Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins (original) (raw)
Abstract
We present the 1.9 Å resolution crystal structure of the catalytic domain of Gyp1p, a specific GTPase activating protein (GAP) for Ypt proteins, the yeast homologues of Rab proteins, which are involved in vesicular transport. Gyp1p is a member of a large family of eukaryotic proteins with shared sequence motifs. Previously, no structural information was available for any member of this class of proteins. The GAP domain of Gyp1p was found to be fully α-helical. However, the observed fold does not superimpose with other α-helical GAPs (e.g. Ras- and Cdc42/Rho-GAP). The conserved and catalytically crucial arginine residue, identified by mutational analysis, is in a comparable position to the arginine finger in the Ras- and Cdc42-GAPs, suggesting that Gyp1p utilizes an arginine finger in the GAP reaction, in analogy to Ras- and Cdc42-GAPs. A model for the interaction between Gyp1p and the Ypt protein satisfying biochemical data is given.
Keywords: GTPase activating protein/Rab protein/vesicular transport/Ypt-GAP domain
Introduction
Membrane vesicle-mediated exchange of substances between different organelles in eukaryotic cells is directional and tightly controlled through the action of a variety of evolutionarily conserved proteins (Jahn and Sudhof, 1999). Among them are the GTPases of the Ypt/Rab family, which are key regulators for transport vesicle targeting to specific membranes of exo- and endocytic compartments (Lazar et al., 1997; Novick and Zerial, 1997; Pfeffer, 1999). Ypt/Rab GTPases form the largest group of the Ras superfamily of small GTP-binding proteins. The common feature in this superfamily is a switching cycle between two functionally distinct conformations controlled by the state of the bound nucleotide. In the active GTP-bound state, they interact in a specific manner with downstream effector proteins, whereas in the GDP-bound state these interactions do not occur as a result of a conformational rearrangement that takes place upon GTP hydrolysis. This functional cycle is tightly controlled through the interaction with guanosine nucleotide exchange factors (GEFs), which stimulate the exchange of GDP against GTP, and GTPase activating proteins (GAPs), which specifically turn off the active state of the Ypt/Rab protein by accelerating the slow intrinsic GTPase activity. Whereas GAPs and GEFs for Ras, Rho and Cdc42p have been studied in great detail, there is only a limited amount of information on the corresponding Ypt/Rab-specific proteins.
The first identified Ypt/Rab-specific GAPs, termed Gyp6p, Gyp7p and Gyp1p, were isolated by high-expression cloning in yeast (Strom et al., 1993; Vollmer and Gallwitz, 1995; Albert et al., 1999; Vollmer et al., 1999). Gyp1p was also recognized by others (Du et al., 1998) by virtue of its relatedness to Gyp7p. As originally noted by Neuwald (1997) based on a sequence database search and a multiple alignment analysis, Gyp1p, Gyp6p and Gyp7p share several related sequence motifs (termed motifs A, B, C, D, E and F) with the yeast spindle pole body protein Bub2p and a variety of other proteins from yeast, Caenorhabditis elegans, Drosophila melanogaster and mammals. These sequence motifs A–F fall within the catalytically active fragments of Gyp1p and Gyp7p (Albert et al., 1999). Several other yeast proteins containing these six motifs were demonstrated to be Ypt/Rab-specific GAPs (Albert and Gallwitz, 1999, 2000). Importantly, a recently identified human Rab6-specific GAP, GAPCenA, displays similar primary structure elements within a central 200 amino acid domain to the Ypt-GAP domain of the Gyp family members (Cuif et al., 1999).
The kinetic characterization of different Ypt-GAPs from Saccharomyces cerevisiae has revealed selectivity and differences in the extent of stimulation towards different Ypt proteins but no absolute specificity for a particular GTPase. For example, Gyp7p has its highest GAP activity towards Ypt7p, and lower activity with Ypt6p and Ypt31p (Albert et al., 1999) while Gyp1p displays its highest specific GTPase activation activity towards Ypt51p and Sec4p (Du et al., 1998; Albert et al., 1999). The GTPase Ypt51p is an important regulator involved in the endocytic membrane traffic of S.cerevisiae (Horazdovsky et al., 1994; Singer-Kruger et al., 1994) and is found to be homologous to the mammalian Rab5 protein (Singer-Kruger et al., 1995). For a structural analysis of Ypt/Rab-GAPs we focused our attention on Gyp1p, since a 1.5 Å high-resolution crystal structure of its substrate Ypt51p in its active conformation has been solved recently (Esters et al., 2000).
A detailed mutational analysis of the catalytic domains of Gyp1p and Gyp7p has revealed a conserved arginine residue in the sequence motif B, which is critical for the catalytic activity (Albert et al., 1999). This could indicate that Gyp proteins exhibit an ‘arginine finger’ mechanism of GTPase activation, as described for Ras- and Cdc42p-GAP [sometimes referred to as Rho-GAP (Lancaster et al., 1994)]. The two key features of this activation mechanism appear to be (i) the positioning of the catalytically essential GTPase glutamine side chain via a hydrogen bonding interaction between the glutamine carbamoyl-NH2 group and the main chain carbonyl group of the GAP arginine, and (ii) the polarization of the γ-phosphate group or the stabilization of charge on it via the interaction with the positively charged side chain guanidinoyl group of the GAP arginine (Rittinger et al., 1997; Scheffzek et al., 1997; Nassar et al., 1998). The charge distribution together with the shape of the GTPase around the active site as well as further specific interactions between the GTPase and the cognate GAP determine the selectivity and specificity of the interaction. As a consequence, the orientation of the arginine finger can be dramatically different in different GTPase–GAP complexes, leading to variations in the ‘arginine finger’ mechanism (Ahmadian et al., 1997).
Here we present the first crystal structure of a Ypt/Rab-specific GAP domain, Gyp1-46p. Together with the structural information available for GAPs specific towards other subfamilies of the Ras superfamily, this structure provides first insights into the determinants of the selective recognition of Ypt proteins. The active site of Gyp1-46p displays structural similarities to the GTPase binding cleft of Ras- and Cdc42p-GAP, suggesting that Ypt-GAPs follow the same GTPase activation mechanism.
