Genomewide Scan in German Families Reveals Evidence for a Novel Psoriasis-Susceptibility Locus on Chromosome 19p13 (original) (raw)
Abstract
Psoriasis is a common chronic inflammatory skin disease with a strong genetic component. Few psoriasis-susceptibility loci have been reported, and only two have been confirmed in independent data sets. This article reports results of a genomewide scan that was performed, using 370 microsatellite markers, for psoriasis-susceptibility loci in 32 German extended families, comprising 162 affected and 195 unaffected individuals. Nonparametric linkage analysis of all families provided strong evidence for a novel psoriasis-susceptibility locus on chromosome 19p (Z _lr_=3.50; _P_=.0002). Parametric analysis revealed a heterogeneity LOD score of 4.06, corresponding to a genomewide significance level of .037, under the assumption of a recessive model with high disease-allele frequency and 66% as the proportion of linked families. This study confirms linkage of psoriasis to the HLA region on chromosome 6p and suggests additional regions on chromosomes 8q and 21q for further investigations.
Psoriasis vulgaris (MIM 177900) is a chronic inflammatory skin disease with a prevalence of ∼2%–3% in white populations (Lomholt 1963; Nevitt and Hutchinson 1996). The hallmarks of psoriasis are a clonal T cell expansion and infiltration of the epidermis, as well as a benign hyperproliferation of keratinocytes. Clinically, the disease is characterized by red, scaly plaques, and it may be associated with severe arthritis. The multifactorial etiology of psoriasis is well established. Although environmental factors, such as streptococcal infections (Boehncke et al. 1997), have been shown to affect the onset of the disease, family studies clearly indicate that psoriasis has a strong genetic component (Abele et al. 1963; Farber et al. 1974; Brandrup et al. 1978). Several psoriasis-susceptibility loci on chromosomes 6p (PSORS1 [MIM 177900] [Trembath et al. 1997; Nair et al. 1997]), 17q (PSORS2 [MIM 602723] [Tomfohrde et al. 1994]), and 4q (PSORS3 [MIM 601454] [Matthews et al. 1996]) have been described, and additional putative psoriasis candidate loci have been reported on 16q, 20p (Nair et al. 1997), 8q (Trembath et al. 1997), 1q (PSORS4 [MIM 603935] [Capon et al. 1999]), and 3q (PSORS5 [MIM 604316] [Enlund et al. 1999]). The genes responsible for susceptibility to psoriasis have not yet been identified.
The inheritance of psoriasis is complex; this complexity arises because variations in the phenotypic expression of the disease, genetic heterogeneity, and interactions either between genetic factors and the environment or among genes do not allow a simple correlation of the disease phenotype with the genotypic constitution (Lander and Schork 1994). As with other genetically complex diseases, the interpretation of linkage data obtained from genome scans for psoriasis susceptibility has been encumbered by many analytical problems. A consensus has therefore been reached that linkage findings in complex diseases require confirmation in independent data sets (Morton 1998). It is noteworthy that only two psoriasis-susceptibility loci, PSORS1 and PSORS2, have been confirmed in replication studies. We have therefore conducted a genomewide scan for psoriasis-susceptibility loci in a cohort of 32 extended, multiply affected pedigrees from northern Germany.
After obtaining approval from the ethics committees of all hospitals involved in the study and after obtaining informed consent from affected individuals, we identified families via an index patient and selected those with at least three affected individuals in two to four generations. Home visits were arranged, and all participating family members underwent a clinical examination, with particular attention to the sites where psoriasis most commonly appears, such as the scalp, elbows, knees, and nails. The diagnosis of psoriasis was made if two or more predilection sites were affected in a characteristic manner or if a single lesion covered >1% of the total body surface area. Unaffected relatives >20 years old, as well as all affected individuals, were enrolled in the study. The age restriction was imposed because the penetrance of psoriasis at age <20 years has been estimated to be only ∼50% (Lomholt 1963). Families with psoriasis pustulosa were excluded from this study.
