TEL, a Putative Tumor Suppressor, Modulates Cell Growth and Cell Morphology of Ras-Transformed Cells While Repressing the Transcription of stromelysin-1 (original) (raw)
Abstract
TEL is a member of the ETS family of transcription factors that interacts with the mSin3 and SMRT corepressors to regulate transcription. TEL is biallelically disrupted in acute leukemia, and loss of heterozygosity at the TEL locus has been observed in various cancers. Here we show that expression of TEL in Ras-transformed NIH 3T3 cells inhibits cell growth in soft agar and in normal cultures. Unexpectedly, cells expressing both Ras and TEL grew as aggregates. To begin to explain the morphology of Ras-plus TEL-expressing cells, we demonstrated that the endogenous matrix metalloproteinase stromelysin-1 was repressed by TEL. TEL bound sequences in the stromelysin-1 promoter and repressed the promoter in transient-expression assays, suggesting that it is a direct target for TEL-mediated regulation. Mutants of TEL that removed a binding site for the mSin3A corepressor but retained the ETS domain failed to repress stromelysin-1. When BB-94, a matrix metalloproteinase inhibitor, was added to the culture medium of Ras-expressing cells, it caused a cell aggregation phenotype similar to that caused by TEL expression. In addition, TEL inhibited the invasiveness of Ras-transformed cells in vitro and in vivo. Our results suggest that TEL acts as a tumor suppressor, in part, by transcriptional repression of stromelysin-1.
The TEL (for “translocation-ETS-leukemia,” also referred to as ETV6) transcription factor is a target for disruption by chromosomal translocations in several forms of acute leukemia (24–27, 38, 50, 51, 54, 57, 63). TEL was originally identified as the gene on chromosome 12 that is disrupted by t(5;12) in patients with chronic myelomonocytic leukemia (25). This translocation fuses the N-terminal homodimerization domain of TEL to the tyrosine kinase domain of the platelet-derived growth factor receptor β. The N terminus of TEL is also fused to the majority of the AML-1B (Runx-1) transcription factor by t(12;21), which is the most frequent translocation in pediatric B-cell acute lymphoblastic leukemias (23, 26, 57, 61).
TEL is a member of the ETS family of transcription factors. ETS factors bind heterogenous sequences centered around a core GGA sequence and cooperate with other transcription factors to regulate the transcription of a diverse set of genes (28, 52, 74). Several ETS factors are downstream effectors of oncogenic Ras proteins and are phosphorylated by mitogen-activated protein kinases (73, 80). Aberrant expression of these ETS factors induces cellular transformation (73, 74). By contrast, TEL acts as a transcriptional repressor. In t(12;21), fusion of the TEL N-terminal domain to AML-1 creates a dominant transcriptional repressor (18, 19, 32). This observation led to the identification of an association between TEL and the mSin3A and SMRT corepressors (13, 20).
The TEL gene maps to chromosome 12 region p13. Loss of heterozygosity in this region of chromosome 12 is found in many types of cancer including leukemias and tumors of the breast and ovary (25, 31, 58, 62, 66, 79). For example, in more than 90% of cases associated with t(12;21), the second TEL allele also is deleted (26, 54, 57, 63). These findings suggest that the widely expressed TEL protein may function as a tumor suppressor (12, 54, 64). However, there is no direct evidence to support this hypothesis, because targeted disruption of the TEL gene in mice is lethal in utero at embryonic day 10.5 (E10.5) (71).
TEL knockout mice die of an inability to maintain the developing vascular network in the yolk sac (71). However, hematopoietic progenitors from these embryos are capable of differentiating along the various blood cell lineages in vitro (71). Therefore, TEL is not intrinsically required for the growth or differentiation of hematopoietic cells. However, in chimeric mice, TEL−/− embryonic stem cells contributed to fetal liver hematopoiesis but not to bone marrow-derived hematopoiesis and were unable to colonize the stromal microenvironment (72). This phenotype was hypothesized to reflect defects in cell adhesion or in pathways responsive to cell adhesion (72).
Matrix metalloproteinases (MMPs) are a family of secreted, zinc-dependent proteinases that degrade various components of the extracellular matrix (ECM). MMPs are required for cell migration, ECM organization, tissue remodeling, and tumor cell invasion (2). Cross talk between the signal transduction pathways that are regulated by cell-cell and cell-ECM adhesion may lead to coordinate regulation of these pathways (9, 17, 30, 37, 40, 47, 48, 60, 76). Consequently, alterations in MMP expression may affect cell-cell interactions as well as cell-ECM adhesion. MMPs are also linked to cell growth. The expression of MMPs is induced by growth factors, and the promoters of the MMP genes contain Ras-responsive elements, which contain binding sites for the ETS family of transcription factors and AP-1 (3, 16, 49, 55). Moreover, MMP expression may directly contribute to tumor development. For example, expression of stromelysin-1 (MMP-3) in transgenic mice promoted spontaneous premalignant cellular changes and stimulated oncogenic transformation in mammary glands (65).
We present evidence showing that the TEL transcriptional repressor affects cell growth and causes cell aggregation. TEL inhibited the growth of Ras-transformed cells both in soft agar and in normal cultures. Expression of TEL in Ras-transformed NIH 3T3 cells resulted in a pronounced aggregation of these cells. The aggregation phenotype coincided with a reduction in the expression of the Ras-responsive gene stromelysin-1 (MMP-3). TEL-mediated repression of stromelysin-1 may be critical for the aggregation phenotype because an MMP-specific chemical inhibitor produced a similar phenotype. Finally, MMPs are required for tumor invasion, and we demonstrated that TEL inhibits tumor invasion. These results add biological support for the role of TEL as a tumor suppressor.
MATERIALS AND METHODS
Plasmids.
The pBabePuro and pCMVTEL constructs have been described elsewhere (32, 46). pCMVTELΔETS was made by replacing nucleotides 1143 to 1179 (25) with a _Bam_HI restriction site. The C-terminal _Bam_HI-_Sal_I fragment was amplified using standard PCR techniques, sequenced for verification, and reinserted into _Bam_HI-_Sal_I-cleaved pCMVTEL. This mutation deletes amino acids 373 to 385, which corresponds to the α2 helix in the ETS DNA-binding domain as described previously (52). The TEL and TELΔETS cDNAs were inserted into pBabePuro as _Eco_RI-_Sal_I fragments. The rat stromelysin-1 reporter from pG7-754TR CAT (43) was inserted into the pGL2 Luciferase vector (Promega) as an _Acc_65I-_Bgl_II fragment. The deletion mutants of the stromelysin-1 promoter were made as follows. pTR334 was made by truncating pGL2-754TR at the unique _Bsr_GI site. pTR182 and pTR127 were made by PCR using oligonucleotides starting at residue −182 or −127 (5′-TAGGTACCCACCATTCGCTTTGCAAA-3′ or 5′-CATGAGCTCTCACTCTTCTGATTT-3′ with 5′-GCAGATCTCGAGCTAGCACGCGTAAG-3′). The fragment was then cloned into the pGL2 vector. pTR89 was made by truncating pGL2-754TR at the unique _Bst_XI site. pTRΔEBS was made by cutting out the _Bsr_GI-_Bst_XI fragment from pGL2-754TR.
Gel mobility shift assay.
Conditions for the gel shift assay were basically as described previously (70). Proteins and radiolabeled probes were incubated in 10 μl of buffer (25 mM HEPES [pH 7.9], 50 mM KCl, 5% glycerol, 0.5 mg of bovine serum albumin per ml, 5 μM zinc sulfate, 0.5 mM dithiothreitol) for 30 min at room temperature and then loaded onto an 8% polyacrylamide gel prepared with Tris-glycine buffer. Poly(dI-dC) was used at 500 ng/ml as a nonspecific competitor. Glutathione _S_-transferase (GST) and GST-TEL fusion proteins were purified with glutathione-Sepharose 4B beads (Pharmacia). Double-stranded consensus ETS factor binding-site oligonucleotides used for the gel shift assay were (sense strand) 5′-AATTCATAAACAGGAAGTGGCTT-3′ (wild type) and 5′-AATTCATAAACACTGAGTGGCTT-3′ (mutant). Oligonucleotides from the stromelysin-1 promoter were as follows: −213 to −183, 5′-CTAAGGCAGGAAGCATTTCCTGGAGATTAA-3′ (wild type) and 5′-CTAAGGCACTGAGCATTGACTGTCGATTAA-3′ (mutant); −111 to −88, 5′-TAATTTTTGGAAATGGTCCCATTT-3′; and −93 to −74, 5′-CCATTTGGATGGAAGCAATT-3′. For the mutant oligonucleotides from the stromelysin-1 promoter, the GGA sequences were changed to TCG in each case.