Results and discussion
Structure determination
The GAP catalytically active domain of Gyp1p (residues 239–638, termed Gyp1-46p) was produced in Escherichia coli as a soluble, C-terminally His6-tagged protein having biochemical properties identical to those of the GAP domain produced in yeast. Its crystal structure was solved using 2.35 Å resolution multiwavelength anomalous diffraction (MAD) data phasing with SeMet-labelled protein (see Table I). The experimental MAD phases were used throughout the refinement and model building process in order to reduce model bias (Pannu et al., 1998). The final free _R_-factor value (Brünger, 1992) for the 2.35 Å MAD data set was 24.9%. The further refinement against a 1.9 Å high-resolution data set from a SeMet Gyp1-46p crystal was performed with molecular replacement methods to position the model and to derive calculated phases. The final model includes 322 residues, lacking one extended and surface exposed region of 61 residues on the convex side of the molecule. The final model is of very good quality with a free _R_-factor of 22.5% for all reflections in the 10% test set with Bragg spacing between 6.0 and 1.9 Å. The quality of the final model was assessed by a number of criteria (see Table II). The excellent statistics suggest that the missing 61 residues are completely disordered and do not contribute to diffraction. Electrospray mass-spectrometry on redissolved crystals confirmed that the protein was intact and no proteolytic cleavage had occurred.
Table I. Crystallographic data and phasing statistics for Gyp1-46p.
| Space group | _P_6(5)22 | | | | | | | | | | | | | | | -------------------------------------------------------------------------------------------------------- | ------------------------------------------------------------------ | ------------------------- | ---------------- | ----------- | ------------- | ---------- | ------ | | | | | | | | | Cell dimensions | a = b = 74.06 Å, c = 277.75 Å (1 mol/AU); 11 Se atoms/mol | | | | | | | | | | | | | | | | _d_min (Å) | No. of reflections | No. of unique reflections | Completeness (%) | I/σa | _R_syma,b (%) | | | | | | | | | | | Se λ1 (0.9790 Å) | 20.0–2.35 | 461 421 | 36 326 | 99.2 (99.9) | 27.5 | 6.8 (19.3) | | | | | | | | | | Se λ2 (0.9795 Å) | 20.0–2.35 | 458 264 | 36 412 | 99.3 (99.5) | 27.0 | 7.1 (19.5) | | | | | | | | | | Se λ3 (0.9200 Å) | 20.0–2.7 | 275 885 | 23 838 | 98.4 (99.8) | 38.8 | 5.5 (10.4) | | | | | | | | | | Se λ4 (0.8428 Å) | 6.0–1.9 | 237 598 | 33 502 | 91.2 (97.1) | 14.4 (6.5) | 9.4 (24.2) | | | | | | | | | | Observed diffraction ratiosc | | | | | | | | | | | | | | | | | λ1 | λ2 | λ3 | | | | | | | | | | | | | | λ1 | 0.0677 | 0.0360 | 0.0470 | | | | | | | | | | | | | λ2 | | 0.0558 | 0.0520 | | | | | | | | | | | | | λ3 | | | 0.0466 | | | | | | | | | | | | | Phasing power,d figure of merit (FOM) and FOM after density modification (DM_FOM) using λ3 as reference | | | | | | | | | | | | | | | | | λ3– > λ3– | λ3– > λ2+ | λ3– > λ2– | λ3– > λ1+ | λ3– > λ1– | FOM | DM_FOM | | | | | | | | | | 20–2.8 Å | 2.3980 | 2.4223 | 3.3621 | 1.4811 | 2.6874 | 0.7712 | 0.9542 | | | | | | | | | 2.8–2.9 Å | 1.5529 | 1.5671 | 2.2353 | 1.1444 | 1.9830 | 0.6684 | 0.9261 | | | | | | | | | Phasing power,d figure of merit (FOM) and FOM after density modification (DM_FOM) using λ1 as reference | | | | | | | | | | | | | | | | | λ1– > λ1– | λ1– > λ2+ | λ1– > λ2– | λ1– > λ3+ | λ1– > λ3– | FOM | DM_FOM | | | | | | | | | | 20–2.35 Å | 3.0609 | 0.7040 | 2.5484 | 1.1987 | 2.3630 | 0.5769 | 0.9588 | | | | | | | | | 2.35–2.45 Å | 2.3321 | 0.4981 | 2.2352 | 1.0491 | 2.1156 | 0.3096 | 0.9415 | | | | | | | |
Table II. Refinement statistics for Gyp1-46p (residues 249–509 and 570–630).
