Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing (original) (raw)
Abstract
The repair of potentially lethal DNA double-stranded breaks (DSBs) by homologous recombination requires processing of the broken DNA into a resected DNA duplex with a protruding 3′-single-stranded DNA (ssDNA) tail. Accordingly, the canonical models for DSB repair require invasion of an intact homologous DNA template by the 3′–end of the ssDNA, a characteristic that the bacterial pairing protein RecA possesses. Unexpectedly, we find that for the eukaryotic homolog, Rad51 protein, the 5′–end of ssDNA is more invasive than the 3′–end. This pairing bias is unaffected by Rad52, Rad54 or Rad55–57 proteins. However, further investigation reveals that, in contrast to RecA protein, the preferred DNA substrate for Rad51 protein is not ssDNA but rather dsDNA with ssDNA tails. This important distinction permits the Rad51 proteins to promote DNA strand invasion using either 3′- or 5′–ends with similar efficiency.
Keywords: DNA strand exchange/homologous recombination/joint molecule formation
Introduction
Spontaneous or damage-induced double-stranded breaks (DSBs) can cause death in all organisms. The repair of such DSBs by homologous recombination requires processing of the broken double-stranded DNA (dsDNA) into a resected DNA duplex with a protruding 3′-single-stranded DNA (ssDNA) tail (Sun et al., 1991; Haber, 1995). Following resection, a homologous pairing protein is loaded onto the ssDNA to form a contiguous nucleoprotein filament. The filament searches for an intact homologous DNA template and pairs with it to commence the replicational repair of the DSB (Haber, 1997). The bacterial RecA protein polymerizes on ssDNA with a 5′→3′ polarity (Register and Griffith, 1985). This directionality ensures that the 3′–end of ssDNA is coated with RecA protein and, hence, is activated for homologous pairing. Indeed, the 3′–end of ssDNA is better protected against exonuclease degradation and is more active than the 5′–end for DNA strand invasion promoted by RecA protein (Konforti and Davis, 1990). This biochemical preference for 3′–end invasion is generally considered advantageous, because this 3′–end can serve as a primer to initiate recombination-dependent replication (Kogoma, 1996) and DSB repair (Szostak et al., 1983) processes.
Despite the sound concordance of biological function and biochemical mechanism in the case of RecA protein, recent findings with eukaryotic proteins seem to question the universality of this canonical view. In contrast to RecA protein, which actively promotes DNA heteroduplex extension in the 5′→3′ direction, the Rad51 protein from Saccharomyces cerevisiae apparently allows DNA heteroduplex extension to occur in either direction (Sung and Robberson, 1995; Namsaraev and Berg, 1998). Another eukaryotic homolog, human Rad51 protein, shows a preferential 3′→5′ polarity for heteroduplex extension, which is opposite to the direction displayed by the RecA protein (Baumann and West, 1999). Given the structural similarities of the nucleoprotein filaments formed by the eukaryotic Rad51 proteins and RecA protein, this difference in the directionality of heteroduplex extension is intriguing. Like RecA protein (Stasiak and Di Capua, 1982), both human and yeast Rad51 proteins form helical filaments with ∼6.2 protein monomers per turn and a pitch of ∼100 Å, stretching the DNA within the filaments by a factor of 1.5 (Ogawa et al., 1993; Benson et al., 1994). Since in the case of RecA protein the polarity of DNA heteroduplex extension reflects the underlying polarity of protein polymerization, as well as the resultant pairing bias displayed with linear ssDNA (Konforti and Davis, 1992), the observations with Rad51 protein raise questions regarding the homologous pairing preference of the eukaryotic homologs. Therefore, we have examined the pairing bias of Rad51 protein-mediated invasion of supercoiled DNA by linear ssDNA.
We show here that, for both yeast and human Rad51 proteins, the 5′–end of ssDNA is more invasive than the 3′–end. For the yeast protein, although the yield of joint molecules is affected by some proteins from the Rad52 epistasis group, the pairing bias is not. However, in contrast to RecA protein, we discovered that the preferred DNA substrate for both Rad51 proteins is not ssDNA but rather DNA composed of dsDNA and ssDNA tails. Thus, tailed dsDNA substrates permit the Rad51 proteins to promote DNA strand invasion of both 3′- and 5′–ends with similar efficiencies.
Results
Homologous pairing by Rad51 protein displays a preference for the 5′–end of ssDNA
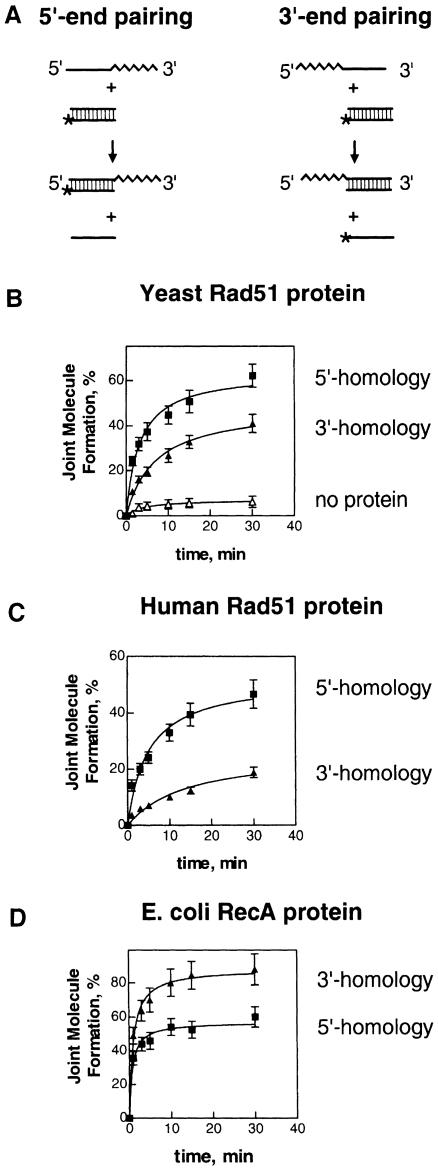
To determine which end of linear ssDNA is more reactive in Rad51 protein-mediated pairing, we measured DNA strand exchange occurring at the opposite ends of nucleoprotein filaments assembled on ssDNA (63mer, #1 or 2) (Figure 1A). The reaction was more efficient when the dsDNA was homologous to the 5′–end of the ssDNA within the S.cerevisiae Rad51 nucleoprotein filament rather than to the 3′–end (Figure 1B). To exclude any possible effect originating from the DNA compositional preference exhibited by RecA-like proteins (Mazin and Kowalczykowski, 1996; Tracy et al., 1997), an oligonucleotide (63mer, #58) was constructed in which the 3′- and 5′–terminal sequences of the oligonucleotide #1 were swapped. DNA strand exchange using this ‘swapped’ oligonucleotide displayed the same bias for the 5′–terminal end (data not shown). Furthermore, the location of the label in the substrates had no effect on the observed bias, because placing the 32P-label in the opposite strand of the dsDNA molecules shown in the Figure 1A had no effect (data not shown).