Cell culture, transfection, retroviral infection, RNA analysis, and cell fractionation.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) supplemented with 10% calf serum (BioWhittaker), 2 mM l-glutamine (BioWhittaker), and 10 μg of gentamicin (Sigma) per ml. v-Harvey Ras-transformed NIH 3T3 cells were the generous gift of Stephen J. Brandt.
Retroviral packaging φNX cells were maintained in DMEM containing 10% fetal calf serum, 2 mM l-glutamine, and 10 μg of gentamicin per ml. Plates (10% confluent) were transfected for 6 to 8 h with 25 μl of Lipofectamine reagent (Gibco) and 5 μg of the pBabePuro retroviral vectors. The medium was replaced 24 h later, and virus-containing supernatants were harvested at 60 h. Supernatants were filtered through a 0.45-μm-pore-size acrodisk (VWR) and added to a plate of NIH 3T3 cells (10% confluent). Ras-expressing cells were infected with retrovirus to eliminate clonal variations. Infected cells were split 1:10 48 h later and selected for 72 h with 1 μg of puromycin (Sigma) per ml. Based on the minimal cell death observed upon selection, we concluded that more than 90% of the cells were infected. The cells were used within the first 10 passages after infection, with the critical experiments being performed within the first few passages. However, we observed the phenotypes (e.g., morphology and soft agar) immediately, and they did not change with cell passage. In addition, we used at least five independent batches of retrovirus and obtained similar results. The expressed proteins were detected by immunoblot analysis as described previously (42). stromelysin-1 mRNA was detected by RNA blot analysis as described previously (1). Affymetrix gene chip analysis was performed by the Duke University gene analysis core using poly(A)+ mRNA from NIH 3T3 cells expressing Ras or Ras plus TEL.
Cell photographs were recorded using a Zeiss Axiophot microscope with a Leaf Lumina digital camera (magnification, ×62.5). Images were recorded as TIFF files in Adobe Photoshop. The images were captured with the camera in either the forward or back position, which yielded subtle differences in magnification.
For cell fractionation, cell pellets were resuspended in Iso-Hi buffer (10 mM Tris-HCl [pH 8.4], 140 mM NaCl, 1.5 mM MgCl2, complete protease inhibitor [Roche], 1 mM EDTA) containing 0.5% NP-40. After 5 min on ice, the nuclei were collected by centrifugation at 2,000 rpm for 5 min in a Beckman GS-6R. Nuclei were reextracted to ensure purity. Equal proportions of the supernatant (cytoplasmic) and nuclear fractions were analyzed by immunoblot analysis with mSin3A, TEL C-terminal, and Ras antisera.
Transcription assays.
Cells and supernatants were harvested 44 to 48 h posttransfection. The cells were washed twice with phosphate-buffered saline and lysed in 100 to 200 μl of reporter lysis buffer (Promega). Aliquots (10 to 80 μl) were assayed for luciferase activity using the luciferase reagent assay (Promega) as specified by the manufacturer. Cytomegalovirus immediate early (CMV)-SEAP (secreted alkaline phosphatase) plasmids were included as internal controls for transfection efficiency (32, 78). SEAP activity was quantitated as described previously (5), except that the incubations were performed at room temperature. Luciferase activities were then normalized with respect to SEAP activity.
Soft-agar assays.
For the bottom agar, 2.5 ml of 1.6% agarose was diluted with 2.5 ml of 2× medium (2× DMEM, 20% calf serum, 4 mM l-glutamine, 20 μg of gentamicin per ml, 2 μg of puromycin per ml). A 4-ml volume of bottom agar was plated in a 60-mm tissue culture dish and allowed to harden. Cells were trypsinized and resuspended at 1.5 × 105 cells/ml in normal medium. The top agar cell suspensions were composed of 300 μl of cell suspension, 3.6 ml of 2× medium, and 2.4 ml of 0.75% agarose. Aliquots (2 ml) of this mixture were overlaid on each of two dishes containing bottom agar. The final plating concentration was 1,500 cells per dish.
Matrigel invasion assay.
Biocoat Matrigel invasion chambers (Becton Dickinson) were used to assess the invasiveness of TEL-infected NIH 3T3 cells as described previously (8). Briefly, 5 × 104 cells were resuspended in 100 μl of serum-free DMEM containing 0.2% bovine serum albumin and added to the cell culture inserts of the invasion chambers. A chemoattractant, 5 μg of human fibronectin (Gibco-BRL) per ml, was added to the lower wells. The cells were incubated at 37°C and allowed to invade through the matrix and the pores (8 μm) of the attached lower membrane for 48 h. The filters were fixed by immersion in 1% glutaraldehyde and stained with 0.2% crystal violet. The upper surface of the membrane was cleaned with a cotton swab to remove all noninvasive cells. The stained, invasive cells were then photographed using a digital camera.
Mouse tumor formation assays.
A single-cell suspension of 5 × 104 cells in phosphate-buffered saline was injected subcutaneously into the right lateral flank of 7-week-old athymic female nude (nu/nu) mice (Harlan Sprague-Dawley). The mice were housed in microisolator cages, given food and water ad libitum, and handled in a sterile laminar-flow hood. Tumor sizes were measured every 3 days using Vernier calipers along two perpendicular axes. The mice were sacrificed after 14 days. Primary tumors were excised, fixed overnight in 4% paraformaldehyde in PBS, and stored in 70% ethanol at 4°C. For histologic testing, the tumors were embedded in paraffin and 5-μm sections were stained with hematoxylin and eosin. Statistical comparison was performed using an analysis of variance followed by a one-tailed t test.
RESULTS
TEL regulates cell growth.
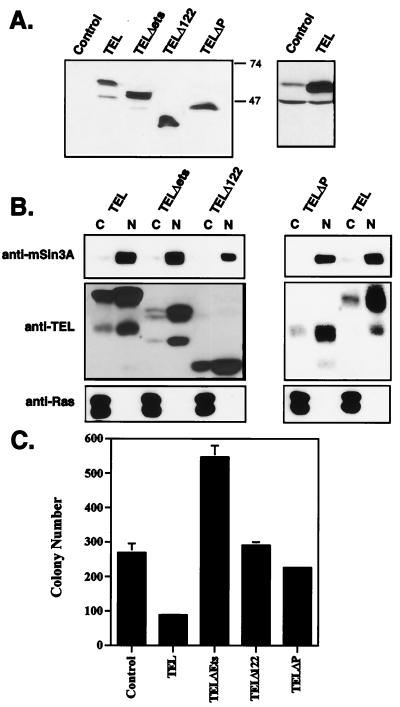
Based on genetic evidence from various cancers, TEL is postulated to be a tumor suppressor in both leukemias and solid tumors (12, 54, 64). Because oncogenic ETS factors are downstream effectors of Ras-signaling pathways (73, 80), we tested whether TEL could directly inhibit the growth of Ras-transformed cells. To express TEL but avoid nonspecific effects of high-level overexpression of ETS domain-containing proteins and clonal variation, we infected normal and Ras-transformed NIH 3T3 cells with recombinant retroviruses expressing TEL. For each assay in this study, several different infected-cell populations were used, to reduce the possibility that the results were affected by viral integration site effects. As controls, we tested TEL mutants lacking the N-terminal pointed domain (Δ122 and ΔP) or containing a small deletion in the ETS domain (TELΔETS) for their ability to inhibit Ras-dependent growth. Immunoblot analysis using a C-terminal TEL antibody confirmed that the mutant proteins were expressed at similar levels (Fig. 1A). To ensure that the mutant proteins were appropriately transported into the nucleus, we performed cell fractionation experiments. Compared to nuclear (mSin3A) and cytoplasmic (Ras) controls, the majority of the proteins expressed for each mutant were found in the nuclear fractions (Fig. 1B).
FIG. 1.