| Resolution range | 6.0–1.9 Å |
|---|---|
| No. of reflections | 33 502 |
| R valuea | 19.9% |
| _R_-free valueb | 22.5% |
| Root-mean-square deviations | |
| bond-length | 0.0050 Å |
| bond-angle | 1.052° |
| improper-angle | 0.7348° |
| dihedrals | 18.68° |
| No. of non-hydrogen atoms | |
| total | 2926 |
| solvent | 166 |
| Average _B_-factor (all atoms) | 31.8 Ų |
| Average _B_-factor (protein atoms) | 31.2 Ų |
| Average _B_-factor (solvent atoms) | 39.0 Ų |
Overall structure
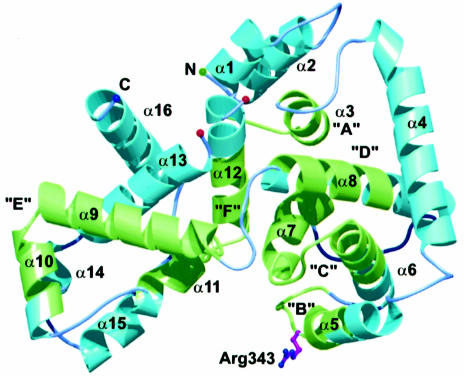
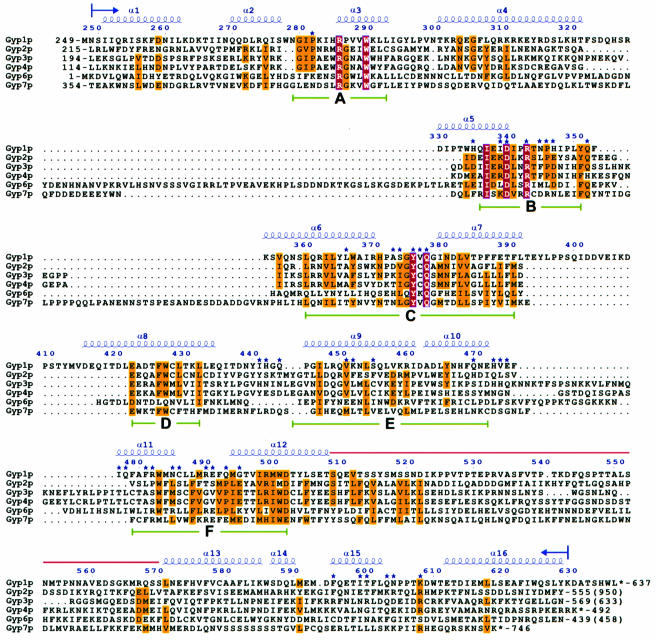
The crystal structure revealed that the Ypt-GAP domain of Gyp1p can be described as a V-shaped molecule consisting of 16 α-helixes and no β-sheet secondary structure elements (see Figure 1). Despite the fact that the Ypt-GAP domain of Gyp1p is purely α-helical, it does not superimpose with the other identified, purely α-helical GAP domains [p120-GAP (Scheffzek et al., 1996), NF1-GAP (Scheffzek et al., 1998), Rho-GAP/Cdc42-GAP (Rittinger et al., 1997; Nassar et al., 1998)]. Using site directed mutagenesis to exchange individually all arginines in the catalytic domain of Gyp1p, two arginine residues were identified to have significant influence on the Gyp1p function. Whereas the first highly conserved arginine in sequence motif A, Arg286, appears to be important for maintaining the structural integrity of Gyp1-46p, the second arginine, Arg343, was found to be essential for the GAP activity of Gyp1-46p (Albert et al., 1999). Arg343 is part of the shared sequence motif B (Neuwald, 1997) and, together with the aspartic acid residue Asp340, is highly conserved among all known Gyp proteins (see Figure 2). It is positioned at the side of the narrow cleft on the concave side of the V-shaped molecule. Therefore, we propose that this cleft is the binding site for the GTPase.
Fig. 1. Three-dimensional structure of the Ypt-GAP domain of Gyp1p. The ribbon diagram displays the secondary structure elements and the active site arginine residue in ball-and-stick representation. Regions of the seven shared sequence motifs A–F are highlighted in green. This figure was generated using the programs BOBSCRIPT (Esnouf, 1997) and raster3D (Merritt and Murphy, 1994).
Fig. 2. Sequence comparison of the catalytically active domains of Gyp1p homologs. The sequence alignment was performed with the program CLUSTAL_W (Thompson et al., 1994) and adjusted manually in the loop regions. Only the residues modelled in the crystal structure of Gyp1-46p were included in the alignment. The residue numbering is based on the Gyp1p full-length sequence. Amino acid residues strictly conserved have a red background, residues (hydrophobic, aromatic, polar, negatively or positively charged) conserved within four or more Gyp proteins are indicated with a yellow background. Secondary structure elements are shown above the aligned sequences. The six shared motifs named A–F, found previously to be common between Ypt-GAPs and proteins involved in spindle checkpoint assembly (Neuwald, 1997), are underlined with solid bars in green. The blue stars indicate residues that are at the surface of the putative active site cleft of Gyp1-46p. The red line indicates the region from residue 510–569 which is not included in the crystal structure. The number for the first and last residue used for the alignment is given together with the overall length of the Gyp protein in parenthesis if the last residue is not the C-terminus. The figure was prepared with the program ESPript (Gouet et al., 1999).
Based on the program TM-pred (Hofmann and Stoffel, 1993), Bi et al. (2000) proposed two possible transmembrane regions for the S.cerevisiae proteins Msb3p (residues 320–337 and 424–440) and its homologue Msb4p (residues 238–255 and 335–351), which are identical to Gyp3p and Gyp4p (Albert and Gallwitz, 1999, 2000). These two regions correspond to the Gyp1p residues 374–391 of the sequence motif C, including most of α-helix α7, and residues 476–492 of α-helix α11, which are part of motif F. These residues are important for maintaining the architecture of the Ypt-GAP domain fold and contribute to the putative binding site for the GTPase. Assuming a similar fold for Gyp3p (Msb3p) and Gyp4p (Msb4p) to that observed for Gyp1-46p, the position of the stretch of hydrophobic amino acids, its content of polar amino acid residues and its location in the tertiary structure excludes the possibility that it is a transmembrane spanning helix. In addition, overexpressed protein could easily be isolated from yeast cytosol (Albert and Gallwitz, 1999), which is rather unlikely for membrane-anchored proteins.
