Reading between the LINEs: Human Genomic Variation Induced by LINE-1 Retrotransposition (original) (raw)
Abstract
The insertion of mobile elements into the genome represents a new class of genetic markers for the study of human evolution. Long interspersed elements (LINEs) have amplified to a copy number of about 100,000 over the last 100 million years of mammalian evolution and comprise ∼15% of the human genome. The majority of LINE-1 (L1) elements within the human genome are 5′ truncated copies of a few active L1 elements that are capable of retrotransposition. Some of the young L1 elements have inserted into the human genome so recently that populations are polymorphic for the presence of an L1 element at a particular chromosomal location. L1 insertion polymorphisms offer several advantages over other types of polymorphisms for human evolution studies. First, they are typed by rapid, simple, polymerase chain reaction (PCR)-based assays. Second, they are stable polymorphisms that rarely undergo deletion. Third, the presence of an L1 element represents identity by descent, because the probability is negligible that two different young L1 repeats would integrate independently between the exact same two nucleotides. Fourth, the ancestral state of L1 insertion polymorphisms is known to be the absence of the L1 element, which can be used to root plots/trees of population relationships. Here we report the development of a PCR-based display for the direct identification of dimorphic L1 elements from the human genome. We have also developed PCR-based assays for the characterization of six polymorphic L1 elements within the human genome. PCR analysis of human/rodent hybrid cell line DNA samples showed that the polymorphic L1 elements were located on several different chromosomes. Phylogenetic analysis of nonhuman primate DNA samples showed that all of the recently integrated “young” L1 elements were restricted to the human genome and absent from the genomes of nonhuman primates. Analysis of a diverse array of human populations showed that the allele frequencies and level of heterozygosity for each of the L1 elements was variable. Polymorphic L1 elements represent a new source of identical–by-descent variation for the study of human evolution.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF242435–AF242451.]
Long interspersed element-1 (LINE-1) sequences are a large family of transposable elements found in the genomes of all mammals (Burton et al. 1986; Xiong and Eickbush 1990). They belong to the poly(A)-containing (also called the non-long-terminal-repeat) class of retrotransposons. The consensus human LINE-1 (L1Hs) is 6.0 kb long, contains two nonoverlapping reading frames, terminates in an A-rich tail, and is surrounded by a short (4–20 bp) duplication of non-LINE-1 (L1) sequence, the target site duplication (Fanning and Singer 1987). The human genome contains an estimated 105 truncated and 4 × 103 full-length L1Hs elements (Adams et al. 1980; Grimaldi et al. 1984; Hwu et al. 1986), which together constitute ∼15% of the genome (Smit 1996).
The majority of L1Hs elements are not capable of transposition because they are truncated or rearranged or contain other significant mutations. Nevertheless, abundant evidence indicates that L1Hs transposition continues to occur. Several examples of recent de novo transposition events have been identified largely as the result of mutations caused by the insertion of new L1Hs elements into functional genes (Kazazian et al. 1988; Woods-Samuels et al. 1989; Miki et al. 1992; Narita et al. 1993; Bleyl et al. 1994; Holmes et al. 1994). All but one of the newly transposed L1Hs sequences in the human genome belongs to a subfamily of L1 elements called Ta (transcribed, subset a). This subfamily was first recognized as a group of expressed elements with a high degree of sequence identity to one another (Skowronski et al. 1988). The Ta subfamily of L1 elements (L1Hs-Ta) are characterized by the presence of the sequence ACA in the 3′ untranslated region at position 5930–5932; numbers refer to the actively transposing element LRE-1 [Dombroski et al. 1991]). Elements with the genomic L1Hs consensus sequence have a GAG sequence at this position (Skowronski et al. 1988). Recent experiments have suggested that the human genome may contain 30–60 active L1Hs retrotransposons (Sassaman et al. 1997).
The de novo insertion of a transposable element into the genome creates a new polymorphic genetic marker with a number of unique properties, as first described for Alu-insertion polymorphisms (Batzer and Deininger 1991; Perna et al., 1992; Deininger and Batzer 1993, 1995, 1999; Batzer et al. 1994, 1996; Stoneking et al. 1997). As with Alu-insertion polymorphisms, each L1Hs insertion represents a unique historic event. This is a result of the large number of potential target sites (theoretically equal to 3 × 109, the number of base pairs in the human genome) for the integration of new mobile elements. Thus, there is an extremely low likelihood that two independent L1Hs insertions would land between the exact same base pairs, and in the unlikely event that this should occur, the two L1 elements would probably differ in length. Accordingly, individual loci bearing the same L1Hs insertion are identical by descent. In addition, the ancestral state of an L1Hs insertion is the absence of the element, because the direction of mutation is the insertion of a new mobile element into the genome. Orthologous loci in nonhuman primates may also be analyzed for the presence of the mobile element insertion to verify the ancestral state. Once inserted, most L1Hs elements are stable over long periods of time (Smit et al. 1995). In rare instances when a transposable element is deleted from the genome, the process is often imperfect, and a “footprint” of the original mobile element is left behind (Edwards and Gibbs 1992). Finally, L1 transposition has been occurring in mammals for millions of years and continues to this day, suggesting that a series of dimorphic L1Hs insertions that have arisen throughout human evolution may be found in the present-day human population; loci that are dimorphic in a population via the presence or absence of an L1 element are called LINE-1 insertion dimorphisms (LIDs).
Most other types of genetic markers do not share these properties (Batzer et al. 1994, 1996; Stoneking et al. 1997). Thus, dimorphic transposable elements, such as Alu or L1, have a number of unique, useful properties for the study of human population genetics. Previously, dimorphic Alu elements have been used to provide insights into human genetic diversity and evolution (Perna et al. 1992; Batzer et al. 1994, 1996; Hammer 1994; Novick et al. 1995, 1998; Tishkoff et al. 1996; Stoneking et al. 1997). Dimorphic L1 elements could present another potentially useful class of genetic polymorphisms if a large number of such dimorphisms could be readily identified. Here, we describe the identification of dimorphic LINE elements from the human genome using a method called L1 display to ascertain the LIDs. Using this approach we have identified six LIDs from six individuals of diverse geographic backgrounds. In addition, we developed PCR-based assays to genotype these six individual LIDs in 850 individuals from 14 worldwide populations. Our results show that evolutionarily young LIDs can be readily identified by the L1 display assay and that these elements are a novel source of genomic variation for the study of human population genetics and forensics.
RESULTS
Identification of LIDs
Our goals in developing the L1 display were to design a method that (1) was capable of efficiently isolating LIDs from the genomic DNA of different individuals and populations, (2) allowed the DNA from many individuals to be compared and processed simultaneously, (3) required minimal preparatory manipulation of the DNA samples, and (4) could tolerate moderately degraded DNA samples. We focused our approach on the L1Hs-Ta subfamily because it includes most of the actively transposing human L1 elements (Sassaman et al. 1997).