TEL inhibits growth in soft-agar assays. (A) Immunoblot analysis showing the expression of TEL and TEL mutant proteins in the stable cell lines used in panel B. The right-hand panel shows the level of the endogenous (Control) and expressed TEL using anti-N-terminal TEL. (B) Cellular localization of TEL and TEL mutants. Cells expressing TEL or the indicated TEL mutants were incubated with 0.5% NP-40 in an isotonic buffer for 5 min before being subjected to low-speed centrifugation to collect the nuclei. Equal amounts of cytoplasmic (C) and nuclear (N) fractions were analyzed by immunoblotting with antibodies directed to mSin3A as a nuclear protein control (upper panel), TEL (middle panel), or Ras as a cytoplasmic protein (bottom panel). (C) TEL inhibits soft-agar colony formation of Ras-transformed cells. Cells were grown for 10.5 days in medium containing 0.375% agarose, as overlays on 0.8% agarose beds. A total of 1,500 cells of the indicated cell lines were plated per dish. Values shown are the mean and range of duplicate samples.
One hallmark of the transformed phenotype is the ability of cells to grow in semisolid medium (56). Ras-transformed NIH 3T3 cells grew efficiently in soft agar (Fig. 1C). TEL expression inhibited the ability of Ras-transformed NIH 3T3 cells to grow in an anchorage-independent manner in these assays (Fig. 1C). This appeared to be due to slower growth, since more colonies became apparent with longer culture times. Deletion of an mSin3-binding domain of TEL, either with a specific internal deletion (ΔP) or by truncation of the N-terminal 122 amino acids, ablated TEL-mediated growth inhibition (Fig. 1C). Thus, simple overexpression of the TEL DNA-binding domain did not inhibit growth, as observed for ETS-2 (21, 34, 39, 75). As a further control, we expressed a TEL mutant with a 12-amino-acid deletion within the DNA-binding domain. Unexpectedly, the TELΔETS mutant cooperated with Ras to stimulate growth in soft agar (Fig. 1C). Because TEL forms homodimers (35, 44), it is possible that this mutant acts as an inhibitory protein to block endogenous TEL functions. In addition, TEL inhibited the growth of Ras-transformed Rat 1A cells in soft agar (data not shown).
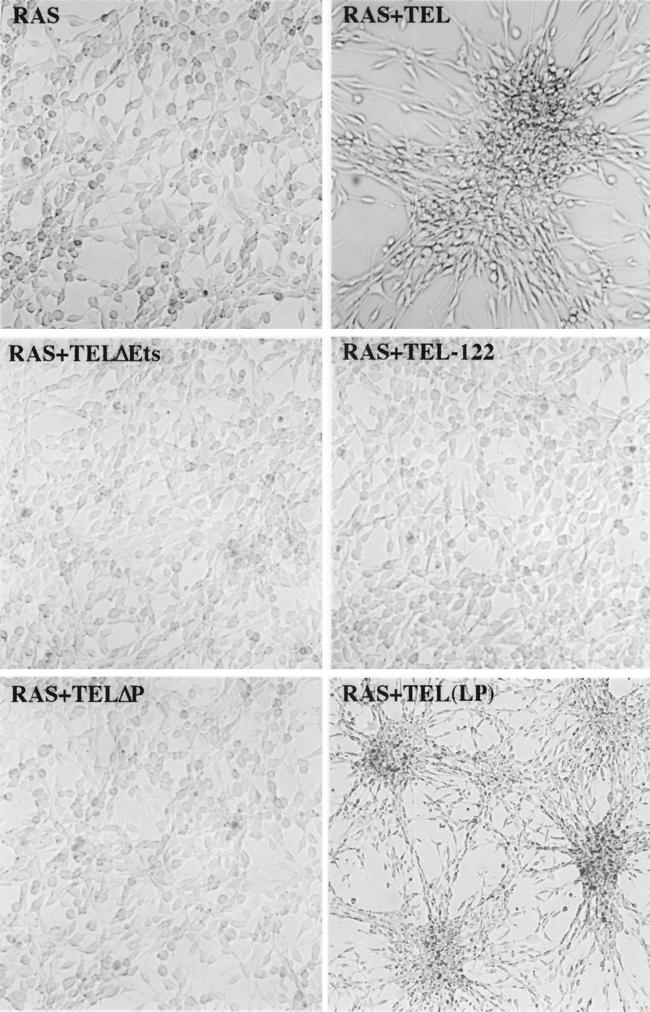
To determine whether TEL could also influence anchorage-dependent cell growth, the growth rates of normally cultured cells were determined. TEL did not affect the growth of control NIH 3T3 cells in culture (Fig. 2A). However, TEL did slow the growth of Ras-transformed NIH 3T3 cells (Fig. 2B). As observed in the soft-agar colony assays, TELΔETS cooperated with Ras to cause Ras-expressing cells to grow faster (Fig. 2B). Although TEL slowed the growth of Ras-transformed cells, there were no signs of apoptosis under normal growth conditions, as determined by morphology or by propidium iodide staining of DNA (data not shown). Also, TEL expression did not dramatically disrupt a specific cell cycle phase as judged by DNA content analysis (data not shown). However, it is possible that the TEL-expressing cells were cycling more slowly but with a proportional lengthening of each phase of the cell cycle.
FIG. 2.
TEL inhibits cell growth. A total of 8 × 105 cells of each cell line were plated in 100-mm tissue culture dishes containing 20 ml of medium. Cells were counted every 24 h for 3 days. Values shown are the mean and standard deviation of quadruplicate samples (A) and the mean and range of duplicate samples (B).
TEL causes Ras-transformed NIH 3T3 cells to grow as aggregates.
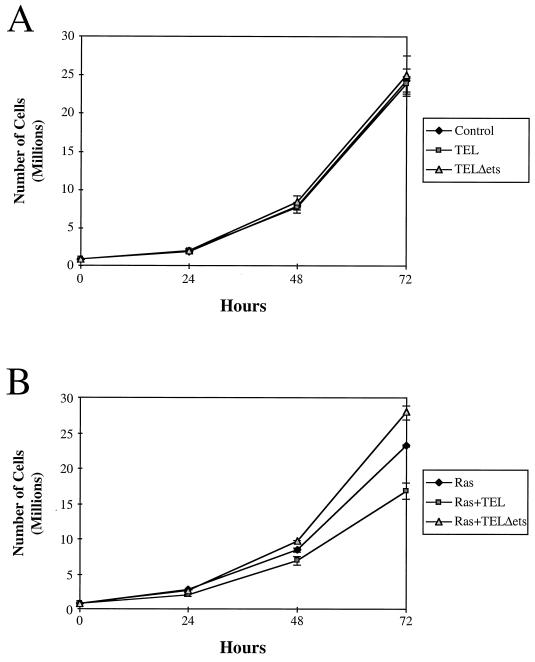
In culturing the TEL-expressing cells, it was readily apparent that TEL was affecting the morphology of these cells. At lower densities (i.e., subconfluent growth), Ras-transformed NIH 3T3 cells grew as monolayers (Fig. 3). TEL did not appear to have any morphological effect on wild-type NIH 3T3 cells (data not shown). By contrast, enforced expression of TEL in Ras-transformed NIH 3T3 cells resulted in an aggregation of these cells under subconfluent conditions (Fig. 3). As Ras-plus TEL-expressing (Fig. 3, Ras+TEL) cells continued to grow, most of them piled up rather than spread over the plate. Distinct lanes of cells could also be seen bridging the large clusters of cells (Fig. 3, Ras+TEL LP). In many cases, these connecting strands consisted of a string of single cells oriented end to end. With extended culturing times, the aggregates grew into large “spheroids” that remained attached to the plate only at the initial point of cell aggregation (data not shown). Unlike control Ras-transformed cells, Ras-plus TEL-expressing cells did not fill the culture dish and did not form transformed “foci” (data not shown). This effect was specific to wild-type TEL, since deletion of the pointed domain or the small deletion in the DNA-binding domain of TEL abolished the phenotype (Fig. 3).
FIG. 3.
TEL causes aggregation of Ras-transformed NIH 3T3 cells. Ras-transformed NIH 3T3 cells infected with recombinant retroviruses expressing TEL and the indicated TEL mutants were plated and allowed to grow for 72 h. Cells were visualized by bright-field microscopy (magnification, ×62.5) and recorded as digital images using Adobe Photoshop. The panel labeled Ras+TEL (LP) contains cell photographed at threefold-lower power (LP, lower power).
TEL represses transcription of the stromelysin-1 gene.