Fig. 1. The pairing bias exhibited by the eukaryotic Rad51 proteins is opposite to that of prokaryotic RecA protein. (A) Scheme of the experiments. Rad51 and RecA nucleoprotein filaments were formed on one of two complementary ssDNA substrates (63mers, #1 or 2). DNA strand exchange was initiated by addition of 32P-labeled dsDNA (31mers, #45 and 55) (12 μM) that was homologous to either the 5′- or 3′–terminal regions of oligonucleotides #1 and 2, respectively. The straight line indicates homology to the dsDNA, the zig-zag line indicates heterology; asterisks denote the 32P-labeled strand of dsDNA. (B, C and D) The kinetics of DNA strand exchange promoted by yeast Rad51 protein, human Rad51 protein and E.coli RecA protein, respectively.
To establish the validity of our assay, RecA protein was also examined. In agreement with previously published data, the bias was reversed and the 3′–terminal end of the RecA nucleoprotein filament was more active in homologous pairing than the 5′–end (Figure 1D). Thus, Rad51 and RecA proteins possess opposite pairing preferences that are intrinsic to these proteins, rather than to the DNA.
Since the pairing bias displayed by the yeast Rad51 protein was opposite to that of RecA protein and, most importantly, was in conflict with the biological expectation that a 3′–end would be more reactive, another eukaryotic homolog, the human Rad51 protein, was examined. We found that the human Rad51 protein displays an even stronger preference for DNA pairing at the 5′-end of ssDNA, when compared with its yeast counterpart (Figure 1C). Thus, this bias for the 5′–end of ssDNA is a common characteristic of these two eukaryotic DNA pairing proteins, which distinguishes them from their prokaryotic counterparts.
The 5′–end of linear ssDNA is more invasive than the 3′–end in Rad51 protein-mediated invasion of supercoiled DNA
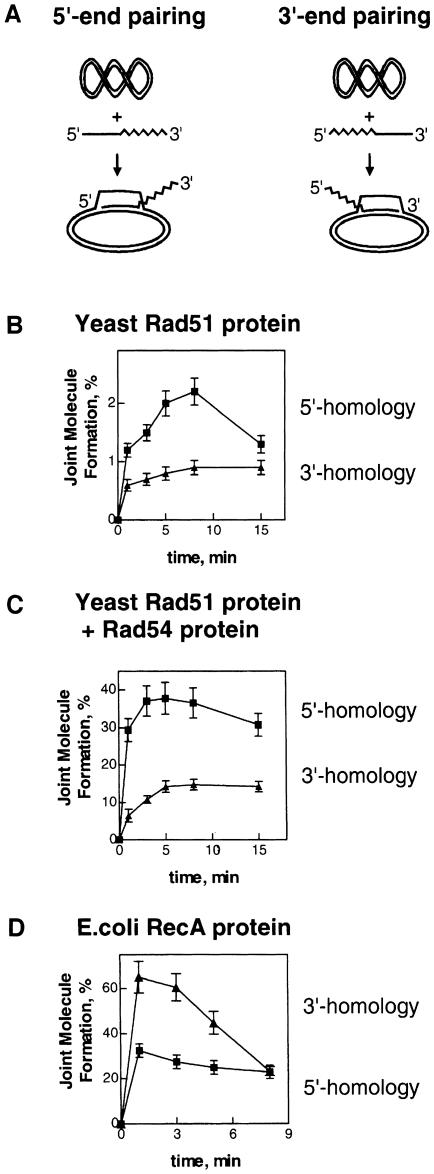
The preferential invasiveness of ssDNA 3′–ends that is mediated by RecA protein was most clearly established using the D–loop pairing assay, an assay that measures joint molecule formation between linear ssDNA and homologous supercoiled DNA (Konforti and Davis, 1990). We used this assay to assess the relative invasiveness of the 3′- and 5′–ends of ssDNA in pairing reactions promoted by Rad51 protein. For this purpose, two 90mer oligonucleotides, SK#3 and SK#5, were synthesized that were partially homologous to pUC19 plasmid DNA. These oligonucleotides contain a homologous region, 63 bases long, which is located at either the 3′- or 5′–end, respectively (Figure 2A).
Fig. 2. In Rad51 protein-mediated invasion of supercoiled DNA, linear ssDNA with homology at the 5′–end is more invasive than ssDNA with homology at the 3′–end. (A) Scheme of the experiments. (B) The kinetics of joint molecule formation promoted by Rad51 protein between supercoiled pUC19 dsDNA and partially homologous ssDNA oligonucleotides SK#3 and SK#5, carrying homology at either the 3′- or 5′–terminal region, respectively. (C) Identical reactions to those in (B), except that they were supplemented with Rad54 protein (0.15 μM). (D) Reactions promoted by RecA protein with the same DNA substrates. The percentage of joint molecule products was expressed relative to the limiting amount of plasmid DNA.