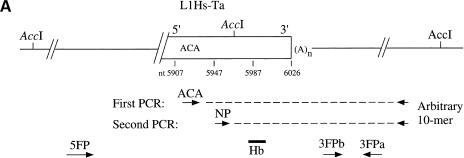
The L1 display method is outlined in Figure 1a. A truncated or full-length L1Hs-Ta is depicted surrounded by flanking DNA. Each DNA sample is amplified by two rounds of polymerase chain reaction (PCR), with multiple samples performed in parallel in each round. Also represented are the products of the PCR amplifications. In the first round, each reaction contains genomic DNA from a single individual (the template), a Ta-specific primer (termed ACA), and a single arbitrary 10-bp primer. In the second round, portions of each of the first-round PCR reactions are reamplified using a nested primer (NP) that hybridizes to a conserved region of the L1Hs 3′ untranslated region (UTR) and the same 10-bp primer used in the first round. The products of this second round of amplification are Southern blotted and hybridized with an oligonucleotide probe (Hb) that is complementary to the L1Hs 3′ UTR. Two patterns of amplification—uniform and variable bands—differentiate fixed and dimorphic L1Hs-Ta elements after such a survey of multiple genomes:— Amplified DNA fragments for a fixed element should be visible in all tested individuals (uniform bands), while amplified DNA fragments from a dimorphic element should be visible in only a fraction of the tested individuals (variable bands).
Figure 1.
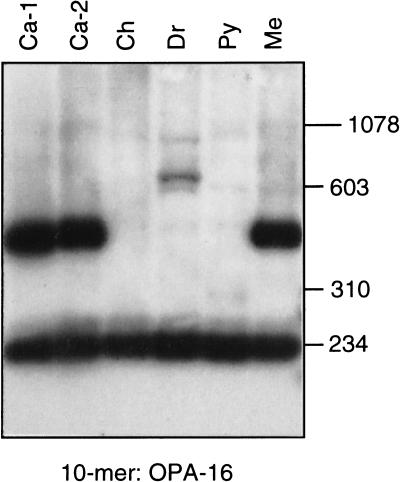
Schematic diagram of the L1Hs display and results. (A) L1 display protocol. A truncated or full-length L1Hs-Ta (rectangle) is depicted surrounded by flanking DNA (solid lines). The relative locations of the ACA and _Acc_I sites are indicated. The dashed lines represent the products of two rounds of PCR amplifications. The arrows below indicate the relative positions and orientations of the arbitrary decamers and the 19- or 20-bp-long flanking primers that were synthesized to match the non-L1 flanking DNA sequences (3FPa, 3FPb, and 5FP). (B) L1 display results. A typical L1 display experiment performed with a single decamer on genomic DNA from six individuals is shown. The figure is an autoradiograph of the gel after Southern blotting and hybridization with oligonucleotide Hb. Ca-1 and 2, European/Caucasian 1 and 2; Ch, Chinese; Dr, Druse; Py, Pygmy; Me, Melanesian. The mobilities of the DNA size markers are indicated.
The L1 display was performed with 14 arbitrary primers on DNA samples isolated from six males with diverse geographic backgrounds (European/Caucasian, Ca-1; Ashkenazi, Ca-2; Chinese, Ch; Druze, Dr; Zaire Pygmy, Py; and Melanesian from the Solomon Islands, Me). With each of the primers, strongly hybridizing bands were evident on the autoradiograms. A typical example of the results obtained using one of the arbitrary primers is shown in Figure 1b. A band of 230 bp is present in all individuals (uniform band) and may represent a fixed L1Hs-Ta locus, while a second band of about 500 bp is present in only three of the samples (variable band) and may represent a LID.
DNA from ten variable bands was isolated from agarose gels, cloned by the TA method (Invitrogen) and sequenced. All ten clones contained sequences of the L1Hs 3′-end and adjacent 3′-flanking unique region. Each of the ten clones was unique as indicated by the unique 3′ flanking sequences. The L1Hs 3′-end sequences contained the terminal 80 bp of a L1Hs 3′ UTR (64 bp of amplified sequence, plus 16 bp from primer NP) and an A-rich region. Only 3 nucleotide differences from the active LRE-1 (also an L1Hs-Ta) sequence were detected among the terminal 64 bp of L1Hs 3′ UTR sequence determined from each of the clones, and 2 of these were present in L1Hs poly(A) addition signals (not shown). This suggests that the cloned L1Hs loci are relatively young and have not had sufficient time to accumulate a large number of random mutations.
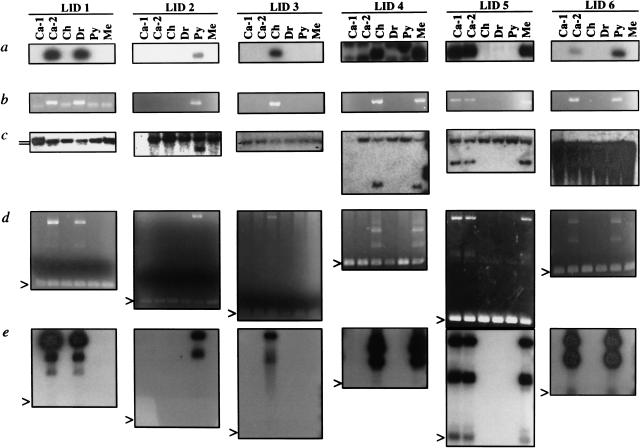
To confirm the dimorphic status of the cloned loci, genomic DNA from each individual was amplified at a higher stringency with primers ACA and 3FPa (Fig. 1a). Each of the 3FPa primers was specific for the non-L1Hs 3′-flanking DNA of one of the cloned variable DNA fragments (Fig. 1a) and each of the amplifications was done with the appropriate 3FPa primer. In six cases, the presence or absence of unique bands of the predicted size matched the pattern seen in L1 display (Figs. 2a,b). We verified the ability of the 3FPa oligonucleotides to prime PCR in all six individuals by amplifying genomic DNA with flanking primers 3FPa and 3FPb (Fig. 1a). Amplified fragments of the expected size were evident in all individuals, confirming that the absence of bands in Fig. 2b did not result from the failure of a 3FPa to prime.
Figure 2.
Identification of LID 1–6 by L1 display and verification of LID dimorphism. (a) L1 display. The products of the second round of PCR amplifications were Southern blotted and probed with oligonucleotide Hb (Fig. 1a). Digital photographs (Kodak DC40) of the sections of the autoradiograms that depict LID 1–6 are shown. Each lane represents the results obtained from one individual. (b) PCR amplification with primers ACA and 3FPa. Digital photographs of ethidium bromide-stained gels (Kodak DC40) are shown. (c) Southern blot of _Acc_I-digested genomic DNA hybridized with 3′-flanking probes. The probes were generated by amplifying the non-L1Hs 3′-flanking DNA of the cloned LIDs with primers 3FPa and 3FPb and tailing the products by the addition of [α-32P]dCTP with terminal transferase. Digital photographs of the autoradiograms of the hybridized blots are depicted. Fragments representing both the empty alleles (slower mobility) and the occupied alleles (faster mobility) can be seen in the blots hybridized with the 3′-flanking probes from LID 1, 2, 4, 5. The two bands in the Ca-2 and Dr samples of the LID-1 blot (positions indicated by short lines) are located extremely close to one another. The absence of fragments for the Ca-1 samples in the LID-2 and LID-4 blots was due to an insufficient loading of DNA. (d,e) PCR amplification of LID 1–6 with 5′- and 3′-flanking primers. Genomic DNA was amplified with primers 3FPa and 5FP. For each LID, 200 ng genomic DNA was amplified with the LID-specific primers 5FP and 3FPa. The arrowheads indicate the location of the amplified products of the empty alleles. The larger bands are the amplified products of the filled alleles. Digital photographs of the ethidium bromide-stained gels (d) or the autoradiograms of the gel after blotting and hybridization with probe Hb (e) are shown.
