Analysis of the Essential Cell Division Gene ftsL of Bacillus subtilis by Mutagenesis and Heterologous Complementation (original) (raw)
Abstract
The ftsL gene is required for the initiation of cell division in a broad range of bacteria. Bacillus subtilis ftsL encodes a 13-kDa protein with a membrane-spanning domain near its N terminus. The external C-terminal domain has features of an α-helical leucine zipper, which is likely to be involved in the heterodimerization with another division protein, DivIC. To determine what residues are important for FtsL function, we used both random and site-directed mutagenesis. Unexpectedly, all chemically induced mutations fell into two clear classes, those either weakening the ribosome-binding site or producing a stop codon. It appears that the random mutagenesis was efficient, so many missense mutations must have been generated but with no phenotypic effect. Substitutions affecting hydrophobic residues in the putative coiled-coil domain, introduced by site-directed mutagenesis, also gave no observable phenotype except for insertion of a helix-breaking proline residue, which destroyed FtsL function. ftsL homologues cloned from three diverse Bacillus species, Bacillus licheniformis, Bacillus badius, and Bacillus circulans, could complement an ftsL null mutation in B. subtilis, even though up to 66% of the amino acid residues of the predicted proteins were different from B. subtilis FtsL. However, the ftsL gene from Staphylococcus aureus (whose product has 73% of its amino acids different from those of the B. subtilis ftsL product) was not functional. We conclude that FtsL is a highly malleable protein that can accommodate a large number of sequence changes without loss of function.
During the 1960s and 1970s, researchers isolated a number of temperature-sensitive Escherichia coli and Bacillus subtilis mutants that formed filaments at the nonpermissive temperature. In this way, several cell division genes were identified and given the designation fts for filamenting temperature sensitive (6). Some B. subtilis genes were also isolated on the grounds of similarity to E. coli cell division genes or by their chromosomal position (2, 9, 14).
In B. subtilis, at least six gene products, FtsZ, FtsA, FtsL, DivIC, DivIB, and PBP 2B, are required for septation, which involves the invagination of the cytoplasmic membrane accompanied by septum-specific peptidoglycan synthesis (3, 4, 5, 8, 9, 23). During vegetative growth, these proteins catalyze the formation of a central septum, which gives rise to two equal-sized daughter cells. In the process of spore formation, asymmetric cell division produces a prespore and a larger mother cell compartment.
Early in the cell cycle, the tubulin-like FtsZ is assembled into a ring structure at the future division site (36). FtsZ recruits other components of the division machinery to mid-cell and is probably directly involved in septal constriction (26). FtsA, which has homology to the ATPase domain found in actin, DnaK, and hexokinases (33), is known to directly interact with FtsZ, but its precise role in septation is still unclear (39). In addition to these two cytoplasmic proteins, there are four bitopic division proteins with their major domains located outside the cytoplasmic membrane, FtsL, DivIC, DivIB, and PBP 2B. PBP 2B belongs to the family of high-molecular-weight penicillin-binding proteins which catalyze the late stages of peptidoglycan biosynthesis. It assembles late at the future division site and is required for the formation of septal peptidoglycan (8, 10). Localization studies of the smaller transmembrane proteins FtsL, DivIC, and DivIB have shown that they are also part of the septator (7, 16, 20, 35). While FtsL and DivIC are essential for division, cells can divide in the absence of DivIB at low temperatures (≤30°C). Thus, DivIB may be involved in stabilizing or promoting the formation of the division complex at higher temperatures (15). Recently, Katis and Wake (21) have shown that only the external C termini of DivIC and DivIB are essential, indicating that these proteins may have a role in septal peptidoglycan synthesis. This group of proteins could therefore form a direct or indirect link between cytokinesis mediated by the Z ring and peptidoglycan synthesis carried out outside the cytoplasmic membrane. Accordingly, these genes appear to be collectively absent from the genomes of bacteria that lack peptidoglycan (e.g., Mycoplasma genitalium and Methanococcus jannaschii), consistent with the idea that they have a role in septal peptidoglycan synthesis or its regulation.
FtsL is a small protein (117 amino acids) with a potential membrane-spanning domain near the N terminus and an external C terminus, which resembles an α-helical leucine zipper. Although FtsL homologues from other organisms have similar structural features, the sequence conservation is relatively weak. Thus, B. subtilis ftsL (yllD) was identified as the homologue of E. coli ftsL (16% identical residues) (9) because of its position upstream of pbpB, homologous to E. coli ftsI. We have shown recently that FtsL interacts with the FtsL-like DivIC protein (35). This probably stabilizes DivIC, which is rapidly degraded when FtsL is depleted (9). In this study, we used a combination of random mutagenesis, site-directed mutagenesis, and substitutions with heterologous genes to probe the sequence of FtsL for amino acid residues that are important for its function.
MATERIALS AND METHODS
Strains, phages, and plasmids.
The strains, phages, and plasmids used in this work are listed in Table 1. All Bacillus strains were isogenic with strain 168.