As shown in Figure 2B, Rad51 protein promotes joint molecule formation more efficiently when the oligonucleotide is homologous at the 5′–end rather than at the 3′–end. Although as expected the overall efficiency of joint molecule formation is low (Petukhova et al., 1998), linear ssDNA with homology at the 5′–end is ∼2-fold more invasive than ssDNA with homology at the 3′–end. However, in contrast to Rad51 protein and in agreement with previously published data, RecA protein forms joint molecules more efficiently with the ssDNA that carries homology at the 3′–end (Figure 2D).
Several proteins encoded by the RAD52 epistasis group of genes are known to increase the pairing activity of Rad51 protein. Hence, we examined whether some of these proteins might alter the observed pairing bias. As reported (Petukhova et al., 1998), Rad54 protein significantly increased the overall efficiency of joint molecule formation by Rad51 protein (Figure 2C). (Rad54 protein alone did not promote joint molecule formation; data not shown.) However, importantly, the Rad54 protein did not alter the observed pairing bias (Figure 2C). Rad52 protein, another protein that stimulates the DNA pairing activity of Rad51 protein (Sung, 1997a; Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998), only slightly stimulated D–loop formation, but it did not change the relative invasiveness of the ssDNA ends (data not shown). Finally, the Rad55–Rad57 heterodimer protein stimulated the extent of D–loop formation by ∼10–20% more than that obtained with Rad51 and Rad54 proteins, but ssDNA with homology confined to its 5′–end retained a higher yield of joint molecule formation (data not shown). Thus, we conclude that the higher reactivity of presynaptic filaments that possess homology at their 5′–end is an intrinsic property of Rad51 protein.
Homologous pairing by Rad51 protein is enhanced with tailed dsDNA substrates
The opposite pairing bias of the eukaryotic and prokaryotic DNA strand exchange proteins raises a number of interesting issues: either (i) the Rad51 proteins are fundamentally different to the prokaryotic RecA proteins with regard to their homologous pairing behavior; (ii) an as yet unknown accessory protein is required in vivo to modify the pairing behavior of the eukaryotic proteins; or (iii) homologous pairing requires a specific type of DNA substrate. Since processing of a DSB in vivo produces a DNA duplex with a protruding 3′–ssDNA tail, we examined the behavior of tailed dsDNA substrates in vitro. Presynaptic filaments with yeast Rad51 protein were formed on either 3′–tailed dsDNA (a 32mer annealed to a 63mer) or ssDNA (a 63mer or 31mer), and DNA strand exchange was initiated by addition of dsDNA that was homologous to the 31 nucleotide region of the ssDNA of the tailed substrate. Interestingly, the tailed dsDNA was more reactive in DNA strand exchange, as measured by the initial reaction rate, than its respective ssDNA counterparts (Figure 3A).
Fig. 3. Duplex DNA with an ssDNA tail is the preferred substrate for DNA strand exchange promoted by Rad51 protein. (A) The kinetics of DNA strand exchange promoted by Rad51 protein. Rad51 nucleoprotein filaments were formed on either ssDNA (63mer, #2 or 31mer, #55) or tailed dsDNA with a 3′ single-stranded end (63mer, #2 and 32mer, #5). DNA strand exchange was initiated by addition of 32P-labeled dsDNA (31mers, #45 and 55) that was homologous to either the 3′–terminal region of both the 63mer ssDNA and 32/63mer tailed dsDNA or to the entire 31mer ssDNA. (B) The kinetics of DNA strand exchange promoted by RecA protein. RecA nucleoprotein filaments were formed on either ssDNA (63mer, #1) or tailed dsDNA with a 5′ single-stranded end (63mer, #1 and 32mer, #6). DNA strand exchange reactions were initiated by addition of 32P-labeled dsDNA (31mers, #45 and 55). In order to measure accurately the initial rates, the reactions with RecA protein were carried out at 24°C. The drawings above each panel illustrate the DNA strand exchange reactions promoted by each protein. The straight lines indicate homology to the dsDNA, the zig-zag lines indicate heterology; asterisks denote the 32P-labeled strand of dsDNA.
Next, we investigated the behavior of tailed dsDNA with homology limited to either the 3′- or 5′–end of the substrate in DNA strand exchange reaction promoted by either yeast or human Rad51 protein. We found that tailed dsDNA with homology at the 5′–end displayed the highest reaction rate for both the yeast and human Rad51 proteins (Figure 4). However, the increase in reaction rate due to the presence of a duplex DNA region, for both eukaryotic proteins, was greater for the 3′–ends than for the 5′–ends when compared with the ssDNA substrates. Thus, the distinction between 5′- and 3′–ends in DNA pairing was diminished. In contrast, DNA strand exchange promoted by RecA protein was equally efficient for either ssDNA or tailed dsDNA with either overhang (Figure 3B; data not shown).
Fig. 4. Both yeast and human Rad51 proteins show a preference for tailed duplex DNA in DNA strand exchange. (A and B) The kinetics of DNA strand exchange promoted by yeast or human Rad51 protein. Rad51 nucleoprotein filaments were formed on either ssDNA (63mers, #1 or 2) or tailed dsDNA with a 5′ single-stranded end (63mer, #1 and 32mer, #6) or a 3′ single-stranded end (63mer, #2 and 32mer, #5). DNA strand exchange reactions were initiated by addition of 32P-labeled dsDNA (31mers, #45 and 55) that was homologous to either the 3′- or 5′–terminal regions of the ssDNA or the tailed dsDNA; the straight line indicates homology to the dsDNA, the zig-zag line indicates heterology. Since only one strand of dsDNA was labeled, DNA strand exchange with the 3′ or 5′ homologous ssDNA substrates produces displaced 32P-labeled 31mer ssDNA or 32P-labeled 32/63mer heteroduplex, respectively (see Figure 1A). In (C), the initial rates of DNA strand exchange were calculated for the first minute of the reactions and were normalized relative to the rate obtained for the 5′–tailed dsDNA (100%) for each protein. Placement of the 32P-label in the opposite dsDNA strand had no effect on the kinetics of joint molecule formation.
