Normal limb development in conditional mutants of Fgf4 (original) (raw)
. Author manuscript; available in PMC: 2007 Jan 22.
Published in final edited form as: Development. 2000 Mar;127(5):989–996. doi: 10.1242/dev.127.5.989
SUMMARY
Fibroblast growth factors (FGFs) mediate multiple developmental signals in vertebrates. Several of these factors are expressed in limb bud structures that direct patterning of the limb. FGF4 is produced in the apical ectodermal ridge (AER) where it is hypothesized to provide mitogenic and morphogenic signals to the underlying mesenchyme that regulate normal limb development. Mutation of this gene in the germline of mice results in early embryonic lethality, preventing subsequent evaluation of Fgf4 function in the AER. A conditional mutant of Fgf4, based on site-specific Cre/loxP-mediated excision of the gene, allowed us to bypass embryonic lethality and directly test the role of FGF4 during limb development in living murine embryos. This conditional mutation was designed so that concomitant with inactivation of the Fgf4 gene by excision of all _Fgf4_-coding sequences, a reporter gene was activated in _Fgf4_-expressing cells, allowing assessment of the site-specific recombination reaction. Although a large body of evidence led us to predict that ablation of Fgf4 gene function in the AER of developing mice would result in abnormal limb outgrowth and patterning, we found that Fgf4 conditional mutants had normal limbs. Furthermore, expression patterns of Shh, Bmp2, Fgf8 and Fgf10 were normal in the limb buds of the conditional mutants. These findings indicate that the previously proposed FGF4-SHH feedback loop is not essential for coordination of murine limb outgrowth and patterning. We suggest that some of the roles currently attributed to FGF4 during early vertebrate limb development may be performed by other AER factors in vivo.
Keywords: Fibroblast growth factor (Fgf), Limb, Mouse
INTRODUCTION
The vertebrate limb provides a powerful system in which to address fundamental questions in development, such as how genes control cell migration, differentiation and the morphology of complex structures. Intercellular signals that pattern the murine limb also function in other regions of the developing embryo and are conserved across species (Cohn and Tickle, 1996; Tabin, 1991). The apical ectodermal ridge (AER) is an epithelial limb bud structure required to support continued outgrowth of the limb bud by maintaining the underlying mesenchyme, the progress zone (PZ), in a highly proliferative, undifferentiated state (Globus and Vethamany-Globus, 1976; Saunders, 1977). Time of residence in the PZ and exposure to AER-derived signals determine cell fate along the proximal-distal limb axis (Summerbell et al., 1973). The AER is also required for initiation and maintenance of the zone of polarizing activity (ZPA), a region of posterior mesenchyme that specifies the anteroposterior axis of the developing limb (Riddle et al., 1993; Todt and Fallon, 1987; Vogel and Tickle, 1993). Similarly, signals from the PZ and ZPA are necessary for survival and function of the AER (Hinchcliffe and Gumpel-Pinot, 1981; Ohuchi et al., 1997; Reiter and Solursh, 1982; Xu et al., 1998)
Members of the fibroblast growth factor gene family (Fgfs) encode secreted signaling proteins (Basilico and Moscatelli, 1992). Subsets of these molecules (FGFs) and their receptors are produced in different regions of the developing vertebrate limb, including the AER and PZ. The importance of FGFs in human skeletal development was confirmed when several dominant human skeletal defects were mapped to genes encoding FGF receptors (Muenke and Schell, 1995). Additionally, mice homozygous for null mutations of Fgf receptor 2 or Fgf10 fail to form limbs (Ohuchi et al., 1997; Sekine et al., 1999; Xu et al., 1998). FGFs can induce and support the outgrowth of ectopic limbs in the absence of an AER and can substitute for the AER after it is surgically ablated from early limb buds (Crossley et al., 1996; Fallon et al., 1994; Niswander et al., 1993). These growth factors have thus been hypothesized to mediate AER activities in vivo.
Fgf4 is first expressed in the developing murine forelimb bud during embryonic day 10 (E10.0, 30-32 somites, stage 3; Wanek et al., 1989); expression is strongest in the posterior AER at E10.5-11.0 and then wanes to undetectable levels by E12.0 (Drucker and Goldfarb, 1993; Niswander and Martin, 1992; Suzuki et al., 1992).
Current models propose that FGF4 mediates mitogenic and morphogenic AER signals. FGF4 may provide a proliferative signal to the posterior PZ to support limb outgrowth (Niswander et al., 1993; Suzuki et al., 1992). Several lines of evidence suggest that expression and function of Sonic Hedgehog (Shh), which mediates the polarizing activity of the ZPA (Riddle et al., 1993), is dependent on FGF4 signaling from the posterior AER (Laufer et al., 1994; Niswander et al., 1994; Vogel and Tickle, 1993). Exogenous FGF4 induces ectopic Shh expression in the presence of retinoic acid and maintains Shh expression in the ZPA in the absence of the AER (Niswander et al., 1994). PZ cells respond to ectopic SHH by asymmetrically expressing Bmp2 and 5′ Hoxd genes only in the presence of the AER; however, FGF4 can substitute for the AER in this assay (Laufer et al., 1994). Additionally, ectopic SHH can induce Fgf4 expression in a more anterior portion of the AER than its endogenous expression domain (Laufer et al., 1994). These observations support the hypothesis that a positive feedback loop between FGF4 in the AER and SHH in the ZPA coordinately regulates limb outgrowth and patterning and thus implicate FGF4 in the generation of the normal limb AP axis (Laufer et al., 1994; Niswander et al., 1994).