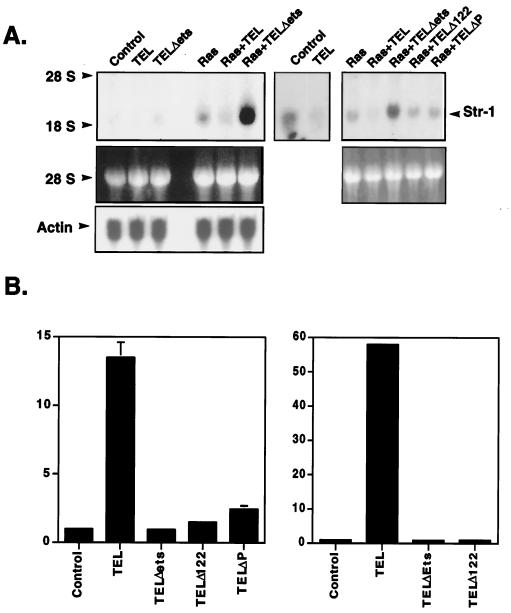
The phenotype caused by TEL overexpression could be due to alterations in cell-cell or cell-ECM adhesion or to a disruption of cell migration due to changes in the signaling pathways that regulate these processes. Because of the large number of genes involved in these processes that could be directly or indirectly affected by TEL, we performed microarray analysis to define changes in gene expression. RNA extracted from NIH 3T3 cells expressing Ras and from cells expressing Ras plus TEL were compared using Affymetrix gene chip arrays. However, of the nearly 50 genes whose expression was significantly altered, none are directly involved in cell adhesion or migration (data not shown). Because microarray analysis may miss changes in gene expression when genes are expressed at low levels, we asked whether genes that are involved in cell adhesion and that are regulated by Ras and oncogenic ETS factors were repressed by TEL (73). MMPs are stimulated by Ras through the activation of ETS and AP-1 factors and repressed by negatively acting ETS factors such as Erg (15, 16, 29, 33, 59, 80). RNA blot analysis indicated that the levels of the MMPs expressed in NIH 3T3 cells, including MMP1, MMP7, and MMP9, were low in these cells and were not affected by TEL (data not shown). While the levels of stromelysin-1 (MMP3) mRNA were also low in control NIH 3T3 cells, these levels were further suppressed by expression of TEL (Fig. 4A, middle panel). As expected, expression of oncogenic Ras increased stromelysin-1 mRNA levels (Fig. 4A). However, coexpression of TEL with Ras reduced the amounts of stromelysin-1 mRNA expressed, whereas TEL mutants that lack an mSin3A binding site failed to repress stromelysin-1 (Fig. 4A). Consistent with the ability of the TELΔETS mutant to cooperate with Ras to stimulate growth, this mutant cooperated with Ras to increase the amount of stromelysin-1 transcript (Fig. 4A). Thus, TEL regulates the level of stromelysin-1 mRNA.
FIG. 4.
TEL inhibits transcription from the stromelysin-1 promoter. (A) Total cellular RNA was isolated from the cell lines described in Fig. 1. Samples (40 μg) were electrophoresed through a 2.5 M formaldehyde–1% agarose gel. (Upper panel) The RNA was transferred to a nylon membrane, UV cross-linked, and probed with a radiolabeled stromelysin-1 cDNA. (Lower panel) RNA stained with ethidium bromide. The third panel (left side) contains the same blot probed for actin expression. (B) NIH 3T3 cells (left panel) or Ras-expressing NIH 3T3 cells (right panel) were transfected with 1 μg of the rat stromelysin-1 luciferase plasmid and 100 ng of the pCMV-TEL expression constructs shown. Luciferase activity was determined 44 h later. Values were normalized for transfection efficiency by using CMV-SEAP activity as an internal control. Values shown are the fold repression (mean and range) of duplicate samples.
To determine whether TEL directly represses the stromelysin-1 promoter, we tested whether TEL could repress expression from a plasmid containing stromelysin-1 regulatory sequences linked to a luciferase reporter gene. As little as 20 ng of TEL-expressing plasmid repressed transcription from the rat stromelysin-1 promoter by more than 15-fold in NIH 3T3 cells (Fig. 4B). This inhibition was specific, since TEL had no effect on the CMV promoter used as an internal control in these experiments. TEL also failed to repress the ETS factor-regulated multidrug resistance gene 1 (MDR-1) promoter (reference 41 and data not shown). In addition, mutations of an mSin3-binding domain (Δ122 and ΔP) or the DNA-binding domain (TELΔETS) impaired TEL-mediated repression (Fig. 4B, left panel). However, at 10-fold-higher levels of input TELΔP expression plasmid, we did observe two- to threefold repression, suggesting competition for DNA-binding sites with endogenous ETS factors (data not shown). When these experiments were repeated in Ras-expressing cells, the basal levels of expression from the stromelysin-1 promoter were higher (data not shown) and TEL-mediated repression was even more dramatic (Fig. 4B, right panel).
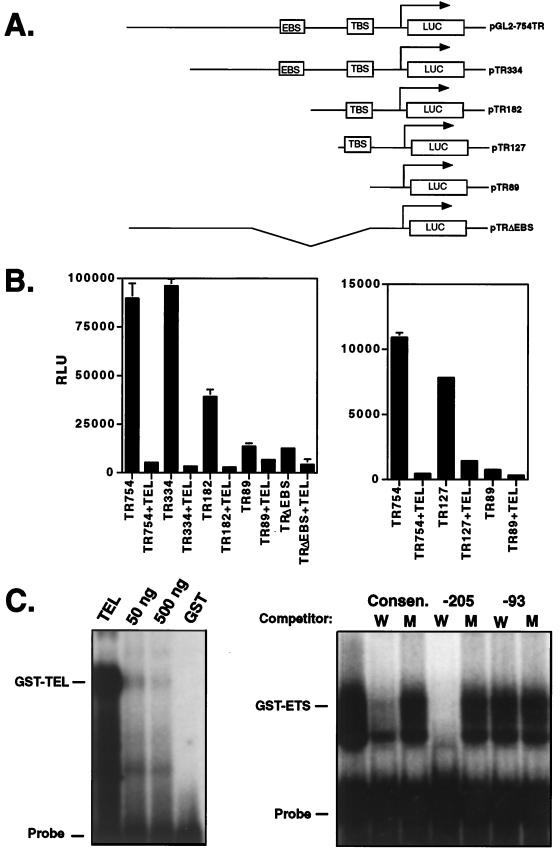
To define the sequences in the stromelysin-1 promoter that are required for TEL-mediated repression, a deletion analysis was performed (Fig. 5A). Deletion of over 400 bp upstream of a previously mapped ETS factor-binding site (3, 10, 11, 16) had no effect on repression, since the −334 construct was repressed by over 15-fold (Fig. 5A and B). However, deletion to nucleotide −182, removing the known ETS-binding site, reduced repression to only five- to sixfold (Fig. 5B, TR182), indicating that these sequences contributed to TEL-mediated repression. TEL-mediated repression was significantly impaired by deletion to residue −89 (Fig. 5B, TR89). In addition, an internal deletion of sequences from −334 to −89 was only marginally responsive to repression by TEL (Fig. 5B, TRΔEBS). Although both of these latter deletions reduced the basal expression from the promoter, each promoter was still active, thus allowing us to determine the effects of TEL.
FIG. 5.
Mapping TEL-responsive elements in the stromelysin-1 promoter. (A) Schematic diagram of the stromelysin-1 promoter and deletion mutants used in panel B. The known ETS factor-binding site is labeled EBS, and a putative TEL-binding site is labeled TBS. (B) Deletion analysis of the stromelysin-1 promoter. The ability of TEL to repress the full-length stromelysin-1 promoter and the deletion mutants shown in panel A is demonstrated. RLU, relative luciferase activity. Assays were performed with NIH 3T3 cells, as in Fig. 4, using 20 ng (left panel) or 100 ng (right panel) of TEL-expressing plasmid. (C) Gel mobility shift assays using GST-TEL and a consensus ETS factor-binding site as the probe or the TEL-binding site (−111 to −88) from the stromelysin-1 promoter as the probe and bacterially produced GST-TEL DNA-binding domain fusion protein (right panel). Oligonucleotides used for competition are designated above each lane (−205, −213 to −183; −93, −93 to −74). The sequences of these oligonucleotides are provided in Materials and Methods. W, wild type; M, mutant. The numbers above the lanes in the left panel are the amounts of the wild-type competitor added to each sample.