DNA strand exchange with both the ssDNA and tailed dsDNA substrates requires a stoichiometric amount of Rad51 protein
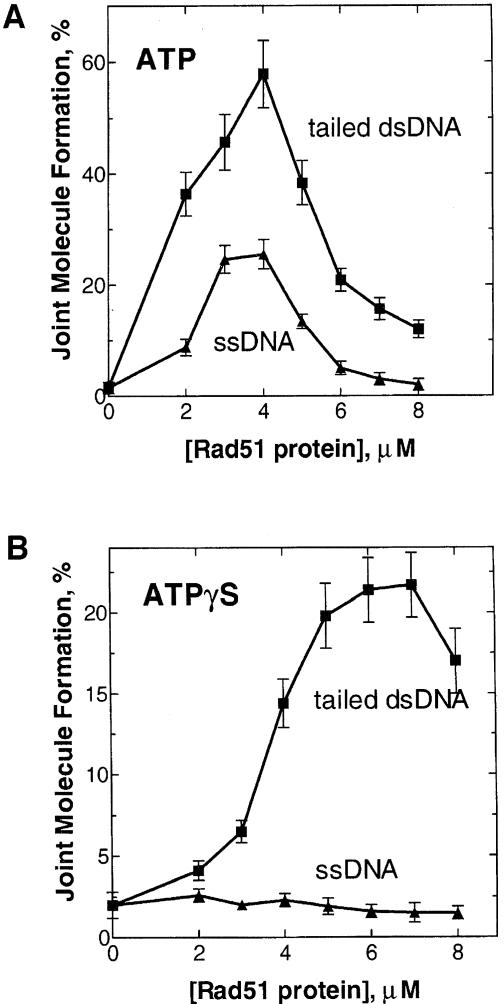
The increased DNA pairing of tailed dsDNA by the eukaryotic Rad51 proteins could be due to a more favorable ratio of Rad51 protein to ssDNA, which would be higher for tailed dsDNA (due to the region of duplex DNA) than for the fully ssDNA substrates. To eliminate this trivial possibility, we measured the efficiency of DNA strand exchange as a function of yeast Rad51 protein concentration for both the ssDNA and 3′–tailed dsDNA substrates.
We found that tailed dsDNA was a more active substrate in DNA strand exchange at all Rad51 protein concentrations examined (Figure 5A). For both the ssDNA and tailed dsDNA substrates, DNA strand exchange was the most efficient at a ratio of one Rad51 protein monomer to three DNA nucleotides (a combination of nucleotides and base pairs in the case of tailed dsDNA). An increase or decrease in Rad51 protein concentration relative to this stoichiometric ratio inhibited DNA strand exchange with either substrate.
Fig. 5. A stoichiometric concentration of Rad51 protein is required for optimal DNA strand exchange activity with either ssDNA or tailed dsDNA substrates. Nucleoprotein filaments were formed by adding yeast Rad51 protein at the indicated concentrations to either ssDNA (63mer, #2) (12 μM) or tailed dsDNA with a 3′ single-stranded end (63mer, #2 and 32mer, #5) (18 μM) in the presence of 2 mM ATP or 1 mM ATPγS. DNA strand exchange was initiated by addition of 32P-labeled dsDNA (31mers, #45 and 55) that was homologous to the 3′–terminal region of the ssDNA or the tailed dsDNA substrate. The reactions were carried out for 5 min. RPA was omitted in both reactions. Reactions were carried out in the presence of ATP (A) and ATPγS (B).
The non-hydrolyzable ATP analog, ATPγS, supports Rad51 protein-promoted DNA strand exchange with φX174 DNA, although higher concentrations of Rad51 protein are required for maximal activity than for equivalent reactions using ATP (Sung and Stratton, 1996). Accordingly, with tailed dsDNA in the presence of ATPγS, we found that the concentration of Rad51 protein required for optimal DNA strand exchange was almost twice as high as in the presence of ATP (Figure 5B). In contrast, the reaction with the ssDNA (63mer) was negligible at all Rad51 protein concentrations.
These results confirm that an intrinsic biochemical property of the tailed dsDNA, rather than a more favorable ratio of Rad51 protein to ssDNA, is responsible for the enhanced DNA pairing activity.
DNA strand exchange involving tailed dsDNA is less susceptible than ssDNA to inhibition by replication protein-A
In vivo data indicate that the ssDNA binding protein, replication protein-A (RPA), is the first to bind ssDNA that is generated during processing of a DSB (Gasior et al., 1998). Rad51 protein subsequently displaces RPA from ssDNA in a process that is mediated by Rad52 protein (Sung, 1997a; New et al., 1998; Shinohara and Ogawa, 1998), and then initiates DNA pairing with the homologous dsDNA. Experiments in vitro show that RPA inhibits DNA strand exchange when it binds to ssDNA prior to Rad51 protein (Sung, 1997b). Hence, we tested whether the tailed dsDNA substrates are less sensitive to this inhibitory effect of RPA than their ssDNA equivalents.
As expected, we found that the prior binding of RPA to the ssDNA (63mer) substrate produced a 10–15 min lag in DNA strand exchange; this lag corresponds to the time required for Rad51 protein to displace the RPA (Sugiyama et al., 1997). In contrast, there was no lag with the tailed dsDNA, demonstrating that RPA can be readily displaced from this substrate (Figure 6). This alleviation was not due to the potentially weaker binding of RPA to the 31 nucleotides of ssDNA in tailed dsDNA than to the 63 nucleotides in the ssDNA, because RPA completely blocked DNA strand exchange with the 31 nucleotide ssDNA.