Shared sequence motifs
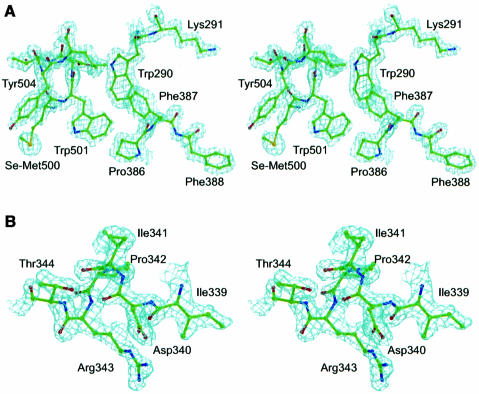
Gyp1p belongs to a group of proteins that share significant sequence similarities in six consecutive regions of the primary structure termed motif A to motif F (Neuwald, 1997). The motifs A, B and C contain three ‘fingerprint’ sequence motifs highly conserved among the members of the Gyp protein family: 286RxxxW (motif A), 337IxxDxxR (motif B) and 376YxQ (motif C) (see Figure 2). The sequence motif A includes the highly conserved residues Arg286 and Trp290 of α-helix α3. Arg286 forms a double salt bridge with Asp502 [d(Arg286-NH2:Asp502-OD2) = 2.8 Å; d(Arg286-NE:Asp502-OD1) = 2.7 Å] of the sequence motif F, which is conserved among all Ypt-GAP domains except for Gyp7p, where a glutamate is at this position and is presumably able to form a similar stabilizing salt bridge. In addition, Asp502 is within hydrogen bonding distance of the side chain of the highly conserved Trp290 [d(Asp502-OD1:Trp290-NE1) = 2.8 Å], which itself is part of a hydrophobic core. Site directed mutagenesis of Arg286 (R286A) in Gyp1p and Gyp7p rendered the protein very unstable during purification and the GAP activity was significantly reduced (Albert et al., 1999). This indicates an important contribution of the conserved Arg286 in the stabilization of the tertiary structure of the Ypt-GAP domain and, therefore, in the formation of the Ypt binding epitope. The highly conserved residue Trp290 of motif A is part of a very hydrophobic region in the core of Gyp1-46p consisting of a number of aromatic residues including Tyr387 and Phe391 at the end of motif C and Trp501 and Tyr504 at the end of motif F (see Figure 3A). The different conserved interactions between the different motifs A, C and F highlight their contribution in maintaining the tertiary structure of the Ypt-GAP fold.
Fig. 3. Central core and active site of the Ypt-GAP domain of Gyp1p. (A) Stereo view of the final 2_F_o – _F_c electron density map contoured at the 2σ level displaying the aromatic core of Gyp1-46p. Displayed are the highly conserved Trp290 of motif A and the surrounding hydrophobic residues. (B) Stereo view of the final 2_F_o – _F_c electron density map contoured at the 2σ level at the active site region of Gyp1-46p. The refined model (ball-and-stick) is superimposed, displaying the residues 339–344 of motif B which includes the highly conserved Asp340 and the catalytically active residue Arg343. This figure was generated using the programs BOBSCRIPT (Esnouf, 1997) and raster3D (Merritt and Murphy, 1994).
Sequence motifs B and C are located close together and also interact with each other to form a well defined and conserved epitope at the edge of the putative GTPase binding cleft. Motif B is composed of the C-terminal end of α-helix α5 and loop L5 between α-helices α5 and α6, whereas motif C consists of α-helices α6 and α7 and the connecting loop L6. The two loops L5 and L6 are tightly packed against each other placing the three absolutely conserved residues Asp340, Arg343 and Gln378, which are exposed to the solvent and form a type of a signature motif (see Figure 3B), at one side of the cleft. The highly conserved residue Asp340 (motif B) forms a salt bridge with the catalytic Arg343 (motif B) and hydrogen bonds to the side chain OH-group of Tyr376 (motif C) and to the side chain of Gln378 (motif C). The presence of hydrophobic residues at the positions of the residues Trp334, Ile337 and Ile341 of α-helix α5 and the residues Gln361, Leu364 and Tyr365 of α-helix α6 is conserved among the different Gyp Ypt-GAP domains, facilitating tight packing of the two α-helices against each other. Due to the localization of the catalytically essential Arg343 and the very rigid packing in this region we propose that this patch at the surface of Gyp1-46p is involved not only in the GTPase activation but also in specific binding of Ypt proteins.
The loop L8 connecting motifs D and E is variable in length and sequence among the different Gyp proteins. However, it is in close proximity to the putative GTPase binding site. Further contributions to this site come from the α-helix α11 and the connecting loop L11 to α-helix α12, both being part of sequence motif F.
The only helices that are not part of any shared motif are α1, α2, α4 and α13–α16. Based on our proposed mode of complex formation between Gyp1-46p and Ypt51p-GppNHp (see below), of theses helices α15 could be involved in the interaction between the two proteins, whereas the other ‘non-motif’ α-helices probably do not contribute to the putative active site.
Active site
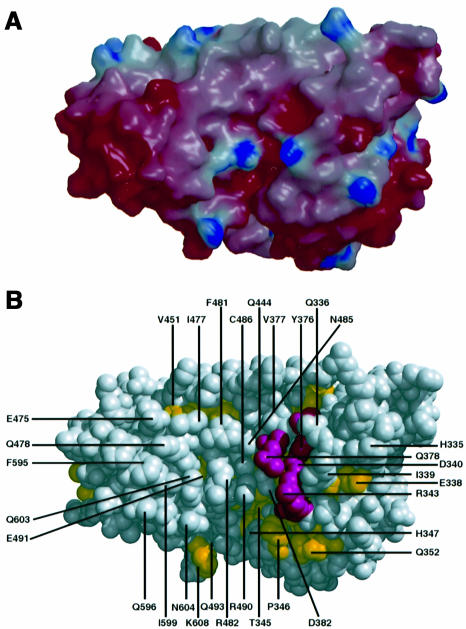
The active site of the Ypt-GAP domain should comprise residues that are highly conserved among different Gyp Ypt-GAP domains as well as protein regions without significant sequence similarity. The conserved residues are important for the mechanism of activation, which is likely to be identical for the different Gyp Ypt-GAP domains and for the specific recognition and binding of conserved Ypt protein motifs. On the other hand the non-conserved, variable amino acids should be partially responsible for rendering the individual Gyp Ypt-GAP domain selective towards the cognate Ypt family member. We postulate that the residues involved in the binding or catalysis of Ypt-GTP should be solvent exposed and invariant among the different Ypt-GAPs. Mapping these residues on the structure of Gyp1-46p reveals that the invariant residues are clustered in two different areas of the Ypt-GAP domain. One cluster is in the central core of the domain (see above) and the other at the surface near the cleft of the concave side of the protein. Four highly conserved residues (Tyr376, Gln378, Asp340 and Arg343) are part of this potential binding cleft (see Figure 4).