The limb phenotypes and gene expression patterns described in naturally occurring vertebrate mutants are also consistent with the existence of an FGF4-SHH feedback loop. Lack of Fgf4 expression in the AER of the murine limb deformity mutant is associated with failure to maintain Shh expression and abnormal patterning of distal limb elements in these animals (Haramis et al., 1995). Recent studies indicate that the Formin gene mutation in ld mutants disrupts a signaling relay from Shh_-expressing mesenchymal cells to the AER (Chan et al., 1995b; Zuniga et al., 1999). Strong's Luxoid polydactylous mice display ectopic, anterior expression of Shh, Fgf4 and 5′_HoxD genes (Chan et al., 1995a). In limbless chicks, the posterior mesoderm does not express Shh nor does the AER express Fgf4. Application of exogenous FGF to limbless buds induces low-level Shh expression in the posterior mesoderm and subsequent asymmetric mesodermal expression of Bmp-2, which is thought to be downstream of SHH (Ros et al., 1996). Chick _talpid_2 and _talpid_3 polydactylous mutants express Shh normally, but Fgf4 is expressed throughout the AER. This has been attributed to abnormal activation of the downstream SHH signaling pathway throughout the anteroposterior axis in the mesoderm (Caruccio et al., 1999; Francis-West et al., 1995; Lewis et al., 1999).
In total, these observations have been interpreted to indicate that FGF4 performs essential AER roles in normal embryos. However, it is possible that some or all of these functions are normally mediated by other AER factors. FGF2 and FGF8 are also produced by the AER, have similar properties in functional assays (Anderson et al., 1993; Cohn et al., 1995; Crossley et al., 1996; Fallon et al., 1994), and activate some of the same receptor isoforms in vitro as FGF4 (Ornitz et al., 1996; Xu et al., 1999). It is unknown how well these assays reflect in vivo FGF activity and receptor usage, or which FGFs are required for specific AER functions during normal limb development.
Fgf4 null embryos die at E4.5 (Feldman et al., 1995), preventing direct evaluation of FGF4 function in the developing limb using conventional methods of inactivation by gene targeting. To bypass embryonic lethality resulting from disruption of Fgf4 in the germline, we generated a system to conditionally inactivate Fgf4 in a developmentally regulated, tissue-restricted manner. Fgf4 conditional mutants, which contain no Fgf4 transcripts in the AER of their forelimb buds, were analyzed with regard to limb structure and gene expression at multiple stages. We found that the limbs of the conditional mutants develop normally and that, in contrast to predictions based on the models of FGF4 function described above, expression of Shh, Bmp2, Fgf10 and Fgf8 was normal in these animals. Our findings suggest that other AER factors may normally perform some of the functions currently attributed to FGF4 or may compensate for the absence of FGF4 in the conditional mutants during limb development.
MATERIALS AND METHODS
Generation of mice bearing targeted Fgf4 alleles and RARCre transgenic lines
To create the Fgf4 conditional allele, F4AP, a loxP site was inserted in the 5′ untranslated region of the gene at an _Fsp_I site located 59 nucleotides upstream of the translation initiation ATG. A cassette was inserted 89 nucleotides 3′ of the polyadenylation signal that contained: the MC1neo gene (Thomas and Capecchi, 1987), followed by a synthetic polyadenylation site (Levitt et al., 1989) and a pause-termination signal derived from the human complement gene C2 (Ashfield et al., 1991), a loxP site in the same orientation as the 5′ loxP, and the entire coding region of the human placental alkaline phosphatase gene.
To produce the F4neo null allele, a deletion of 141 nucleotides was introduced between an _Xma_III site at nucleotide 286 of exon 1 and an _Xma_III site at position 113 of the first intron. The MC1neo gene was inserted in place of the deleted genomic sequences. The deletion in exon 1 and translational stops in all three reading frames of the MC1 promoter led us to predict that this would be a null allele of Fgf4. This was confirmed by failure to detect any F4neo/neo homozygotes among the progeny of heterozygous intercrosses at E9.5, 10.5 and adult (_n_=85, expected 22 homozygotes).
Targeting vectors were electroporated into RI ES cells and cell lines carrying the desired alterations to the Fgf4 gene were used to generate chimeras that transmitted the targeted allele.
The RARCre transgene was constructed by placing a 3.5 kb murine RARβ2 promoter fragment (Charité et al., 1994) upstream of coding sequences for Cre recombinase (Gu et al., 1993). The RARCre fragment was microinjected into zygotes by standard methods (Hogan et al., 1986) to generate two transgenic lines.
Genotype determination
DNA was prepared from tails or yolk sacs of adult and embryonic mice, respectively. Southern blot analysis was applied to identify and characterize mice bearing targeted alleles of Fgf4 or the RARCre transgene. Subsequently, PCR assays were utilized to determine the genotype of embryos and adult mice. PCR strategies used for the Fgf4 conditional and neo alleles are diagrammed in Fig. 1.
Fig. 1.
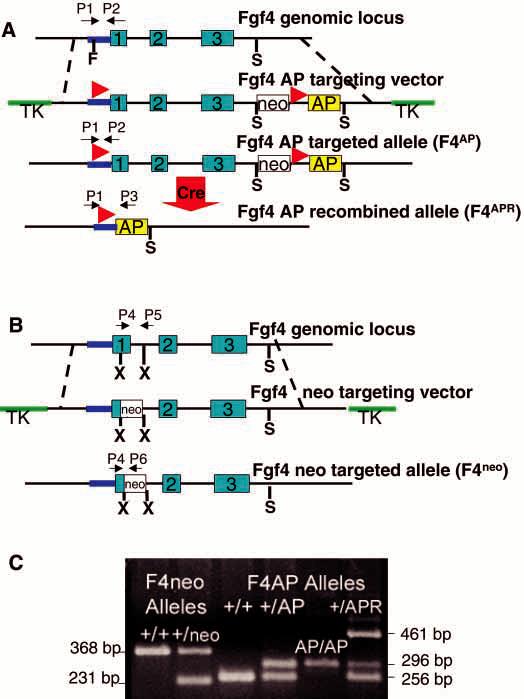
Conditional mutagenesis of the Fgf4 locus. (A) The wild-type (WT) Fgf4 locus is depicted on the first line; exons are represented by blue boxes; untranslated regions are shown as lines (purple, Fgf4 promoter; black, introns and 3′UT). The F4AP targeting vector (second line) was constructed by insertion of a loxP site (red arrowheads) 59 bp upstream of the translation start site. A cassette containing: MC1neor (white box), another loxP site in the same orientation as that in the 5′ untranslated region and, human placental alkaline phosphatase-coding sequences and the SV40 polyadenylation signal (yellow box), was inserted 89 bp downstream of the polyadenylation signal. Thymidine kinase (TK, green) genes flank the Fgf4 homology to permit negative selection against random integrants. Cre-mediated recombination (large red arrow) of the F4AP-targeted allele deletes all Fgf4 sequences between the loxP sites, generating F4APR. (B) The F4neo targeting vector contains MC1neor between _Xma_III (X) restriction sites at nucleotides 286 of exon 1 and 113 of intron 1. (C) Genotyping at the Fgf4 locus. PCR primer sets are shown in A and B and fragment sizes are as shown in C. The F4AP and F4APR alleles contain WT sequences in the region examined by the F4neo primer set and vice versa. Each animal was analyzed with both primer sets to obtain a complete genotype. PCR products obtained with P1 and P2 on the WT and F4AP alleles are 256 base pairs (bp) and 296 bp, respectively. The P1/P3 primer set amplifies a 461 bp region of the recombined allele, F4APR. The P4/P5 set detects a 368 bp WT product; while P4/P6 detects a 231 bp product from the F4neoallele.
