Metabolic Analysis of Escherichia coli in the Presence and Absence of the Carboxylating Enzymes Phosphoenolpyruvate Carboxylase and Pyruvate Carboxylase (original) (raw)
Abstract
Fermentation patterns of Escherichia coli with and without the phosphoenolpyruvate carboxylase (PPC) and pyruvate carboxylase (PYC) enzymes were compared under anaerobic conditions with glucose as a carbon source. Time profiles of glucose and fermentation product concentrations were determined and used to calculate metabolic fluxes through central carbon pathways during exponential cell growth. The presence of the Rhizobium etli pyc gene in E. coli (JCL1242/pTrc99A-pyc) restored the succinate producing ability of E. coli ppc null mutants (JCL1242), with PYC competing favorably with both pyruvate formate lyase and lactate dehydrogenase. Succinate formation was slightly greater by JCL1242/pTrc99A-pyc than by cells which overproduced PPC (JCL1242/pPC201, ppc+), even though PPC activity in cell extracts of JCL1242/pPC201 (ppc+) was 40-fold greater than PYC activity in extracts of JCL1242/pTrc99a-pyc. Flux calculations indicate that during anaerobic metabolism the pyc+ strain had a 34% greater specific glucose consumption rate, a 37% greater specific rate of ATP formation, and a 6% greater specific growth rate compared to the ppc+ strain. In light of the important position of pyruvate at the juncture of NADH-generating pathways and NADH-dissimilating branches, the results show that when PPC or PYC is expressed, the metabolic network adapts by altering the flux to lactate and the molar ratio of ethanol to acetate formation.
In mixed-acid fermentation of glucose, succinate is formed via the reductive arm of the tricarboxylic acid cycle, a pathway which includes the fixation of 1 mol of carbon dioxide per mol of succinate generated. The key reaction in this sequence is the carboxylation of three-carbon intermediates such as phosphoenolpyruvate (PEP) to four-carbon oxaloacetate. The principal PEP-carboxylating enzyme found in Escherichia coli is PEP carboxylase (PPC). In E. coli PEP may also be converted to pyruvate, which during anaerobic growth leads to the formation of lactate, formate, acetate, and ethanol. In other prokaryotes and many eukaryotes during glucose metabolism, oxaloacetate is synthesized by carboxylation of pyruvate by pyruvate carboxylase (PYC) (3, 24, 25), an enzyme that is absent in E. coli. PEP is also required for glucose consumption via the PEP-phosphotransferase system (PEP-PTS) and for the synthesis of aromatic amino acids (7, 14). Because of its central position in glucose metabolism, PEP partitioning is highly regulated by cellular mechanisms.
In order to affect the metabolic rigidity of the biochemical network at the PEP branch point, several metabolic engineering approaches have been proposed. As one would expect, increased succinate production has been shown to result from overexpression of PPC from E. coli (20) or overexpression of PYC via the Rhizobium etli pyc gene (13). Similarly, the expression of malic enzyme in E. coli strains lacking the enzymes pyruvate formate lyase (PFL) and lactate dehydrogenase (LDH) yielded succinate as the major fermentation product (28). Each of these genetic perturbations directly affects the central metabolic network and therefore impacts the carbon flow through the metabolic branches. Prior metabolic engineering efforts to affect succinate production have not included detailed flux analysis. Such changes in metabolic fluxes could be determined using flux analysis methodologies (8, 26, 33, 34). Flux analysis of a metabolic system typically involves calculation of the intracellular fluxes based on measured excretion fluxes and the stoichiometry of the reactions involved in the metabolic network. Previous studies have applied this technique to a wide variety of fermentations (1, 8, 12, 22, 26, 33, 34, 36).
In order to improve our understanding of anaerobic succinate production in E. coli, we analyzed carbon flux distributions in response to genetic perturbations affecting the activities of PPC and PYC. In this study, we investigated the metabolic shifts in E. coli resulting from a null mutation in the ppc gene, overexpression of PPC and overexpression of PYC.
MATERIALS AND METHODS
Strains and plasmids.
E. coli strains VJS676 [F− λ− Δ(argF-lac)U169] (provided by V. J. Stewart, Cornell University) and JCL1242 [F− λ− Δ(argF-lac)U169 ppc:Km] (6) were used in this study. Strain JCL1242 is a derivative of VJS676 and has a null mutation in the ppc gene. The plasmids used in this study are shown in Table 1. The native E. coli ppc gene was expressed using the pPC201 plasmid, in which the expression of ppc is under the control of the artificial tac promoter (6). The pyc gene from R. etli (9) was expressed using the pTrc99A-pyc plasmid, in which the expression of pyc is controlled by the artificial trc promoter. Since both the pPC201 and pTrc99A-pyc plasmids contained the lac operator, expression of the ppc and pyc genes was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). Plasmids pJF118EH (11) and pTrc99A (2), the parental vectors of pPC201 and pTrc99A-pyc, were used as controls.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference |
|---|---|---|
| pPC201 | E. coli ppc bla lacI_q_tac ColE1 | 7 |
| pJF118EH | _bla lacI_q tac ColE1 | 11 |
| pTrc99A | _bla lacI_q trc ColE1 | 2 |
| pTrc99A-pyc | _R. etli pyc bla lacI_q trc ColE1 | Laboratory plasmid |
Medium and fermentation conditions.