Fig. 4. The putative Ypt binding cleft. (A) Electrostatic surface representations viewed into the concave side of the molecule. The figure was generated using the program GRASP (Nicholls et al., 1993) and rendered with the program raster3D (Merritt and Murphy, 1994). Red indicates negatively charged (–7 kT) and blue positively charged regions (+7 kT). (B) Surface CPK representation of the Gyp1p Ypt-GAP domain shown in the same orientation as in (A). Residues that are highly conserved in the different Gyp Ypt-GAP domains are coloured pink; well conserved residues are coloured yellow (see sequence alignment, Figure 2). Residues that are solvent accessible and form the surface of the cleft are labelled.
The surface of the cleft accounts for ∼20% of the solvent-accessible surface area with dimensions of ∼30 × 25 Å and a depth of ∼10 Å (see Figure 4). The inner part of this putative Ypt binding site is dominated by charged or polar amino acid residues and only at the edge of the binding site is a hydrophobic patch located around residues Phe481 and Phe595 (see Figure 4). The contribution of this non-polar surface patch to the binding of the Ypt protein can explain the experimental observation that in the presence of detergents such as Triton X-100 the GAP activity of Ypt-GAPs is significantly reduced (S.Albert, unpublished results).
The solvent-exposed residues of α-helix α15 are directed towards the putative Ypt binding pocket. However, its primary structure displays no sequence similarity between the different identified Gyp Ypt-GAP domains. Since C-terminally truncated Gyp proteins lacking α-helix α15 display no GAP activity (Albert et al., 1999) we assume that this α-helix α15 is either important for maintaining the tertiary architecture of Gyp1-46p or that the solvent-exposed residues are involved in the binding of the Ypt protein and might determine selectivity for members of the Ypt protein family. This conclusion is further supported by comparison with the related proteins Bub2p from S.cerevisiae and Cdc16p from Schizosaccharomyces pombe. Based on sequence alignment both proteins are significantly shorter than Gyp1p and do not include the complete Ypt-GAP domain, since the last three α-helices are missing. For Cdc16p it could be shown that in association with Byr4p it is a GAP for the GTPase Spg1p but without Byr4p it is inactive (Furge et al., 1998). Cdc16p and Byr4p form a tight complex and it can be hypothesized that Byr4p contributes the missing α-helices, thus completing the GTPase binding site for building the active GAP protein. Furthermore, GAPCenA, a GTPase activating protein for Rab6, was identified using a yeast two-hybrid screen with the GTPase-defective mutant Rab6(Q72L) as bait (Cuif et al., 1999). The isolated region interacting with Rab6(Q72L) is located downstream of the sequence motif F and therefore could overlap with the C-terminal region of Gyp1p reaching from α-helix α13 to α16. However, in contrast to Gyp1p for the Rab6 binding site of GAPCenA, a high propensity for forming coiled-coil helices is predicted (Lupas, 1996) within this region, which might indicate a different architecture of GAPCenA in this part of the Ypt-GAP fold.
Diversities in the Ypt-GAP fold
The sequence alignment of different identified Ypt-GAP domains reveals several regions of the Ypt-GAP fold with significant heterogeneity in sequence and size of loops. These variations are probably determinants for selectivity and specificity for interaction with other proteins, particularly towards substrate GTPases in addition to sequence-specific regions in Ypt proteins themselves (e.g. Ypt51p and Ypt52p, see below). Significant differences can be highlighted for loops L4 (connecting α4 and α5), L5 (connecting α5 and α6), L7 (connecting α7 and α8), L10 (connecting α10 and α11) and to some extent loop L8 (connecting α8 and α9). Loop L4 has an especially large insertion for Gyp6p, loop L5 for Gyp7p, loop L7 for Gyp1p and loop L10 for Gyp3p, Gyp4p and Gyp6p. In our proposed model of the complex between Gyp1-46p and Ypt51p, loop L7 does not participate in complex formation and we assume its involvement in the interaction with a Gyp1-specific protein.
Missing region of amino acids 510–569
Whereas most parts of the Gyp Ypt-GAP domain are very well defined by electron density, the structure composed of residues 510–569 is not defined and could not be included into the structural model. Using the secondary structure prediction server Jpred (Cuff et al., 1998) to predict the secondary structure elements of the Gyp1-46p domain all of the observed α-helices were predicted, however, sometimes with some minor differences in length. For the missing region three further α-helices and the start of α-helix α13 at around residue Thr555 were predicted. Since the two easily visible ends of this flexible part are very close to each other [distance (Ser509-C–Ser570-N) = 8.9 Å], a prediction of its localization taking into account the predicted secondary structure is not possible.