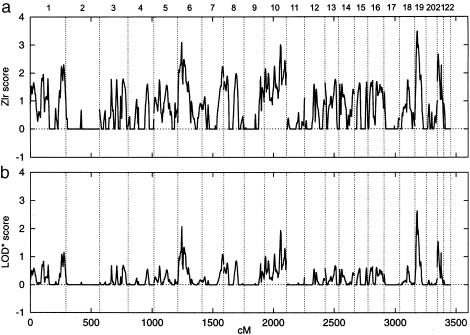
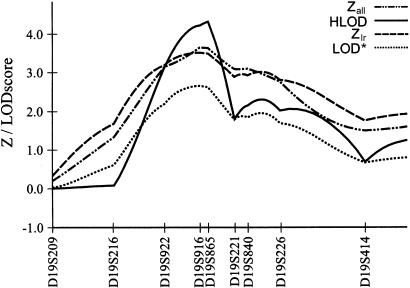
A genomewide scan for linkage was performed, using 370 microsatellite markers with an average marker interval of 10 cM and a mean heterozygosity of .8. In chromosomal regions of interest, the study population was genotyped with additional markers to further define those loci. All marker genotypes were checked for Mendelian inheritance by use of the PedCheck software (O'Connell and Weeks 1998). Nonparametric multipoint linkage analysis was performed, using uniform allele frequencies, with the GENEHUNTER-PLUS (Kong and Cox 1997) modification of GENEHUNTER (Kruglyak et al. 1996). One family was split to ensure that the program retained all affected individuals in the analysis. GENEHUNTER uses the nonparametric _Z_all statistic to estimate the significance of excess sharing of alleles identical by descent among all affected relatives (Kruglyak et al. 1996). Because the GENEHUNTER method produces conservative estimates when the descent information is incomplete, the GENEHUNTER-PLUS software uses the modified _Z_lr statistic that is well approximated by a normal distribution. GENEHUNTER-PLUS provides an accurate likelihood-ratio test to assess evidence for linkage and also allows the calculation of a nonparametric LOD score that is based on allele sharing and that can be interpreted in the same way as a traditional LOD score. The results of the nonparametric analysis are summarized in figure 1. The most significant allele sharing among affected relatives in families was detected at a novel locus on chromosome 19p13, near marker D19S916 (Z _lr_=3.50; _P_=.0002). This finding provided suggestive evidence for linkage. Because the nonparametric analysis includes information only from affected individuals, its power to detect linkage is expected to be lower than the power of parametric approaches that use information from all available relatives. However, the power to detect linkage by parametric LOD score analysis is sensitive to misspecification of the genetic model. It has been shown that maximization of LOD scores over multiple genetic models (maximized LOD [MOD] score) increases the power to detect linkage when the true mode of inheritance of the disease is unknown (Hodge et al. 1997). We have therefore maximized parametric multipoint LOD scores over penetrance models, using GENEHUNTER version 2.0 (Kruglyak et al. 1996). First, the penetrances were altered by a grid space of 0.1. The model leading to the highest LOD score was then varied, using a finer grid. In addition, the phenocopy rate was varied. Finally, we optimized the MOD score by changing allele frequencies. Under the assumption of locus heterogeneity, multipoint LOD scores (heterogeneity [HLOD]) were maximized for varying fractions of linked families (α), by use of GENEHUNTER version 2.0. On chromosome 19p, a MOD score of 2.38 was obtained at marker D19S865 (31.9 cM from pter), under the assumption of a recessive inheritance model with high disease-allele frequency. The HLOD score on 19p was 4.06, with α=66% of linked families (figure 2). To address the concern about the magnitude of the type 1 error after multiple markers and multiple genetic models were tested, the significance level of the maximum HLOD score was assessed by simulation. With the use of information from all 33 families, genotype data were generated by SIMULATE (Ott 1989), under the assumption of no linkage. Maximum HLOD scores were computed for the models used in the actual analysis by use of MSIM and ELODHET of the SLINK software (Ott 1989; Weeks et al. 1990). An empirical significance level was calculated as the proportion of replicates for which the maximum HLOD for any model was greater than the maximum obtained in the real data set and was corrected for testing multiple markers. The genomewide probability of obtaining an HLOD of 4.06 by chance in the present set of families and for all models tested in the actual analysis was estimated, using 20,000 replicates, to be <.037. This finding provides significant evidence, based on stringent criteria, for a novel psoriasis-susceptibility locus on 19p (Lander and Kruglyak 1995).