Fermentations (2.0 liters in volume) were carried out in 2.5-liter BioFlo III bench top fermentors (New Brunswick Scientific, Edison, N.J.). The medium contained the following (in grams per liter): Luria-Bertani Miller broth, 25; glucose, 10; Na2HPO4 · 7H2O, 3; KH2PO4, 1.5; NH4Cl, 1; MgSO4 · 7H2O, 0.25; and CaCl2 · 2H2O, 0.02. Inocula for each fermentation were started from a single colony grown on a Luria-Bertani–glucose plate. A 3-ml aerobic culture grown 6 to 8 h was transferred into 50 ml of fresh medium prepared anaerobically under an atmosphere of carbon dioxide. This culture was grown 12 h in sealed serum bottles at 37°C, and 20 ml was used to inoculate a fermentor. Each fermentor operated at 150 rpm, 0% oxygen saturation (as determined with a Polarographic oxygen sensor [Mettler-Toledo Process Analytical, Inc., Wilmington, Mass.]), 37°C, and a constant pH of 7.0, which was maintained by the automatic addition of 2 M Na2CO3. Anaerobic conditions were maintained by flushing the fermentor headspace with oxygen-free carbon dioxide. To maintain initial selective pressure for strains carrying plasmids, media were supplemented initially with 100 μg of ampicillin per ml. The induction of the ppc or the pyc gene in the E. coli strains was achieved by adding 1 mM IPTG. The fermentation of each strain was performed in duplicate, and statistical significance was determined by using Student's t test.
Analytical methods.
During the course of a fermentation, samples were anaerobically withdrawn at regular intervals for measurement of glucose, products, and biomass concentrations. Cell growth was monitored by measuring the optical density (with a DU-650 spectrophotometer [Beckman Instruments, San Jose, Calif.]) at 600 nm, and this measurement was used to correlate with the dry cell concentration by using the following equation: dry cell concentration (in grams per liter) = 0.48 × optical density. A portion of each sample was centrifuged (8,000 × g for 15 min at 4°C), and the supernatant was stored at −20°C for subsequent chromatographic analyses.
Glucose and fermentation products were analyzed by high-pressure liquid chromatography as previously described (10), with a Coregel 64-H ion-exclusion column (Interactive Chromatography, San Jose, Calif.). Glucose, succinate, lactate, acetate, formate, and ethanol were simultaneously detected with a differential refractive index detector (model 410; Waters, Milford, Mass.).
Enzyme assays.
Cell-free extracts of E. coli strains were prepared by first withdrawing 50 ml of mid-log-phase culture from the fermentor and then harvesting the cells by centrifugation (8,000 × g for 15 min at 4°C). Cells were washed with 10 ml of 100 mM Tris-HCl buffer (pH 8.0) and then resuspended in 2 ml of the same buffer. Cell disruption was achieved by sonication (with a Sonifier II apparatus [Branson Ultrasonics, Danbury, Conn.]), and cell debris were removed by centrifugation at 4°C (20,000 × g for 20 min). The supernatant was stored on ice until further use. The total protein concentration of the cell extract was determined using bovine serum albumin as the standard (18).
The activities of PYC, PPC, and LDH were spectrophotometrically measured at 37°C. PYC activity was measured by the method of Payne and Morris (24), in which the oxaloacetate produced by PYC is converted to citrate by adding citrate synthase in the presence of acetyl coenzyme A (acetyl-CoA) and 5,5′-dithio-bis(2-nitrobenzoate). The rate of increase in absorbance at 412 nm due to the presence of CoA-dependent formation of the thionitrobenzoate was monitored, first after the addition of pyruvate and then after the addition of ATP. The difference between these two rates was taken as the ATP-dependent PYC activity. To determine the saturation constant of PYC, we performed initial rate studies using the same assay but varied the pyruvate concentration (0.0 to 6.0 mM) while maintaining other substrates in excess (5.0 mM ATP, 50 mM HCO3−, and 5.0 mM Mg2+). The value for the pyruvate saturation constant (Km) was determined from a Lineweaver-Burk plot. PPC activity was assayed by monitoring the decrease in absorbance of NADH at 340 nm using malate dehydrogenase as a coupling enzyme (31). LDH activity was also measured by monitoring the disappearance of NADH at 340 nm (5).
Intracellular flux determination.
The methodology followed in this study to calculate intracellular fluxes has been detailed elsewhere (8, 23, 34). Molar balance equations were formulated from the biochemical pathways of E. coli shown in Fig. 1. The PEP carboxylation flux (_J_5) was fixed to zero for the strains which lacked PPC, while the pyruvate carboxylation flux (_J_PYC) was fixed to zero for the strains which lacked PYC. The resulting networks for the E. coli strains (details are given in the Appendix) were usually composed of nine fluxes, except for JCL1242, for which _J_11 and _J_PYC were both equal to zero and the number of fluxes was eight. Also, for each of the strains, balance equations were written for each of the five biochemical intermediates or nodes. For example, for the acetyl-CoA node, the balance equation was as follows: rate of accumulation of acetyl-CoA = _J_7 − _J_8 − _J_9. Application of the pseudo-steady-state assumption permitted all of the accumulation terms to be assigned a value of zero, yielding five linear equations with nine fluxes (eight for JCL1242). Those six fluxes (five for JCL1242) which involved glucose or the measured products (_J_1, _J_5 or _J_PYC, _J_6, _J_7, _J_8, and _J_9) were directly calculated from experimental results. These fluxes were calculated by dividing the net change in the concentration of the metabolite during the exponential growth phase (about 4 to 7 h) by the duration of that phase, providing units of millimoles per liter · hour. For the calculation of specific rates, such as the specific glucose consumption rate, these volumetric rates were divided by the average biomass concentration during the same time interval. For the calculation of flux values, these volumetric rates were normalized by the specific glucose consumption rate (so that _J_1 = 100). Each system of five equations ultimately contained three unknown fluxes (_J_2, _J_3, and _J_4), which caused each to be mathematically overdetermined. The least-squares estimates for both the measured and unknown fluxes were calculated using the method of Tsai and Lee (32).