In four of the ten cases, amplification of genomic DNA with primers ACA and 3FPa resulted in bands of identical length in all six individuals (not shown). In these cases, it is possible that the L1 display was in error and the clones may actually represent monomorphic L1Hs insertions. It is important to note that two of the four potentially false-positive clones were obtained using a single arbitrary primer in L1 display. Other experiments confirm that some arbitrary primers are not suitable for L1 display (F-m. Sheen and G.D. Swergold, unpubl.). Careful selection of arbitrary primers that yield reproducible banding patterns in L1 display reactions can substantially reduce the false-positive rate. Alternatively, transduction of 3′-flanking DNA may have occurred during the insertion of these four elements. In this situation, the 3FPa primers, which were designed to prime near the L1Hs insertion sites, may be expected to amplify both the new (dimorphic) L1Hs insertions, as well as the progenitor L1Hs elements. As a result, the amplification of genomic DNA from different individuals with primers ACA and 3FPa may falsely indicate the presence of a single insertion with a high gene frequency. In contrast, performing PCR with the ACA and the arbitrary primers would amplify only the younger (dimorphic) L1Hs insertion because the arbitrary primer annealing site is farther from the insertion site and outside of the transduced region.
We employed several methods to confirm that the dimorphic amplified DNA fragments labeled LID 1–6 derived from the insertion of L1Hs-Ta elements on some of the sample chromosomes. First, genomic DNA from each individual was digested with _Acc_I, blotted, and hybridized with non-L1Hs 3′-flanking probes generated by PCR amplification of the cloned DNA with primers 3FPa and 3FPb (Fig. 1a). L1Hs elements contain two conserved _Acc_I sites located near the 5′- and 3′-ends of the elements. Chromosomes containing the LID-occupied alleles are expected to display shorter DNA fragments than chromosomes containing the empty alleles (Fig. 1a.). In four cases (LID 1, 2, 4, and 5), the patterns of _Acc_I fragments detected by the 3′-flank probes were also consistent with the L1 display (Fig. 2c). We were unable to confirm the dimorphic status of the other two insertion loci by Southern blotting. Hybridization with the LID-3 flanking probe revealed only a single weakly hybridizing DNA fragment, which was present in each individual, while hybridization with the LID 6 probe resulted in a smear (Fig. 2c).
Subsequently, we obtained 5′-flanking sequence from each of the insertion sites (see Methods) and amplified genomic DNA from each of the six samples using the LID-specific flanking sequence primers 5FP and 3FPa (Figs. 1a, 2d,e). In this experiment, amplification of empty alleles is expected to result in DNA fragments that are shorter than amplification of the occupied alleles by a size that depends on the length of the L1Hs insertions (Batzer and Deininger 1991). In each case, both the empty alleles (arrowheads) and the filled alleles were visible and the pattern of bands were as expected. The ethidium bromide-stained gels also contained shadow bands (Fig. 2d) that probably resulted from the formation of heterodimers between the filled and empty alleles. We confirmed the identity of the LID PCR products by blotting the gels and hybridizing with oligonucleotide Hb (Fig. 2e).
These data also indicate that LIDs 1–6 were heterozygous in each of the six tested individuals (Figs. 2c,d). DNA sequencing of the LID PCR products revealed that LID 1–6 all contained the ACA subfamily specific sequence, indicating that they indeed are members of Ta subfamily of L1 elements. Two of the LIDs (2 and 3) were present in only a single subject and each of the six subjects was unique on the basis of the presence or absence of LID 1–6 (Fig. 2).
L1Hs-Ta Subfamily Quantification
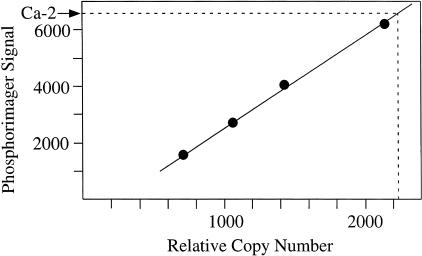
To estimate the number of L1Hs-Ta dimorphisms present in a typical genome, we first determined the total number of L1Hs-Ta 3′ UTRs in the haploid genome that contains about 100,000 L1Hs elements. Quantitative Southern blotting revealed that 2250 L1Hs-Ta 3′ UTRs were present (Fig. 3). This result compares favorably to the previous estimate, also based on quantitative Southern blotting, that 2% (80/4000) of full-length L1Hs elements belong to subset Ta (Sassaman et al. 1997). It may, however, be artificially elevated by the inappropriate hybridization of the probe to elements with sequences other than ACA at position 5930–5932. Although we did establish that the probe does not hybridize to GAG sequences (not shown) the large number of alternative sequences present in the human genome made it impractical to rule out inappropriate hybridization to a different subset of elements. We estimated the minimum frequency of Ta element dimorphisms by counting the L1 display bands obtained using the 14 arbitrary primers. An average of ten variable and 20 uniform bands were evident per individual (not shown). Only six of the ten cloned variable bands represent confirmed dimorphic L1 loci, indicating that roughly 20% (0.6 × 10/30) of Ta sites in an individual are dimorphic. We then estimated that ∼500 dimorphic loci exist in an average diploid human genome (see Methods for the calculation). L1 display cannot distinguish between individuals who are homozygous or heterozygous for a LID. This is because individuals with both of these genotypes will yield an L1 display band when the appropriate arbitrary primer is used. For a LID to be detected, at least one individual with one or more occupied alleles and at least one individual with two empty alleles must be present in the L1 display panel. Accordingly, dimorphic L1Hs insertions with high gene frequencies will be difficult to detect using either small population samples, as used here, or samples drawn from populations with high gene frequencies for the insertion. This suggests that our estimate of the number of dimorphic L1Hs insertions in the human population is lower than the actual value. A greater number of LIDs should be identifiable by performing L1 display on a larger diverse set of DNA samples.
Figure 3.
Quantification of L1Hs-Ta 3′ UTRs in the human genome. Southern blot quantification. Genomic DNA from (1) individual Ca-2, (2) mouse LMTK-cells, and (3) LMTK-cells to which plasmid pL1.2A, which contains a subset Ta L1Hs), was added were digested with _Sau_3AI and _Acc_I to release the L1Hs 3′ UTRs. Samples 2 and 3 were mixed in varying ratios to represent 0, 700, 1050, 1400, and 2100 relative copies of L1Hs per haploid genome. Samples (1 μg) of each were Southern blotted and hybridized to oligomer C, a L1Hs-Ta-specific probe. The relative activity of the hybridized bands was measured on a PhosphorImager (Molecular Dynamics). Results indicate a relative copy number of 2250 for the Ca-2 band and a linear relationship of copy number to signal in the standard lanes.
LID Localization and Human Genomic Variation
To facilitate the rapid determination of the LID genotypes of untested individuals, we developed a PCR-based assay to determine the presence or absence of individual LIDs. A schematic diagram of the assay and the expected results is depicted in Figure 4. In the assay, two PCR reactions are used to genotype each LID insertion. The first reaction utilizes 5′- and 3′-flanking unique DNA-sequence primers to ascertain genomic sites that are not occupied by L1 elements. In the second reaction, an L1Hs-Ta specific internal primer is used along with a 3′-flanking unique DNA-sequence primer to amplify genomic sites that are occupied by L1 elements. This assay was used not only to genotype individuals, but also to determine the chromosomal location of each LID element. To accomplish this, we performed a series of LID specific PCR reactions on a set of human/rodent monochromosomal hybrid cell line DNA samples (Coriell Institute) using the LID-specific PCR primers shown in Table 1. Each of the cell lines contains a full complement of rodent chromosomes, along with an individual human chromosome. Therefore, a PCR product from the LID-occupied site or the LID-empty site will be generated from a single DNA sample within the panel indicating that the LID element or preintegration site resides on that human chromosome, respectively. With this approach, we were able to map each of the LID elements to the chromosome on which they reside; the results of these experiments are shown in Table 1. Two of the six LIDs (LID 2,6) reside on human chromosome 17.