Structural model of the binary complex formed between Gyp1p and Ypt51p-GppNHp
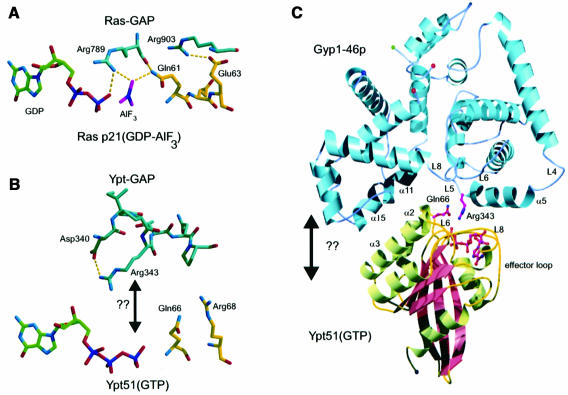
Based on the common features observed in the crystal structures for different GAP–GTPase complexes (Figure 5A), we have manually docked the active form of Ypt51p on to the structure of Gyp1-46p. The main consideration for complex formation was the possibility of forming a hydrogen bonding interaction between the side chain carbamoyl group of Ypt51-Gln66 and the main chain carbonyl oxygen atom of Gyp1-Arg343 without major spatial overlaps between the two proteins (Figure 5B). Since Arg343 is at the C-terminal end of α-helix α5, which is unlikely to rearrange dramatically upon complex formation, this is a very tight constraint and leaves only one possibility for docking. Based on these structural requirements for complex formation we manually oriented and ‘docked’ the structure of Ypt51p-GppNHp (Esters et al., 2000) at the active site cleft of Gyp1-46p as shown in Figure 5C. However, we have not performed energy minimization or adjustment of side chains for this complex. Two further aspects were mainly of importance in reaching the modelled complex. First, the Ypt51p structure can be described as a wedge with the nucleotide binding site at its thin end. There therefore only appear to be two general ways of fitting this wedge into what appears to be the sterically corresponding V-shaped groove in Gyp1-46p, these two possibilities being related by a 180° rotation. While one of them allows the important groups to come near enough to each other as described above, the other possibility does not allow this without departing significantly from the basic feature of a wedge fitting into a pre-existing V-shaped groove. In the orientation chosen, the rearrangements outlined above can take place and only a few other side chains have to adopt new orientations. Certain new and feasible side chain interactions between polar residues from the two proteins can be formulated. Importantly, in this proposed orientation of the two proteins, the hydrogen bond between Ypt51-Gln66-NE2 and Gyp1-Arg343-O would not be blocked by a side chain at position 16 of the GTPase P-loop (16Ala-Ala-Val in Ypt51p). In H-Ras p21 any amino acid substitution of the corresponding glycine (Gly12) blocks the GTPase stimulation by Ras-GAP even if the complex can be formed. This is explained by the fact that Ras-Gln61 cannot reach a favourable position since the hydrogen bond between Ras-Gln61 and Ras-GAP-Arg789 cannot be formed (Scheffzek et al., 1997)
Fig. 5. Docking approach for the complex between Gyp1p and Ypt51p-GTP, modelled manually on the basis of known GAP–GTPase structures. (A) Active site of the complex formed between p120-GAP and H-Ras p21(GDP-AlF3). The hydrogen bonds between GAP residues Arg789 and Arg903 and H-Ras p21 residues Gln61 and Glu63 as well as with AlF3 are indicated. This specific hydrogen bond network and the orientation of the side chains were used as a model for manual docking of Ypt51-GTP to Gyp1-46p. (B) Close-up view of the active site in the putative Gyp1-46p–Ypt51-GTP complex. For the interaction between the side chain of Arg343 and the γ-phosphate group, the salt bridge formed between Arg343 and Asp340 has to be broken. Gln66 of Ypt51p is well oriented to become positioned closer to the γ-phosphate by forming a hydrogen bond between its side chain and the main chain carbonyl group of Arg343 of Gyp1p. (C) Ribbon representation of the putative complex. The orientation of Gyp1-46p is the same as in Figure 1. The essential arginine Arg343 of Gyp1p, the active site glutamine Gln66 of Ypt51p and the bound nucleotide GTP are shown in ball-and-stick representation. This figure was generated using the programs BOBSCRIPT (Esnouf, 1997) and raster3D (Merritt and Murphy, 1994).
In this model the following regions are in close contact and therefore would determine binding selectivity. The N-terminal parts of the Gyp1-46p α-helices α11 and α15 are in close contact with the Ypt51p α-helix α3 (residues 93–103). The C-terminal half of the Gyp1-46p α-helix α15 is in proximity to the N-terminal half of Ypt51p α-helix α2 (residues 68–71), which is at the end of the switch II region of small GTP binding proteins. In addition, residues in the middle of this putative switch II region would further interact with residues of the Gyp1-46p loop L11, e.g. Ypt51-Glu67 can probably form a salt bridge with Gyp1-Arg490. Loop L6 of Ypt51p (residues 89–93) is deeply buried in the cleft of the Ypt-GAP domain near the Gyp1-46p loops L6 (part of motif C around residue 378) and loop L8 (around residue 443). Ypt51p loop L8 (residues 127–132) is surrounded by the N-terminal part of α-helix α5, residues of loop L6 (around residue 375) and of loop L8 (around residue 442) from Gyp1-46p. This close interaction via loop L8 of Ypt51p is probably significant, since the isoform Ypt52p has an insertion of 17 residues in this loop region, which could be an explanation for it not being a target protein of Gyp1p. Of further importance could be the observation that the so-called effector loop region of Ypt51p (residues 34–46) is only at the periphery of the complex interface next to α-helix α5. This could be an explanation for the observation that most Gyp proteins can interact with a variety of GTPases, which display quite high diversity in the effector loop region.
From the structure and the previous mutational and biochemical characterization of the Gyp1p catalytic domain we propose a GTPase activation mechanism similar to that of H-Ras p21 GAP (Scheffzek et al., 1997) and Rho-GAP/Cdc42-GAP (Rittinger et al., 1997; Nassar et al., 1998): the side chain of the conserved glutamine residue (Gln66) of Ypt51p is suggested to be repositioned upon interaction with Gyp1p through the formation of a hydrogen bond with the main chain carbonyl group of Gyp1-Arg343, in direct analogy to that seen for the known Ras- and Cdc42-GAP–GTPase complexes (see Figure 5A); the side chain of Gyp1-Arg343 reorients in order to interact with the γ-phosphate group of the Ypt51p-bound GTP; the salt bridges formed between Gyp1-Arg343 and Gyp1-Asp340 on the Gyp side and the tight interaction between Ypt51-Gln66 and Ypt51-Arg68 on the Ypt side have to be broken and replaced by new interactions, presumably via the formation of salt bridges or hydrogen bonds between the two proteins.
From the evolutionary point of view, it is a remarkable finding that Ras-, Rho-, Cdc42p- and Ypt/Rab-GTPases, important regulators of many cellular activities, are significantly related in primary and tertiary structure whereas the corresponding GTPase-activating proteins are not at all related in primary structure and display distinct folds. However, their overall structures are nearly exclusively α-helical and their catalytic activities are based on the same mechanistic principle.