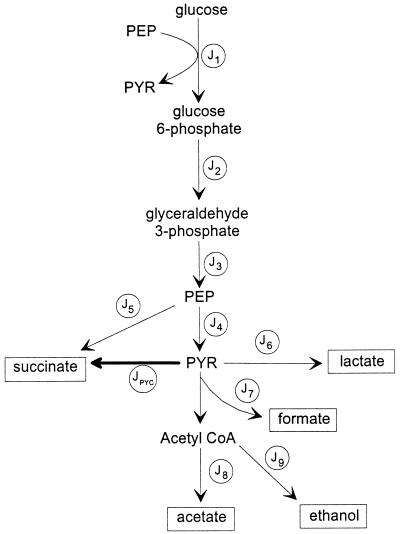
FIG. 1.
Fermentative pathways of E. coli grown in a glucose-limited rich medium. This figure depicts the principal branches of glucose metabolism by E. coli under anaerobic conditions. PYR, pyruvate.
RESULTS
Fermentations of strains with and without two carboxylating enzymes.
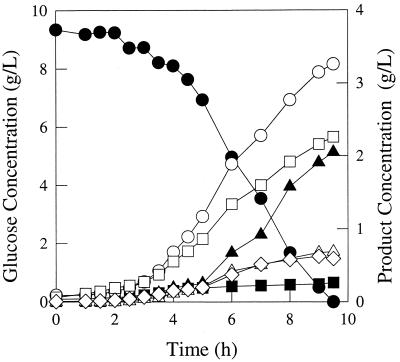
Anaerobic fermentations were performed under controlled conditions in order to assess the metabolic consequences of a null mutation in the ppc gene and/or of the expression of ppc or pyc on overall growth, glucose consumption, and formation of products. Representative fermentation time profiles for the E. coli strains VJS676 (which contains native ppc), JCL1242 (ppc null mutant), JCL1242/pPC201 (ppc+), and JCL1242/pTrc99A-pyc are shown in Fig. 2 to 5, respectively. (Similar fermentation results with JCL1242 carrying the control plasmids pJF118EH or pTrc99A are not shown.) Several significant results can readily be noted from these fermentations. Compared with the strain having a single chromosomal copy of ppc (VJS676), the fermentation with the strain having a null mutation in the ppc gene (JCL1242) resulted in a significant decrease in the final succinate concentration (1.0 g/liter versus 0.2 g/liter; P < 0.001). The small quantity of succinate detected with the three ppc mutant strains may be a consequence of the presence of aspartate and glutamate in the rich medium. The observed decrease in succinate formation in the ppc mutant strains was accompanied by an increase in lactate production (P < 0.10). As shown in Fig. 4 and 5, expression of either PPC or PYC in JCL1242 led to a significant decrease in lactate production compared to that in the ppc mutant JCL1242 (P < 0.05).
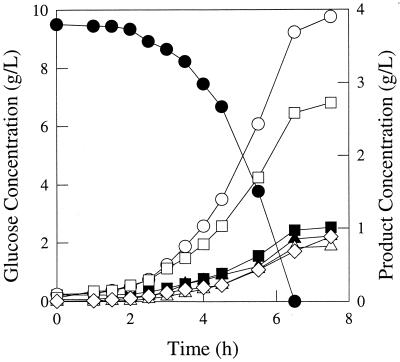
FIG. 2.
Fermentation pattern of E. coli strain VJS676 growing in glucose-limited rich media. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol; ◊, biomass.
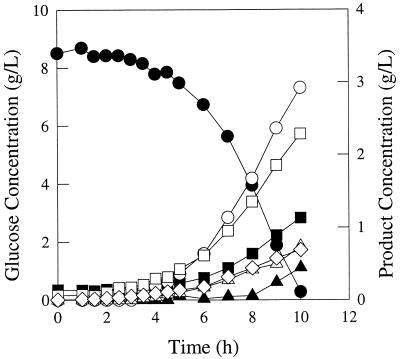
FIG. 5.
Fermentation pattern of E. coli strain JCL1242/pTrc99A-pyc growing in glucose-limited rich medium. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol; ◊, biomass.
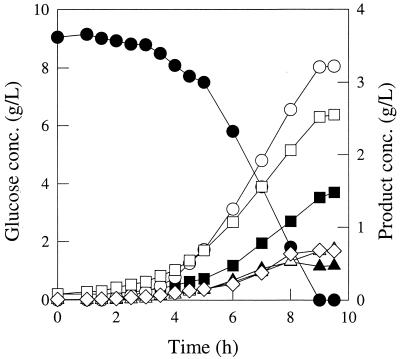
FIG. 4.
Fermentation pattern of E. coli strain JCL1242/pPC201 (ppc+) growing in glucose-limited rich medium. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol, and ◊, biomass.
The average product yields for the four principal strains (Table 2) indicate the overall stoichiometric differences in the carbon metabolism of these strains. Significant differences in succinate and lactate yields occur among these strains. In the wild-type strain VJS676, the succinate yield was 0.10 g/g of glucose consumed. The absence of PPC significantly decreased the succinate yield (P < 0.001). Succinate formation was restored in JCL1242 by the expression of either PPC or PYC. Indeed, the fermentations using JCL1242/pPC201 (ppc+) resulted in a succinate yield which surpassed by 44% that of fermentations using VJS676. The pyc gene from R. etli was similarly able to compensate for the lack of PPC activity. Fermentations using JCL1242/pTrc99A-pyc resulted in a 66% increase in succinate yield compared to fermentations using VJS676. The strain JCL1242 with the pyc gene showed a succinate yield 19% greater than that of the same strain with multiple copies of the ppc gene (P < 0.01). Lactate was also sensitive to the absence of PPC activity, with the lactate yield being significantly higher in JCL1242 than in any of the strains with PPC or PYC activity (P < 0.001). Carbon recovery based on glucose consumed was calculated from final molar product concentrations, including the carbon dioxide fixation and evolution steps but excluding biomass synthesis. That the carbon recovery was close to 100% for each of the fermentations suggests that biomass was largely derived from the precursor metabolites, such as amino acids, fatty acids, and amino sugars available in the rich fermentation medium.