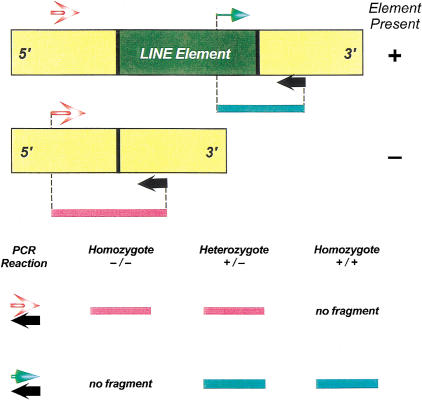
Figure 4.
Schematic diagram of the LID-insertion PCR assay. The diagram displays the LI-insertion dimorphism assay. The L1 element is in dark green, with the flanking unique sequence regions in yellow. The 5′- and 3′-flanking unique sequence primers are in red stripe and black, respectively. The internal Ta subfamily specific primer is shown in light green. The PCR amplicons generated from the L1-occupied and empty alleles are shown as green and red lines. In the assay, two PCR reactions are utilized to genotype each L1 insertion. In the first PCR reaction, 5′- and 3′-flanking unique sequence oligonucleotide primers are used to assay individual loci for empty alleles that do not contain Ta L1 elements. In the second PCR reaction, the 3′-flanking unique sequence oligonucleotide is used for the PCR, along with Ta L1 element subfamily specific primer ACA. With this approach, the size of the PCR-based amplicons generated from L1-occupied alleles is minimized and individual loci are tested for L1-occupied sites. The expected results of the PCR reactions are shown for the three potential genotypes at the bottom of the figure.
Table 1.
LID Element Primers, Annealing Temperatures, Chromosomal Locations, and PCR Amplicon Sizes
| Name | 5′ Primer sequence (5′-3′) | 3′ Primer sequence (5′-3′) | Chromosomal location | Annealing temp | Product sizes | |
|---|---|---|---|---|---|---|
| filled | empty | |||||
| LID 1 | CTCCTGACCTTGGATCTCAG | GTCCCTAATCTCTGCACTAC | 7 | 59 | 180 | 300 |
| LID 2 | AGGAAGTCTTGTAAATGTATCC | GCCTTCAGATGAGTTTTGAGATCAGAGC | 17 | 59 | 120 | 300 |
| LID 3 | TCTACAGATGTTTGAGTGCC | TGACGTAGGCTTGGATGATG | 8 | 59 | 407 | 500 |
| LID 4 | CGAATTCAGGAGGCAGAG | TAACGCCACTCTTTAAGCAG | 4 | 59 | 506 | 550 |
| LID 5 | AGGCCATGAAAACACTGAGCTTGGC | AGCCAGCGAATAGCAGGTGAAAAACAC | 5 | 59 | 600 | 470 |
| LID 6 | CGTTCTGGTATGCAGTCCAC | CCCTGAGTGTGCTTTGTACT | 17 | 59 | 505 | 350 |
| ACA | CCTAATGCTAGATGACACA | 61 |
We also used this PCR genotyping assay to determine the phylogenetic distribution of each of the LID elements within the human genome. We performed a series of LID-specific PCR reactions using DNA from 15 nonhuman primates as templates. In these experiments, we expected a preintegration-site PCR product from the genomes that do not contain the L1 element and an L1 element-specific PCR product from the genomes that do. The preintegration sites of LIDs 3, 4, and 6 were successfully amplified in all of the great-ape, old- and new-world monkey species (see Methods for a list of the samples used), while the preintegration sites for LIDs 1, 2, and 5 were successfully amplified in the chimpanzee, gorilla, and orangutan samples but not in the old- or new-world monkeys. Occupied LID alleles were not amplified from any of the nonhuman primate samples. These data are indicative of the relatively recent origin of LIDs 1–6 within the human genome.
Finally, we performed a survey of the human genomic variation associated with the six LID loci in 850 DNA samples from 14 worldwide populations. The results of this survey are shown in Table 2. Each of the LID elements was dimorphic in a number of diverse populations, with allele frequencies that ranged from 0.56 for LID 5 in the Hispanic American population to a frequency of 0 for LID1–LID6 in a number of cases. The average heterozygosity values for each locus also varied from 0.298 for LID 1 to 0.013 for LID 2. This is not surprising, for these are bi-allelic loci with a maximum heterozygosity of 0.5, or 50%. Only one of the 84 individual tests for Hardy Weinberg equilibrium were significant at the 0.01 level. We would expect one test to be significant at this level based upon chance alone, suggesting that this departure may be due to random statistical fluctuation. The between-population differentiation for each LID locus was determined using Wright's Fst statistic (Wright 1921). The amount of between-population differentiation for each LID locus ranged from 0.035 for LID3 to 0.253 for LID 5. These data indicate that 3.5%–25.3% of the variation in the data was between populations.
Table 2.
Survey of Human Genomic Variation for LIDs 1–6
| Population | LID 1 | LID 2 | LID 3 | LID 4 | LID 5 | LID 6 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | (+) freq | h | n | (+) freq | h | n | (+) freq | h | n | (+) freq | h | n | (+) freq | h | n | (+) freq | h | |
| African American | 72 | 0.08 | 0.142 | 72 | 0.02 | 0.041 | 72 | 0.07 | 0.130 | 72 | 0.38 | 0.475 | 72 | 0.17 | 0.289 | 72 | 0.13 | 0.220 |
| Armenian | 43 | 0.22 | 0.348 | 43 | 0.00 | 0.000 | 36 | 0.13 | 0.222 | 42 | 0.13 | 0.230 | 41 | 0.28 | 0.409 | 42 | 0.04 | 0.070 |
| !Kung | 40 | 0.00 | 0.000 | 41 | 0.01 | 0.024 | 41 | 0.00 | 0.000 | 32 | 0.00 | 0.000 | 40 | 0.04 | 0.073 | 42 | 0.06 | 0.113 |
| African Bantu speakers | 50 | 0.06 | 0.114 | 49 | 0.00 | 0.000 | 47 | 0.06 | 0.121 | 45 | 0.17 | 0.281 | 49 | 0.01 | 0.020 | 49 | 0.01 | 0.020 |
| Syrian | 70 | 0.17 | 0.286 | 70 | 0.01 | 0.014 | 70 | 0.05 | 0.096 | 69 | 0.28 | 0.402 | 69 | 0.22 | 0.351 | 70 | 0.04 | 0.083 |
| Turkish Cypriot | 59 | 0.21 | 0.337 | 59 | 0.00 | 0.000 | 58 | 0.03 | 0.051 | 60 | 0.15 | 0.257 | 47 | 0.02 | 0.042 | 59 | 0.00 | 0.000 |
| French | 72 | 0.22 | 0.348 | 70 | 0.00 | 0.000 | 71 | 0.08 | 0.144 | 72 | 0.22 | 0.348 | 69 | 0.01 | 0.029 | 72 | 0.06 | 0.106 |
| Alaska natives | 50 | 0.23 | 0.358 | 48 | 0.00 | 0.000 | 46 | 0.22 | 0.344 | 50 | 0.07 | 0.132 | 50 | 0.46 | 0.502 | 50 | 0.01 | 0.020 |
| Greenland natives | 50 | 0.27 | 0.398 | 50 | 0.00 | 0.000 | 46 | 0.12 | 0.213 | 50 | 0.02 | 0.040 | 59 | 0.53 | 0.503 | 50 | 0.00 | 0.000 |
| Breton | 69 | 0.24 | 0.367 | 72 | 0.00 | 0.000 | 72 | 0.14 | 0.241 | 72 | 0.16 | 0.270 | 57 | 0.02 | 0.035 | 70 | 0.11 | 0.193 |
| Germans | 72 | 0.22 | 0.348 | 70 | 0.00 | 0.000 | 71 | 0.10 | 0.179 | 68 | 0.13 | 0.231 | 64 | 0.01 | 0.016 | 72 | 0.08 | 0.154 |
| Swiss | 65 | 0.38 | 0.473 | 71 | 0.02 | 0.042 | 71 | 0.08 | 0.156 | 71 | 0.15 | 0.264 | 62 | 0.10 | 0.176 | 68 | 0.02 | 0.043 |