Alkaline phosphatase staining
Embryos were dissected in PBS+2mM MgCl2 and fixed overnight in 4% paraformaldehyde in PBS+2 mM MgCl2. After three rinses in PBS+2mM MgCl2, embryos were heated at 65°C for 90 minutes to destroy endogenous alkaline phosphatase activity. Embryos were rinsed once for 15 minutes in 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween 20, 0.5 mg/ml levamisole and stained at room temperature in the dark in BM purple alkaline phosphatase substrate (Boehringer Mannheim Biochemicals) for 2 hours. Embryos to be sectioned were stained overnight, refixed and embedded in paraffin by standard methods. 10 μm sections were counterstained with nuclear fast red.
Whole-mount in situ hybridization and X-gal staining
In situ hybridization on intact embryos was carried out as described (Carpenter et al., 1993) with the following minor modifications: embryos were treated with proteinase K at 10 μg/ml for 5 minutes at 25°C; the anti-digoxigenin alkaline phosphatase-conjugated antibody (Boehringer Mannheim Biochemicals) was used at a dilution of 1:5000; 0.5 mg/ml levamisole was only added to the reaction buffer used to rinse the embryos prior to the enzyme reaction; the substrate was BM purple (Boehringer Mannheim Biochemicals). All experiments were repeated a minimum of three times; littermates were used when feasible and control and mutant embryos were hybridized in the same vial.
X-gal staining was performed as described (Mansour et al., 1993), except that the reactions were carried out at 30°C. Fixation time was 5 to 15 minutes depending on gestational age of embryos.
Skeletal analysis
Skeleton preparations of newborn mice were carried out as described (Mansour et al., 1993).
RESULTS
Elements of the Fgf4 conditional mutagenesis system
Our system for conditional mutagenesis of Fgf4 employs three components. Mice carrying either a conditional or a null allele of Fgf4 were created by gene targeting in ES cells (Thomas and Capecchi, 1987) (Fig. 1) and a transgenic ‘switch’ was generated to inactivate the conditional allele (Fig. 2). ES clones and chimera offspring containing the Fgf4 mutant alleles were initially identified by Southern analysis (data not shown). Subsequently, animals were genotyped using the PCR strategy illustrated in Fig. 1.
Fig. 2.
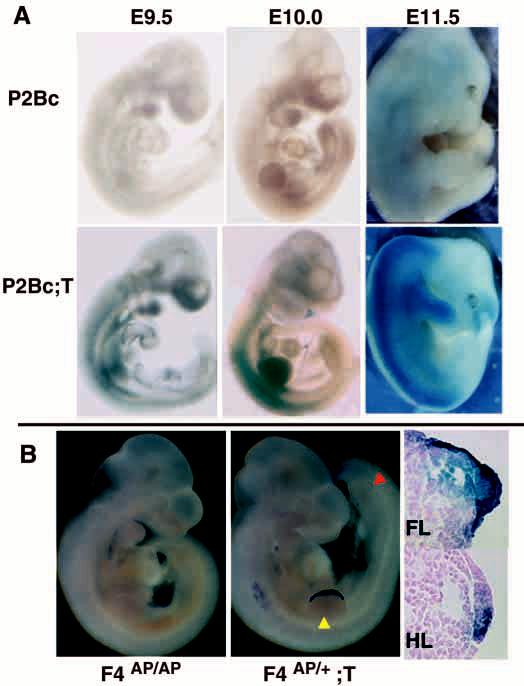
Activity of the RARCre transgene. (A) β-galactosidase activity in offspring of RARCre X P2Bc reporter mice. E9.5, E10.0 and E11.5 embryos were stained with X-gal; there is no activity in embryos with only the P2Bc reporter allele at any stage. Littermates that also carry the RARCre transgene (P2Bc/+;T) have detectable β-galactosidase activity, resulting from Cre mediated recombination of the reporter gene, in the forelimb by E 9.5 and in the AER by E 10.0. Note absence of significant hindlimb staining. (B) Alkaline phosphatase (AP) activity requires Cre-mediated recombination of the F4AP conditional allele. E10.0, 30-32 somite, forelimb stage 3 littermates, were stained for AP activity. There is no staining in F4AP/AP mice in the absence of RARCre. Cre-mediated recombination of the F4AP allele results in uniform staining of the entire AER in the forelimb (FL, yellow arrowhead) but only partial staining in the hindlimb (HL, red arrowhead) AER. The difference in recombination between fore- and hindlimb is confirmed by examining the AERs in cross-section. The appearance of AP signal in cells outside the AER could be a technical artifact or reflect actual low-level activity of the Fgf4 promoter in these cells.
The F4AP conditional allele contains loxP sites in the 5′ untranslated (UTR) and 3′ flanking regions of Fgf4 (Fig. 1A). Sequences encoding human placental alkaline phosphatase (AP) were placed downstream of the loxP site in the untranscribed 3′ flanking region. This allele is functional prior to inactivation by the Cre-mediated ‘switch’; F4AP/AP homozygotes are born at predicted Mendelian frequencies and survive to be fertile adults with normal limbs. Cre-mediated recombination (Baubonis and Sauer, 1993) of this allele deletes all _Fgf4_-coding sequences and generates a null allele of Fgf4 (F4APR). This recombination/deletion event also repositions the AP-coding sequences so that they are expressed and regulated by the Fgf4 promoter. Thus, AP activity labels those cells in which Cre-mediated recombination has occurred and the Fgf4 promoter is active.