Fig. 6. RPA inhibits DNA strand exchange with tailed dsDNA less than with ssDNA substrates. (A) The kinetics of DNA strand exchange promoted by yeast Rad51 protein when RPA was pre-bound to either ssDNA or 3′–tailed dsDNA. Reactions were identical to those described in the legend to Figure 1, except that RPA (3 μM) was added to either ssDNA (63mer, #2 or 31mer, #55) or 3′–tailed dsDNA (63mer, #2 and 32mer, #5), and was followed by a 7 min incubation prior to addition of yeast Rad51 protein. Homologous dsDNA was added 3 min after addition of Rad51 protein. For comparison, (B) shows the kinetics of identical DNA strand exchange reactions except that RPA was omitted.
Previously, it was described that several auxiliary proteins facilitate the displacement of RPA from ssDNA by Rad51 protein (Sung, 1997a,b; New et al., 1998; Shinohara and Ogawa, 1998). Our results demonstrate that tailed dsDNA substrates also enable Rad51 protein to compete more efficiently with RPA, thereby increasing the efficiency of presynaptic complex formation and the subsequent DNA strand exchange process.
Tailed dsDNA is the favored substrate for Rad51 protein binding
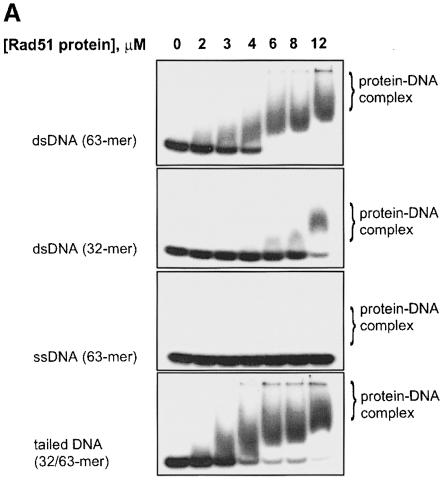
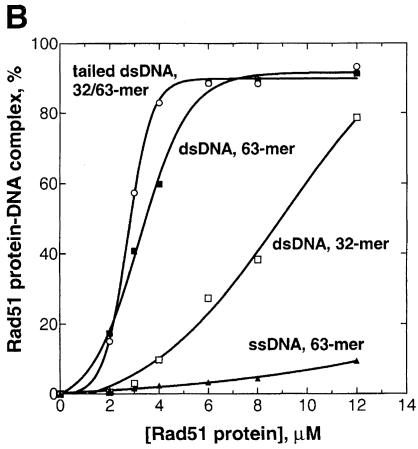
To understand why tailed dsDNA was the preferred substrate for DNA strand exchange promoted by Rad51 protein, we analyzed the affinity of Rad51 protein for ssDNA, dsDNA and tailed dsDNA using a gel mobility-shift assay that was used earlier to study RecA nucleoprotein complex formation (Mazin and Kowalczykowski, 1996, 1998).
In the presence of ATPγS, Rad51 protein forms complexes with dsDNA (63mer), but fails to form a sufficiently stable complex with ssDNA (63mer) to be detected in the gel (Figure 7). Binding of Rad51 protein to dsDNA appears to be length dependent, since higher Rad51 protein concentrations were required for binding to the shorter dsDNA (32mer) (Figure 7). However, when short dsDNA (32 bp long) was equipped with an ssDNA tail (31 nucleotides), the resultant tailed dsDNA (3′–tailed, 32/63mer) interacted effectively with Rad51 protein (Figure 7). A protein titration showed that the binding stoichiometry is 2.5–3 bp and/or bases of dsDNA (63mer) or tailed dsDNA (32/63mer) per protein monomer (Figure 7B). A similar hierarchy and stoichiometry was observed in the presence of ATP, although the apparent affinities were higher (A.V.Mazin and S.C.Kowalczykowski, unpublished observation). We present the data obtained with ATPγS here because they provided a ready distinction between most of the different DNA substrates. In the presence of ATP, Rad51 protein binds sufficiently tightly to ssDNA to obscure any differences in affinity compared with the tailed dsDNA (data not shown); the competition experiments presented below permit a clear comparison when ATP is used.
Fig. 7. Rad51 protein displays a hierarchy of complex formation with different DNA substrates. DNA substrates, dsDNA 63mer (63mers, #1 and 2) (24 μM), dsDNA 31mer (31mers, #45 and 55) (24 μM), tailed dsDNA 32/63mers (63mer, #2 and 32mer, #5) (18 μM) and ssDNA 63mer (63mer, #1) (12 μM), were mixed with increasing concentrations of yeast Rad51 protein and analyzed. (A) shows images of the gels; (B) represents quantitation of the PhosphorImager scans.
Because the direct binding assay could not readily differentiate the apparent affinities for all of the DNA substrates (Figure 7), competition experiments were performed. Labeled dsDNA (63mer) or 3′–tailed dsDNA (32/63mer) was pre-mixed with unlabeled heterologous dsDNA (63mer), and Rad51 protein was bound in the presence of either ATP or ATPγS. The results demonstrate that in the presence of ATP the percentage of Rad51 protein bound to the tailed dsDNA was greater than that bound to the dsDNA at all concentrations of competitor (Figure 8); similar results were obtained with ATPγS (inset). Thus, Rad51 protein binds preferentially to DNA with single-stranded tails. Therefore, we suggest that the binding preference of Rad51 protein for tailed duplex DNA is responsible for the increased efficiency of DNA strand exchange promoted by Rad51 protein with these DNA substrates.