Within the region from −182 to −89, there are two putative ETS factor-binding sites at −103 and −95 (labeled TBS in Fig. 5A). Therefore, we deleted sequences upstream of residue −127 to further isolate the second TEL-responsive sequence. The −127 promoter was nearly as active as the full-length −754 promoter, and it was repressed five- to sixfold by TEL expression (Fig. 5B, right panel). Once again, the −89 promoter was only weakly inhibited by TEL. Thus, two regions of the stromelysin-1 promoter that contain putative ETS factor-binding sites are responsive to TEL.
To determine whether TEL could directly interact with the TEL-responsive sites in the stromelysin-1 promoter, we asked if TEL could bind these sequences in gel mobility shift assays. A GST-TEL DNA-binding domain fusion protein formed a specific complex with an oligonucleotide encompassing the two ETS factor-binding sites (GGAA at residue −103 and TCC at residue −95; the probe contains residues −111 to −88 [Fig. 5C]). This complex was competed by oligonucleotides containing a wild-type but not a mutant consensus ETS factor-binding site that TEL recognizes (52, 53, 77) (Fig. 5C). Wild-type double-stranded oligonucleotides containing the known ETS-binding sites from the stromelysin-1 promoter (at nucleotides −205 and −196) also competed for TEL binding, whereas oligonucleotides containing mutations in the GGA sequences did not compete (Fig. 5C, lanes −205 W and −205 M). Although the TR89 promoter was slightly repressed at higher levels of input TEL expression plasmid (Fig. 5B, right panel), the two GGA putative ETS factor-binding sites that remain in this promoter failed to compete for TEL binding (nucleotides −93 to −74, labeled −93 in Fig. 5C). However, we cannot rule out the possibility that TEL binds these sequences weakly to mediate the reduction in expression observed in Fig. 5B. Therefore, TEL can bind at least two sites in the stromelysin-1 promoter.
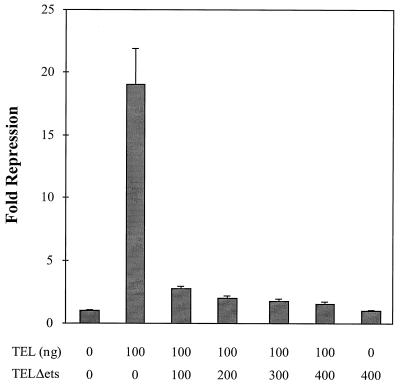
In several of the assays described above, the expression of TELΔETS had the opposite effect to the expression of TEL. Because NIH 3T3 cells express endogenous TEL, TELΔETS might act to inhibit endogenous TEL activity or might sequester or titrate a TEL cofactor. However, in these transient-expression assays, the ratio of reporter plasmid to endogenous TEL is likely to be too high for the endogenous TEL to repress transcription and thus allow TELΔETS to “activate” transcription by blocking its action. Consequently, we asked whether TELΔETS could inhibit TEL function when TEL was expressed exogenously in the transcription assays (Fig. 6). An equimolar amount of TELΔETS plasmid was sufficient to block TEL-mediated repression, but the mutant did not further activate transcription. Taken together with the biological data (Fig. 1 to 3), these results suggest that TELΔETS can act as an inhibitor of TEL function.
FIG. 6.
TELΔETS is an inhibitor of TEL-mediated repression. NIH 3T3 cells were transfected with the rat stromelysin-1 luciferase plasmid and the pCMV-TEL or pCMV-TELΔETS expression constructs shown. The numbers below the graph are the amounts of each expression plasmid transfected. Luciferase activity was determined 44 h later. Values were normalized for transfection efficiency using CMV-SEAP as an internal control. Values shown are the mean and standard deviation of triplicate samples. Fold repression represents the promoter activity from cells transfected with expression plasmids compared to those transfected with the empty vector.
An MMP inhibitor yields the same phenotype as TEL expression.
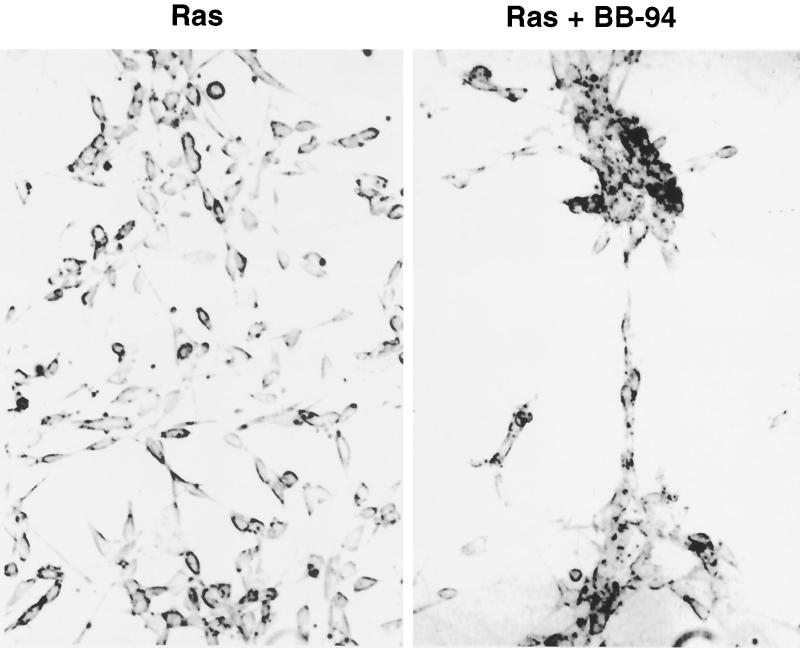
Although we thought it unlikely that a single target gene could result in the observed TEL-induced aggregation phenotype, MMPs are critical regulators of cell adhesion and metastasis (14). Therefore, we used a specific MMP inhibitor, BB-94 (67), to determine whether inhibition of MMPs is important for TEL-mediated cellular aggregation. BB-94 (2 μM) was added to the culture medium at the time when the cells were plated, and the cells were allowed to grow for 3 days. BB-94 had no effect on NIH 3T3 morphology (data not shown). By contrast, when added to the culture medium of Ras-transformed NIH 3T3 cells, BB-94 caused the cells to grow in clusters (Fig. 7), with lanes of cells extending from one cluster to another. This morphology was similar to that observed when TEL was expressed (Fig. 3).
FIG. 7.
An MMP inhibitor produces a phenotype similar to TEL expression. Ras-transformed NIH 3T3 cells were cultured in the presence or absence of BB-94, a specific MMP inhibitor (67). Cells were grown for 72 h and photographed as described in Materials and Methods.
TEL inhibits Ras-dependent tumor cell invasion.
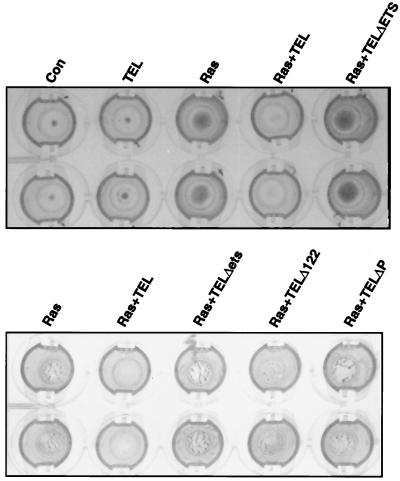
MMPs are critical regulators of cell migration and adhesion. For tumor growth, MMPs are required for angiogenesis and for invasion of surrounding tissue (other than adipose tissue). Therefore, we asked whether TEL expression in Ras-transformed cells could inhibit tumor cell invasion in vitro and in vivo. First, we compared the ability of cells to invade through a reconstituted three-dimensional basement membrane gel (Matrigel) (45). Control NIH 3T3 cells or TEL-expressing cells invaded the reconstituted matrix only poorly (Fig. 8 upper panel). Cells expressing oncogenic Ras or cells expressing Ras and TELΔETS, TELΔP, or TELΔ122 invaded through and adhered to the underside of the polycarbonate filter (the invading cells are indicated as dark regions in these stained cultures in Fig. 8). By contrast, when TEL was coexpressed with Ras, it inhibited the ability of Ras-transformed cells to invade the three-dimensional matrix (Fig. 8).
FIG. 8.
