, Humphrey M B, Cooper T A, Berget S M. A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol Cell Biol. 1998;18:343–352. doi: 10.1128/mcb.18.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 21.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallego M E, Balvay L, Brody E. cis-Acting sequences involved in exon selection in the chicken β-tropomyosin gene. Mol Cell Biol. 1992;12:5415–5425. doi: 10.1128/mcb.12.12.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallego M E, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gattoni R, Chebli K, Himmelspach M, Stevenin J. Modulation of alternative splicing of adenoviral E1A transcripts: factors involved in the early-to-late transition. Genes Dev. 1991;5:1847–1858. doi: 10.1101/gad.5.10.1847. [DOI] [PubMed] [Google Scholar]
- 26.Gattoni R, Schmitt P, Stevenin J. In vitro splicing of adenovirus E1A transcripts: characterization of novel reactions and of multiple branch points abnormally far from the 3′ splice site. Nucleic Acids Res. 1988;16:2389–2409. doi: 10.1093/nar/16.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge H, Manley J L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 28.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graveley B R, Hertel K J, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 31.Harper J E, Manley J L. Multiple activities of the human splicing factor ASF. Gene Expr. 1992;2:19–29. [PMC free article] [PubMed] [Google Scholar]
- 32.Hedley M L, Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- 33.Heinrichs V, Baker B S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinrichs V, Ryner L C, Baker B S. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol Cell Biol. 1998;18:450–458. doi: 10.1128/mcb.18.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 36.Hertel K J, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 37.Hibbert C S, Gontarek R R, Beemon K L. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA. 1999;5:333–343. doi: 10.1017/s1355838299981347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Himmelspach M, Cavaloc Y, Chebli K, Stevenin J, Gattoni R. Titration of serine/arginine (SR) splicing factors during adenoviral infection modulates E1A pre-mRNA alternative splicing. RNA. 1995;1:794–806. [PMC free article] [PubMed] [Google Scholar]
- 39.Jamison S F, Pasman Z, Wang J, Will C, Luhrmann R, Manley J L, Garcia-Blanco M A. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 41.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 42.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 43.Krainer A R, Conway G C, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 44.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 45.Lim L P, Sharp P A. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lou H, Neugebauer K M, Gagel R F, Berget S M. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 49.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 50.Mayeda A, Screaton G R, Chandler S D, Fu X D, Krainer A R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNally L M, McNally M T. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol Cell Biol. 1998;18:3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muro A F, Iaconcig A, Baralle F E. Regulation of the fibronectin EDA exon alternative splicing. Cooperative role of the exonic enhancer element and the 5′ splice site. FEBS Lett. 1998;437:137–141. doi: 10.1016/s0014-5793(98)01201-0. [DOI] [PubMed] [Google Scholar]
- 54.Nagoshi R N, Baker B S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- 55.Popielarz M, Gattoni R, Stevenin J. Contrasted cis-acting effects of downstream 5′ splice sites on the splicing of a retained intron: the adenoviral E1A pre-mRNA model. Nucleic Acids Res. 1993;21:5144–5151. doi: 10.1093/nar/21.22.5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan K J, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaal T D, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaal T D, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitt P, Gattoni R, Keohavong P, Stevenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987;50:31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- 62.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Selvakumar M, Helfman D M. Exonic splicing enhancers contribute to the use of both 3′ and 5′ splice site usage of rat beta-tropomyosin pre-mRNA. RNA. 1999;5:378–394. doi: 10.1017/s1355838299981050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Si Z H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stark J M, Bazett-Jones D P, Herfort M, Roth M B. SR proteins are sufficient for exon bridging across an intron. Proc Natl Acad Sci USA. 1998;95:2163–2168. doi: 10.1073/pnas.95.5.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephens C, Harlow E. Differential splicing yields novel adenovirus 5 E1A mRNAs that encode 30 kd and 35 kd proteins. EMBO J. 1987;6:2027–2035. doi: 10.1002/j.1460-2075.1987.tb02467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 70.Tacke R, Chen Y, Manley J L. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tacke R, Tohyama M, Ogawa S, Manley J L. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 73.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ulfendahl P J, Linder S, Kreivi J P, Nordqvist K, Sevensson C, Hultberg H, Akusjarvi G. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J. 1987;6:2037–2044. doi: 10.1002/j.1460-2075.1987.tb02468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Xiao S H, Manley J L. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z, Hoffmann H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 78.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 79.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 80.Xiao J H, Davidson I, Matthes H, Garnier J M, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 81.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 83.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang W J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zuo P, Manley J L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]