Fig. 8. Rad51 protein binds preferentially to duplex DNA with ssDNA tails. DNA substrates, dsDNA (63mers, #1 and 2) (24 μM) or tailed dsDNA (63mer, #2 and 32mer, #5) (18 μM) (both 32P-labeled), were mixed with various concentrations of unlabeled dsDNA (63mers, #41 and 49). These mixtures were incubated for 15 min with yeast Rad51 protein (7.8 μM) in the presence of ATP. Rad51 protein–DNA complexes were analyzed by gel electrophoresis and quantified. The inset shows the data from an identical experiment, except that it was performed in the presence of ATPγS instead of ATP.
Discussion
Our results establish a novel property of the eukaryotic DNA strand exchange proteins: namely, these proteins show a pronounced affinity for dsDNA with ssDNA tails, the presumptive physiological substrate of both DSB repair and recombination-dependent replication. This finding, at least partially, resolves the potential dilemma that despite the demonstrated presence of 3′–ssDNA tailed duplex DNA as a physical intermediate of DSB repair, both the yeast and human proteins favor pairing of the 5′–end of ssDNA. We find that the pairing activity at the 3′–end is potentiated by incorporation of the ssDNA into a tailed duplex, allowing nearly equivalent pairing by either a 3′- or 5′–end of ssDNA; thus, the nature of the cellular nucleolytic processing of DSBs will determine which end is presented to Rad51 protein in vivo. The basis for this enhanced pairing activity resides in a preferential affinity of the eukaryotic proteins for tailed dsDNA, a preference that is not shared by its prokaryotic counterpart.
The prokaryotic RecA protein assembles on ssDNA with a 5′→3′ polarity of polymerization (Register and Griffith, 1985). This directionality ensures that the 3′–end of ssDNA is coated with RecA protein and, hence, is both protected against exonucleases (Konforti and Davis, 1991) and activated for homologous pairing (Konforti and Davis, 1990). Our results demonstrate that binding of Rad51 protein activates preferentially the 5′–end of ssDNA for homologous pairing. By inference, we suggest that the eukaryotic Rad51 protein may assemble on ssDNA with a 3′→5′ polarity of polymerization. Such a directionality would result in a higher probability of assembly at the 5′–end of linear ssDNA and, hence, in the observed pairing bias. Although additional physical evidence will be required to affirm our hypothesis, recent observations with human Rad51 protein clearly establish that nucleoprotein filament formation protects the 5′–end of ssDNA better than the 3′–end against exonuclease degradation (Gupta et al., 1998); this result is opposite to that seen for RecA protein (Konforti and Davis, 1991), and is consistent with our interpretation.
Since the polarity of RecA protein polymerization on ssDNA is the same as the polarity of DNA heteroduplex extension during DNA strand exchange (Konforti and Davis, 1992), one might expect that Rad51 protein and RecA protein should also have opposite polarities of DNA heteroduplex extension. However, although the 5′→3′ polarity of DNA heteroduplex extension is clear for RecA protein (Cox and Lehman, 1981a), the situation for Rad51 protein is quite complicated. It was found that Rad51 protein-promoted DNA strand exchange can initiate at either end of dsDNA, provided that this end has a short ssDNA overhang, and that it can proceed in either direction (Namsaraev and Berg, 1998). Although this finding seems to highlight an important difference in behavior between RecA and Rad51 proteins, it should be noted that none of the published studies on DNA heteroduplex extension by Rad51 protein demonstrate that the extension phase is in fact protein mediated, as was done for RecA protein (Cox and Lehman, 1981b). Thus, rather than promoting DNA heteroduplex formation in either direction, Rad51 protein may simply promote joint molecule formation at either end of the linear dsDNA substrate. The subsequent extension of the DNA heteroduplex region could be entirely thermal in nature. The apparent directionality could arise from a bias introduced by the presence of Rad51 protein bound to the nascent region of DNA heteroduplex, forcing thermal branch migration in the opposite direction. Recently, it was also reported that Rad51 protein promotes extension of DNA heteroduplex with the same polarity as RecA protein (Gupta et al., 1998). However, the experimental system did not necessarily distinguish between the effects of preferential invasiveness of the ssDNA end and the polarity of DNA branch migration. In fact, the bias observed in that work can be explained by the interpretation for Rad51 protein that we offer here.
In summary, we found that for the eukaryotic DNA pairing proteins, the 5′–end of ssDNA is more invasive than the 3′–end. This is in contrast with the behavior of the prokaryotic homolog, RecA protein, which shows an opposite bias. This finding poses a bit of a paradox, since 3′–ssDNA tailed duplex DNA is a well documented physical intermediate of DSB repair (Cao et al., 1990). We offer two simple considerations that help to resolve this potential problem. The first is that, as we established here, the presence of a dsDNA region attached to the ssDNA enhances its reactivity to a degree that the distinction between 5′- and 3′–ends is minimized. Although the precise molecular details are not yet known, the presence of a ssDNA–dsDNA junction apparently stabilizes the nucleoprotein complex. Our results raise the possibility that the ssDNA–dsDNA junctions can alter the assembly or stability of the Rad51 nucleoprotein filament to prompt additional polymerization toward the 3′–end of ssDNA. The second consideration is that within the cell, the binding of Rad51 protein to ssDNA may be initiated by some as yet unknown protein(s) that binds to the 3′–end of ssDNA and promotes the loading of Rad51 protein onto ssDNA presumably via protein–protein interactions; in general terms, this possibility has as a prototype, which is the loading of RecA protein onto ssDNA by RecBCD enzyme (Anderson and Kowalczykowski, 1997). To form a contiguous Rad51 nucleoprotein filament on ssDNA starting from the 3′–end, this hypothetical loading mechanism would, in fact, require a 3′→5′ polarity of Rad51 protein polymerization on ssDNA, a property that is consistent with our results. Although further work is needed to refine our understanding of this process, it is clear that the behavior of the Rad51 family of proteins is unique with regard to its DNA pairing bias.