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant characteristic(s) | Source or reference |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| 2001 | trpC2 (φ105J506) cat Pxyl-ftsL Ω(ftsL::neo)1401 | 35 |
| 2003 | trpC2 (φ105J506) cat Pxyl-ftsL Ω(ftsL::neo)1401 Ω(amyE::spc Pspac-ftsL lacI) | This study |
| 2005 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) | Strain 168 transformed to spectinomycin resistance with strain 2003 DNA |
| 2006 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 | Strain 2005 transformed to kanamycin resistance with strain 2000 DNA |
| 2007 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125) | Strain 2006 lysogenized with φ105J125 |
| 2030 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL26) | This study |
| 2031 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL27) | This study |
| 2032 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL28) | This study |
| 2033 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL29) | This study |
| 2034 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL30) | This study |
| 2036 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL31) | This study |
| 2037 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL32) | This study |
| 2038 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL33) | This study |
| 2039 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL34) | This study |
| 2040 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL37) | This study |
| 2041 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL38) | This study |
| 2042 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL36) | This study |
| 2043 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsL35) | This study |
| 2044 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsLSa) | This study |
| 2045 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsLBl) | This study |
| 2046 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsLBb) | This study |
| 2047 | trpC2 Ω(amyE::spc Pspac-ftsL lacI) Ω(ftsL::neo)1401 (φ105J125::cat Pxyl-ftsLBc) | This study |
| B. licheniformis 6346 | NCIMBa | |
| B. badius S33 | F. G. Priestb | |
| B. circulans DSMII | F. G. Priestb | |
| S. aureus RN4420 | S. J. Fosterc | |
| E. coli DH5α | F−endA1 hsdR17 supE44 thi-1 λ− recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 φ80 d_lacZΔM15_ | Gibco BRL |
| Plasmids | ||
| pRD96 | bla cat Pxyl | 9 |
| pRD99 | bla cat Pxyl-ftsL | 9 |
| pSG1405 | bla cat Pxyl-ftsL26 | This study |
| pSG1406 | bla cat Pxyl-ftsL27 | This study |
| pSG1407 | bla cat Pxyl-ftsL28 | This study |
| pSG1408 | bla cat Pxyl-ftsL29 | This study |
| pSG1409 | bla cat Pxyl-ftsL30 | This study |
| pSG1411 | bla cat Pxyl-ftsL31 | This study |
| pSG1412 | bla cat Pxyl-ftsL32 | This study |
| pSG1413 | bla cat Pxyl-ftsL33 | This study |
| pSG1414 | bla cat Pxyl-ftsL34 | This study |
| pSG1415 | bla cat Pxyl-ftsL37 | This study |
| pSG1416 | bla cat Pxyl-ftsL38 | This study |
| pSG1417 | bla cat Pxyl-ftsL36 | This study |
| pSG1418 | bla cat Pxyl-ftsL35 | This study |
| pSG1436 | bla cat Pxyl-ftsLSa | This study |
| pSG1437 | bla cat Pxyl-ftsLBl | This study |
| pSG1438 | bla cat Pxyl-ftsLBb | This study |
| pSG1439 | bla cat Pxyl-ftsLBc | This study |
| Phages | ||
| φ105J125 | ind-125 cts-23 sal-104 mcs-23 Δ(DI:lt) (cloning vector) | 37 |
| φ105J506 | φ105J125::cat Pxyl-ftsL | 9 |
General methods.
B. subtilis strains were transformed by the method described by Anagnostopoulos and Spizizen (1), as modified by Jenkinson (18), or by the method described by Kunst and Rapoport (22), except that 20 min after addition of DNA the transformed cultures were supplemented with 0.66% Casamino Acids solution. Transformants were selected on Oxoid nutrient agar containing, as necessary, kanamycin (5 μg ml−1), spectinomycin (50 μg ml−1), or chloramphenicol (5 μg ml−1).
DNA manipulations and E. coli transformations were carried out as described by Sambrook et al. (32). All cloning was done in E. coli DH5α (Gibco BRL). Methods for the manipulation and growth of φ105 ind cts derivatives were essentially as described by Errington (11).
Construction of a strain containing two inducible ftsL genes.
The xylose-dependent strain 2001 is lysogenic for the recombinant bacteriophage φ105J506, which contains a copy of ftsL under the control of the xylose-inducible Pxyl promoter. The natural copy of ftsL has been disrupted by a neo resistance cassette, which provides sufficient transcription for the essential pbpB gene downstream. A second copy of ftsL under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Pspac promoter was inserted into the amyE locus of strain 2001, giving strain 2003. Full details of the complicated constructions of strains and plasmids are available on request. Strain 2003 grew normally in the presence of IPTG or xylose but failed to divide and formed elongated filaments in unsupplemented medium.
Strain 168 was transformed with chromosomal DNA from strain 2003 with selection for spectinomycin resistance to give strain 2005, containing the Pspac-ftsL construct inserted at the amyE site. The genomic copy of ftsL in 2005 was disrupted by transformation with chromosomal DNA from strain 2000 with selection for kanamycin resistance, resulting in the IPTG-dependent strain 2006. This strain was lysogenized with the bacteriophage φ105J125 to give strain 2007.
ftsL expression experiments.
The effects of the expression of different ftsL alleles were studied in strain 2007 and its derivatives. The strains were grown on plates containing xylose (0.5%), IPTG (1 mM), or no inducer. To examine vegetatively growing cells, an overnight culture grown in Difco antibiotic medium 3 (PAB) containing 1 mM IPTG was diluted 50-fold and incubated at 37°C until it reached an optical density at 600 nm (OD600) of 0.8. The cells were centrifuged for 1 min, washed once with prewarmed, unsupplemented PAB, and then resuspended in this medium. The culture was split into two or three aliquots and supplemented with xylose (0.5%), IPTG (1 mM), and no inducer, respectively. These aliquots were incubated for another 1.5 h at 37°C (or at 30 or 45°C to test for temperature dependence) and then fixed with ethanol for microscopic analysis (see below).
Light microscopy.
Cell morphology was analyzed by phase-contrast light microscopy after fixation with ethanol as described previously (17). The images obtained were processed with IPlab Spectrum 3.1a software (Signal Analyticals, Vienna, Va.), and final images were assembled with Adobe Photoshop version 3.0.5.
Hydroxylamine mutagenesis of ftsL and isolation of mutants.