The F4neo allele is a null allele created by a deletion in Exon 1 of Fgf4 and insertion of a neo r gene (Fig. 1B). No F4neo/neo homozygotes were detected among the progeny of heterozygote intercrosses, a finding consistent with the embryonic lethal phenotype of Fgf4 null homozygotes previously described (Feldman et al., 1995). F4neo/+ heterozygotes were present at expected Mendelian frequencies and were phenotypically normal, fertile adults. We performed conditional mutagenesis in compound heterozygotes carrying an F4 AP and an F4 neo allele to limit the requirement for Cre-mediated recombination to only one of the Fgf4 alleles. Use of the F4neo allele also avoids potential problems that could arise from Cre-mediated recombination between F4AP alleles on separate chromosomes since the F4neo allele does not contain loxP sites.
The ‘switch’ used to regulate inactivation of the conditional Fgf4 allele was the Cre-recombinase gene expressed under the control of the Retinoic Acid Receptor-_B_2 promoter (RARCre). The function of this promoter has been well characterized in transgenic mice, and includes expression in the intermediate and lateral plate mesoderm as early as E8.5, as well as in the forelimb buds at E9.5 and the AER as it develops at E10.0-10.5 (Charité et al., 1994; Mendelsohn et al., 1991; Reynolds et al., 1991). Thus, RARCre should be expressed in all of the relevant tissues that potentially require Fgf4 for patterning of the forelimb. Two RARCre transgenic lines were generated. Expression of RARCre was the same in both lines, similar to that previously described in the literature, and consistent throughout our experiments.
Activity of the RARCre transgenic ‘switch’ in murine embryos
To characterize the production of functional Cre recombinase by the RARCre transgene, we mated RARCre transgenic animals to reporter mice carrying the lacZ gene targeted to the RNA polymerase II large subunit locus (P2Bc). The polII promoter is ubiquitously active but, in these reporter mice, β-galactosidase (β-gal) activity is dependent on Cre-mediated recombination of the locus (O'Gorman et al., 1997). As expected, no β-gal activity was detected in embryos carrying only the reporter P2Bc allele (Fig. 2A). Embryos that carry both P2Bc and RARCre alleles have readily detectable β-gal activity at E9.5 in the forelimb bud and in the developing AER at E10.0. Note that there is very little β-gal activity in the hindlimb at any stage, indicating that few cells express Cre in the hindlimb.
The F4AP conditional allele provides a very sensitive means of assessing the function of RARCre (Fig. 2B). AP activity is undetectable in E10.0 F4AP/AP embryos, confirming that there is no ‘leaky’ expression of the AP gene from the unrecombined F4APallele. In a 31- to 32-somite E10.0 embryo carrying an F4AP allele and RARCre (F4AP/+; T [RARCre transgene]), AP activity is detectable only at those sites where the Fgf4 promoter is active and Cre has recombined the F4AP allele; it labels those cells in which functionally relevant recombination has occurred. As expected, only faint staining of the hindlimb AER is evident, whereas staining of the forelimb AER is dark and uniform (Fig. 2B). Cross-sections through the forelimb (Fig. 2B, FL) of the F4AP/+;T embryo confirm staining of every cell in the AER, indicating that Cre has recombined the F4AP allele throughout the AER by E10.0. In contrast, the hindlimb AER (Fig. 2B, HL) contains many unstained cells that possess the unrecombined F4AP allele. These results indicate that the F4AP allele is already recombined (and thus a null allele) in RARCre-expressing cells at the time of earliest Fgf4 expression in the forelimb AER.
Additionally, the AP expression and lacZ reporter data demonstrate that the hindlimbs of conditionally mutant animals (where RARCre is not uniformly expressed) function as an internal control for the effects of a complete ‘knock-out’ of Fgf4 in the forelimb AER.
Fgf4 conditional mutants are viable and fertile
Conditional mutagenesis of Fgf4 was accomplished by mating mice containing the F4AP allele (genotypes: F4AP/+ or F4AP/AP) to mice bearing RARCre and either null allele (F4APR or F4neo, hereafter noted as F4Null) in their germline (genotype: F4Null/+;T). The results were the same with the use of either null allele. Conditional mutants (F4AP/Null; T) were obtained at the expected Mendelian frequencies as embryos and adults, confirming that embryonic lethality had been circumvented. For example, among the 74 progeny of F4AP/AP X F4Null/+;T matings examined at E10.0-E11.5, we expected 18 and observed 21 conditional mutants. We generated 50 adults with this mating strategy; we expected 13 and saw 14 conditional mutants. The conditional mutant adults were fertile and produced healthy offspring.
Fgf4 transcripts are absent in the forelimb AER of Fgf4 conditional mutants
To demonstrate that RARCre activity disrupted Fgf4 gene function throughout the forelimb AER of Fgf4 conditional mutant embryos, we performed RNA in situ hybridization on embryos using an antisense Fgf4 riboprobe (Fig. 3). As expected, at E10.5, there was a strong Fgf4 signal in the hindlimb AER of the F4AP/Null;T conditional mutants, but no Fgf4 transcripts were detected in the forelimb AER (Fig. 3A,C). Also, as expected, E10.5 F4AP/Null animals without RARCre had normal patterns of Fgf4 expression in the AER of both limbs. The earliest stage that we and others have detected Fgf4 transcripts is E10.0, or the 30-somite stage when the AER is morphologically distinct (Drucker and Goldfarb, 1993; Niswander and Martin, 1992; Suzuki et al., 1992). When we examined the forelimb AER of Fgf4 conditional mutants at this stage (Fig. 3B), no Fgf4 transcripts were detected. Thus, in embryos bearing RARCre, the F4AP conditional allele and a null allele, F4AP is recombined throughout the AER prior to the onset of normal Fgf4 transcription resulting in an AER with no functional Fgf4 alleles.
