AT-Rich Palindromes Mediate the Constitutional t(11;22) Translocation (original) (raw)
Abstract
The constitutional t(11;22) translocation is the only known recurrent non-Robertsonian translocation in humans. Offspring are susceptible to der(22) syndrome, a severe congenital anomaly disorder caused by 3:1 meiotic nondisjunction events. We previously localized the t(11;22) translocation breakpoint to a region on 22q11 within a low-copy repeat termed “LCR22” and within an AT-rich repeat on 11q23. The LCR22s are implicated in mediating different rearrangements on 22q11, leading to velocardiofacial syndrome/DiGeorge syndrome and cat-eye syndrome by homologous recombination mechanisms. The LCR22s contain AT-rich repetitive sequences, suggesting that such repeats may mediate the t(11;22) translocation. To determine the molecular basis of the translocation, we cloned and sequenced the t(11;22) breakpoint in the derivative 11 and 22 chromosomes in 13 unrelated carriers, including two de novo cases and der(22) syndrome offspring. We found that, in all cases examined, the reciprocal exchange occurred between similar AT-rich repeats on both chromosomes 11q23 and 22q11. To understand the mechanism, we examined the sequence of the breakpoint intervals in the derivative chromosomes and compared this with the deduced normal chromosomal sequence. A palindromic AT-rich sequence with a near-perfect hairpin could form, by intrastrand base-pairing, on the parental chromosomes. The sequence of the breakpoint junction in both derivatives indicates that the exchange events occurred at the center of symmetry of the palindromes, and this resulted in small, overlapping staggered deletions in this region among the different carriers. On the basis of previous studies performed in diverse organisms, we hypothesize that double-strand breaks may occur in the center of the palindrome, the tip of the putative hairpin, leading to illegitimate recombination events between similar AT-rich sequences on chromosomes 11 and 22, resulting in deletions and loss of the palindrome, which then could stabilize the DNA structure.
Introduction
The chromosome 22q11 region is susceptible to rearrangements associated with congenital anomaly disorders, including velocardiofacial syndrome/DiGeorge syndrome (VCFS/DGS [MIM 192430/MIM 188400]; DiGeorge 1965; Shprintzen et al. 1978), cat-eye syndrome (CES [MIM 115470]; Guanti et al. 1981; Reiss et al. 1985) and der(22) syndrome (Fraccaro et al. 1980; Zackai and Emanuel 1980; Schinzel et al. 1981). Most patients with VCFS/DGS have 3-Mb hemizygous deletions of 22q11; a subset have a nested distal deletion endpoint, resulting in a 1.5-Mb deletion, and a few rare patients have unique deletions (Lindsay et al. 1995; Morrow et al. 1995; Carlson et al. 1997). Most cases occur sporadically in the population, suggesting that this region is prone to chromosome rearrangements. Physical-mapping studies revealed the presence of low-copy repeat clusters containing genes or pseudogenes, >200 kb in size, at the common 3-Mb deletion endpoints and at a third endpoint, mapping in between the two common ones, containing the 1.5-Mb distal deletion endpoint (Edelmann et al. 1999_a;_ Funke et al. 1999; Shaikh et al. 1999). To determine the mechanism causing the deletion, haplotypes were generated in three generations of individuals. Both inter- and intrachromosomal homologous recombination events between the LCR22s mediated the 3-Mb deletion (Baumer et al. 1998; Edelmann et al. 1999_b_). A family with an interstitial duplication of the same 3-Mb region was identified last year, and haplotype analysis was consistent with an interchromosomal homologous recombination event between the LCR22s (Edelmann et al. 1999_b_). In addition to these breakpoints, the CES duplication breakpoints resulting in a supernumerary bisatellited chromosome 22 occurred in the same LCR22s as the 3-Mb deletion, resulting in two CES categories, CES-I and CES-II (McTaggart et al. 1998; Edelmann et al. 1999_b_). These results demonstrated that the LCR22s mediate a number of distinct rearrangements on 22q11 by homologous recombination mechanisms leading to congenital anomaly disorders.