φ105J506 phage DNA was mutagenized in a 100 mM potassium phosphate buffer (pH 4.0) containing 1 mM EDTA. A 1 M hydroxylamine solution (pH 6.0) was added to a final concentration of 0.4 M. Samples were incubated for 1 h at 75°C and then diluted 10-fold with water to stop the reaction. The DNA was recovered by ethanol precipitation. The mutagenesis was repeated once as described above. The phage DNA was transformed into strain 2007 with selection for chloramphenicol in the presence of IPTG (1 mM). Transformants were then transferred to plates containing 0.5% xylose but no IPTG. To sequence mutations in ftsL, primers xyl(promoter)-f (5′-GGGCAACAAACTAATGTGCAAC-3′) and 3860 (5′-TTTTGGATCCTTTGAATCATTCCTGTATG-3′) were used to amplify Pxyl-ftsL by PCR. Following purification of the PCR product, using a QIAquick PCR purification kit (Qiagen), the same primers were used for sequencing.
DNA sequence analysis.
Plasmids and PCR products were sequenced on both strands using the ABI PRISM BigDye terminator cycle sequencing kit (Perkin-Elmer). Custom-synthesized oligonucleotides were used as primers, and gels were run on an ABI PRISM 377 DNA sequencer.
Site-directed mutagenesis of ftsL and isolation of mutants.
Most of the ftsL mutations were generated by PCR-based mutagenesis using Pfu DNA polymerase (Stratagene), as recommended by the manufacturer. Each 100-μl PCR mixture in 1× Pfu buffer consisted of 35 ng of pRD99 DNA as the template, primers (one primer carrying the desired base changes; primer sequences are available on request) at a final concentration of 0.5 μM, deoxynucleotide triphosphates (dNTPs; final concentration of 200 μM for each dNTP) and 5 U of Pfu DNA polymerase. Each reaction cycle, repeated 25 times, included the following: 96°C for 45 s, 50°C for 45 s, and 72°C for 11 min. The PCR product was separated on an 0.8% agarose gel and purified using the QIAquick gel extraction kit (Qiagen). The purified PCR product was treated with T4 polynucleotide kinase and ligated in a simultaneous reaction (30 μl) carried out in 1× ligation buffer (Roche Molecular), containing ATP (3.3 mM), 1 U of T4 DNA ligase, and 1 U of T4 polynucleotide kinase. This mixture was incubated for 24 h at room temperature, and 15 μl of this reaction mixture was transformed into E. coli DH5α.
All plasmids were checked for the desired mutations in ftsL by DNA sequence analysis. Plasmids were digested with _Sma_I, ligated to _Sma_I-digested φ105J125 phage DNA at a high DNA concentration, and directly transformed into strain 2007 with selection for chloramphenicol in the presence of IPTG (1 mM). This resulted in the random insertion of the Pxyl-ftsL cassette between _Sma_I sites in the φ105J125 prophage. Transformants were transferred to plates containing 0.5% xylose but no IPTG. The alleles ftsL35 and ftsL36 were generated by amplification errors of Taq DNA polymerase (constructs not shown) and subcloned into pRD96 to give pSG1418 and pSG1417, respectively. ftsL37 was amplified from strain 168 by PCR using primers ftsL.fw2 (5′-CGAAACGCGAACTCTAGATTAAAAGGAGG-3′) and ftsL.rv3 (5′-GAATCATTCCTGCAGGTTTTACACGTTTTACACTTTTTTATC-3′), and ftsL38 was amplified using primers ftsL.fw2 and ftsL.rv4 (5′-TTCTGCAGCTTATAAATTCAAGCCGTTCTTTTTCG-3′), introducing _Xba_I and _Pst_I sites, respectively. The _Xba_I-_Pst_I fragments were inserted between these sites in pRD96 to give pSG1415 and pSG1416, respectively. These ftsL alleles were introduced into the chromosome of strain 2007 as described above.
Isolation of ftsL genes from Bacillus licheniformis, Bacillus badius, and Bacillus circulans.
Two peptide sequences in YllC and in PBP 2B and PBP 2X, respectively, conserved in B. subtilis and Streptococcus pneumoniae, were chosen for use in the design of degenerate primers. The peptide TFHSLED (residues 241 to 247 in B. subtilis YllC) was used to design the forward primer pool yllC.degen.fw [5′-GGAATTC AC(A/C/G/T) TT(C/T) CA(C/T) TC(A/C/G/T) CT(A/C/G/T) GA(A/G) G-3′]. The reverse primer pool pbpB.degen.rv2 [5′-(C/T)TT CAT IGT I(C/G)(A/T) ICC IGG (C/T)TC-3′] is based on EPGSTMK (residues 309 to 315 in B. subtilis PBP 2B), which includes the common motif of PBPs, SXXK. The 3′ primer also contained deoxyinosine (I) to reduce the degeneracy. The ftsL gene was amplified by PCR from strains B. licheniformis 6346, B. badius S33, and B. circulans DSMII using the degenerate primer pools and Taq DNA polymerase. The corresponding gene products of 1.5 kb were directly sequenced and (nondegenerate) primers were constructed to allow gene amplification using the more accurate Pfu DNA polymerase (Stratagene). B. licheniformis ftsL was amplified using primers Bl.fw1 (5′-CCTCACGGACTGCCCGTGATCCCC-3′) and Bl.rv1 (5′-TTTCCCGTCGCTTGAATAAACGC-3′). Primers for B. badius ftsL were Bb.fw1 (5′-CCTCCGGGGCTGCCGGTCATTCC-3′) and Bb.rv1 (5′-CCAGTTATTTGTATCATCAAAAAACGG-3′), and primers for B. circulans ftsL were Bc.fw1 (5′-CCGCCAGGGCTACCATTTATTCC-3′) and Bc.rv1 (5′-CCTGTCGCCTGAATATAGACAACCCG-3′). To reduce any sequencing errors resulting from PCR amplification artefacts, at least three independent PCR products were sequenced.