Figure 1.
Summary of the genome scan for psoriasis-susceptibility loci in 32 German extended families. The figure shows nonparametric multipoint GENEHUNTER PLUS _Z_lr (A) and LOD* score (B), across all autosomes, from pter to qter. Dotted lines indicate chromosome boundaries.
Figure 2.
Linkage analysis on chromosome 19, showing GENEHUNTER nonparametric LOD* score (_Z_all) and HLOD score, assuming a recessive model with high disease-allele frequency, as well as GENEHUNTER PLUS _Z_lr and nonparametric LOD score. Markers are arranged in map order, according to the final Généthon human linkage map (Dib et al. 1996).
The maximum HLOD was detected under the assumption of a recessive model with high disease-gene frequency. Although our parametric analysis does not allow conclusions about the true underlying mode of inheritance, it is interesting to note that the best model is consistent with the recessive-gene hypothesis proposed by Swanbeck et al. (1994). Epidemiological observations suggest that a recessive mode of inheritance of very common disease alleles would result in a pseudodominant inheritance pattern (Swanbeck et al. 1994). The new psoriasis locus on 19p13 coincides with a susceptibility locus for inflammatory bowel disease (Rioux et al. 2000). This finding supports the hypothesis proposed by Nair et al., on the basis of linkage findings on 16q for both psoriasis (Nair et al. 1997) and Crohn disease (Hugot et al. 1996), that common genetic factors may influence susceptibility to both systemic inflammatory conditions. The candidate interval on 19p is still quite large, comprising ⩾15 cM. Numerous genes and expressed sequence tags have been localized to this region. It is noteworthy that intercellular adhesion molecule-1 (ICAM-1) maps to the interval between D19S413 (31.9 cM from pter) and D19S221 (35.5 cM from pter) (Deloukas et al. 1998). ICAM-1, a glycoprotein of the immunoglobulin superfamily, is expressed on leukocytes, endothelium, and fibroblasts and is a ligand for lymphocyte function–associated (LFA) antigens. Through the interaction with LFA-1, ICAM-1 functions as a major cell-adhesion molecule, mediating leukocyte migration into sites of inflammation (Springer 1994), T cell activation (Siu et al. 1989), and T cell–effector function (Ybarrondo et al. 1994). It has been proposed that keratinocytes participate in the immunohomeostasis of the skin and in the regulation of T cell activation (Nickoloff et al. 1995). In psoriasis, ICAM-1 is expressed on the surface of keratinocytes, and Nickoloff et al. (1993) have suggested that ICAM-1 has a major role in the keratinocyte-mediated costimulation of T cell proliferation. ICAM-1 therefore represents an interesting positional and functional candidate, on 19p, for psoriasis susceptibility. Transmission disequilibrium studies on an extended sample are under way to further elucidate the role of the ICAM-1 gene in psoriasis.
In addition, suggestive evidence of linkage was detected on chromosomes 6p, 21q, and 8q (table 1). Of 20,000 replicates, 16 had an HLOD of 2.5. Because we tested 370 markers, an HLOD of 2.5 corresponds to a genomewide P value of .296, according to our simulation. Increased allele sharing was observed at marker D6S422 in the HLA region where studies involving two independent genome scans have reported linkage to psoriasis (Nair et al. 1997; Trembath et al. 1997). A MOD score of 2.46 was obtained, under the assumption of a recessive mode of inheritance. Analogous to other investigators (Leder et al. 1998), we found no significant evidence for heterogeneity on 6p. Given that the replication of previously reported loci requires less stringent criteria for significance (Lander and Kruglyak 1995), our finding confirms the presence of a major psoriasis locus in this region. Additional investigations will be required to confirm the relevance of the chromosomal regions on 21q and 8q for psoriasis susceptibility.
Table 1.