| Hispanic | 72 | 0.17 | 0.289 | 72 | 0.01 | 0.014 | 72 | 0.22 | 0.340 | 70 | 0.27 | 0.398 | 71 | 0.56 | 0.497 | 72 | 0.03 | 0.054 |
| European American | 72 | 0.24 | 0.371 | 72 | 0.02 | 0.041 | 72 | 0.05 | 0.093 | 70 | 0.32 | 0.439 | 62 | 0.19 | 0.315 | 72 | 0.08 | 0.142 |
| Average frequency and h | 0.194 | 0.298 | 0.006 | 0.013 | 0.095 | 0.166 | 0.176 | 0.269 | 0.187 | 0.233 | 0.047 | 0.087 | ||||||
| Total frequency and h | 856 | 0.199 | 0.319 | 859 | 0.007 | 0.014 | 845 | 0.095 | 0.172 | 843 | 0.192 | 0.311 | 812 | 0.193 | 0.311 | 860 | 0.051 | 0.096 |
| Fst | 0.065 | 0.092 | 0.035 | 0.134 | 0.253 | 0.094 |
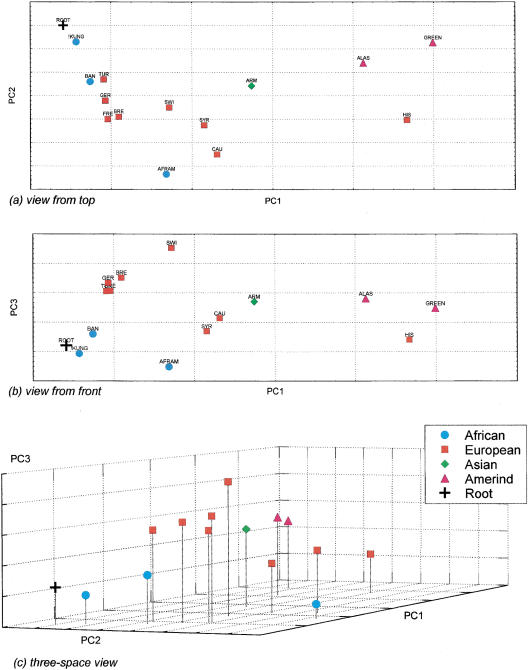
To determine the utility of the LID elements for the study of human population genetics, we performed a principal-components analysis of the distribution of the six LID elements in a series of 14 human populations (Figure 5; Harpending et al. 1996). The clustering of populations within the plot shows a good concordance with the geographic proximity of the populations. Within the front view, there are groups of populations that contain Africans, Asians, Europeans, Amerinds, and Hispanic Americans. Because the ancestral state is the absence of the LINE elements from a particular chromosomal location (as shown above), we added a hypothetical ancestral population that did not contain the LINE elements into the analysis to determine the origin. This is identical to the way that plots of population relationships derived from Alu-insertion polymorphisms are analyzed (Batzer et al. 1994, 1996; Stoneking et al. 1997). In the PC plot, the hypothetical ancestral population is denoted (root) and is closest to the African populations in the first, second, and third principal components of the analysis. This supports an African origin for our species based upon the analysis of the six LID elements reported here.
Figure 5.
Principal components analysis of LID elements in humans. A principal coordinate (PC) genetic map of 14 human populations as defined by variation in six LINE elements is presented in three views. The top two panels (a,b) show two-dimensional views of the data by plotting PC1 against PC2 and PC3, respectively. The lower panel (c) shows a three-dimensional view of the genetic distances. The first, second, and third PC axes account for 59.1%, 20.6%, and 11.4% of the variation in the samples. Thus, panel a captures 79.7% of the sample variation, panel b 70.5%, and panel c 91.1%. Population classifications–African: Bantu (BAN), African American (AFRAM), !Kung (!Kung); Asian: Armenian (ARM); European/Caucasian: Syrian (SYR), Turkish Cypriot (TUR), French (FRE), Breton (BRE), German (GER), Swiss (SWI), European-American (CAU), Hispanic American (HIS); Native American: Greenland Native (GREEN), Alaska Native (ALAS). A hypothetical ancestral population (ROOT) with a frequency of 0.0 for all LINE insertions was added into the analysis and serves as a point of initial dispersion for all other points on the map.
DISCUSSION
L1 insertion polymorphisms offer several advantages over other autosomal DNA polymorphisms for human evolution studies. First, they are typed by rapid, simple PCR-based assays. This rapid nonradioactive approach to genotyping the loci makes it possible to quickly screen large numbers of DNA samples derived from a variety of different sources. In contrast, many other types of polymorphisms are much more time consuming to analyze and often require radioactivity or automated DNA sequencers for analysis (e.g., Bowcock et al. 1994; Deka et al. 1995, 1996).
LINE elements are also stable polymorphisms that rarely undergo deletion. Even when the deletion of a mobile-element fossil occurs within the genome, a partial fossil relic is typically left behind in the genome, as previously reported for Alu elements (Edwards and Gibbs 1992). It is also important to note that the rate of L1 element mobilization in the human genome has been faster than that of Alu elements but is still relatively slow, with only about 4500 L1H-Ta elements integrated in the genome since the radiation of African apes. This is important because the presence of an L1 element represents identity by descent, for the probability that two different Ta L1 repeats would integrate independently in the same chromosomal location is negligible. This means that the L1 elements are similar to the previously reported mobile element insertion polymorphisms (Batzer et al. 1991, 1994, 1995; Hammer 1994; Zietkiewicz et al. 1994; Arcot et al. 1995a,b, 1996, 1998; Novick et al. 1995; Tishkoff et al. 1996; Stoneking et al. 1997; Boissinot et al. 2000; Jorde et al. 2000; Santos et. al. 2000). In addition, this makes the L1-insertion dimorphisms and other mobile-element insertion polymorphisms (e.g., Alu-insertion polymorphisms) unique as compared to other genomic polymorphisms, such as single nucleotide polymorphisms (Sherry et al. 2000), simple sequence repeats (Nakamura et al. 1987), or restriction site polymorphisms (Botstein et al. 1980), which may arise numerous times within a population and, hence, are merely identical by state.
We have also shown that the ancestral state of L1-insertion polymorphisms is the absence of the L1 element through PCR-based analysis of orthologous positions within the genomes of several nonhuman primates. This information concerning the ancestral state can be used to root trees/plots of population relationships derived from the analysis of L1-insertion dimorphisms. Unambiguous knowledge of the ancestral state of these and other mobile-element-insertion polymorphisms (e.g., Alu-insertion polymorphisms in the human genome), as well as other insertion/deletion polymorphisms makes these types of polymorphic markers unique (Batzer et al. 1994, 1996; Stoneking et al. 1997). The insertion of L1 elements into the human genome is also an ongoing process, resulting in a wide array of L1-based insertion dimorphisms that have arisen at different times during human evolution and are shared within a population or between different populations or may be unique to a single individual or family.