Complementation studies.
ftsL genes were PCR-amplified from chromosomal DNA of the appropriate strains using the following primers: ftsLSa.fw (5′-TGCTCTAGAAAGGAGGTCATCAGCCTATGGCTGTAGAAAAAGTGTACCAACC-3′) and ftsLSa.rv (5′-TTTAAGCTTAATTTTTTGCTTCGCCATTAC-3′) for S. aureus ftsL, ftsLBl.fw (5′-GGAAAAAGCTCTAGAGTGAGCTGTACG-3′) and ftsLBl.rv (5′-TTTAAGCTTTTTGCATCATTCCTGTATGTC-3′) for B. licheniformis ftsL, ftsLBb.fw (5′-AATCTAGAAAAGGAGGTCATCAGCCTATGAGCAATCTAGCCAGAAAACAAC-3′) and ftsLBb.rv (5′-CATAAGCTTTTCATTCATTGCGGCTGC-3′) for B. badius ftsL, and ftsLBc.fw (5′-AATC TAGAAAAGGAGGTCATCAGCCTATGAGCAACTTAGCAAGAAAAAT GC-3′) and ftsLBc.rv (5′-CGGATCAAAGCTTAATGCACAACC-3′) for B. circulans ftsL, introducing _Xba_I and _Hin_dIII sites, in order respective to the listed primers for each gene. The DNA regions upstream of Staphylococcus aureus ftsL, B. badius ftsL, and B. circulans ftsL were replaced by the sequence upstream of B. subtilis ftsL, including its ribosome-binding site (RBS). The _Xba_I-_Hin_dIII fragments were inserted between these sites in pRD96. Plasmids and φ105J125 phage DNA were digested with _Sst_I and _Hin_dIII, ligated, and directly transformed into strain 2007 with selection for chloramphenicol resistance in the presence of IPTG. Transformants were patched out on plates containing IPTG or xylose. To make derivatives in which the Pspac-ftsL construct located in the amyE gene was eliminated, each strain was transformed with a plasmid containing a tet cassette flanked by segments from the amyE locus (J. Sievers, unpublished data), with selection for resistance to tetracycline.
Nucleotide sequence accession numbers.
The EMBL accession numbers of the nucleotide sequences of B. licheniformis, B. badius, and B. circulans ftsL determined in this work are AJ271356, AJ271357, and AJ271358, respectively.
RESULTS AND DISCUSSION
Construction of a strain with two inducible alleles of ftsL.
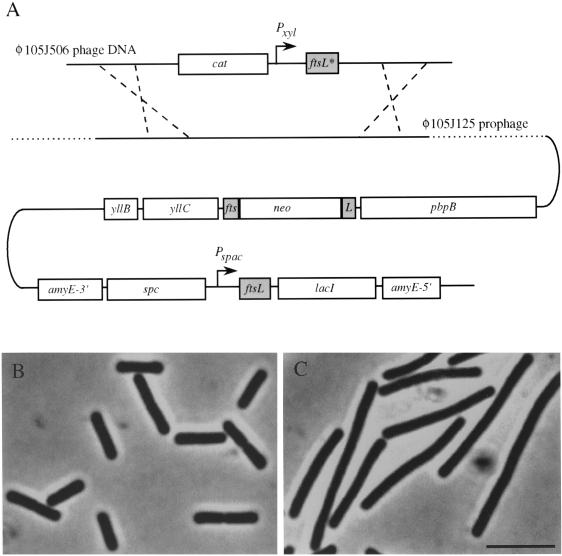
We constructed a conditional B. subtilis mutant with two copies of ftsL, each under the control of a different inducible promoter. This allowed us to mutagenize one copy of ftsL, study the effects of mutations on division, and maintain strains by expression of the other (wild-type) allele. In strain 2007, one allele of ftsL under the control of the IPTG-inducible Pspac promoter was inserted at the amyE locus (Fig. 1A). The natural ftsL gene has been disrupted with a kanamycin resistance cassette (neo), which maintains transcription of the likewise essential pbpB gene downstream of ftsL (Fig. 1A). This strain formed elongated aseptate filaments when grown in the absence of IPTG, but growth was indistinguishable from that of strain 168 in IPTG-supplemented PAB as judged by phase-contrast microscopy (Fig. 1B and C). Strain 2007 was also lysogenized with the φ105J125 phage. Hence, a second allele of ftsL under the control of the xylose-inducible Pxyl promoter, placed in the recombinant φ105J506 phage (9), could be integrated into the chromosome of 2007 by a double-crossover event (indicated by dashed lines in Fig. 1A). We previously showed that the Pxyl promoter can be used to control ftsL expression and that division is normal in the presence of xylose and completely blocked in the absence of xylose (9).
FIG. 1.
Strain 2007 constructed for mutagenesis of ftsL. (A) Schematic representation of the genetic organization of strain 2007. The chromosomal pbpB operon contains four genes, yllB, yllC, ftsL, and pbpB. The chromosomal copy of ftsL is disrupted by a neo cassette, whose promoter also provides transcription of the downstream pbpB gene. An IPTG-inducible wild-type copy of ftsL is placed in the amyE locus. The φ105J125 prophage, integrated elsewhere in the chromosome, allows transformation with φ105J506 phage DNA and the insertion by a double-crossover event (dashed lines) of a second copy of ftsL (ftsL*) under the control of the xylose-inducible Pxyl promoter. (B and C) Phase-contrast images of 2007 grown in the presence of the inducer IPTG (B) and after ca. three generations in its absence (C). Bar, 5 μm.
Random mutagenesis of the ftsL gene.