Fig. 3.
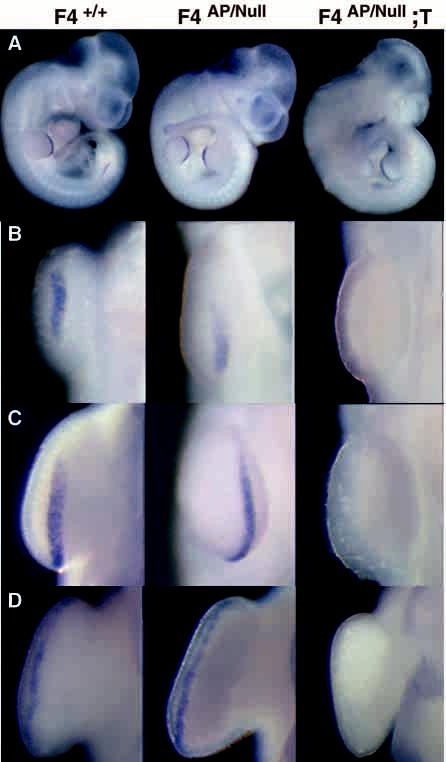
Fgf4 expression in conditional mutants and their littermates. Whole-mount RNA in situ hybridization with an Fgf4 antisense riboprobe containing exon 3 and 3′UTR sequences. (A) E10.5 whole-mount embryos; transcripts are present in hindlimb, but not forelimb AER of conditional mutants (F4AP/Null;T), confirming that Cre-mediated recombination of the conditional allele in the context of a null allele completely disrupts Fgf4 gene function. (B) E10.0 (30 somite) embryos. Close-up view of forelimb AER. Absence of Fgf4 transcripts correlates with presence of RARCre and confirms disruption of Fgf4 gene function at the earliest stage of normal Fgf4 transcription. (C,D) Close-up views of the forelimb AERs of E10.5 and E11.5 embryos, respectively.
Fgf4 conditional mutants have normal forelimbs
Current models of FGF4 function in the developing limb led us to hypothesize that disruption of Fgf4 in the AER of embryonic mice would result in defective outgrowth of posterior limb elements and altered anteroposterior patterning secondary to disruption of the proposed FGF4-SHH feedback loop. We examined the skeletons of conditional mutants (_n_=21) in E15.5 embryos, newborns (Fig. 4) and adults, and were extremely surprised to find that patterning and outgrowth of the forelimbs of Fgf4 conditional mutants were normal, as were external appearance and function in living animals. Embryonic conditional mutants (E10.0-11.5) were also indistinguishable from their littermates with regard to initiation, location or size of the limb bud and appearance of the AER.
Fig. 4.
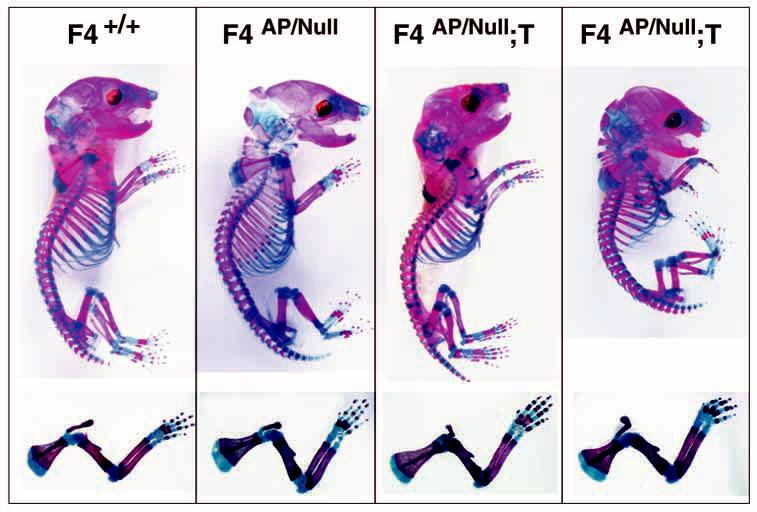
Skeletal phenotypes of newborns. Alizarin red stains ossified structures and Alcian blue stains cartilage in these preparations. Note that the limbs of Fgf4 conditional mutants are indistinguishable from wild-type controls.
Sonic Hedgehog is expressed normally in the limb mesenchyme of Fgf4 conditional mutants
If FGF4 normally maintains Shh expression in the ZPA, one would expect decreased Shh expression in the forelimbs of Fgf4 conditional mutants, particularly by E11.5. However, no changes in Shh expression were detected (Fig. 5A). Because Bmp2 is thought to act in opposition to Fgf4 to control limb outgrowth (Niswander and Martin, 1993) and to be regulated by the FGF4-SHH feedback loop (Laufer et al., 1994), we also expected altered expression of Bmp2 in the absence of FGF4 (Fig. 5C). Instead, the expression pattern was normal in the mutants. Similarly, there were no detectable alterations in timing, location or intensity of signal when we examined Fgf8 expression in the AER (Fig. 5B) or Fgf10 expression in the mesenchyme (data not shown).
Fig. 5.