Summary of Nonparametric and Parametric Linkage Analysis for Regions on Chromosomes 6, 8, 19, and 21 with Suggestive Evidence for Linkage
| Nonparametric Statistics | Parametric Statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome and Marker | Distance from pter (cM)a | _Z_allb | _Z_lrc | Pd | LODc | MOD | PenetranceVector | Disease AlleleFrequency | HLOD | α |
| 6: | ||||||||||
| D6S260 | 29.6 | 2.03 | 2.30 | .012 | 1.15 | |||||
| D6S422 | 35.7 | 3.07 | 3.10 | .001 | 2.08 | 2.46 | .02, .1, .6 | .01 | 2.93 | .75 |
| 8: | ||||||||||
| D8S286 | 93.5 | 1.78 | 1.52 | .064 | .50 | 3.13 | .03, .15, .3 | .001 | 3.15 | .85 |
| D8S270 | 102.1 | 2.02 | 1.97 | .024 | .84 | |||||
| 19: | ||||||||||
| D19S922 | 26.0 | 3.12 | 3.17 | .0008 | 2.18 | |||||
| D19S916 | 30.8 | 3.62 | 3.50 | .0002 | 2.64 | |||||
| D19S865 | 31.9 | 3.60 | 3.46 | .0003 | 2.60 | 2.38 | .03, .1, 1.0 | .04 | 4.06 | .66 |
| D19S221 | 35.5 | 3.06 | 2.86 | .0021 | 1.78 | |||||
| D19S840 | 37.3 | 3.07 | 2.90 | .0019 | 1.82 | |||||
| D19S226 | 41.7 | 2.74 | 2.78 | .0027 | 1.68 | |||||
| 21: | ||||||||||
| D21S236 | 2.6 | 2.06 | 2.43 | .0075 | 1.28 | |||||
| D21S1256 | 8.6 | 2.02 | 2.48 | .0066 | 1.33 | 2.84 | .03, .1, .95 | .98, .02 | 3.35 | .73 |
The present study in a large cohort of German extended families with high rates of psoriasis provides significant evidence for a novel psoriasis-susceptibility locus on 19p, which awaits replication in independent data sets. In addition, linkage to the HLA region on 6p was confirmed, demonstrating the importance of this region for psoriasis susceptibility.
Acknowledgments
The kind cooperation of all participating families is gratefully acknowledged. We are grateful to Madeleine Skorna, Johanna Harder-d'Heureuse, and Alexandra Forster for excellent technical assistance. This project was supported by Deutsche Forschungsgemeinschaft grants Re 679/10-1, Tr228/5-1, and Wi155/1-1. The Gene Mapping Center is funded by a grant from the German Human Genome Project (to A.R. and T.F.W).
Electronic-Database Information
The accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for PSORS1 [MIM 177900], PSORS2 [MIM 602723], PSORS3 [MIM 601454], PSORS4 [MIM 603935], and PSORS5 [MIM 604316], respectively)
- GeneMap'99, http://www.ncbi.nlm.nih.gov/genemap/
References
- Abele DC, Dobson RL, Graham JB (1963) Heredity and psoriasis: study of a large family. Arch Dermatol 88:38–47 [DOI] [PubMed] [Google Scholar]
- Boehncke WH, Zollner TM, Dressel D, Kaufmann R (1997) Induction of psoriasiform inflammation by a bacterial superantigen in the SCID-hu xenogeneic transplantation model. J Cutan Pathol 24:1–7 [DOI] [PubMed] [Google Scholar]
- Brandrup F, Hauge M, Henningsen K, Eriksen B (1978) Psoriasis in an unselected series of twins. Arch Dermatol 114:874–878 [PubMed] [Google Scholar]
- Capon F, Novelli G, Semprini S, Clementi M, Nudo M, Vultaggio P, Mazzanti C, Gobello T, Botta A, Fabrizi G, Dallapiccola B (1999) Searching for psoriasis susceptibility genes in Italy: genome scan and evidence for a new locus on chromosome 1. J Invest Dermatol 112:32–35 [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, Bentolila S, Bihoreau M, Birren BB, Browne J, Butler A, Castle AB, Chiannilkulchai N, Clee C, Day PJ, Dehejia A, Dibling T, Drouot N, Duprat S, Fizames C, Bentley DR (1998) A physical map of 30,000 human genes. Science 282:744–746 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Enlund F, Samuelsson L, Enerback C, Inerot A, Wahlstrom J, Yhr M, Torinsson A, Riley J, Swanbeck G, Martinsson T (1999) Psoriasis susceptibility locus in chromosome region 3q21 identified in patients from southwest Sweden. Eur J Hum Genet 7:783–790 [DOI] [PubMed] [Google Scholar]