φ105J506 phage DNA was mutagenized with hydroxylamine (HA) in vitro. As a hydroxyl donor, HA deaminates cytosine to uracil, or a stable intermediate in the deamination of cytosine, hydroxyaminocytosine, is formed. Both mechanisms give rise to C-G to T-A transition mutations (38). The mutagenized DNA was transformed into strain 2007 with selection for chloramphenicol resistance in the presence of IPTG (therefore maintaining expression only of the nonmutagenized copy of ftsL). Transformants were then streaked on plates containing xylose but no IPTG to screen for mutations affecting the ftsL allele under the control of the Pxyl promoter. Out of some 2,500 transformants in several separate experiments, 22 independently isolated mutants were obtained that showed impaired growth (cell filamentation) in the presence of xylose (Table 2). None of these mutations turned out to be dominant to the wild type, as tested by growth in the presence of both IPTG and xylose (not shown). DNA sequence analysis revealed that we had obtained 12 distinct point mutations in the ftsL gene, which fell into two clear classes. The major class comprised nonsense mutations. Given the effect of HA, nine possible stop codons could be generated by single base changes; in each case a glutamine codon, CAA or CAG, would become a stop codon, TAA or TAG, respectively. Substitutions of six of these codons, yielding Q7, Q11, Q12, Q72, Q79, and Q94 of the FtsL protein, were obtained. A nonsense mutation in a seventh codon, encoding Q116, which is only one residue from the C terminus, would have no phenotypic effect because an FtsL protein truncated of the last five residues is functional (see below; Table 2). Thus, only two potential stop codons were missed. An additional TAG mutation (ftsL4) resulted from an unusual transversion in TCG, which encodes S54. In only one of 117 residues did we obtain a missense mutation, L37→F in the ftsL11 product, but this allele also carried a TAG mutation (Q11→amber). Four of the seven nonsense mutations were isolated independently more than once.
TABLE 2.
Sequences of new ftsL mutations and their phenotypic effects
| Allele(s) | Amino acid change(s) | Base change(s) | Phenotype |
|---|---|---|---|
| Random mutagenesis | |||
| ftsL2, ftsL15, ftsL18, ftsL23 | RBS | GGAGG→GAAGG | Filamentous |
| ftsL3, ftsL22 | Q94→amber | CAG→TAG | Filamentous |
| ftsL4 | S54→amber | TCG→TAG | Filamentous |
| ftsL5, ftsL7 | RBS | GGAGG→GGAAG | Filamentous |
| ftsL6, ftsL10, ftsL16 | RBS | GGAGG→AGAGG | Filamentous |
| ftsL8, ftsL25 | Q12→amber | CAG→TAG | Filamentous |
| ftsL11 | Q11→amber, L37→F | CAG→TAG, CTT→TTT | Filamentous |
| ftsL12, ftsL13 | Q11→amber | CAG→TAG | Filamentous |
| ftsL14, ftsL24 | Q72→ochre | CAA→TAA | Filamentous |
| ftsL17 | S74→ochre | TCA→TAA | Filamentous |
| ftsL19 | Q79→ochre | CAA→TAA | Filamentous |
| ftsL21 | Q7→ochre, Y6→Y | CAA→TAA, TAC→TAT | Filamentous |
| Site-directed mutagenesis | |||
| ftsL26 | L69→A | CTT→GCT | Wild type |
| ftsL27 | L83→A | CTC→GCT | Wild type |
| ftsL28 | L83→D | CTC→GAC | Wild type |
| ftsL29 | L83→F | CTC→TTT | Wild type |
| ftsL30 | L83E84K85→AAA | CTCGAAAAA→GCTGCGGG | Wild type |
| ftsL31 | E70→A | GAG→GCG | Wild type |
| ftsL32 | K102→A | AAG→GCG | Wild type |
| ftsL33 | L69E70→AA | CTTGAG→GCTGCG | Wild type |
| ftsL34 | L69E70E71→AAA | CTTGAGGAG→GCTGCGGG | Wild type |
| ftsL35 | I99→T | ATT→ACT | Wild type |
| ftsL36 | L69→P | CTT→CCT | Filamentous |
| ftsL37 | K113→ochre | AAA→TAA | Wild type |
| ftsL38 | K108→ochre | AAA→TAA | Filamentous |
The second class of mutations weakened the ftsL RBS, which strongly resembles the consensus sequence (28). Again, all of these mutations were obtained at least twice, and three of the four conserved G residues in the site were affected. It seems reasonable to assume that these mutations work by reducing the level of synthesis of FtsL to the point where it no longer functions. We conclude, from the abundance of mutations that severely truncate or reduce synthesis of FtsL, that the chemical mutagenesis was efficient and almost saturating for C-G to T-A transitions. Hence, many other amino acid substitutions in FtsL must have been generated but with no phenotypic effect. In principle, 86 amino acid substitutions affecting 67 different amino acids (57% of the total) could have been generated. Although most of the transitions would generate relatively conservative substitutions, the results suggest that relatively few residues in the FtsL sequence are critical for its function. Another group has recently reported a disproportionate number of nonsense mutations in B. subtilis, in experiments studying the spoIIR gene (19).
The repeating leucine heptad motif in B. subtilis FtsL is dispensable.
To probe more directly for mutations affecting FtsL function, we utilized site-directed mutagenesis to make defined missense mutations in ftsL. As for the random mutagenesis, the mutated ftsL alleles were placed in the φ105J125 phage under the control of the Pxyl promoter and introduced into the chromosome of strain 2007.