TEL inhibits Ras-dependent cell invasion in vitro. NIH 3T3 cells (control) or cells expressing TEL, Ras, Ras plus TEL, or Ras plus the indicated TEL mutants (lower panel) were cultured in Matrigel chambers for 3 days using fibronectin in the lower chamber as a chemoattractant. The filters were fixed by immersion in 1% glutaraldehyde and then stained with 0.2% crystal violet. The upper surface of the membrane was cleaned with a cotton swab to remove all noninvasive cells. The stained, invasive cells were then photographed using a digital camera. The two rows are duplicate samples.
Based on these in vitro results, we injected Ras-expressing NIH 3T3 cells and Ras-plus TEL-expressing cells into nude mice to determine if TEL could inhibit tumor invasion in vivo. The Ras-transformed cells formed tumors readily, and nearly every tumor invaded the surrounding muscle tissue (Table 1), as determined by gross morphology and by histologic analysis (data not shown). In general, the Ras-plus TEL-expressing cells formed somewhat smaller tumors (Table 1). However, when tumors of similar size were compared, the TEL-expressing tumors failed to invade the surrounding muscle. At necropsy, three of the TEL-expressing tumors showed the first signs of invasion and were scored as partially positive (Table 1). This small number of partially invading tumors could be due to our use of populations of cells that express different amounts of TEL rather than using single-cell clones expressing only large amounts of TEL. However, we conclude that TEL inhibits Ras-mediated tumor cell invasion in vitro and in vivo.
TABLE 1.
Tumor formation in nude micea
| Type of tumor | No. of tumors/no. of mice | |
|---|---|---|
| Ras | Ras-TEL | |
| Total | 14/14 | 15/15 |
| Invasiveb | 13/14 | ±3d/15 |
| Metastaticc | 2/14 | 0/15 |
DISCUSSION
Loss of heterozygosity in tumor samples suggests that TEL acts as a tumor suppressor in both leukemias and solid tumors. We have provided biochemical and biological evidence that supports this hypothesis. Expression of TEL inhibited the growth of Ras-transformed cells in soft agar and slowed their growth in normal cultures (Fig. 1 and 2). The ability of TEL to induce cell aggregation suggested that TEL-mediated growth inhibition may be due to alterations in cell adhesion (Fig. 3). Analysis of stromelysin-1 mRNA levels and the ability of TEL to regulate the stromelysin-1 promoter suggested that TEL acted as a transcriptional repressor to regulate MMP levels. The aggregation phenotype appeared to be partially due to the repression of stromelysin-1 because an MMP inhibitor also caused these cells to grow as aggregates (although the phenotype was not identical to that of TEL-expressing cells [compare Fig. 2 and 7]). Finally, TEL was shown to inhibit tumor invasion. Therefore, we propose that at least a portion of the action of TEL as a tumor suppressor is to regulate cell growth and tumor cell invasion by repressing target genes such as stromelysin-1.
The repression of both the stromelysin-1 promoter and the endogenous gene suggests that stromelysin-1 is a direct target of TEL-mediated repression. TEL binds a canonical ETS factor-binding site (52, 77), and it also binds the previously identified consensus ETS-binding site and at least one other site in the stromelysin-1 promoter (Fig. 5). Our promoter analysis identified two regions containing TEL-binding sites as being important for TEL-mediated repression. Although a link between modulation of stromelysin-1 transcription and Ras-dependent transformation has been established by expressing wild-type Ets-2 or dominant inhibitory mutants of ETS factors (21, 34, 39, 75), the morphological phenotype associated with TEL expression is distinct. Although the morphological effects of an MMP inhibitor are similar to the effects observed with TEL, TEL probably represses the transcription of other genes involved with growth control.
When an mSin3-binding domain of TEL was deleted, TEL was transcriptionally and biologically inactivated (Fig. 1 to 4 and 8). Given that these TEL mutants retain the ETS domain, the biological effects of TEL are due to active repression and not competition for DNA binding with endogenous ETS factors. This is in contrast to the effects of a dominant inhibitor of ETS-2 or the expression of isolated ETS factor-binding domains, which competed for ETS factor-binding sites to inhibit Ras-dependent transformation (21, 34, 39, 75). Interestingly, the TEL mutants lacking an mSin3A-binding motif retain a domain capable of interacting with the SMRT corepressor (13). However, in transcription assays, the mSin3A-binding mutant failed to repress transcription (Fig. 4), suggesting that multiple corepressor contacts are required for TEL-mediated repression.
Although the TELΔETS mutant was designed to be a nonfunctional protein, its overexpression had some physiological consequences. When expressed alone, TELΔETS had little or no effect on cell aggregation, cell growth, or transcriptional regulation of stromelysin-1. However, in conjunction with Ras, TELΔETS stimulated cell growth both in exponentially growing cultures and in soft-agar assays. TELΔETS also cooperated with oncogenic Ras to activate stromelysin-1 expression. The effects of this mutant on cell growth but not cell adhesion suggest that TEL also regulates growth independently of its effects on cell adhesion. We will need to define other TEL target genes to fully understand how TEL regulates these complex cell phenotypes.
In chimeric mice derived using TEL−/− embryonic stem cells, the TEL-null cells failed to contribute to the developing bone marrow, possibly due to a defect in stem cell migration or adhesion (72). In NIH 3T3 cells, TEL altered the morphology of the cells, possibly by affecting cell adhesion (Fig. 3 and 7). The results obtained with these two experimental systems suggest that the regulation of stromelysin-1 levels by TEL may be important during development. In addition, TEL null mice die in utero at E10.5 due to defects in angiogenesis and MMPs are critical for the tissue remodeling required for angiogenesis (6, 14). Tumors formed by injecting Ras-plus TEL-expressing cells into nude mice (Table 1) appeared to have less vasculature than did tumors derived from Ras-transformed cells, but this was difficult to quantitate (data not shown). Further work must be performed to establish a direct role for TEL in regulating angiogenesis.
There are several precedents for tumor suppressors that regulate cell adhesion. Perhaps the best characterized is E-cadherin. E-cadherin mediates cell-cell interactions and is frequently deleted in metastatic tumors (4, 22, 69). In fact, altering the expression of stromelysin-1 changed the proteolytic cleavage of E-cadherin (40). The observation that TEL can inhibit tumor invasion (Fig. 8 and Table 1) may suggest that TEL acts at a late stage of tumor development and that its loss may be associated with metastasis. In at least one case, loss of TEL was a late event in the progression of a t(12;21)-containing leukemia (36). For leukemogenesis, the ability of immature hematopoietic progenitors to survive in the absence of stromal cell contacts is critical (7, 68). It is possible that loss of TEL may promote leukemogenesis by affecting cell growth and/or by altering cell adhesion. Future investigations will be necessary to identify additional targets of TEL that may directly or indirectly affect cell adhesion and growth.
ACKNOWLEDGMENTS
Randy Fenrick and Lilin Wang contributed equally to this paper.
Peter Brown, British Biotech, Ltd., Oxford, England, kindly supplied the BB-94. We also thank Helena Abushamaa, Duke University Genome Core Facility, for assistance with the Affymetrix GeneChip analysis. We thank Yue Hou and Jonathon Sheehan for technical assistance and members of the Hiebert, Kinch, and Matrisian laboratories for helpful discussions.
The experiments described here were performed in part through the use of the VUMC Cell Imaging Resource and the VCC sequencing facility, which are supported by NIH grants CA68465 and DK20593.R01. This work was also supported by NIH/NCI grants RO-1 CA46843 (to L.M.M.) and RO1-CA64140 and RO1-CA77274 (to S.W.H.), by American Cancer Society grants JFRA-591 (to S.W.H.) and RPG CSM-86522 (to M.S.K.), by a Center grant from the National Cancer Institute (CA68485), and by the Vanderbilt Cancer Center. R.F. and J.A. were funded by NIH training grant T32 DK07186-22. J.N. is a Special Fellow of the Leukemia Society of America (3827-99).