TABLE 2.
Product yields and carbon recovery in fermentations using E. coli strainsa
| Strain | Yield (SD) | Carbon recovery (SD) | ||||
|---|---|---|---|---|---|---|
| Succinate | Lactate | Formate | Acetate | Ethanol | ||
| VJS676 | 0.10 (0.00) | 0.09 (0.01) | 0.40 (0.00) | 0.28 (0.00) | 0.20 (0.01) | 0.99 (0.02) |
| JCL1242 | 0.03 (0.00) | 0.21 (0.00) | 0.35 (0.01) | 0.24 (0.01) | 0.19 (0.01) | 0.97 (0.03) |
| JCL1242/pPC201 (ppc+) | 0.12 (0.00) | 0.04 (0.01) | 0.35 (0.01) | 0.27 (0.01) | 0.20 (0.03) | 0.93 (0.09) |
| JCL1242/pTrc99A-pyc | 0.15 (0.00) | 0.08 (0.05) | 0.33 (0.04) | 0.26 (0.01) | 0.18 (0.01) | 0.94 (0.00) |
From cell extracts of each of the six strains, the activities of PPC, PYC, and LDH were determined (Table 3). These measurements indicate that the expression of PPC in JCL1242/pPC201 (ppc+) resulted in about a 25-fold increase in PPC activity over that in strain VJS676. As expected, no endogenous PYC activity was observed in E. coli. Also, the cell extracts from strains in which both PPC and PYC were absent exhibited the greatest LDH activity.
TABLE 3.
Activities of enzymes in cell extracts of E. coli strains during exponential growtha
| Strain | Activity (μmol/mg of protein · min) of: | ||
|---|---|---|---|
| PPC | PYC | LDH | |
| VJS676 | 0.05 | 0.00 | 0.42 |
| JCL1242 | 0.00 | 0.00 | 0.67 |
| JCL1242/pJF118EH | 0.00 | 0.00 | 0.61 |
| JCL1242/pTrc99A | 0.00 | 0.00 | 0.56 |
| JCL1242/pPC201 (ppc+) | 1.25 | 0.00 | 0.42 |
| JCL1242/pTrc99A-pyc | 0.00 | 0.03 | 0.23 |
Flux analysis.
Metabolic flux analysis is now routinely used to gain insight into metabolic changes caused by genetic perturbations or changes in physiological states. The carbon recovery from fermentations of the strains used in this study (Table 2) demonstrated that glucose was not used in biomass synthesis. Therefore, carbon flux from glucose to biomass was excluded from the analysis, and the role of the pentose pathway was assumed to be negligible (14). Also, succinate formation via the glyoxylate bypass was neglected, since in the presence of glucose and under anaerobic conditions the transcription of glyoxylate bypass genes is repressed (16, 19). Since the observed carbon recoveries were about 100%, decomposition of formate to hydrogen and carbon dioxide was also neglected. The biochemical network used for the flux analysis is shown in Fig. 1.
The results of the flux analysis are summarized in Table 4, showing fluxes during the exponential growth phase normalized with respect to glucose flux (i.e., _J_1 = 100). The genetic perturbations that were studied occurred below PEP in the pathway, and therefore the normalized fluxes before PEP (_J_1 through _J_3) were very similar in each of the strains. The flux from PEP toward pyruvate via pyruvate kinase (_J_4) was affected by the presence or absence of PPC activity, with greater fluxes naturally observed in strains without PPC activity (JCL1242 and JCL1242/pTrc99A-pyc). In JCL1242, this relatively high flux to pyruvate was accompanied by an increased flux to lactate (_J_6). In JCL1242/pTrc99A-pyc, the increased carbon flowing to pyruvate was diverted about equally between oxaloacetate (_J_PYC) and lactate.
TABLE 4.
Flux distributions from fermentations of E. coli strains during exponential growth on glucose and rich mediuma
| Parameter | Flux (SD) in strain: | |||
|---|---|---|---|---|
| VJS676 | JCL1242 | JCL1242/pPC201 (ppc+) | JCL1242/pTrc99A-pyc | |
| _J_1 | 100 | 100 | 100 | 100 |
| _J_2 | 88 (1) | 96 (2) | 92 (1) | 94 (1) |
| _J_3 | 169 (1) | 188 (3) | 180 (2) | 184 (10) |
| _J_4 | 51 (1) | 80 (5) | 59 (1) | 81 (12) |
| _J_5 | 11 (0) | 0 | 17 (1) | 0 |
| _J_6 | 14 (6) | 32 (1) | 2.7 (3.1) | 20 (11) |
| _J_7 | 131 (7) | 148 (2) | 153 (1) | 137 (4) |
| _J_8 | 67 (4) | 68 (5) | 73 (3) | 70 (1) |
| _J_9 | 64 (3) | 81 (3) | 79 (3) | 67 (5) |
| _J_PYC | 0 | 0 | 0 | 21 (1) |
Comparison of specific growth rates and glucose consumption rates.
The flux analysis was performed during exponential growth (approximately 4 to 7 h for these fermentations). We calculated specific growth rates and specific glucose consumption rates for each of the four strains during that same time interval (Table 5), but these calculations do not rely on the flux model. Both of these rates were affected by the genetic perturbations studied. Fermentations with VJS676 yielded the greatest specific growth rates and specific glucose consumption rates. Compared to JCL1242, the growth rate of JCL1242/pPC201 (ppc+) was unchanged. However, the growth rate of JCL1242/pTrc99A-pyc was 6% greater than the growth rate of JCL1242/pPC201 (ppc+) (P < 0.0025). Compared to JCL1242, the strain with PPC activity (JCL1242/pPC201) showed a decreased specific glucose consumption rate (P < 0.01). In contrast, overexpression of PYC increased the specific glucose consumption rate to 15.7 mmol/g of cells · h, which is significantly greater than the values observed in either JCL1242 or JCL1242/pPC201 (ppc+) (P < 0.025).