Materials and methods
Bacterial expression and purification of Gyp1-46p
The catalytically active GAP domain of Gyp1p (Albert et al., 1999) was synthesized in E.coli as a 46 kDa C-terminally His6-tagged version. A DNA fragment encoding the N-terminally truncated protein was amplified by PCR using the cloned GYP1 gene as template and two primers with different restriction sites for cloning into the bacterial expression vector pET19b (Novagen). The 5′ primer #1 (5′-ATACTGCAGGATCCATGGGTAACTCCATCATCCAGCG-3′) contained a _Bam_HI (underlined) and an _Nco_I restriction site (bold), the latter overlapping the ATG translation initiation codon. The 3′ primer #2 (5′-TATAGATCTCTGCAGTTAGTGATGGTGATGGTGATGCAGCCAGTGCGACGTAGC-3′) contained a _Pst_I (underlined) and a _Bgl_II recognition sequence (bold) downstream of the translation stop codon. PCR amplification under stringent conditions was performed as described (Albert et al., 1999). The amplified fragment encoded amino acids 249–637 of Gyp1p with an additional Met-Gly sequence at the N-terminus and six histidines at the C-terminus. It was cleaved with _Nco_I and _Bgl_II and inserted into pET19b to obtain pET19b-GYP1-46, which was transformed into E.coli BL21 (DE3) (Novagen).
For isolation of His6-tagged Gyp1-46p, 1 l of prewarmed LB medium was inoculated with an overnight culture of transformed E.coli strain BL21(DE3) to an OD600 of 0.1 and shaken at 30°C until an OD600 of 0.5–0.6 was reached. Isopropyl-β-d-galactopyranoside (IPTG) was added to 1 mM final concentration, and incubation continued for 5 h. Pelleted cells were washed with cold lysis buffer (50 mM NaH2PO4 pH 8.0, 0.3 M NaCl, 10 mM imidazole, 1 mM Pefabloc and protease inhibitor mix) and resuspended in 30 ml of this buffer per liter of bacterial culture. Cells were disintegrated using a microfluidizer (Microfluidics Corp.) and a cleared lysate was obtained by ultracentrifugation. The supernatant was loaded on to Ni-NTA–agarose (Qiagen) and after washing the beads, bound proteins were eluted with a gradient of imidazole 25–250 mM in phosphate buffer. For further purification, affinity purified Gyp1p protein was subjected to gel filtration using Superdex 75 (Pharmacia) equilibrated with 20 mM NaCl, 4 mM MgCl2, 10 mM Tris–HCl pH 7.5. From 1 l of culture, ∼15 mg of Gyp1-46p-His6 protein with a purity of >98% could be obtained. Selenomethionine (SeMet)-substituted Gyp1-46p was overexpressed in the same E.coli strain BL21(DE3) (a met+ strain) by downregulating methionine biosynthesis (Van Duyne et al., 1993; Doublie, 1997) using media supplemented with l-SeMet (60 mg/l). Selenomethionyl Gyp1-46p was purified using the same protocol as used for native protein, except that 20 mM dithiothreitol was used in all buffers. Electrospray mass spectrometry of purified selenomethionyl Gyp1-46p indicated 100% replacement of methionine with SeMet.
Crystallization
Purified Gyp1-46p in 20 mM NaCl, 4 mM MgCl2, 10 mM Tris–HCl pH 7.5 was concentrated in Centricon centrifugal filters (Millipore, Amicone). Two crystal types of Gyp1p protein were produced by the hanging drop vapour diffusion method. Triclinic crystals were grown at 20°C from 10% PEG 3350, 75 mM KOAc, 2 mM MgCl2, 70 mM Bicine pH 9, using a protein concentration of 12 mg/ml. Hexagonal bipyramid crystals were grown from 5% PEG MME 5000, 5–10 mM spermine–HCl, 20 mM Tris pH 7.5, at 20°C, using 10 mg/ml protein concentration in the drops. The triclinic crystals belonged to space group _P_1, with unit cell dimensions a = 152.5, b = 197.8, c = 296.8 Å, α = 77°, β = 89°, γ = 89° and ∼24 molecules per asymmetric unit as predicted based on the Matthews coefficient (Matthews, 1968), whereas the hexagonal form belongs to space group _P_6522 with a = b = 75.8, c = 278.5 Å with one molecule per asymmetric unit. Isomorphous crystals of the hexagonal form were also grown from SeMet-substituted protein under the same conditions.
Data collection, structure determination and refinement
MAD Se-Met Gyp1-46p data were collected at the MPG beamline BW6 and a second, high-resolution data set (maximum Bragg spacing at 1.8 Å resolution) at the EMBL beamline BW7B at HASYLAB/DESY, Hamburg using a 165 mm MarCCD in both cases. The MAD data set was processed and scaled using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). The high-resolution data set was processed with the programs XDS and XSCALE (Kabsch, 1993). Eight of the expected 11 selenium sites were identified in a Patterson search using CNS version 0.9 (Brünger et al., 1998). Site refinement, phase determination and density modification were performed using CNS version 0.9. The enantiomorph of the space group was selected by inspection of the respective electron density maps. Since the data sets collected at the white line (λ1 = 0.9790 Å) and at the inflection point (λ2 = 0.9795 Å) had the highest resolution the structure determination was performed with the two-wavelength-derived electron density map calculated with data up to 2.35 Å resolution. The map was of excellent quality and >80% of the model could be built using the program O (Jones et al., 1991). Subsequent rounds of refinement were carried out with CNS using experimental phases only and were monitored using a 10% free-R factor sample (Brünger, 1992). Further refinement against the second, high-resolution data set (cut-off at 1.9 Å resolution) was carried out after rigid-body positioning of the model. Model quality and secondary structure were determined with the program PROCHECK (Laskowski et al., 1993). The coordinates and structure factors have been deposited with the Protein Data Bank (Bernstein et al., 1977) (accession code 1FKM).