Materials and methods
Proteins and DNA
Escherichia coli RecA, single-stranded DNA-binding (SSB) protein, S.cerevisiae and human Rad51 proteins and RPA, were purified as described (LeBowitz, 1985; Sugiyama et al., 1997; P.Sung, unpublished). The oligonucleotides used in this study were: #1, 63mer, ACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA; #2, 63mer, TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGT; #5, 32mer, CCATCCGCAAAAATGACCTCTTATCAAAAGGA; #6, 32mer, TCCTTTTGATAAGAGGTCATTTTTGCGGATGG; #41, 63mer, TCTGCTCCTGTTTCTGCTTTTTTGCTCTTTGCCTTCCGCTTTTTTGTCCTCTTTTCTTTTGGT; #45, 31mer ACAGCACCAGATTCAGCAATTAAGCTCTAAG; #49, 63mer, ACCAAAAGAAAAGAGGACAAAAAAGCGGAAGGCAAAGAGCAAAAAAGCAGAAACAGGAGCAGA; #55, 31mer, CTTAGAGCTTAATTGCTGAATCTGGTGCTGT; #58, 63mer, CCATCCGCAAAAATGACCTCTTATCAAAAGGAACAGCACCAGATTCAGCAATTAAGCTCTAAG; SK#3, 90mer, AATTCTCATTTTACTTACCGGACGCTATTAGCAGTGGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGC- ACAGATGCGT; SK#5, 90mer, CGCGTGTCGGGGGTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTTAGAAGCGTTATTCTCTGGGCGAATAGAGTGCTATC. Oligonucleotides were purified as described (Mazin and Kowalczykowski, 1996). The concentrations of the oligonucleotides were determined spectrophotometrically using an extinction coefficient (ɛ260) of 9833 M–1cm–1 (1 OD260 = 33 μg/ml). DNA concentrations are expressed in moles of nucleotides. Both oligonucleotide labeling and dsDNA preparation were as described (Sambrook et al., 1989). Oligonucleotides were stored at –20°C.
DNA strand exchange
Rad51 nucleoprotein filaments were formed by incubation of human or yeast Rad51 protein (4 μM) with ssDNA (12 μM) or with an equivalent molar (molecule) amount of tailed dsDNA in the standard buffer containing 33 mM HEPES pH 7.0, 2 mM dithiothreitol (DTT), 100 μg/ml bovine serum albumin, 2 mM ATP, 1.2 mM magnesium acetate at 37°C for 5 min, followed by an increase in the magnesium acetate concentration to 20 mM, and an additional 5 min incubation. When indicated, ATP was substituted with 1 mM ATPγS. Human or yeast RPA proteins (0.5 μM) were added to the corresponding nucleoprotein filaments and incubation was continued for another 5 min. RecA nucleoprotein filaments were assembled at slightly different optimal conditions by incubating RecA protein (4 μM) with ssDNA (12 μM) in standard buffer containing 11.2 mM magnesium acetate, 2 mM DTT, 100 μg/ml bovine serum albumin, 1 mM ATP, 3 mM phosphoenolpyruvate, 10 U/ml pyruvate kinase and 1 μM SSB protein at 37°C for 12 min. DNA strand exchange was initiated by addition of homologous dsDNA to the nucleoprotein filaments. Aliquots were withdrawn from the reaction mixture, deproteinized by the addition of EDTA to 50 mM, SDS to 1% and proteinase K to 500 μg/ml followed by incubation for 5 min at 37°C, mixed with a 1/10 vol of loading buffer (20% ficoll, 0.1% bromophenol blue), and loaded onto a 10% polyacrylamide gel.
Products of DNA strand exchange were quantitated using a Storm 840 PhosphorImager (Molecular Dynamics).
D–loop assay
To form Rad51 nucleoprotein filaments, yeast Rad51 protein (0.3 μM) was incubated with ssDNA (0.9 μM) in buffer containing 25 mM Tris–acetate pH 7.5, 10 mM magnesium acetate, 1 mM DTT, 2 mM ATP, 3 mM phosphoenolpyruvate, 20 U/ml pyruvate kinase, 100 μg/ml bovine serum albumin at 37°C for 5 min. Unless otherwise indicated, RPA protein (0.11 μM) was added to the Rad51 nucleoprotein filaments and incubation was continued for another 3 min. Where indicated, Rad54 protein (0.23 μM) (a generous gift of W.-D.Heyer, University of California, Davis) and Rad52 protein (0.04–0.32 μM) were added to the reaction mixtures. In reactions containing the Rad55–Rad57 heterodimer, presynaptic filaments were formed by incubating ssDNA (SK#3 or SK#5) first with RPA (0.04 μM) for 1 min followed by addition of Rad51 protein (0.3 μM) and the Rad55–Rad57 heterodimer (0.04 μM). After 10 min of incubation, Rad54 protein (0.23 μM) was added to increase the yield of joint molecules (when Rad54 protein was omitted, the yield of D–loops was too low to permit accurate measurement). In control experiments, both RPA and Rad51 protein were omitted and Rad54 protein was added to ssDNA prior to addition of dsDNA. RecA nucleoprotein filaments were assembled by incubation of RecA protein (0.3 μM) with ssDNA (0.9 μM) at 37°C for 5 min in the same buffer except that the ATP concentration was 1 mM and both phosphoenolpyruvate and pyruvate kinase were omitted. SSB protein (0.09 μM) was then added to the RecA nucleoprotein filaments followed by incubation for 3 min. Pairing reactions were initiated by addition of pUC19 supercoiled plasmid dsDNA (9 μM) to the nucleoprotein filaments. Aliquots were withdrawn from the reaction mixture, deproteinized by the addition of EDTA to 50 mM, SDS to 1% and proteinase K to 500 μg/ml followed by incubation for 5 min at 37°C, mixed with a 1/10 vol of loading buffer (20% ficoll, 0.1% bromophenol blue), and loaded onto a 1% agarose gel. Joint molecule formation was quantitated using a Storm 840 PhosphorImager (Molecular Dynamics). Since the ssDNA is in excess, the yield of joint molecule products is expressed as a percentage of the limiting plasmid DNA concentration.