TABLE 5.
Metabolic data from fermentations of E. coli strains during exponential growth on glucose and rich mediuma
| Parametera | Value for strain (SD): | |||
|---|---|---|---|---|
| VJS676 | JCL1242 | JCL1242/pPC201 (ppc+) | JCL1242/pTrc99A-pyc | |
| qS | 23.2 (2.1) | 13.6 (0.0) | 11.7 (0.1) | 15.7 (0.1) |
| μ | 0.509 (0.003) | 0.405 (0.018) | 0.399 (0.002) | 0.424 (0.001) |
| R/Ob | 1.02 (0.00) | 0.98 (0.02) | 0.92 (0.01) | 0.95 (0.05) |
| _q_ATPb | 48.9 (2.9) | 32.6 (1.1) | 27.7 (0.6) | 37.9 (1.1) |
| _Y_ATPb | 2.11 (0.07) | 2.39 (0.09) | 2.38 (0.05) | 2.42 (0.17) |
Redox balance.
Flux analysis permits the estimation of several key fermentation parameters during the exponential growth phase. The selected flux network and model should result in a redox balance. The redox balance (R/O) can be calculated by dividing the flux which generates NADH by the sum of all the fluxes which generate NAD (and FAD). For the current flux model, the redox balance is calculated as follows: R/O = _J_3/(_J_6 + 2_J_5 + 2_J_PYC + 2_J_9). Because the redox balance is a ratio, the value of R/O may be calculated by the normalized fluxes shown in Table 4. The results of these calculations (Table 5) indicate that the redox balance is close to 1 on the basis of the flux model for fermentations of the four principal strains.
Rate of ATP formation and ATP yield.
During anaerobic glucose metabolism, ATP is consumed by some reactions and generated by others. Since the flux model includes these reactions, it may be used to calculate the specific rates of ATP formation due to glucose metabolism (_q_ATP) using non-normalized fluxes. That is, the fluxes as scaled to _J_1 = 100 must be multiplied by the factor q S/100 to result in units of millimoles of ATP formed per gram of cells · hour. So, the values of _q_ATP may be calculated for each strain by the following equation: _q_ATP = (_J_3 + _J_4 + _J_5 + _J_8 − _J_2)(q S/100). This equation includes 1 mol of ATP generated by electron transport phosphorylation during the reduction of 1 mol of fumarate to succinate. Therefore, the flux from pyruvate to succinate via PYC has no net generation or consumption of ATP, while the flux from PEP to succinate via PPC has a net generation of 1 mol of ATP per mol of succinate formed. Another important measurement of ATP is the ATP yield, which is the number of moles of ATP formed during exponential growth by glucose metabolism per mole of glucose consumed. The value of ATP yield may be calculated by dividing _q_ATP by qS.
We calculated the specific formation of ATP and the ATP yield for each of the four strains during the same time interval as the flux analysis (Table 5). The parent strain VJS676, which had the least genetic perturbation, had the greatest rate of ATP formation (48.9 mmol/g of cells · h). Compared to JCL1242, the strain with PPC activity (JCL1242/pPC201) showed a 15% decreased rate of ATP formation, from 32.6 to 27.7 mmol/g of cells · h (P < 0.05). In contrast, overexpression of PYC increased the rate of ATP formation to 37.9 mmol/g of cells · h, which is significantly greater than the values observed in either JCL1242 or JCL1242/pPC201 (ppc+) (P < 0.025). Although the yield of ATP was lowest in the wild-type strain VJS676, the ATP yields in the three other strains were identical.
Flux partitioning.
The effects of these genetic perturbations may be further elucidated by studying flux partitioning at key metabolic branch points, or nodes. By comparing the fractional output of carbon at each node, flux partitioning is a means to compare the steady-state competition of multiple enzymes for a single substrate in response to genetic perturbations. We calculated the flux partitioning at the PEP, pyruvate, and acetyl-CoA nodes for the four principal strains (Table 6).
TABLE 6.
Flux partitioning at nodes for E. coli fermentations during exponential growth on glucose and rich mediuma
| Strain | Flux at node | ||||||
|---|---|---|---|---|---|---|---|
| PEP | Pyruvate | Acetyl-CoA | |||||
| _J_1 + _J_4 | _J_5 | _J_PYC | _J_6 | _J_7 | _J_8 | _J_9 | |
| VJS676 | 93 | 7.1 | 0 | 9.7 | 90 | 51 | 49 |
| JCL1242 | 100 | 0 | 0 | 18 | 82 | 46 | 54 |
| JCL1242/pPC201 (ppc+) | 90 | 9.8 | 0 | 1.7 | 98 | 48 | 52 |
| JCL1242/pTrc99A-pyc | 100 | 0 | 12 | 11 | 77 | 51 | 49 |
At the PEP node, flux toward pyruvate is the dominant branch for all strains (by the combined effects of pyruvate kinase and the PTS). Compared to VJS676 (which has a chromosomal copy of ppc), overexpression of PPC in JCL1242/pPC201 (ppc+) resulted in a relatively minor difference in the fraction of carbon flowing toward oxaloacetate (from 7.1 to 9.8%).