It has previously been suggested that the C-terminal domains of FtsL proteins (e.g., E. coli) and of the FtsL-like DivIC protein of B. subtilis have features of an α-helical leucine zipper (13, 23). In B. subtilis, they could be involved in the heterodimer formation of FtsL and DivIC (35). However, the characteristic repeating leucine heptad motif with four or five leucine residues spaced seven amino acids apart, is not fully conserved in B. subtilis FtsL, as the second leucine is replaced by a glutamate residue (E76). First, we altered two of the leucine residues of this motif, L69 and L83, by site-directed mutagenesis (see Material and Methods). Single substitutions of L69 for alanine and of L83 for alanine, aspartate, or phenylalanine gave no observable phenotype (Table 2). Thus, the introduction of a charged residue or a bulky amino acid like phenylalanine was tolerated. Further, single changes of other amino acid residues, E70 and K102 with alanine and I99 with threonine, also yielded the wild-type phenotype. Even substitutions of two or three residues, L69E70, L69E70E71, and L83E84K85, for alanine residues did not cause cell filamentation (Table 2). Ultimately, we found a mutation that generated a null phenotype, as a spontaneous PCR-related base substitution (ftsL36) (Table 2). The resulting amino acid change involved the substitution of a leucine residue, L69, with proline (L69P). This mutation was highly significant because proline residues are not compatible with a helical structure. According to the program COILS 2.1, which predicts coiled coils from protein sequences (24, 25), this mutant protein is unlikely to form a coiled-coil structure, whereas all of the other mutations introduced by site-directed mutagenesis seem to have no or little effect on this structural motif (data not shown). Thus, the phenotypic effect of this mutation would be consistent with the C-terminal domain of FtsL comprising a helical coiled-coil structure in which the structure itself is important but the precise identities of the hydrophobic core residues (Fig. 2) are variable.
FIG. 2.
Comparison of the FtsL proteins of B. subtilis (FtsLBs), B. licheniformis (FtsLBl), B. badius (FtsLBb), B. circulans (FtsLBc), and S. aureus (FtsLSa). The alignment was performed using the CLUSTAL W method, with residues identical to the B. subtilis sequence shown in gray boxes. The membrane-spanning domain and the a and d positions of the heptad repeat of the putative coiled-coil domain are shown above the alignment.
To test whether FtsL protein could tolerate loss of residues from its extreme C terminus, we made two defined nonsense mutations that would truncate FtsL 5 (ftsL37) or 10 (ftsL38) residues from its normal C terminus. Removal of 5 amino acids had no detectable phenotypic effect. However, when 10 amino acids were removed, FtsL function was abolished. Thus, the extreme C terminus of FtsL is essential for its function or stability.
The two alleles with clear division phenotypes, ftsL38 and ftsL36 (L69P), were tested further, first by examining their possible temperature dependence. Reducing or increasing the growth temperature had no observable effect on the division block. The mutant alleles were then expressed in parallel with a wild-type ftsL gene in strains 2041 and 2042. Both alleles were complemented fully by the wild-type protein. It was also interesting to examine the effects of the mutations on the interaction with DivIC. We have recently shown that DivIC interacts with FtsL protein in vitro (35) and that in the absence of FtsL, DivIC is destabilized in vivo (9). Western blotting experiments showed that in both of the new mutants, DivIC was undetectable (as in ftsL null mutants), whereas for example in strains 2036 and 2039, which express the phenotypically silent alleles ftsL30 and ftsL34, respectively, DivIC was present at normal levels (data not shown).
Cloning and sequencing of ftsL homologues from B. licheniformis, B. circulans, and B. badius.
As an alternative means of identifying regions of FtsL important for its function, we attempted to isolate ftsL genes from other Bacillus species. We assumed that the genetic organization would be similar to the pbpB operons of B. subtilis and other bacteria (10, 31). Consequently, we designed degenerate primers that bound to sequences in yllC and pbpB, upstream and downstream of ftsL, respectively (see Materials and Methods). We succeeded in PCR amplifying and sequencing the ftsL genes of three species, B. licheniformis, B. circulans, and B. badius. As expected, translation of partial DNA sequences upstream and downstream of the ftsL genes revealed the presence of yllC and pbpB homologues, respectively (data not shown).
Figure 2 shows the alignment of the primary sequences with the B. subtilis homologue (FtsLBs). As for B. subtilis FtsL, they show a predicted bitopic topology, and the coiled-coil prediction for parts of the C termini of these proteins, using COILS 2.1 (24, 25), is high (100%) for all three protein sequences (data not shown). The positions of the predominantly hydrophobic residues of the heptad repeat characteristic of coiled-coil proteins are shown in Fig. 2. These residues form the helix interface of coiled-coil proteins (24). The ftsL gene from the closely related species B. licheniformis (30) encodes a predicted protein of 117 amino acids (FtsLBl), 64% of which are identical to those of FtsLBs; a similar level of conservation is found for the DivIB proteins of these two species (14). Unlike DivIB, there are no major differences in the conservation of the N and C termini, nor in the hydrophobic segment of the FtsL homologues. Most of the amino acid residues in FtsLBs that were exchanged by site-directed mutagenesis (e.g., L69 and L83) are conserved in FtsLBl. B. badius ftsL codes for a protein of 118 amino acids (FtsLBb), which is 37% identical to FtsLBs. Interestingly, the characteristic leucine heptad motif of FtsL proteins is virtually absent in FtsLBb; only L83 is present. Further, the residues conserved in each pairwise comparison were quite dissimilar. For example, of 46 identical residues conserved between FtsLBs and FtsLBb, 13 are not conserved between FtsLBs and FtsLBl. This gives evidence for a rapid and fairly random evolution of the FtsL protein, including residues involved in its likely structure. As for FtsLBb, the predicted protein product of the B. circulans ftsL gene (FtsLBc) is moderately conserved (34% identical to the B. subtilis homologue), although the identity to FtsLBb is higher (47%). Three residues in FtsLBs—E70, L83, and K113—conserved in all four homologues, are not essential, as shown by site-directed mutagenesis (Table 2). Thus, the consensus sequence is likely to be of only limited use for identifying any essential residues.
Complementation of a B. subtilis ftsL null mutation.