REFERENCES
- 1.Ball L A, Amann J M, Garrett B K. Replication of nodamura virus after transfection of viral RNA into mammalian cells in culture. J Virol. 1992;66:2326–2334. doi: 10.1128/jvi.66.4.2326-2334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basbaum C B, Werb Z. Focalized proteolysis: spatial and temporal regulation of extracellular matrix degradation at the cell surface. Curr Opin Cell Biol. 1996;8:731–738. doi: 10.1016/s0955-0674(96)80116-5. [DOI] [PubMed] [Google Scholar]
- 3.Basuyaux J P, Ferreira E, Stehelin D, Buttice G. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J Biol Chem. 1997;272:26188–26195. doi: 10.1074/jbc.272.42.26188. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Ze'ev A. Cytoskeletal and adhesion proteins as tumor suppressors. Curr Opin Cell Biol. 1997;9:99–108. doi: 10.1016/s0955-0674(97)80158-5. [DOI] [PubMed] [Google Scholar]
- 5.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 6.Bergers G, Javaherian K, Lo K M, Folkman J, Hanahan D. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science. 1999;284:808–812. doi: 10.1126/science.284.5415.808. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia R, Verfaillie C M. The effect of interferon-alpha on beta-1 integrin mediated adhesion and growth regulation in chronic myelogenous leukemia. Leuk Lymphoma. 1998;28:241–254. doi: 10.3109/10428199809092680. [DOI] [PubMed] [Google Scholar]
- 8.Brodt P, Reich R, Moroz L A, Chambers A F. Differences in the repertoires of basement membrane degrading enzymes in two carcinoma sublines with distinct patterns of site-selective metastasis. Biochim Biophys Acta. 1992;1139:77–83. doi: 10.1016/0925-4439(92)90085-2. [DOI] [PubMed] [Google Scholar]
- 9.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 10.Buttice G, Duterque-Coquillaud M, Basuyaux J P, Carrere S, Kurkinen M, Stehelin D. Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–2306. [PubMed] [Google Scholar]
- 11.Buttice G, Kurkinen M. A polyomavirus enhancer A-binding protein-3 site and Ets-2 protein have a major role in the 12-O-tetradecanoylphorbol-13-acetate response of the human stromelysin gene. J Biol Chem. 1993;268:7196–7204. [PubMed] [Google Scholar]
- 12.Cave H, Gerard B, Martin E, Guidal C, Devaux I, Weissenbach J, Elion J, Vilmer E, Grandchamp B. Loss of heterozygosity in the chromosomal region 12p12-13 is very common in childhood acute lymphoblastic leukemia and permits the precise localization of a tumor-suppressor gene distinct from p27KIP1. Blood. 1995;86:3869–3875. [PubMed] [Google Scholar]
- 13.Chakrabarti S R, Nucifora G. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem Biophys Res Commun. 1999;264:871–877. doi: 10.1006/bbrc.1999.1605. [DOI] [PubMed] [Google Scholar]
- 14.Chambers A F, Matrisian L M. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 15.Cosgaya J M, Recio J A, Aranda A. Influence of Ras and retinoic acid on nerve growth factor induction of transin gene expression in PC12 cells. Oncogene. 1997;14:1687–1696. doi: 10.1038/sj.onc.1200997. [DOI] [PubMed] [Google Scholar]
- 16.Crawford H C, Matrisian L M. Mechanisms controlling the transcription of matrix metalloproteinase genes in normal and neoplastic cells. Enzyme Protein. 1996;49:20–37. doi: 10.1159/000468614. [DOI] [PubMed] [Google Scholar]
- 17.Edwards D R, Murphy G. Cancer. Proteases—invasion and more. Nature. 1998;394:527–528. doi: 10.1038/28961. [DOI] [PubMed] [Google Scholar]
- 18.Fears S, Gavin M, Zhang D E, Hetherington C, Ben-David Y, Rowley J D, Nucifora G. Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter Proc. Natl Acad Sci USA. 1997;94:1949–1954. doi: 10.1073/pnas.94.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenrick R, Hiebert S W. The role of histone deacetylases in acute leukemia J. Cell Biochem Suppl. 1999;30-31:194–202. [PubMed] [Google Scholar]
- 20.Fenrick R, Amann J M, Lutterbach B, Wang L, Westendorf J J, Downing J, Hiebert S W. Both Tel and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol Cell Biol. 1999;19:6566–6574. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foos G, Garcia-Ramirez J J, Galang C K, Hauser C A. Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J Biol Chem. 1998;273:18871–18880. doi: 10.1074/jbc.273.30.18871. [DOI] [PubMed] [Google Scholar]
- 22.Frixen U H, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golub T, McLean T, Stegmaier K, Carroll M, Tomasson M, Gilliland D G. The TEL gene and human leukemia. Biochim Biophys Acta. 1996;1288:7–10. doi: 10.1016/0304-419x(96)00015-7. [DOI] [PubMed] [Google Scholar]
- 24.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golub T R, Barker G F, Lovett M, Gilliland D G. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 26.Golub T R, Barker G F, Stegmaier K, Gilliland D G. Involvement of the TEL gene in hematologic malignancy by diverse molecular genetic mechanisms. Curr Top Microbiol Immunol. 1996;211:279–288. doi: 10.1007/978-3-642-85232-9_28. [DOI] [PubMed] [Google Scholar]
- 27.Golub T R, Goga A, Barker G F, Afar D E, McLaughlin J, Bohlander S K, Rowley J D, Witte O N, Gilliland D G. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 29.Gum R, Lengyel E, Juarez J, Chen J H, Sato H, Seiki M, Boyd D. Stimulation of 92-kDa gelatinase B promoter activity by ras is mitogen-activated protein kinase kinase 1-independent and requires multiple transcription factor binding sites including closely spaced PEA3/ets and AP-1 sequences. J Biol Chem. 1996;271:10672–10680. doi: 10.1074/jbc.271.18.10672. [DOI] [PubMed] [Google Scholar]
- 30.Gumbiner B M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 31.Hatta Y, Takeuchi S, Yokota J, Koeffler H P. Ovarian cancer has frequent loss of heterozygosity at chromosome 12p12.3-13.1 (region of TEL and Kip1 loci) and chromosome 12q23-ter: evidence for two new tumour-suppressor genes. Br J Cancer. 1997;75:1256–1262. doi: 10.1038/bjc.1997.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussell M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription Mol. Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himelstein B P, Lee E J, Sato H, Seiki M, Muschel R J. Transcriptional activation of the matrix metalloproteinase-9 gene in an H-ras and v-myc transformed rat embryo cell line. Oncogene. 1997;14:1995–1998. doi: 10.1038/sj.onc.1201012. [DOI] [PubMed] [Google Scholar]
- 34.Jin D I, Jameson S B, Reddy M A, Schenkman D, Ostrowski M C. Alterations in differentiation and behavior of monocytic phagocytes in transgenic mice that express dominant suppressors of ras signaling Mol. Cell Biol. 1995;15:693–703. doi: 10.1128/mcb.15.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jousset C, Carron C, Boureux A, Quang C T, Oury C, Dusanter-Fourt I, Charon M, Levin J, Bernard O, Ghysdael J. A domain of TEL conserved in a subset of ETS proteins defines a specific oligomerization interface essential to the mitogenic properties of the TEL-PDGFR beta oncoprotein. EMBO J. 1997;16:69–82. doi: 10.1093/emboj/16.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D H, Moldwin R L, Vignon C, Bohlander S K, Suto Y, Giordano L, Gupta R, Fears S, Nucifora G, Rowley J D, et al. TEL-AML1 translocations with TEL and CDKN2 inactivation in acute lymphoblastic leukemia cell lines. Blood. 1996;88:785–794. [PubMed] [Google Scholar]
- 37.Kinch M S, Clark G J, Der C J, Burridge K. Tyrosine phosphorylation regulates the adhesions of ras-transformed breast epithelia. J Cell Biol. 1995;130:461–471. doi: 10.1083/jcb.130.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacronique V, Boureux A, Della Valle V, Poirel H, Quang C T, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 39.Langer S J, Bortner D M, Roussel M F, Sherr C J, Ostrowski M C. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol Cell Biol. 1992;12:5355–5362. doi: 10.1128/mcb.12.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell M J. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutterbach B, Sun D, Scheutz J, Hiebert S W. The MYND motif is required for repression of basal transcription from the multidrug resistance-1 promoter by the t(8;21) fusion protein Mol. Cell Biol. 1998;18:3604–3611. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Glass C K, Rosenfeld M G, Seto E, Hiebert S W. ETO, a target of the t(8;21) in acute leukemia, interacts with N- CoR, mSin3, and histone deacetylases. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonnell S E, Kerr L D, Matrisian L M. Epidermal growth factor stimulation of stromelysin mRNA in rat fibroblasts requires induction of proto-oncogenes c-fos and c-jun and activation of protein kinase C. Mol Cell Biol. 1990;10:4284–4293. doi: 10.1128/mcb.10.8.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLean T W, Ringold S, Neuberg D, Stegmaier K, Tantravahi R, Ritz J, Koeffler H P, Takeuchi S, Janssen J W, Seriu T, Bartram C R, Sallan S E, Gilliland D G, Golub T R. TEL/AML-1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood. 1996;88:4252–4258. [PubMed] [Google Scholar]