At the pyruvate node, PFL was the dominant branch in all strains, with 77 to 98% of carbon flowing toward formate. The presence of PPC activity greatly affected flux to lactate. In JCL1242, which has no PPC activity, the fraction of the flux going to lactate was 18%, whereas in the strain with a chromosomal copy of ppc this fraction was only 9.7%. Overexpression of PPC in JCL1242/pPC201 (ppc+) resulted in only 1.7% of the pyruvate carbon flowing towards lactate. Thus, in the three strains in which PYC activity was absent, flux partitioning to lactate was inversely correlated to PPC activity. In the presence of PYC activity in JCL1242/pTrc99A-pyc, 11% of the carbon flowing into the pyruvate node was diverted to lactate, a result indistinguishable from the fraction observed with VJS676. Compared to the result with VJS676, the presence of PYC activity had the effect of decreasing the flux partition to formate by 13% without affecting the flux partition to lactate. Compared to the result with JCL1242, the addition of the pyc gene in JCL1242/pTrc99A-pyc had the effect of reducing each of the two other competing fluxes, _J_6 and _J_7.
Genetic perturbations with ppc or pyc also affected the partitioning of carbon at the acetyl-CoA node. Compared to the result with JCL1242, the presence of PYC activity in JCL1242/pTrc99A-pyc served to increase the fraction of carbon at this node that was diverted to acetate (P < 0.10).
DISCUSSION
In this study we have examined the metabolic alterations in a model organism, E. coli, as a result of the presence or absence of carboxylating enzymes PPC or PYC. The synthesis of oxaloacetate is a key step in the formation of four-carbon compounds, such as malate, fumarate, and succinate. Possible routes used by various organisms for the synthesis of oxaloacetate include carboxylation of PEP by PPC, PEP carboxykinase, or PEP carboxytransphosphorylase; glyoxylate shunt enzymes; and the ATP-dependent carboxylation of pyruvate by PYC. Of these enzymes, PPC and PYC are most commonly employed for synthesis of oxaloacetate during glucose metabolism. Even though E. coli synthesizes PEP carboxykinase, an enzyme employed by several anaerobes in the formation of oxaloacetate (27, 35), this enzyme was unable to complement the absence of PPC in E. coli. Similar to results reported previously (20), our results demonstrate that E. coli relies principally on PPC for anaerobic diversion of carbon toward succinate production. Previously, PYC has been reported to compensate for the absence of PPC in aerobic systems, such as in lysine production (23). We report here the ability of PYC to restore the (anaerobic) succinate-producing ability of E. coli ppc null mutants. This restoration of succinate production by PYC in ppc null mutants moreover comes with an increase in glucose consumption rate compared to strains with elevated PPC activity.
E. coli adapts to alterations in the activities of PYC and PPC principally through the production of lactate. For sustained anaerobic fermentation, an organism must regenerate the reducing equivalent (NAD) consumed during glycolysis. E. coli accomplishes this task by the formation of ethanol, lactate, or succinate and could conceivably alter the formation of these products to maintain reducing equivalents. In the absence of the carboxylating enzyme PPC in JCL1242, the carbon from glucose flows exclusively from PEP to pyruvate. Pyruvate is known to be an allosteric effector of LDH (29, 30). Our observations of increased flux partitioning to lactate in the absence of PPC support the hypothesis that removing PPC activity increases the pool of intracellular pyruvate available to activate LDH and increase flux toward lactate in these strains. Moreover, increased PPC activity beyond the native activity would tend to reduce pyruvate availability and reduce flux toward lactate, a result we observed in fermentations of JCL1242/pPC201 (ppc+). Although overexpression of PPC in JCL1242/pPC201 (ppc+) dramatically decreased lactate partitioning, overexpression of PYC in JCL1242/pTrc99A-pyc caused lactate partitioning to decrease much less compared to that in JCL1242. Since E. coli could accomplish growth by generating only succinate and ethanol or acetate, one might speculate at the role LDH serves. Considering the response of the lactate flux to the genetic perturbations studied, LDH might be a means to afford the cell some metabolic flexibility in adapting readily to redox demands.
Ethanol synthesis also is affected in response to alterations in PPC and PYC activities. Indeed, even though lactate partitioning at the pyruvate node was greatest in JCL1242, this strain, which is deficient in both PPC and PYC activities, also showed the greatest ethanol partitioning at the acetyl-CoA node (Table 6). Previous studies have shown that NADH plays a role in inducing the gene coding for alcohol dehydrogenase (17). Since the route of NADH recycling via succinate is absent in JCL1242, perhaps increased NADH levels in this strain also caused slightly greater ethanol synthesis. However, fermentation of the strain with elevated PPC activity also led to slightly elevated (though not significantly different) ethanol flux during exponential growth compared to that in the strain with native PPC activity.
Increased levels of PPC activity in E. coli serve to decrease the specific rate of glucose consumption and the specific rate of ATP formation. For organisms which use the PEP-PTS, such as E. coli, 1 mol of PEP is required for each mole of glucose transported into the cell, thus limiting the quantity of PEP which might be diverted to oxaloacetate by PPC. Also, PPC must compete with ATP-generating pyruvate kinase. These limitations may explain why the succinate production by JCL1242/pPC201 (ppc+) is lower than that by JCL1242/pTrc99A-pyc even though the activity of PPC expressed via plasmid pPC201 in cell extracts was about 40 times greater than PYC activity from plasmid pTrc99A-pyc. These limitations also provide an explanation for the relatively small increase in flux partitioning to succinate in JCL1242/pPC201 (ppc+) compared to that in the wild-type VJS676. Increasing the activity of PPC beyond the native level reduced the specific rate of glucose consumption, perhaps through competition with the PEP-PTS for PEP. With a lowered glucose consumption rate, such cells would likely synthesize ATP more slowly. Our results with JCL1242/pPC201 (ppc+) demonstrate that a 14% decrease in the specific glucose consumption rate compared to that for JCL1242 was sufficient to reduce the specific rate of ATP formation through glucose metabolism by 15%. In E. coli, overexpression of native ppc appears to be a self-defeating means of increasing the yield of succinate. The enzyme PPC might more competitively be able to divert carbon to oxaloacetate in fermentations with organisms not having the PEP-PTS, a hypothesis which requires additional studies.