To explore the utility of L1-based insertion dimorphisms for the study of human population relationships, we analyzed the distribution of six LID elements in 14 human populations. In the PC analysis, the populations cluster in a manner that shows good concordance with geographic proximity between populations. In addition, the hypothetical ancestral population, or root of the PC plot, resided in Africa, suggesting an African origin of our species. This result is in agreement with a growing body of literature involving the analysis of a variety of genetic systems (reviewed in Jorde et al. 1998). However, these results should be interpreted with caution, given the small number of populations and loci involved.
A comparison of the levels of genetic variation associated with the LID elements reported here and those of previously reported Alu-insertion polymorphisms (Batzer et al. 1994, 1996; Stoneking et al. 1997) reveals that both the levels of heterozygosity and the average allele frequencies of the LID elements reported in this study are lower than those previously reported for Alu-insertion polymorphisms. Although this may partially reflect the limited set of populations that have been surveyed for L1-element-based genetic variation, we believe that this potential source of bias is minor. Rather, the major reason for the difference probably resides in the method by which the LID and Alu elements were originally identified. Most of the previously identified Alu-insertion polymorphisms have been identified by screening total genomic libraries (Batzer et al. 1990, 1991, 1995; Batzer and Deininger 1991; Arcot et al. 1995a,b, 1996, 1998) or data mining (Roy et al. 1999). Because these elements were identified by first screening the entire genome or database for the presence of subfamily specific repetitive elements and then testing each element for polymorphism, the frequency distribution of these ascertained elements is biased toward very common elements, for the element must be present in the individual whose genome is being analyzed. By contrast, the direct identification of mobile elements that are polymorphic by PCR-based display as reported here for L1 elements shifts the frequency spectrum of the ascertained elements toward the less common or more recently integrated elements within the genome. The higher frequency elements are not identified as polymorphic because they are more likely to be shared between genomes in heterozygous or homozygous states. This difference makes the data-mining and genome-screening approaches for the identification of polymorphic mobile-element insertions complementary to the PCR-based displays reported previously for Alu repeats (Roy et al. 1999) and in our study for L1 elements. Alu and L1 elements have also previously been shown to have different physical distributions within the human genome (Soriano et al. 1983; Manuelidis and Ward 1984; Korenberg and Rykowski 1988; Moyzis et al. 1989). Therefore, the combination of L1-insertion and Alu-insertion polymorphisms should provide a genome-wide assortment of mobile-element-based polymorphisms composed of 2000 or more elements for the analysis of human evolutionary history.
One previous limitation of the use of transposable element markers for the study of population genetics has been the difficulty in discovering new markers using laborious library screening procedures (Batzer et al. 1991, 1995; Arcot et al. 1995a,b, 1998) and the related difficulty of discovering mobile element insertion events that have occurred during recent human evolutionary history. For example, previously reported L1Hs dimorphisms were identified either by the chance discovery of insertions in genes being investigated for other reasons or by the screening of genomic libraries for elements belonging to the Ta subclass (Dombroski et al. 1993; Bleyl et al. 1994; Sassaman et al. 1997). Given the large background of mobile elements that have amplified in the past within our genomes, the identification of the more recent events becomes the genomic equivalent of the identification of needles in a haystack. In our study, we have described an efficient method, called L1 display, that is designed for discovering LINE-1 insertion dimorphisms from diverse human populations. The L1 display can be performed simultaneously on many genomic DNA samples, thereby greatly increasing the likelihood of discovering both recent and ancient insertions. This assay should also prove useful for determining the rate of L1 transposition in somatic and germ-line tissues and to investigate a possible role for transposition in nondisjunction and oncogenesis (Bratthauer and Fanning 1992, 1993; Bratthauer et al. 1994; Hawley et al. 1994).
METHODS
Cell Lines and DNA Samples
The human DNA samples used for the L1 display were as follows: Melanesian (Me), Pygmy (Py), Druze (Dr), and Caucasian-1 (Ca-1) DNA were isolated from tissue culture cell lines GM10540, GM10492, GM11522, and GM05386 (Coriell Cell Repository), respectively; the Caucasian-2 (Ca-2) and Chinese (Ch) DNA were isolated from blood samples donated by the authors using standard protocols (Sambrook et al. 1989). All of the samples used for the display analyses were derived from males. Human (Homo sapiens), HeLa (ATCC CCL2); chimpanzee (Pan troglodytes), Wes (ATCC CRL1609); gorilla (Gorilla gorilla), Ggo-1 (primary gorilla fibroblasts), were provided by Stephen J. O'Brien (National Cancer Institute, Frederick, Maryland, USA). Additional nonhuman primate DNA samples from five chimpanzees, one gorilla, three orangutans (Pongo pygmaeus), one macaque (Macaca fascicularis), and one tamarin (Saguinus oedipus) were obtained from BIOS Laboratories (New Haven, Connecticut, USA). Cell lines were maintained as directed by the source and DNA isolations were performed using Wizard genomic DNA purification (Promega). Human DNA samples from geographically diverse populations were either isolated from peripheral blood lymphocytes using Wizard genomic DNA purification kits (Promega) or were available from previous studies (Stoneking et al. 1997).
Oligonucleotides
Arbitrary oligonucleotide decamers were purchased from Operon (Li et al. 1996). The other oligonucleotides used in this study were prepared by the Center for Biologics Evaluation and Research core facility, or purchased from Life Technologies. The sequence of the L1 display oligonucleotides were as follows: ACA 5′-CTAATGCTAGATGACACA-3′NP 5′-GCACCAGCATGGCACA-3′ Hb 5′-CCTGCACAATGTGCACATGTACCC-3′.The sequences of the oligonucleotides used in the analyses of individual LID elements are shown in Table 1.
LID Identification Polymerase Chain Reaction
Three different types of PCR reactions were performed as follows. (1) L1 display PCR. The first round of L1 display PCR reactions were carried out with 25 ng genomic DNA, 0.5 μM primer ACA, and 0.3 μΜ decamer primer in 20 mM Tris pH 8.4, 1.5 mM MgCl2, 50 mM CaCl2, 0.2 mM deoxynucleotides, and 2.5 U Taq DNA polymerase for 40 cycles of 94°C for 30 sec, 36°C for 30 sec, and 72°C for 30 sec. The second round of reactions were carried out under the same conditions, except that the primer NP was substituted for primer ACA, and the template consisted of 2.5 μl of the products of the first-round reactions. (2) 3′-flanking PCR. Amplifications with primer ACA and primer 3FPa (Fig. 2) were performed with 200 ng genomic DNA, 0.2 μΜ of each primer and an annealing temperature of 50°C. (3) 5′- and 3′-flanking PCR. To amplify the LID insertion sites using primers 5FP and 3FPa (Fig. 1), we utilized BIO-X-ACT DNA polymerase (GeneMate), 200 ng genomic DNA, and 0.2 μM of each primer in OptiBuffer (GeneMate). Reactions were carried out for 31–36 cycles of 59°C for 30 sec and of 68°C for 3–6 min, depending on the length of the expected products.