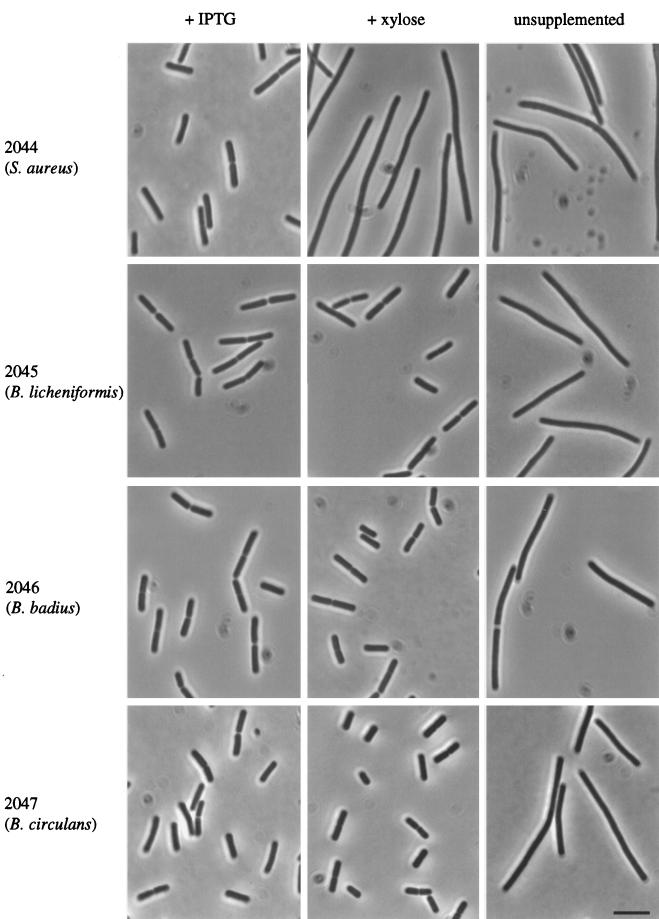
According to the notion that the FtsL primary sequence is highly flexible, it was possible that the highly divergent genes from other Bacillus spp. might nevertheless be able to substitute functionally for FtsL in B. subtilis. To test this, we introduced the heterologous genes into the chromosome of a B. subtilis strain with a conditional native ftsL gene. All three of the new strains, 2045, 2046, and 2047, producing FtsL of B. licheniformis, B. badius, and B. circulans, respectively, were able to grow and divide in the absence of native ftsL expression (Fig. 3). All of the heterologous FtsL proteins also allowed these strains to grow at high temperature (45°C) and to sporulate at levels similar to the wild type (data not shown). It was possible that septation in the three new strains was dependent on residual expression of the native ftsL gene (in the absence of inducer). To exclude this possibility, the conditional B. subtilis ftsL gene was eliminated by replacing it with a tetracycline resistance casette. All three derivative strains still grew and divided normally, as judged by phase-contrast microscopy (data not shown). The absence of the repeating leucine heptad motif in FtsLBb confirms that this motif is not necessary for FtsL function in B. subtilis. The results also showed that many other residues can be replaced without loss of function, because, for example, FtsLBb and FtsLBc, with identities as low as 37% and 34%, respectively, could restore septation in a null mutant.
FIG. 3.
Complementation of a B. subtilis ftsL null mutation by ftsL alleles of S. aureus, B. licheniformis, B. circulans, and B. badius. Shown are phase-contrast images of strains 2044, 2045, 2046, and 2047 grown at 37°C in PAB supplemented with either IPTG (left panel) or xylose (central panel) or not supplemented with any inducer (right panel). IPTG induces the expression of the wild-type gene, whereas the foreign ftsL alleles are transcribed in the presence of xylose. As expected, all four strains were filamented in unsupplemented medium (i.e., when FtsL is depleted). Bar, 5 μm.
To test whether even more distantly related ftsL homologues could substitute for B. subtilis ftsL, we cloned the equivalent gene from S. aureus, whose gene product shows 27% identity to B. subtilis FtsL (31). When the S. aureus gene, including the B. subtilis ftsL RBS (see Materials and Methods), was introduced into strain 2007 it did not complement the neo insertion in ftsL, even at a reduced growth temperature (strain 2044 [Fig. 3]). It is possible that the S. aureus protein is nonfunctional in B. subtilis because of its low identity and/or its inability to interact with the DivIC protein (35), which S. aureus seems to lack. Alternatively, S. aureus FtsL may not be stable or properly incorporated into the membrane in B. subtilis.
Consequences for FtsL structure and function.
It has previously been suggested that the repeating leucine motifs in E. coli FtsL (13) and B. subtilis DivIC (23) might constitute key residues in a protein dimerization domain. Such motifs, leucine zippers, have been described for many eukaryotic and also prokaryotic transcription factors (27, 34). Pairs of leucine zippers cooperate to form a DNA-binding domain through a helical coiled-coil structure (29). Accordingly, both E. coli FtsL (12) and FtsL and DivIC of B. subtilis (35) have subsequently been shown to undergo protein-protein interactions. However, our data show convincingly that the leucine residues themselves are not essential for FtsL function in B. subtilis. Several were removed from the B. subtilis protein by mutagenesis without a detectable phenotype, and the B. badius homologue is functional even though it retains only one of the leucines. However, it seems likely that FtsL does have an α-helical structure and that hydrophobic residues, although not necessarily leucines, are needed at appropriate positions in the structure, presumably to participate in hydrophobic interactions at the protein-protein interface. Certainly, introduction of a helix-breaking proline residue in place of one of the leucines abolished FtsL function.
It was interesting that the S. aureus ftsL did not function in B. subtilis even though the protein product is not much more divergent that the B. badius gene. The most likely explanation for this lack of complementation is the lack of a divIC homologue in S. aureus. Thus, FtsL might form homodimers in this organism, as it does in E. coli (12), and cannot interact with B. subtilis DivIC. So far, database searches have revealed homologues of divIC only in bacteria closely related to B. subtilis: Bacillus anthracis, Bacillus stearothermophilus, and Listeria monocytogenes (a related non-spore former) (21).