Meiotic nondisjunction events in carriers of the constitutional t(11;22) translocation result in the severe congenital anomaly disorder, referred to as “der(22) syndrome,” caused by 3:1 meiotic nondisjunction events (Fraccaro et al. 1980; Zackai and Emanuel 1980). Der(22) syndrome patients carry a der(22) chromosome and are therefore trisomic for 11q23-qter and 22pter-q11. The main clinical findings of der(22) syndrome are moderate mental retardation, mild craniofacial anomalies, and congenital heart defects (Zackai and Emanuel 1980; Lin et al. 1986). To determine the molecular basis of the reciprocal t(11;22) translocation, the breakpoint intervals on 11q23 and 22q11 were defined. In all carriers examined, the constitutional t(11;22) breakpoint occurred in the same LCR22 as the distal 1.5-Mb deletion in patients with VCFS/DGS (Edelmann et al. 1999_b;_ Funke et al. 1999; Shaikh et al. 2000; Tapia-Paez et al. 2000) referred to as “LCR22-3a” (Dunham et al. 1999). To narrow the region on 11q23, we generated somatic hybrid cell lines from two unrelated carriers and found that the site of strand exchange occurred in an AT-rich repeat between the genetic markers D11S1340 and APOC3 (Edelmann et al. 1999_c_). The LCR22s also harbor AT-rich repetitive sequence motifs, which we hypothesized could be the region of meiotic exchange. A few years ago, an AT-rich motif was implicated in the novel, reciprocal t(17;22)(q11;q11) translocation in a family with neurofibromatosis type 1 (NF1) (Kehrer-Sawatzki et al. 1997). The exchange event occurred between an AT-rich repeat within intron 31 of the NF1 gene on chromosome 17 and a similar AT-rich repeat on 22q11 (Kehrer-Sawatzki et al. 1997), suggesting that there was a common mechanism underlying both the t(11;22) and the t(17;22) translocations. Interestingly, the sequences of the normal chromosomes 17 and 22 surrounding this region consisted of an inverted repeat and therefore was palindromic.
Recently, the constitutional t(11;22) translocation was cloned and sequenced (Hill et al. 2000; Kurahashi et al. 2000_a,_ 2000_b_). In one report (Hill et al. 2000), the t(11;22) breakpoint in both chromosomes, in five carriers, was mapped to an Alu repeat, and inter-Alu homologous recombination events were the proposed mechanism leading to the translocation. The Alu sequence on chromosome 11 mapped 350 bp proximal to the AT-rich repeat. This result was in conflict with our earlier findings, as well as with the data we present in this report. Two additional reports are in agreement with our finding that the breakpoints occur in palindromic AT-rich repeats (Kurahashi et al. 2000_a,_ 2000_b_); however, we differ in our interpretation of the data as to the exact region of chromosome breakage and exchange. Characterizing the precise sites of exchange between chromosomes is paramount to understanding the molecular mechanism leading to the translocation. On the basis of studies of palindromic sequences in prokaryotes through mammalian species, we propose that the chromosomal breaks occur in the center of the palindromes, at the tip of the putative hairpins, resulting in illegitimate recombination events producing overlapping staggered deletions.
Patients and Methods
Patients
Carriers of the t(11;22) translocation and patients with der(22) syndrome with “BM” codes were ascertained and their conditions diagnosed as described elsewhere (Funke et al. 1999), under an internal review board–approved program. Cell lines obtained for the t(11;22) translocation carriers included GM06229b, GM04403, GM03847, GM03372, and GM06275a, as well as der(22) syndrome patients GM06228, GM04370a, GM03371, and GM00084a, and were purchased from the National Institute of General Medical Science cell repository (Coriell Cell Repositories). Carrier cell lines purchased from the European Collection of Cell Cultures (ECACC) included DD3474, DD0185 (de novo), DD1990, and DD0329. The carrier, BM737—representing a de novo case of the constitutional t(11;22) translocation, as suggested by FISH mapping studies using the N25 probe (Oncor)—was ascertained in our laboratory. To generate haplotypes to rule out nonpaternity, genomic DNA (50 ng) from the carrier (BM737) and the parents (BM738 and 739) was genotyped using the markers D22S427, D22S1638, D22S941, and D22S264, mapping to 22q11, as well as D11S1885, D11S1340, and D11S4127, mapping to 11q23, as described elsewhere (Edelmann et al. 1999_c;_ Funke et al. 1999). The carrier, V1, was ascertained by the Obstetrics and Gynecology Department at the Albert Einstein College of Medicine.