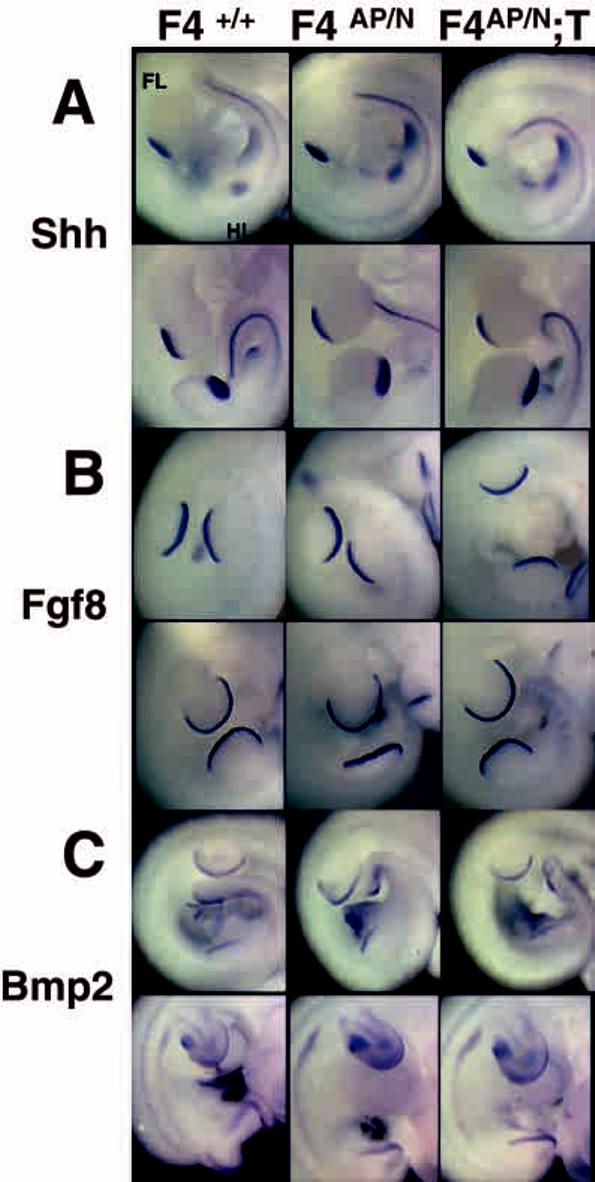
Molecular phenotypes as assessed by RNA in situ hybridization; normal expression of Shh in Fgf4 conditional mutants. For each probe, top row are E10.5 embryos; bottom row, E11.5. Littermates were used when possible. (A) Shh expression in the ZPA. (B) Fgf8 expression in the AER. (C) Bmp2 expression in the AER and mesenchyme. Note that each of these genes is expressed normally in Fgf4 conditional mutants.
DISCUSSION
By applying a system to conditionally mutate Fgf4 in the forelimbs of developing mouse embryos, we have successfully bypassed the early embryonic lethality that results from mutation of this gene in the germline. A large body of experimental evidence led us to predict that Fgf4 conditional mutants would have alterations in forelimb bud outgrowth and anteroposterior patterning as a result of altered PZ-AER interactions and disruption of the proposed FGF4-SHH feedback loop. However, our results indicate that Fgf4 is not required for normal limb development or Shh expression in the ZPA. In the absence of FGF4, Shh and Bmp2 are appropriately induced and maintained, consistent with correct anteroposterior forelimb patterning seen in the conditional mutants; Fgf 8 and 10 are expressed normally in the AER and PZ, a finding consistent with the continued normal forelimb outgrowth observed in these animals.
Besides demonstrating that Fgf4 gene function is not required for normal limb development, this work also provides new insight into Fgf4 regulatory elements. Based on expression of Fgf4 gene fragments linked to lacZ in transgenic mice, it has been proposed that the Fgf4 promoter has minimal _cis_-regulatory activity and that specific sequences in the 3′UTR direct Fgf4 gene expression in particular expression domains (i.e. AER versus myotome) (Curatola and Basilico, 1990; Fraidenraich et al., 1998). Additionally, in the transgenic system, restriction of Fgf4 expression to cells in the posterior AER required the presence of ‘inhibitor’ sequences in the Fgf4 3′UTR. However, Cre-mediated recombination of the F4AP allele deletes the entire 3′UTR and an additional 89 nucleotides and yet we have found that the Fgf4 promoter, in its normal 5′ chromasomal context, is active and contains sufficient information to direct AP expression in the AER, myotomes and pharyngeal pouches in the absence of any of the proposed enhancer or _cis_-regulatory sequences contained in the 3′UTR (Fig. 2B and data not shown). The Fgf4 5′ promoter fragment used in the transgenic constructs was 1.2 kb, ended 83 bp upstream of the translation start site and may not have contained all the regulatory elements located 5′ of the coding sequences. Notably, the F4APR allele is expressed throughout the AER (Fig. 2B). The broader expression domain is likely the consequence of the AP enzymatic assay being more sensitive than in situ hybridization as a means of detecting low level Fgf4 expression in the anterior AER at E10.0 (note that wild-type E11.5 embryos have detectable Fgf4 mRNA throughout the AER, Fig. 3B).
The observation that limbs develop normally in the absence of FGF4 in the AER of Fgf4 conditional mutants suggests that some of the functions currently ascribed to FGF4 may be performed by other AER factors in vivo. The simplest hypothesis is that another FGF compensates for the absence of FGF4 in the conditional mutants and provides a functionally equivalent signal to the underlying mesenchyme to support continued limb outgrowth and patterning. FGF1, FGF 2 and FGF 8 can each support limb outgrowth in the absence of the AER (Cohn et al., 1995; Crossley et al., 1996; Fallon et al., 1994). FGF2 maintains ZPA signaling in mouse limb microdissectate cultures (Anderson et al., 1993). Similarly, FGF8 can induce and maintain SHH expression in the absence of the AER (Crossley et al., 1996). FGF2, FGF4 and FGF8 display high levels of mitogenic activity when bound to the mesenchymally expressed FGF receptor isoforms 1c and 2c in vitro (Ornitz et al., 1996).
It is extremely unlikely that the normal skeletal and molecular phenotypes of the conditional mutants result from early or persistent Fgf4 gene function. We used several different methods to prove that Fgf4 was already disrupted in the forelimb AER of conditional mutants prior to the time that it is normally expressed in this structure: we did not detect Fgf4 transcripts in the forelimb AER of conditional mutants at E10.0-11.5; AP activity, which requires Cre-mediated deletion of all Fgf4-coding sequences from the F4AP conditional allele, is detectable throughout the forelimb AER by E10.0, indicating that not only has recombination of the allele occurred by this stage, but also transcription and translation of the AP reporter. Furthermore, we have confirmed that Cre is expressed in the limb bud ectoderm of E9.5 embryos by detecting AP expression from an allele of Fgf8 that expresses the AP gene under the control of the Fgf8 promoter (which is expressed this early in the ventral ectoderm and developing AER) after Cre-mediated recombination of the allele. The limb phenotype in these Fgf8 conditional mutants is obvious at E10.0. These findings demonstrate that Cre is expressed and active in the ectoderm of the limb bud prior to formation of the AER and prior to the onset of normal Fgf4 expression in the AER. (A. M. M. and M. R. C., unpublished results). In summary, our data confirm that RARCre is active sufficiently early and in the appropriate location to recombine F4AP prior to the onset of Fgf4 expression in the limb and to ensure that the conditional knockout of Fgf4 is both timely and complete in the forelimb AER.