Cloning and Sequencing of LID PCR Products
L1 display DNA fragments were isolated from agarose gels with the QIAquick gel extraction kit (Qiagen) and cloned by the TA method (Invitrogen). DNA sequences were determined with either the SequiTherm EXCEL kit (Epicentre Technologies) using 32P-labeled primers or with the Thermo Sequanase kit (Amersham) using 33P-labeled dideoxynucleotides. Sequence analyses and database searching were performed with the MacVector program version 6.0 (Oxford Molecular Group). The sequence of the insertion sites of LID 1–4, 6 were obtained by amplifying empty alleles using the Genome Walker kit (Clontech) and LID-specific primers. The accession numbers for the sequences from the LID elements re as follows LID 1, AF242438-AF242441; LID 2, AF242442-AF242444; LID 3, AF242445-AF242448; LID 4, AF242449-AF242451; LID 5, AF242452; LID 6, AF242453-AF242455. The 5′ flanking sequence of LID 5 was obtained from GenBank (accession AC002122)
Southern Blotting and Ta Subfamily Quantification
PCR products were separated electrophoretically in a 3% 3:1 NuSieve agarose gel (FMC) and alkaline blotted onto a Nytran Plus membrane (Schleicher & Schuell). Hybridizations were performed with 32P-end-labeled oligonucleotides (109 cpm/μg) in 5 × SSPE/0.3% SDS/10 μg/ml salmon sperm DNA at 42°C overnight. The membranes were washed in 2 × SSPE at 25°C for 15 min, 2 × SSPE/0.1% SDS at 25°C for 45 min and twice in 0.5 × SSPE/0.1% SDS at 42°C for 15 min. Southern analysis of genomic DNA was performed as described (Church and Gilbert 1984). To quantify the number of Ta 3′ UTRs in an average genome, 6 ng pL1.2A (Dombroski et al. 1991) DNA was added to 4 μg of mouse LMTK- DNA and digested with _Sau_3AI and _Acc_I. After digestion, the DNA was extracted with phenol/chloroform, ethanol precipitated, redissolved and the concentration again determined spectrophotometrically. The DNA was then diluted with a similarly prepared sample of mouse LMTK-DNA (into which no plasmid DNA had been added) at relative copy numbers of 700, 1050, 1400, and 2100 copies of pL1.2A per haploid genome. A 1 μg sample of each and 1 μg of similarly digested genomic DNA from individual Ca-2 were Southern blotted and hybridized to the L1Hs-Ta specific oligonucleotide (oligomer C [5′-TGCTAGATGACACATTAGTG-3′] from Sassaman et al. 1997). Hybridization and washing conditions were as listed above. The relative activity of the hybridized bands was measured on a PhosphoIimager (Molecular Dynamics). Results indicate a relative copy number of 2250 for the Ca-2 band and a linear relationship of copy number to signal in the standard lanes. Calculation of the number of LIDs was performed as follows:
Variable bands = 10
Total bands = 30
True dimorphic bands = (10/30) × (6 dimorphic bands)/(10 putative dimorphic bands) = 0.2
X = number of monomorphic L1Hs-Ta loci
Y = number of dimorphic L1Hs-Ta loci
4500 = total number of Ta loci/diploid genome
Total number of L1 display bands = X + Y
Equation 1 4500 = 2(X) + Y
Equation 2 0.2 = Y/(X + Y)
Solve for Y: = 500 dimorphic loci /diploid genome.
LID Genotyping and Human Genomic Diversity
Nucleotide sequences flanking individual Ta L1 elements were screened against the GenBank nonredundant database for the presence of repetitive elements using the basic local alignment search tool (BLAST program) from the National Center for Biotechnology Information (Altschul et al. 1990). PCR primers for each locus were designed either manually or using the software PRIMER (Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA). PCR amplification was carried out in 25 μl reactions under exact conditions as described previously for Alu-insertion polymorphisms (Stoneking et al. 1997). Individual LINE insertion dimorphisms were genotyped by direct inspection of agarose gels after amplification. The sequences of the primers for each locus and their annealing temperatures are shown in Table 1. The observed numbers of each genotype for each locus and population are available upon request from the authors.
Acknowledgments
We thank Dr. Maxine Singer for helpful discussions. This research was supported by National Science Foundation SBR-9610147 (MAB) and award number 1999-IJ-CX-K009 from the Office of Justice Programs, National Institute of Justice, Department of Justice (MAB). Points of view in this document are those of the authors and do not necessarily represent the official position of the U.S. Department of Justice. S.T.S. was supported by an National Research Service Award from the National Institutes of Health (GM19110).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gs314@columbia.edu; FAX (212) 342-5316.
REFERENCES
- Adams JW, Kaufman RE, Kretschmer PJ, Harrison M, Nienhuis AW. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980;8:6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Fontius JJ, Deininger PL, Batzer MA. Identification and analysis of a “young” polymorphic Alu element. Biochem Biophys Acta. 1995a;1263:99–102. doi: 10.1016/0167-4781(95)00080-z. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995b;29:136–144. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Adamson AW, Lamerdin JE, Kanagy B, Deininger PL, Carrano AV, Batzer MA. Alu fossil relics—distribution and insertion polymorphism. Genome Res. 1996;6:1084–1092. doi: 10.1101/gr.6.11.1084. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Adamson AW, Risch G, LaFleur J, Lamerdin JE, Carrano AV, Batzer MA. High-resolution cartography of recently integrated chromosome 19-specific Alu fossils. J Mol Biol. 1998;281:843–855. doi: 10.1006/jmbi.1998.1984. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. A human-specific subfamily of Alu sequences. Genomics. 1991;9:481–487. doi: 10.1016/0888-7543(91)90414-a. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Kilroy GE, Richard PE, Shaikh TH, Desselle TD, Hoppens CL, Deininger PL. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 1990;18:6793–6798. doi: 10.1093/nar/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Gudi VA, Mena JC, Foltz DW, Herrera RJ, Deininger PL. Amplification dynamics of human-specific (HS) Alu family members. Nucleic Acids Res. 1991;19:3619–3623. doi: 10.1093/nar/19.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Stoneking M, Allegria-Hartman M, Bazan H, Kass DH, Shaikh TH, Novick GE, Ioannou PA, Scheer WD, Herrera RJ, et al. African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci. 1994;91:12288–12292. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Rubin CM, Hellmann-Blumberg U, Alegria-Hartman M, Leeflang EP, Stern JD, Bazan HA, Shaikh TH, Deininger PL, Schmid CW. Dispersion and insertion polymorphism in two small subfamilies of recently amplified human Alu repeats. J Mol Biol. 1995;247:418–427. doi: 10.1006/jmbi.1994.0150. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Arcot SS, Phinney JW, Alegria-Hartman M, Kass DH, Milligan SM, Kimpton C, Gill P, Hochmeister M, Ioannou PA, et al. Genetic variation of recent Alu insertions in human populations. J Mol Evol. 1996;42:22–29. doi: 10.1007/BF00163207. [DOI] [PubMed] [Google Scholar]
- Bleyl S, Ainsworth P, Nelson L, Viskochil D, Ward K. An ancient Ta subclass L1 insertion results in an intragenic polymorphism in an intron of the NF1 gene. Hum Mol Genet. 1994;3:517–518. doi: 10.1093/hmg/3.3.517. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick MH, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Amer J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Bratthauer GL, Fanning TG. Active LINE-1 retrotranposons in human testicular cancer. Oncogene. 1992;7:507–510. [PubMed] [Google Scholar]