Somatic Cell–Hybrid Cell Lines
The method used to generate hamster-human somatic hybrid cell lines from the t(11;22) carriers, BM114 and GM06229b, has been described elsewhere (Edelmann et al. 1999_c_). For the PCR-based genotyping experiments, 50 ng of template DNA from each sample was used, under standard reaction conditions, as described elsewhere (Carlson et al. 1997).
Genomic Southern Blot Hybridization
Genomic DNA was purified from Epstein-Barr–transformed human lymphoblastoid cell lines, human fibroblast cell lines, or hamster-human somatic hybrid cell lines and 10 ug from each sample was digested with the restriction endonucleases, _Sac_I or _Hin_dIII. The digested DNA was separated electrophoretically, according to size, on 0.8% agarose gels in TAE buffer and then was transferred onto Hybond N nytran membranes (Amersham). Probes were generated from the PCR products, b1030-4 for the der(11) chromosomes and b1030-6 for the der(22) chromosomes. Primer sequences for these PCR products were as defined elsewhere (Edelmann et al. 1999_c_). The radiolabeled PCR products were used as probes and were hybridized to the membranes at 65°C overnight in standard buffer conditions. Membranes then were washed under stringent conditions in 0.2 × SSC, 0.1% SSC at 65°C for 15 min, with a total of four washes. Autoradiograms were exposed with MS film (Kodak) overnight.
PCR Amplification of the Breakpoint Junction Fragments
A nested PCR approach was used to amplify specific junction fragment products from the der(11) and der(22) chromosomes. To amplify a 1.6-kb product containing the der(11) breakpoint junction fragment, the Expand Long Template PCR system (Roche) was used. Initially, outer primers b1030-5F (Edelmann et al. 1999_c_) and LCR22-1R (5′-GTCGGGAGAACAAAGACACA-3′) were used, under the following cycling conditions: 94°C for 2 min, followed by30 cycles of 94°C for 30 s, 60°C for 1 min, and 68°C for 2 min, followed by a final extension of 68°C for 7 min. The first-round PCR product was diluted 50-fold and was subjected to a second round of nested PCR. Inner primers b1030-9F (5′-GGGAGAGCATGTAGAGATTG-3′) and AT(26)R1 (Kehrer-Sawatski et al. 1997) were used to generate an 800-bp PCR product, using the following cycling conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 58°C for 1 min, and 68°C for 2 min, followed by a final extension of 68°C for 7 min. The resulting PCR products were isolated from 1% agarose gels (Ultra-Free DA columns, Millipore) and were purified using the Qiagen PCR Purification Kit. To amplify a 1.2-kb product containing the der(22) breakpoint junction fragment, the Expand Long Template PCR system (Roche) was used. Outer primers AT(26)F3 (5′-GGTGTAGTCCCAGTGTGAATT-3′) and b1030-6R (Edelmann et al. 1999_c_) were used, under the following cycling conditions: 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, 58°C for 45 sec, and 68°C for 1.5 min, followed by a final extension of 68°C for 10 min. The first-round PCR product was diluted 50-fold and was subjected to a second round of nested PCR. Inner primers AT(26)F1a.2 (5′-TCCCAGTGTGAGTTGGGATT-3′) and b1030-6Frev (5′-CTGAAAGGGCAGAAGTCTTG-3′) were used to generate an 800-bp PCR product, using Platinum Taq polymerase (Gibco) and the supplied buffer, under the following cycling conditions: 94°C for 10 min, followed by 35 cycles of 94°C for 30 sec, 58°C for 45 sec, and 72°C for 45 sec, followed by a final extension of 72°C for 10 min. The resulting PCR products were isolated and were purified as described above for the der(11) products. The purified PCR products were then subjected to automated sequence analysis on ABI 377 sequencing machines.