Binding of Rad51 protein to DNA
Yeast Rad51 protein was incubated with DNA in buffer containing 33 mM HEPES pH 7.0, 20 mM magnesium acetate, 2 mM DTT, 100 μg/ml bovine serum albumin and 3 mM ATP at 37°C for 15 min. Where indicated, ATP was substituted with 1 mM ATPγS. Rad51 nucleoprotein complexes were mixed with a 1/10 vol of loading buffer (30% glycerol, 0.1% bromophenol blue), loaded onto a 10% polyacrylamide gel, and subjected to electrophoresis at 9 V/cm for 2 h in TBE (90 mM Tris–borate pH 8.3 and 0.5 mM EDTA) (Mazin and Kowalczykowski, 1996). Complexes were detected by a shift of mobility and were quantitated using a Storm 840 PhosphorImager (Molecular Dynamics).
Acknowledgments
Acknowledgements
We are grateful to Wolf Heyer and Andrei Kuzminov for their comments on the manuscript. We would like to thank the following members of the laboratory for both discussion of our results and critical reading of the manuscript: Dan Anderson, Piero Bianco, Joel Brockman, Frederick Chedin, Deana Haddox, Frank Harmon, Noriko Kentake, Julie Kleiman, James New, Erica Seitz, Tomohiko Sugiyama and Eugene Zaitsev. A.V.M. is grateful to the Institute of Cytology and Genetics, Russian Academy of Science, Novosibirsk, for granting him a leave of absence. This work was supported by grants from the NIH (AI-18987) and from the Human Frontiers Science Program (RG63) to S.C.K.
References
- Anderson D.G. and Kowalczykowski, S.C. (1997) The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell, 90, 77–86. [DOI] [PubMed] [Google Scholar]
- Baumann P. and West, S.C. (1999) Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J. Mol. Biol., 291, 363–374. [DOI] [PubMed] [Google Scholar]
- Benson F.E., Stasiak, A. and West, S.C. (1994) Purification and characterization of the human Rad51 protein, an analogue of E.coli RecA. EMBO J., 13, 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F.E., Baumann, P. and West, S.C. (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani, E. and Kleckner, N. (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S.cerevisiae. Cell, 61, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Cox M.M. and Lehman, I.R. (1981a) Directionality and polarity in recA protein-promoted branch migration. Proc. Natl Acad. Sci. USA, 78, 6018–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.M. and Lehman, I.R. (1981b) RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc. Natl Acad. Sci. USA, 78, 3433–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S.L., Wong, A.K., Kora, Y., Shinohara, A. and Bishop, D.K. (1998) Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev., 12, 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.C., Golub, E.I., Wold, M.S. and Radding, C.M. (1998) Polarity of DNA strand exchange promoted by recombination proteins of the RecA family. Proc. Natl Acad. Sci. USA, 95, 9843–9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. (1995) In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. BioEssays, 17, 609–620. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1997) A super new twist on the initiation of meiotic recombination. Cell, 89, 163–166. [DOI] [PubMed] [Google Scholar]
- Kogoma T. (1996) Recombination by replication. Cell, 85, 625–627. [DOI] [PubMed] [Google Scholar]
- Konforti B.B. and Davis, R.W. (1990) The preference for a 3′ homologous end is intrinsic to RecA-promoted strand exchange. J. Biol. Chem., 265, 6916–6920. [PubMed] [Google Scholar]
- Konforti B.B. and Davis, R.W. (1991) DNA substrate requirements for stable joint molecule formation by the RecA and single-stranded DNA-binding proteins of Escherichia coli. J. Biol. Chem., 266, 10112–10121. [PubMed] [Google Scholar]
- Konforti B.B. and Davis, R.W. (1992) ATP hydrolysis and the displaced strand are two factors that determine the polarity of RecA-promoted DNA strand exchange. J. Mol. Biol., 227, 38–53. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. (1985) Biochemical mechanism of strand initiation in bacteriophage λ DNA replication. PhD thesis, Johns Hopkins University, Baltimore, MD. [Google Scholar]
- Mazin A.V. and Kowalczykowski, S.C. (1996) The specificity of the secondary DNA binding site of RecA protein defines its role in DNA strand exchange. Proc. Natl Acad. Sci. USA, 93, 10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A.V. and Kowalczykowski, S.C. (1998) The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J., 17, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namsaraev E.A. and Berg, P. (1998) Branch migration during Rad51-promoted strand exchange proceeds in either direction. Proc. Natl Acad. Sci. USA, 95, 10477–10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J.H., Sugiyama, T., Zaitseva, E. and Kowalczykowski, S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu, X., Shinohara, A. and Egelman, E.H. (1993) Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science, 259, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Petukhova G., Stratton, S. and Sung, P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- Register J.C. III and Griffith, J. (1985) The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem., 260, 12308–12312. [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shinohara A. and Ogawa, T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- Stasiak A. and Di Capua, E. (1982) The helicity of DNA in complexes with recA protein. Nature, 299, 185–186. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Zaitseva, E.M. and Kowalczykowski, S.C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem., 272, 7940–7945. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco, D. and Szostak, J.W. (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell, 64, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997a) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997b) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- Sung P. and Robberson, D.L. (1995) DNA strand exchange mediated by a RAD51–ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–461. [DOI] [PubMed] [Google Scholar]
- Sung P. and Stratton, S.A. (1996) Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem., 271, 27983–27986. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver, T.L., Rothstein, R.J. and Stahl, F.W. (1983) The double-strand break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Tracy R.B., Baumohl, J.K. and Kowalczykowski, S.C. (1997) The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev., 11, 3423–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]