- Bratthauer GL, Fanning TG. LINE-1 retrotransposon expression in pediatric germ cell tumors. Cancer. 1993;71:2383–2386. doi: 10.1002/1097-0142(19930401)71:7<2383::aid-cncr2820710733>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Bratthauer GL, Cardiff RD, Fanning TG. Expression of LINE-1 retrotransposons in human breast cancer. Cancer. 1994;73:2333–2336. doi: 10.1002/1097-0142(19940501)73:9<2333::aid-cncr2820730915>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Burton FH, Loeb DD, Voliva CF, Martin SL, Edgell MH, Hutchinson CAI. Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA Long Interspersed Sequence One. J Mol Biol. 1986;187:291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic Sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Evolution of retroposons. Evol Biol. 1993;27:157–196. [Google Scholar]
- ————— . Evolution of retroposons. In: Maraia RJ, editor. The Impact of Short Interspersed Elements (SINEs) on the Host Genome. Georgetown, Texas: R.G. Landes; 1995. pp. 43–60. [Google Scholar]
- ————— Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Deka R, Jin L, Shriver MD, Yu LM, DeCroo S, Hundrieser J, Bunker CH, Ferrell RE, Charaborty R. Population genetics of dinucleotide (dC-dA)n.(dG-dT)n polymorphisms in world populations. Am J Hum Genet. 1995;56:461–474. [PMC free article] [PubMed] [Google Scholar]
- Deka R, Jin L, Shriver MD, Yu LM, Saha N, Barrantes R, Chakraborty R, Ferrell RE. Dispersion of human Y chromosome haplotypes based on five microsatellites in global populations. Genome Res. 1996;6:1177–1184. doi: 10.1101/gr.6.12.1177. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an Active Human Transposable Element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Scott aF, Kazazian HH., Jr Two additional potential retrotranspsosons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MC, Gibbs RA. A Human dimorphism resulting from loss of an Alu. Genomics. 1992;14:590–597. doi: 10.1016/s0888-7543(05)80156-9. [DOI] [PubMed] [Google Scholar]
- Fanning TG, Singer MF. LINE-1: A mammalian transposable element. Biochim Biophys Acta. 1987;910:203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of the KpnI family segments. EMBO J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF. A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol. 1994;11:749–761. doi: 10.1093/oxfordjournals.molbev.a040155. [DOI] [PubMed] [Google Scholar]
- Harpending HC, Relethford J, Sherry ST. Methods and models for understanding human diversity. In: Boyce AJ, Mascie-Taylor CGN, editors. Molecular biology and human diversity. Cambridge, UK: Cambridge University Press; 1996. pp. 283–299. [Google Scholar]
- Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST. Genetic trances of ancient demography. Proc Natl Acad Sci. 1998;95:1961–1967. doi: 10.1073/pnas.95.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RS, Frazier JA, Rasooly R. Commentary: Separation anxiety: The etiology of nondisjunction in flies and people. Hum Mol Genet. 1994;3:1521–1528. doi: 10.1093/hmg/3.9.1521. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–8. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Hwu HR, Roberts JW, Davidson EH, Britten RJ. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci. 1986;83:3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Bamshad M, Rogers AR. Using mitochondrial and nuclear DNA markers to reconstruct human evolution. BioEssays. 1998;20:126–136. doi: 10.1002/(SICI)1521-1878(199802)20:2<126::AID-BIES5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA. The distribution of human genetic diversity: A comparison of mitochondrial, autosomal, and Y chromosome data. Amer J Hum Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HHJ, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis S. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature (London) 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Rykowski MC. Human genome organization: Alu, Lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- Li S-W, Nembhard KM, Prockop DJ, Khillan JS. Identification and cloning of integration site DNA by PCR. BioTechniques. 1996;20:356–358. doi: 10.2144/19962003356. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Ward DC. Chromosomal and nuclear distribution of the HindIII 1.9-kb human DNA repeat segment. Chromosoma. 1984;91:28–38. doi: 10.1007/BF00286482. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomia J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Canc Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Torney DC, Meyne J, Buckingham JM, Wu J-R, Burks C, Sirotkin KM, Goad WB. The distribution of interspersed repetitive DNA sequences in the human genome. Genomics. 1989;4:273–289. doi: 10.1016/0888-7543(89)90331-5. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Leppert M, O'Connell P, Wolff R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E, White R. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987;235:1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Narita N, Nishio H, Kitoh Y, Ishikawa Y, Ishikawa Y, Minami R, Nakamura H, Matsuo M. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest. 1993;9:1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick GE, Novick C, Yunis J, Yunis E, Martinez K, Duncan GG, Troup GM, Deininger PL, Stoneking M, Batzer MA, et al. Polymorphic human specific Alu insertions as markers for human identification. Electrophoresis. 1995;16:1596–1601. doi: 10.1002/elps.11501601263. [DOI] [PubMed] [Google Scholar]
- Novick GE, Novick C, Yunis J, Yunis E, Antunez De Mayolo P, Scheer WD, Deininger PL, Stoneking M, York DS, et al. Polymorphic Alu insertions and the Asian origin of Native American populations. Hum Biol. 1998;70:23–29. [PubMed] [Google Scholar]
- Perna NT, Batzer MA, Deininger PL, Stoneking M. Alu insertion polymorphism: A new type of marker for human population studies. Hum Biol. 1992;64:641–648. [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AM, Carroll ML, Kass DH, Nguyen SV, Salem A, Batzer MA, Deininger PL. Recently integrated human Alu repeats: Finding needles in the haystack. Genetica. 1999;107:149–161. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Press; 1989. [Google Scholar]
- Santos FR, Pandya A, Kayser M, Mitchell RJ, Liu A, Singh L, Destro-Bisol G, Novelletto A, Qamar R, Mehdi SQ, et al. A polymorphic L1 retroposon insertion in the centromere of the human Y chromosome. Hum Mol Genet. 2000;9:421–30. doi: 10.1093/hmg/9.3.421. [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nature Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. Use of molecular variation in the NCBI dbSNP database. Hum Mutat. 2000;15:68–75. doi: 10.1002/(SICI)1098-1004(200001)15:1<68::AID-HUMU14>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Toth G, Riggs AD, Jurka J. Ancestral mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Soriano P, Meunier-Rotival M, Bernardi G. The distribution of interspersed repeats in nonuniform and conserved in the mouse and human genomes. Proc Natl Acad Sci. 1983;80:1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M, Fontius JJ, Clifford S, Soodyall H, Arcot SS, Saha N, Jenkins T, Tahir MA, Deininger PL, Batzer MA. Alu insertion polymorphisms and human evolution: Evidence for a larger population size in Africa. Genome Res. 1997;7:1061–1071. doi: 10.1101/gr.7.11.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Ruano G, Kidd JR, Kidd KK. Distribution and frequency of a polymorphic Alu insertion at the plasminogen activator locus in humans. Hum Genet. 1996;97:759–764. doi: 10.1007/BF02346186. [DOI] [PubMed] [Google Scholar]
- Wright S. Systems of mating. Genetics. 1921;16:97–159. [Google Scholar]
- Woods-Samuels P, Wong C, Mathias SL, Scott AF, Kazazian HH, Jr, Antonarakis SE. Characterization of a nondeleterious L1 insertion in an intron of the human factor VIII gene and further evidence of open reading frames in functional L1 elements. Genomics. 1989;4:290–296. doi: 10.1016/0888-7543(89)90332-7. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E, Richer C, Makalowski W, Jurka J, Labuda D. A young Alu subfamily amplified independently in human and African great apes lineages. Nucleic Acids Res. 1994;22:5608–5612. doi: 10.1093/nar/22.25.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]