Acknowledgments
Acknowledgements
The authors thank Andrea Beste and Georg Holtermann (Dortmund) and Ursula Welscher-Altschäffel (Göttingen) for technical assistance and our colleagues at the Max-Planck-Institut für molekulare Physiologie for helpful suggestions and discussions. We thank Gleb P.Bourenkov and Hans D.Bartunik for their assistance during the collection of the MAD data set at beamline BW6 (DESY, Hamburg, Germany), Alexander Popov and Paul Tucker for their assistance during the collection of the high-resolution data set at beamline BW7B (DESY, Hamburg, Germany) and the BM30 beamline staff (ESRF, Grenoble, France). We gratefully acknowledge support by a Long Term Fellowship from Human Frontier Science Program (A.R.) and grants to A.J.S. (SCH545/1-2) and D.G. (SFB 523) from the Deutsche Forschungsgemeinschaft.
References
- Ahmadian M.R., Stege,P., Scheffzek,K. and Wittinghofer,A. (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature Struct. Biol., 4, 686–689. [DOI] [PubMed] [Google Scholar]
- Albert S. and Gallwitz,D. (1999) Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem., 274, 33186–33189. [DOI] [PubMed] [Google Scholar]
- Albert S. and Gallwitz,D. (2000) Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol. Chem., 381, 453–456. [DOI] [PubMed] [Google Scholar]
- Albert S., Will,E. and Gallwitz,D. (1999) Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J., 18, 5216–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle,T.F., Williams,G.J.B., Meyer,E.T.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The protein data bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- Bi E., Chiavetta,J.B., Chen,H., Chen,G.C., Chan,C.S. and Pringle,J.R. (2000) Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell, 11, 773–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system—a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cuff J.A., Clamp,M.E., Siddiqui,A.S., Finlay,M. and Barton,G.J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics, 14, 892–893. [DOI] [PubMed] [Google Scholar]
- Cuif M.H. et al. (1999) Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J., 18, 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Du L.L., Collins,R.N. and Novick,P.J. (1998) Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J. Biol. Chem., 273, 3253–3256. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph., 15, 133–138. [DOI] [PubMed] [Google Scholar]
- Esters H., Alexandrov,K., Constantinescu,A.T., Goody,R.S. and Scheidig,A.J. (2000) High-resolution crystal structure of S.cerevisiae Ypt51(Delta C15)-GppNHp, a small GTP-binding protein involved in regulation of endocytosis. J. Mol. Biol., 298, 111–121. [DOI] [PubMed] [Google Scholar]
- Furge K.A., Wong,K., Armstrong,J., Balasubramanian,M. and Albright,C.F. (1998) Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol., 8, 947–954. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Stoffel,W. (1993) A database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler, 374, 166. [Google Scholar]
- Horazdovsky B.F., Busch,G. and Emr,S. (1994) VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J., 13, 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R. and Sudhof,T.C. (1999) Membrane fusion and exocytosis. Annu. Rev. Biochem., 68, 863–911. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr., 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Lancaster C.A., Taylor-Harris,P.M., Self,A.J., Brill,S., van Erp,H.E. and Hall,A. (1994) Characterization of rhoGAP. J. Biol. Chem., 269, 1137–1142. [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lazar T., Gotte,M. and Gallwitz,D. (1997) Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem. Sci., 22, 468–472. [DOI] [PubMed] [Google Scholar]
- Lupas A. (1996) Prediction and analysis of coiled-coil structures. Methods Enzymol., 266, 513–525. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. (1968) Solvent content of protein crystals. J. Mol. Biol., 33, 491–495. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Nassar N., Hoffman,G.R., Manor,D., Clardy,J.C. and Cerione,R.A. (1998) Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nature Struct. Biol., 5, 1047–1052. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F. (1997) A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem. Sci., 22, 243–244. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Bharadwaj,R. and Honig,B. (1993) GRASP graphical representation and analysis of surface properties. Biophys. J., 64, A166. [Google Scholar]
- Novick P. and Zerial,M. (1997) The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol., 9, 496–504. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enyzmol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pannu N.S., Murshudov,G.N., Dodson,E.J. and Read,R.J. (1998) Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr. D, 54, 1285–1294. [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. (1999) Motivating endosome motility. Nature Cell Biol., 1, E145–E147. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker,P.A., Eccleston,J.F., Smerdon,S.J. and Gamblin,S.J. (1997) Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature, 389, 758–762. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Lautwein,A., Kabsch,W., Ahmadian,M.R. and Wittinghofer,A. (1996) Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature, 384, 591–596. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian,M.R., Kabsch,W., Wiesmüller,L., Lautwein,A., Schmitz,F. and Wittinghofer,A. (1997) The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science, 277, 333–338. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian,M.R., Wiesmuller,L., Kabsch,W., Stege,P., Schmitz,F. and Wittinghofer,A. (1998) Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J., 17, 4313–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B., Stenmark,H., Dusterhoft,A., Philippsen,P., Yoo,J.S., Gallwitz,D. and Zerial,M. (1994) Role of three rab5-like GTPases, Ypt51p, Ypt52p and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol., 125, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B., Stenmark,H. and Zerial,M. (1995) Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J. Cell Sci., 108, 3509–3521. [DOI] [PubMed] [Google Scholar]
- Strom M., Vollmer,P., Tan,T.J. and Gallwitz,D. (1993) A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature, 361, 736–739. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Vollmer P. and Gallwitz,D. (1995) High expression cloning, purification and assay of Ypt-GTPase-activating proteins. Methods Enzymol., 257, 118–128. [DOI] [PubMed] [Google Scholar]
- Vollmer P., Will,E., Scheglmann,D., Strom,M. and Gallwitz,D. (1999) Primary structure and biochemical characterization of yeast GTPase-activating proteins with substrate preference for the transport GTPase Ypt7p. Eur. J. Biochem., 260, 284–290. [DOI] [PubMed] [Google Scholar]