In conclusion, our results strongly support the idea that FtsL is a highly malleable protein that can tolerate many amino acid changes without loss of function. One way to explain this would be that its function involves formation of multiple, relatively weak interactions with DivIC and possibly other cell division proteins. How such a low degree of sequence specificity allows the FtsL protein to carry out its essential function in cell division is a challenging question.
ACKNOWLEDGMENTS
This work was supported by grants from the Biotechnology and Biological Research Council and the BIOTECH programme of the European Community. J.S. was the recipient of a Boehringer Ingelheim Fonds postgraduate fellowship. J.E. is the recipient of a BBSRC Senior Research Fellowship.
We thank Armajit Bhomra and Alice Taylor for help with the DNA sequencing.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall B, Lowe M, Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988;170:4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall B, Lutkenhaus J. Nucleotide sequence and insertional activation of a Bacillus subtilis gene that affects cell division, sporulation and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5:447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- 5.Beall B, Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992;174:2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 7.Daniel R A, Errington J. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis, and a role for DivIB protein in FtsL turnover. Mol Microbiol. 2000;36:278–289. doi: 10.1046/j.1365-2958.2000.01857.x. [DOI] [PubMed] [Google Scholar]
- 8.Daniel R A, Harry E J, Errington J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol Microbiol. 2000;36:278–289. doi: 10.1046/j.1365-2958.2000.01724.x. [DOI] [PubMed] [Google Scholar]
- 9.Daniel R A, Harry E J, Katis V L, Wake R G, Errington J. Characterisation of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- 10.Daniel R A, Williams A M, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Errington J. Gene cloning techniques. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. pp. 175–220. [Google Scholar]
- 12.Ghigo J-M, Beckwith J. Cell division in Escherichia coli: role of FtsL domains in septal localization, function, and oligomerization. J Bacteriol. 2000;182:116–129. doi: 10.1128/jb.182.1.116-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman L, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 14.Harry E J, Partridge S R, Weiss A S, Wake R G. Conservation of the 168 divIB gene in Bacillus subtilis W23 and B. licheniformis, and evidence for homology to ftsQ of Escherichia coli. Gene. 1994;147:85–89. doi: 10.1016/0378-1119(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 15.Harry E J, Stewart B J, Wake R G. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol Microbiol. 1993;7:611–621. doi: 10.1111/j.1365-2958.1993.tb01152.x. [DOI] [PubMed] [Google Scholar]
- 16.Harry E J, Wake R G. The membrane-bound cell division protein DivIB is localised to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- 17.Hauser P M, Errington J. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3923–3931. doi: 10.1128/jb.177.14.3923-3931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkinson H F. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J Gen Microbiol. 1983;129:1945–1958. doi: 10.1099/00221287-129-6-1945. [DOI] [PubMed] [Google Scholar]
- 19.Karow M L, Rogers E J, Lovett P S, Piggot P J. Suppression of TGA mutations in the Bacillus subtilis spoIIR gene by prfB mutations. J Bacteriol. 1998;180:4166–4170. doi: 10.1128/jb.180.16.4166-4170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katis V L, Harry E J, Wake R G. The Bacillus subtilis division protein DivIC is a highly abundant membrane-bound protein that localises to the division site. Mol Microbiol. 1997;26:1047–1055. doi: 10.1046/j.1365-2958.1997.6422012.x. [DOI] [PubMed] [Google Scholar]
- 21.Katis V L, Wake R G. Membrane-bound division proteins DivIB and DivIC of Bacillus subtilis function solely through their external domains in both vegetative and sporulation division. J Bacteriol. 1999;181:2710–2718. doi: 10.1128/jb.181.9.2710-2718.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunst F, Rapoport G. Salt stress is an environmental signal affecting digestive enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin P A, Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994;176:1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 25.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 26.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 27.Maxon M E, Wigboldus J, Brot N, Weissbach H. Structure-function studies on Escherichia coli MetR protein, a putative prokaryotic leucine zipper protein. Proc Natl Acad Sci USA. 1990;87:7076–7079. doi: 10.1073/pnas.87.18.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mountain A. Gene expression systems for Bacillus subtilis. In: Harwood C R, editor. Bacillus. New York, N.Y: Plenum Press; 1989. pp. 73–114. [Google Scholar]
- 29.O'Shea E K, Rutkowski R, Kim P S. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 30.Priest F G. Systematics and ecology of Bacillus. In: Sonenshein A L, Hoch A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 3–16. [Google Scholar]
- 31.Pucci M J, Thanassi J A, Discotto L F, Kessler R E, Dougherty T J. Identification and characterization of cell wall-cell division gene clusters in pathogenic gram-positive cocci. J Bacteriol. 1997;179:5632–5635. doi: 10.1128/jb.179.17.5632-5635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanchez M, Valencia A, Ferrandiz M-J, Sander C, Vincente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasse-Dwight S, Gralla J D. Role of the eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor ς54. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 35.Sievers J, Errington J. The Bacillus subtilis cell division protein FtsL localises to sites of cell division and interacts with DivIC. Mol Microbiol. 2000;36:846–855. doi: 10.1046/j.1365-2958.2000.01895.x. [DOI] [PubMed] [Google Scholar]
- 36.Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornewell S J, East A K, Errington J. An efficient expression and secretion system based on Bacillus subtilis phage φ105 and its use for the purification of B. cereus β-lactamase I. Gene. 1993;133:47–53. doi: 10.1016/0378-1119(93)90223-p. [DOI] [PubMed] [Google Scholar]
- 38.Walton C, Booth R K, Stockley P G. Random chemical mutagenesis and the non-selective isolation of mutated DNA sequences in vitro. In: McPherson M J, editor. Directed mutagenesis. Oxford, United Kingdom: Oxford University Press; 1991. pp. 135–162. [Google Scholar]
- 39.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]