Increased levels of PYC activity in E. coli serve to increase the specific growth rate, the specific rate of glucose consumption, and the specific rate of ATP formation. In contrast to overexpression of ppc, expression of pyc increased the specific glucose consumption rate by 15%. The presence of PYC activity might reduce the pool of pyruvate, a result which would favorably support increased fluxes through the PEP-PTS and pyruvate kinase, which would favorably impact the glucose consumption rate and cells' ability to synthesize ATP. Paralleling the increase in glucose consumption, the specific rate of ATP formation was 16% greater in JCL1242/pTrc99A-pyc than in JCL1242. Consistent with these observations is an increased specific growth rate for JCL1242/pTrc99A-pyc over that for JCL1242.
An important result is that neither pyc nor ppc genetic perturbations affected the yield of ATP. This observation can be interpreted by considering the fluxes below the PEP node which involve ATP generation or consumption. The only fluxes that were significantly influenced by the presence or absence of the two carboxylating enzymes and that are involved in the net generation of ATP are _J_4 and _J_5. In the absence of PPC activity (such as with JCL1242), no energy is generated through _J_5. However, this decrease in ATP generation was compensated for in JCL1242 by an increase in _J_4. Conversely, in the presence of elevated PPC activity in JCL1242/pPC201 (ppc+), the heightened flux _J_5 was offset by a decrease in _J_4. In the presence of PYC activity in JCL1242/pTrc99A-pyc, the new flux from pyruvate to succinate (_J_PYC) had no net effect on ATP consumption. Even though PYC consumes ATP, production of succinate via PPC is energetically equivalent to the production of succinate via pyruvate kinase (_J_4) and PYC.
PYC is able to compete with PFL and LDH. At the pyruvate node, the presence of PYC activity introduced a third competitor for the substrate pyruvate. The presence of PYC activity reduced the fraction of carbon flowing through each of the two other paths, indicating that PYC competed well with each enzyme. We measured an enzyme saturation constant (Km) for PYC of 0.24 mM, which is much lower than reported values of 2.0 mM for PFL (15) and 7.2 mM for LDH (30, 31). Although these measured (i.e., in vitro) saturation constants suggest PYC should compete even more favorably with PFL and LDH, our results with JCL1242/pTrc99A-pyc suggest different in vivo apparent Km values for these three enzymes. PYC may be limited by the availability of ATP. Also, PYC may be limited by the known allosteric behavior of the PYC. Specifically, the intracellular metabolites aspartate and malate are inhibitors of PYC, while PYC requires acetyl-CoA for its activation (9). In the fermentations in the present study, the PYC activity could readily be limited in vivo by the presence of aspartate and malate, since the presence of PYC itself would tend to increase the pools of aspartate and malate (from oxaloacetate). One possible means to overcome this drawback would be to express an α4β4-type PYC, which has been shown not to be affected by aspartate, malate, or acetyl-CoA (21).
FIG. 3.
Fermentation pattern of E. coli strain JCL1242 (ppc null mutant) growing in glucose-limited rich medium. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol; ◊, biomass.
ACKNOWLEDGMENTS
We express particular thanks to J. C. Liao, V. J. Stewart, and M. F. Dunn for strains and plasmids and to V. Hatzimanikatis for helpful discussions.
We also acknowledge the University of Georgia Research Foundation, the University of Georgia Experiment Stations, Applied Carbo Chemicals, Inc., and the Consortium for Plant Biotechnology Research for financial support.
Appendix
The flux for the PEP-PTS, _J_1, is defined as follows: glucose + PEP = glucose 6-phosphate + pyruvate.
The fluxes for the Embden-Meyerhof-Parnas pathway are defined as follows: for _J_2, glucose 6-phosphate + ATP = 2 glyceraldehyde 3-phosphate + ADP; for _J_3, glyceraldehyde 3-phosphate + ADP + NAD = NADH + ATP + PEP + H2O; and for _J_4, PEP + ADP = ATP + pyruvate.
The fluxes for pyruvate dissimilation are defined as follows: for _J_6, pyruvate + NADH = lactate + NAD, and for _J_7, pyruvate + CoA = acetyl-CoA + formate.
The fluxes for acetyl-CoA dissimilation are defined as follows: for _J_8, acetyl-CoA + ADP = acetate + CoA + ATP, and for _J_9, acetyl-CoA + 2 NADH = ethanol + 2 NAD + CoA.
The fluxes for carboxylation of PEP and pyruvate are defined as follows: for _J_5, PEP + CO2 + NADH + FADH + ADP = succinate + NAD + FAD + H2O + ATP, and for _J_PYC, pyruvate + CO2 + NADH + FADH = succinate + NAD + FAD + H2O.