An important feature of the conditional mutagenesis system that we have described is that it provides a means for evaluating the extent of Cre-mediated recombination in the relevant cells. Since the marker (AP) is expressed under the control of the Fgf4 promoter only after Cre-mediated disruption of Fgf4, labeling is limited to the subset of cells that express Fgf4 and have a recombined allele. This provides an independent and extremely sensitive assay for the extent of Cre-mediated recombination, which is critical to proper evaluation of the phenotypic consequences.
The power of conditional mutagenesis may not be restricted to modulating a single locus at a time; it should be possible to conditionally disrupt multiple genes simultaneously. Our experience suggests that use of the conditional allele in a null background was not necessary; if Cre is present in a cell, it appears to operate to completion. Recombination between loxP sites at homologous positions on homologous chromosomes also does not appear to be problematic. Thus, simultaneous conditional elimination of more than one Fgf in the AER may allow us to examine the potential for functional redundancy among these factors and thus to determine their relative contribution to normal AER activity and limb development.
Acknowledgments
We thank Kirk Thomas, vivarium staff and members of the Capecchi laboratory for advice and assistance, and C. L. Peterson and L. Hager for their work on the F4neo targeting vector. We are grateful to Julie Tomlin for excellent technical assistance. We thank A. McMahon, B. Hogan, C. Deng and D. Ornitz for generously providing in situ probes. For the AP plasmid and protocols, we thank C. Neville and N. Rosenthal. N. Proudfoot kindly provided the C2 pause and synthetic poly(A) sequences used in the targeting vectors. We thank S. O'Gorman for the P2Bc reporter mice and J. Deschamps for the RARβ2 promoter fragment. A. M. M. is supported by grants from the Primary Children's Medical Center Foundation and an HHMI Fellowship to Faculty award.
REFERENCES
- Anderson R, Landry M, Muneoka K. Maintenance of ZPA signaling in cultured mouse limb bud cells. Development. 1993;117:1421–1433. doi: 10.1242/dev.117.4.1421. [DOI] [PubMed] [Google Scholar]
- Ashfield R, Enriquez-Harris P, Proudfoot NJ. Transcriptional termination between the closely linked human complement genes C2 and Factor B: common termination factor for C2 and C-myc? EMBO J. 1991;10:4197–4207. doi: 10.1002/j.1460-2075.1991.tb04998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv. Cancer Res. 1992;59:115–163. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- Baubonis W, Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Research. 1993;21:2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EM, Goddard JM, Chisaka O, Manley NR, Capecchi MR. Loss of hox-A1 function results in the reorganization of the murine hindbrain. Development. 1993;118:1063–1075. doi: 10.1242/dev.118.4.1063. [DOI] [PubMed] [Google Scholar]
- Caruccio NC, Martinez-Lopez A, Harris M, Dvorak L, Bitgood J, Simandl K, Fallon JF. Constitutive activation of Sonic Hedgehog signaling in the chicken mutant talpid2: Shh-independent outgrowth and polarizing acitivity. Dev. Biol. 1999;212:137–149. doi: 10.1006/dbio.1999.9321. [DOI] [PubMed] [Google Scholar]
- Chan DC, Laufer E, Tabin C, Leder P. Polydactylous limbs in Strong's Luxoid mice result from ectopic polarizing activity. Development. 1995a;121:1971–1978. doi: 10.1242/dev.121.7.1971. [DOI] [PubMed] [Google Scholar]
- Chan DC, Wynshaw-Boris A, Leder P. Formin isoforms are differentially expressed in the mouse embryo and are required for normal expression of fgf-4 and shh in the limb bud. Development. 1995b;121:3151–3162. doi: 10.1242/dev.121.10.3151. [DOI] [PubMed] [Google Scholar]
- Charité J, de Graaff W, Shen S, Deschamps J. Ectopic expression of Hoxb-8 causes duplication of the ZPA in the forelimb and homeotic transformation of axial structures. Cell. 1994;78:589–601. doi: 10.1016/0092-8674(94)90524-x. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Izpisúa-Belmonte JC, Abud H, Heath JK, Tickle C. Fibroblast growth factors induce additional limb development from the flank of chick embryos. Cell. 1995;80:739–746. doi: 10.1016/0092-8674(95)90352-6. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. Limbs: a model for pattern formation within the body plan. Trends in Genetics. 1996;12:253–257. doi: 10.1016/0168-9525(96)10030-5. [DOI] [PubMed] [Google Scholar]
- Crossley HP, Minowada G, MacArthur CA, Martin GR. Roles for Fgf-8 in the induction, initiation and maintenance of chick limb development. Cell. 1996;84:127–136. doi: 10.1016/s0092-8674(00)80999-x. [DOI] [PubMed] [Google Scholar]
- Curatola AM, Basilico C. Expression of the K-fgf protooncogene is controlled by 3′ regulatory elements which are specific for embryonal carcinomal cells. Molec. Cell. Biol. 1990;10:2475–2484. doi: 10.1128/mcb.10.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker BJ, Goldfarb M. Murine FGF-4 gene expression is spatially restricted within embryonic skeletal muscle and other tissues. Mech. Dev. 1993;40:155–163. doi: 10.1016/0925-4773(93)90073-7. [DOI] [PubMed] [Google Scholar]
- Fallon JF, Lopez A, Ros MA, Savage MP, Olwin BB, Simandl BK. Fgf-2: apical ectodermal ridge growth signal for chick limb development. Science. 1994;264:104–107. doi: 10.1126/science.7908145. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaipoannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Fraidenraich D, Lang R, Basilico C. Distinct regulatory elements govern Fgf4 gene expression in the mouse blastocyst, myotomes, and developing limbs. Dev. Biol. 1998;204:197–209. doi: 10.1006/dbio.1998.9053. [DOI] [PubMed] [Google Scholar]