- Farber EM, Nall ML, Watson W (1974) Natural history of psoriasis in 61 twin pairs. Arch Dermatol 109:207–211 [PubMed] [Google Scholar]
- Hodge SE, Abreu PC, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Leonard-Jones JE, Cortot A, Colombel JF, Thomas G (1996) Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature 379:821–823 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed] [Google Scholar]
- Leder RO, Mansbridge JN, Hallmayer J, Hodge SE (1998) Familial psoriasis and HLA-B: unambiguous support for linkage in 97 published families. Hum Hered 48:198–211 [DOI] [PubMed] [Google Scholar]
- Lomholt G (1963) Psoriasis: prevalence, spontaneous course, and genetics. GEL Gad, Copenhagen [Google Scholar]
- Matthews D, Fry L, Powles A, Weber J, McCarthy M, Fisher E, Davies K, Williamson R (1996) Evidence that a locus for familial psoriasis maps to chromosome 4q. Nat Genet 14:231–233 [DOI] [PubMed] [Google Scholar]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62:690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Henseler T, Jenisch S, Stuart P, Bichakjian CK, Lenk W, Westphal E, Guo SW, Christophers E, Voorhees JJ, Elder JT (1997) Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 6:1349–1356 [DOI] [PubMed] [Google Scholar]
- Nevitt GJ, Hutchinson PE (1996) Psoriasis in the community: prevalence, severity and patients' beliefs and attitudes towards the disease. Br J Dermatol 135:533–537 [PubMed] [Google Scholar]
- Nickoloff BJ, Mitra RS, Green J, Zheng XG, Shimizu Y, Thompson C, Turka LA (1993) Accessory cell function of keratinocytes for superantigens: dependence on lymphocyte function–associated antigen-1/intercellular adhesion molecule–1 interaction. J Immunol 150:2148–2159 [PubMed] [Google Scholar]
- Nickoloff BJ, Turka LA, Mitra RS, Nestle FO (1995) Direct and indirect control of T-cell activation by keratinocytes. J Invest Dermatol Suppl 105:25S–29S [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci U S A 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB, Bitton A, Williams CN, Greenberg GR, Cohen Z, Lander ES, Hudson TJ, Siminovitch KA (2000) Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet 66:1863–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu G, Hedrick SM, Brian AA (1989) Isolation of the murine intercellular adhesion molecule 1 (ICAM-1) gene: ICAM-1 enhances antigen-specific T cell activation. J Immunol 143:3813–3820 [PubMed]
- Springer TA (1994) Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76:301–314 [DOI] [PubMed] [Google Scholar]
- Swanbeck G, Inerot A, Martinsson T, Wahlstrom J (1994) A population genetic study of psoriasis. Br J Dermatol 131:32–39 [DOI] [PubMed] [Google Scholar]
- Tomfohrde J, Silverman A, Barnes R, Fernandez-Vina MA, Young M, Lory D, Morris L, Wuepper KD, Stasmy P, Menter A (1994) Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 264:1141–1145 [DOI] [PubMed] [Google Scholar]
- Trembath RC, Clough RL, Rosbotham JL, Jones AB, Camp RD, Frodsham A, Browne J, Barber R, Terwilliger J, Lathrop GM, Barker JN (1997) Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet 6:813–820 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Ybarrondo B, O'Rourke AM, Brian AA, Mescher MF (1994) Contribution of lymphocyte function-associated-1/intercellular adhesion molecule-1 binding to the adhesion/signaling cascade of cytotoxic T lymphocyte activation. J Exp Med 179:359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]