REFERENCES
- 1.Aiba S, Matsuoka M. Identification of metabolic model: citrate production from glucose by Candida lipolytica. Biotechnol Bioeng. 1979;21:1373–1386. [Google Scholar]
- 2.Amann E, Ochs B, Abel K-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 3.Attwood P V. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 4.Blaschkowski H P, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in E. coli. Eur J Biochem. 1980;123:563–569. [PubMed] [Google Scholar]
- 5.Bunch P K, Mat-Jan F, Lee N, Clark D P. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143:187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 6.Chao P-Y, Liao J C. Alteration of growth yield by overexpression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in Escherichia coli. Appl Environ Microbiol. 1993;59:4261–4265. doi: 10.1128/aem.59.12.4261-4265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark D P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;63:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Ricci J C, Tsu M, Bailey J E. Influence of expression of the pet operon on intracellular metabolic fluxes of Escherichia coli. Biotechnol Bioeng. 1992;38:1318–1324. doi: 10.1002/bit.260390110. [DOI] [PubMed] [Google Scholar]
- 9.Dunn M F, Encarnacion G, Araiza G, Vargas M C, Davalos A, Peralta H, Mora Y, Mora J. Pyruvate carboxylase from Rhizobium etli: mutant characterization, nucleotide sequence, and physiological role. J Bacteriol. 1996;178:5960–5970. doi: 10.1128/jb.178.20.5960-5970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiteman M A, Chastain M J. Optimization of the ion-exchange analysis of organic acids from fermentation. Anal Chim Acta. 1997;338:69–75. [Google Scholar]
- 11.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdagarian M, Landa E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 12.Goel A, Ferrance J, Jeong J, Ataai M M. Analysis of metabolic fluxes in batch and continuous cultures of Bacillus subtilis. Biotechnol Bioeng. 1993;42:686–696. doi: 10.1002/bit.260420603. [DOI] [PubMed] [Google Scholar]
- 13.Gokarn R R, Eiteman M A, Altman E. Expression of pyruvate carboxylase enhances succinate production in Escherichia coli without affecting glucose uptake. Biotechnol Lett. 1998;20:795–798. [Google Scholar]
- 14.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer; 1985. [Google Scholar]
- 15.Knappe J, Blaschkowski H P, Grobner P, Schmitt T. Pyruvate-formate lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem. 1974;50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 16.Kornberg H. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J. 1966;99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonardo M R, Dailly Y, Clark D P. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Maloy S R, Nunn W D. Genetic regulation of glyoxylate shunt in Escherichia coli K-12. J Bacteriol. 1982;149:173–180. doi: 10.1128/jb.149.1.173-180.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millard C S, Chao Y-P, Liao J C, Donnelly M I. Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl Environ Microbiol. 1996;62:1808–1810. doi: 10.1128/aem.62.5.1808-1810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milrad de Forchetti S R, Cazzulo J J. Some properties of the pyruvate carboxylase from Pseudomonas fluorescens. J Gen Microbiol. 1976;93:75–81. doi: 10.1099/00221287-93-1-75. [DOI] [PubMed] [Google Scholar]
- 22.Papoutsakis E T, Meyer C L. Equations and calculations of product yields and preferred pathways for butanediol and mixed acid fermentations. Biotechnol Bioeng. 1985;27:56–66. doi: 10.1002/bit.260270108. [DOI] [PubMed] [Google Scholar]
- 23.Park S M, Sinskey A J, Stephanopoulos G. Metabolic and physiological studies of Corynebacterium glutamicum mutants. Biotechnol Bioeng. 1997;55:864–879. doi: 10.1002/(SICI)1097-0290(19970920)55:6<864::AID-BIT5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 24.Payne J, Morris J G. Pyruvate carboxylase in Rhodopseudomonas spheroides. J Gen Microbiol. 1969;59:97–101. doi: 10.1099/00221287-59-1-97. [DOI] [PubMed] [Google Scholar]
- 25.Peters-Wendisch P G, Wendisch V F, Paul S, Eikmanns B J, Sahm H. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology. 1997;143:1095–1103. doi: 10.1099/00221287-143-4-1095. [DOI] [PubMed] [Google Scholar]
- 26.Reardon K F, Scheper T H, Bailey J E. Metabolic pathway rates and culture fluorescence in batch fermentations of Clostridium acetobutylicum. Biotechnol Prog. 1987;3:153–167. [Google Scholar]
- 27.Samuelov N S, Lamed R, Lowe S, Zeikus J G. Influence of CO2-HCO3− levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl Environ Microbiol. 1991;57:3013–3019. doi: 10.1128/aem.57.10.3013-3019.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stols L, Kulkarni G, Harris B G, Donnelly M I. Expression of Ascaris suum malic enzyme in a mutant Escherichia coli allows production of succinic acid from glucose. Appl Biochem Biotechnol. 1997;63/65:153–158. doi: 10.1007/BF02920421. [DOI] [PubMed] [Google Scholar]
- 29.Tarmy E M, Kaplan N O. Kinetics of Escherichia coli B d-lactate dehydrogenase and evidence for pyruvate-controlled change in conformation. J Biol Chem. 1968;243:2587–2596. [PubMed] [Google Scholar]
- 30.Tarmy E M, Kaplan N O. Chemical characterization of d-lactate dehydrogenase from Escherichia coli B. J Biol Chem. 1968;243:2587–2596. [PubMed] [Google Scholar]
- 31.Terada K, Murata T, Izui K. Site directed mutagenesis of phosphoenolpyruvate carboxylase from E. coli; the role of his-579 in the catalytic and regulatory functions. J Biochem. 1991;109:49–54. [PubMed] [Google Scholar]
- 32.Tsai S P, Lee Y H. Application of metabolic pathway stoichiometry to statistical analysis of bioreactor measurement data. Biotechnol Bioeng. 1988;32:713–715. doi: 10.1002/bit.260320517. [DOI] [PubMed] [Google Scholar]
- 33.Vallino J J, Stephanopoulos G. Flux determination in cellular bioreaction networks. Application to lysine fermentations. In: Sikdar S, Bier M, Todd P, editors. Frontiers in bioprocessing. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 205–219. [Google Scholar]
- 34.Vallino J J, Stephanopoulos G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol Bioeng. 1993;41:633–646. doi: 10.1002/bit.260410606. [DOI] [PubMed] [Google Scholar]
- 35.Van der Werf M J, Guettler M V, Jain M K, Zeikus J G. Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch Microbiol. 1997;167:332–334. doi: 10.1007/s002030050452. [DOI] [PubMed] [Google Scholar]
- 36.Van de Walle M, Shiloach J. Proposed mechanism of acetate accumulation in two recombinant Escherichia coli strains during high density fermentation. Biotechnol Bioeng. 1998;57:71–78. doi: 10.1002/(sici)1097-0290(19980105)57:1<71::aid-bit9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]