- Francis-West PH, Robertson KE, Ede DA, Rodriguez C, Ipisua-Belmonte JC, Houston B, Burt DW, Brickell PM, Tickle C. Expression of genes encoding bone morphogenetic proteins and Sonic Hedgehog in Talpid (ta3) limb buds: their relationships in the signalling cascade involved in lamb patterning. Dev. Dynam. 1995;203:187–197. doi: 10.1002/aja.1002030207. [DOI] [PubMed] [Google Scholar]
- Globus M, Vethamany-Globus S. An in vitro analog of early chick limb bud outgrowth. Differentiation. 1976;6:91–96. doi: 10.1111/j.1432-0436.1976.tb01474.x. [DOI] [PubMed] [Google Scholar]
- Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Haramis AG, Brown JM, Zeller R. The limb deformity mutation disrupts the SHH/FGF-4 feedback loop and regulation of 5′Hox -D genes during limb pattern formation. Development. 1995;121:4237–4245. doi: 10.1242/dev.121.12.4237. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe JR, Gumpel-Pinot M. Control of maintenance and anteroposterior skeletal differentiation of the anterior mesenchyme of the chick wing by its posterior margin (the ZPA) J. Embryol. Exp. Morph. 1981;62:63–82. [PubMed] [Google Scholar]
- Hogan B, Constantini F, Lacy E. Manipulating the Mouse Embryo. Cold Spring Harbor Laboratory; New York: 1986. [Google Scholar]
- Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Levitt N, Briggs D, Hil A, Proudfoot NJ. Definition of an efficient synthetic poly (A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Drossopoulou G, Paton IR, Morrice IR, Robertson KE, Burt DW, Ingham PW. Expressiono f ptc and gli genes in talpid3 suggests bifurcation in Shh pathway. Development. 1999;126:2397–2407. doi: 10.1242/dev.126.11.2397. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Developmental analysis of the retinoic acid-inducible RAR-B2 promoter in transgenic animals. Development. 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- Muenke M, Schell U. Fibroblast growth factor receptor mutations in human skeletal disorders. Trends Genet. 1995;11:308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- Niswander L, Jeffery S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–614. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. FGF-4 and BMP-2 have opposite effects on limb growth. Nature. 1993;361:68–71. doi: 10.1038/361068a0. [DOI] [PubMed] [Google Scholar]
- Niswander L, Tickle C, Vogel A, Booth I, Martin GR. FGF-4 replaces the apical ectodermal ridge and directs outgrowth and patterning of the limb. Cell. 1993;75:579–587. doi: 10.1016/0092-8674(93)90391-3. [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y,H,Y, Kuwana T, Nohno T, Yamasaki M, et al. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- Ornitz D, Xu J, Colvin J, McEwen D, MacArthur C, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Reiter RS, Solursh M. Mitogenic property of the apical ectodermal ridge. Dev. Biol. 1982;93:28–35. doi: 10.1016/0012-1606(82)90235-4. [DOI] [PubMed] [Google Scholar]
- Reynolds K, Mezey E, Zimmer A. Activity of the B-retinoic acid receptor promoter in transgenic mice. Mech. Dev. 1991;36:15–29. doi: 10.1016/0925-4773(91)90068-h. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic Hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Ros MA, Lopez-Marinez A, Simandl B, Rodriguez C, Izpisua-Belmonte JC, Dahn R, Fallon JF. The limb field mesoderm determines initial limb bud anteroposterior asymmetry and budding independent of sonic hedgehog or apical ectodermal gene expressions. Development. 1996;122:2319–2330. doi: 10.1242/dev.122.8.2319. [DOI] [PubMed] [Google Scholar]
- Saunders JW. The experimental analysis of chick limb bud development. In: Ede DA, Hinchcliffe JR, Balls M, editors. Vertebrate limb and somite morphogenesis. Cambridge University Press; Cambridge: 1977. pp. 1–24. [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Soto T, Yagishita N, Matsui D, Koga Y, Itoh N, et al. Fgf10 is essential for limb and lung formation. Nature Genetics. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Summerbell D, Lewis JH, Wolpert L. Positional information in chick limb morphogenesis. Nature. 1973;224:492–496. doi: 10.1038/244492a0. [DOI] [PubMed] [Google Scholar]
- Suzuki HR, Sakamoto H, Yoshida T, Sugimura T, Terada M, Solursh M. Localization of Hst1 transcripts to the Apical Ectodermal ridge in the Mouse Embryo. Dev. Biol. 1992;150:219–222. doi: 10.1016/0012-1606(92)90020-h. [DOI] [PubMed] [Google Scholar]
- Tabin CJ. Retinoids, homeoboxes and growth factors: toward molecular models of limb development. Cell. 1991;66:199–217. doi: 10.1016/0092-8674(91)90612-3. [DOI] [PubMed] [Google Scholar]
- Thomas KT, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Todt WL, Fallon JF. Posterior apical ectodermal ridge removal in the chick wing bud triggers a series of events resulting in defective anterior pattern formation. Development. 1987;101:501–515. doi: 10.1242/dev.101.3.501. [DOI] [PubMed] [Google Scholar]
- Vogel A, Tickle C. Fgf-4 maintains polarizing activity of posterior limb bud cells in vivo and in vitro. Development. 1993;119:199–206. doi: 10.1242/dev.119.1.199. [DOI] [PubMed] [Google Scholar]
- Wanek N, Muneoka K, Holler-Dinsmore G, Burton R, Bryant SV. A staging system for mouse limb development. J. Exp. Zool. 1989;249:41–49. doi: 10.1002/jez.1402490109. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Cuiling L, Naski M, Cohen RI, Ornitz D, Leder P, Deng C. Fibroblast growth factor receptor 2-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Deng C. Fibroblast growth factor receptors (FGFRs) and their roles in limb development. Cell Tissue Res. 1999;296 doi: 10.1007/s004410051264. [DOI] [PubMed] [Google Scholar]
- Zuniga A, Haramis AG, McMahon A, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]