- 45.Melchiori A, Carlone S, Allavena G, Aresu O, Parodi S, Aaronson S A, Albini A. Invasiveness and chemotactic activity of oncogene transformed NIH/3T3 cells. Anticancer Res. 1990;10:37–44. [PubMed] [Google Scholar]
- 46.Morgenstern J P, Land H. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newgreen D F, Minichiello J. Control of epitheliomesenchymal transformation. II. Cross-modulation of cell adhesion and cytoskeletal systems in embryonic neural cells. Dev Biol. 1996;176:300–312. doi: 10.1006/dbio.1996.0135. [DOI] [PubMed] [Google Scholar]
- 48.Novak A, Hsu S C, Leung-Hagesteijn C, Radeva G, Papkoff J, Montesano R, Roskelley C, Grosschedl R, Dedhar S. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and beta-catenin signaling pathways. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owen R D, Ostrowski M C. Transcriptional activation of a conserved sequence element by ras requires a nuclear factor distinct from c-fos or c-jun. Proc Natl Acad Sci USA. 1990;87:3866–3870. doi: 10.1073/pnas.87.10.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papadopoulos P, Ridge S A, Boucher C A, Stocking C, Wiedemann L M. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. [PubMed] [Google Scholar]
- 51.Peeters P, Raynaud S D, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor-associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- 52.Pio F, Kodandapani R, Ni C Z, Shepard W, Klemsz M, McKercher S R, Maki R A, Ely K R. New insights on DNA recognition by ets proteins from the crystal structure of the PU.1 ETS domain-DNA complex. J Biol Chem. 1996;271:23329–23337. doi: 10.1074/jbc.271.38.23329. . (Erratum, 271:33156.) [DOI] [PubMed] [Google Scholar]
- 53.Poirel H, Oury C, Carron C, Duprez E, Laabi Y, Tsapis A, Romana S P, Mauchauffe M, Le Coniat M, Berger R, Ghysdael J, Bernard O A. The TEL gene products: nuclear phosphoproteins with DNA binding properties. Oncogene. 1997;14:349–357. doi: 10.1038/sj.onc.1200829. [DOI] [PubMed] [Google Scholar]
- 54.Raynaud S, Cave H, Baens M, Bastard C, Cacheux V, Grosgeorge J, Guidal-Giroux C, Guo C, Vilmer E, Marynen P, Grandchamp B. The 12;21 translocation involving TEL and deletion of the other TEL allele: two frequently associated alterations found in childhood acute lymphoblastic leukemia. Blood. 1996;87:2891–2899. [PubMed] [Google Scholar]
- 55.Reddy M A, Langer S J, Colman M S, Ostrowski M C. An enhancer element responsive to ras and fms signaling pathways is composed of two distinct nuclear factor binding sites. Mol Endocrinol. 1992;6:1051–1060. doi: 10.1210/mend.6.7.1324418. [DOI] [PubMed] [Google Scholar]
- 56.Rizzino A. Soft agar growth assays for transforming growth factors and mitogenic peptides. Methods Enzymol. 1987;146:341–352. doi: 10.1016/s0076-6879(87)46035-7. [DOI] [PubMed] [Google Scholar]
- 57.Romana S P, Poirel H, Leconiat M, Flexor M A, Mauchauffe M, Jonveaux P, Macintyre E A, Berger R, Bernard O A. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 58.Sato Y, Suto Y, Pietenpol J, Golub T R, Gilliland D G, Davis E M, Le Beau M M, Roberts J M, Vogelstein B, Rowley J D, et al. TEL and KIP1 define the smallest region of deletions on 12p13 in hematopoietic malignancies. Blood. 1995;86:1525–1533. [PubMed] [Google Scholar]
- 59.Schorpp M, Mattei M G, Herr I, Gack S, Schaper J, Angel P. Structural organization and chromosomal localization of the mouse collagenase type I gene. Biochem J. 1995;308:211–217. doi: 10.1042/bj3080211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwartz M A, Schaller M D, Ginsberg M H. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 61.Shurtleff S A, Buijs A, Behm F G, Rubnitz J E, Raimondi S C, Hancock M L, Chan G C, Pui C H, Grosveld G, Downing J R. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 62.Spirin K S, Simpson J F, Takeuchi S, Kawamata N, Miller C W, Koeffler H P. p27/Kip1 mutation found in breast cancer. Cancer Res. 1996;56:2400–2404. [PubMed] [Google Scholar]
- 63.Stegmaier K, Pendse S, Barker G F, Bray-Ward P, Ward D C, Montgomery K T, Krauter K S, Reynolds C, Sklar J, Donnelly M, et al. Frequent loss of heterozygosity at the TEL gene locus in acute lymphoblastic leukemia of childhood. Blood. 1995;86:38–44. [PubMed] [Google Scholar]
- 64.Stegmaier K, Takeuchi S, Golub T R, Bohlander S K, Bartram C R, Koeffler H P. Mutational analysis of the candidate tumor suppressor genes TEL and KIP1 in childhood acute lymphoblastic leukemia. Cancer Res. 1996;56:1413–1417. [PubMed] [Google Scholar]
- 65.Sternlicht M D, Lochter A, Sympson C J, Huey B, Rougier J P, Gray J W, Pinkel D, Bissell M J, Werb Z. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeuchi S, Bartram C R, Miller C W, Reiter A, Seriu T, Zimmerann M, Schrappe M, Mori N, Slater J, Miyoshi I, Koeffler H P. Acute lymphoblastic leukemia of childhood: identification of two distinct regions of deletion on the short arm of chromosome 12 in the region of TEL and KIP1. Blood. 1996;87:3368–3374. [PubMed] [Google Scholar]
- 67.Talbot D C, Brown P D. Experimental and clinical studies on the use of matrix metalloproteinase inhibitors for the treatment of cancer. Eur J Cancer. 1996;32A:2528–2533. doi: 10.1016/s0959-8049(96)00398-x. [DOI] [PubMed] [Google Scholar]
- 68.Verfaillie C M, McCarthy J B, McGlave P B. Mechanisms underlying abnormal trafficking of malignant progenitors in chronic myelogenous leukemia. Decreased adhesion to stroma and fibronectin but increased adhesion to the basement membrane components laminin and collagen type IV. J Clin Investig. 1992;90:1232–1241. doi: 10.1172/JCI115985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 70.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 71.Wang L C, Kuo F, Fujiwara Y, Gilliland D G, Golub T R, Orkin S H. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L C, Swat W, Fujiwara Y, Davidson L, Visvader J, Kuo F, Alt F W, Gilliland D G, Golub T R, Orkin S H. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–2402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- 74.Wasylyk B, Hahn S L, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. . (Erratum, 215:907, 1993.) [DOI] [PubMed] [Google Scholar]
- 75.Wasylyk C, Maira S M, Sobieszczuk P, Wasylyk B. Reversion of Ras transformed cells by Ets transdominant mutants. Oncogene. 1994;9:3665–3673. [PubMed] [Google Scholar]
- 76.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91:439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 77.Werner M H, Clore M, Fisher C L, Fisher R J, Trinh L, Shiloach J, Gronenborn A M. The solution structure of the human ETS1-DNA complex reveals a novel mode of binding and true side chain intercalation. Cell. 1995;83:761–771. doi: 10.1016/0092-8674(95)90189-2. . (Erratum, 87:355, 1996.) [DOI] [PubMed] [Google Scholar]
- 78.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocyte differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wlodarska I, Marynen P, La Starza R, Mecucci C, Van den Berghe H. The ETV6, CDKN1B and D12S178 loci are involved in a segment commonly deleted in various 12p aberration in different hematological malignancies Cytogenet. Cell Genet. 1996;72:229–235. doi: 10.1159/000134197. [DOI] [PubMed] [Google Scholar]
- 80.Yang B S, Hauser C A, Henkel G, Colman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2 Mol. Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]