Results
PCR Mapping
To delineate the chromosomal breakpoints in the somatic hybrid–cell lines created from the unrelated carriers BM114 and GM06229b, we previously performed PCR mapping studies and found that the breakpoints on 22q11 occurred in LCR22-3a, between markers D22S1623 and D22S264, and on 11q23, between the markers D11S1340 and APOC3 (Edelmann et al. 1999_c_). This finding was confirmed recently (Shaikh et al. 1999; Tapia-Páez et al. 2000). We used a PCR-based approach to systematically divide and narrow the region in the somatic hybrid lines from BM114 and GM 06229b, using the available genomic sequence in the region on 11q23 (GenBank accession number AC007707) (Edelmann et al. 1999_c_). The interval that contained the breakpoint, between the two PCR markers, b1030-5 and b1030-6, was 190 bp and consisted almost entirely of an AT-rich repeat (Edelmann et al. 1999_c_).
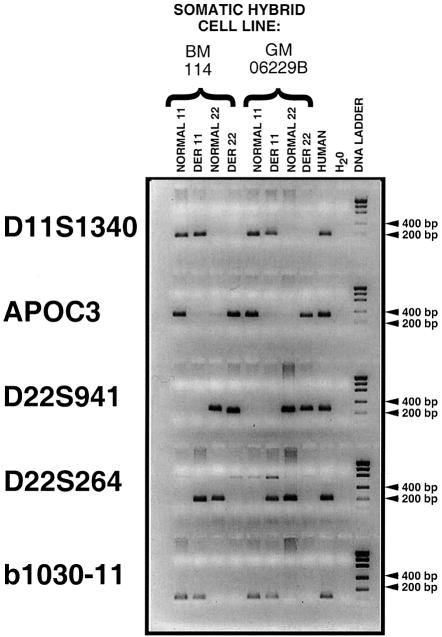
In a recent report by Hill and colleagues (2000), it was proposed that the t(11;22) breakpoint in five carriers occurred in Alu sequences on chromosomes 11 and 22. To demonstrate that the breakpoint was in the AT-rich repeat sequence on 11q23 and not in Alu sequences, PCR mapping studies were performed on the derivative chromosomes and with one of the same carriers, GM06229b, described in the second study (Hill et al. 2000). To demonstrate the integrity of the derivative and normal chromosomes, we performed PCR mapping studies with D11S1340, APOC3, D22S941, and D22S264, as shown in figure 1. As expected, the marker D11S1340 was present on the normal chromosome 11 and der(11) chromosome, APOC3 was present on the normal chromosome 11 and der(22) chromosome, D22S941 was present on the normal chromosome 22 and der(22) chromosome, and D22S264 was present on the normal chromosome 22 and der(11) chromosome. To determine the position of the t(11;22) breakpoint with respect to the AT-rich repeat and the Alu sequence, PCR amplification was performed with primers b1030-11F (5′-CAATCTCTACATGCTCTCCC-3′) and b1030-11R (5′-GACCTTCACAGAATAACAATAA-3′). If the breakpoint in the two carriers occurred in the Alu sequence, the PCR product would be present on the der(22) chromosome, and, if it was telomeric to the Alu sequence, the PCR product would be present on the der(11) chromosome. The presence of the PCR product in the der(11) chromosome in both carriers, including GM06229b, that is shared in both studies, definitively shows that the t(11;22) breakpoint occurred distal to the Alu sequences, in the vicinity of AT-rich repeat. We conclude that the t(11;22) in the two carriers demonstrates that our original observation was correct.
Figure 1.
Definition of the t(11;22) breakpoint interval by PCR analysis. PCR amplification was performed on the separated normal and derivative chromosomes 11 and 22 in somatic hybrid cell lines from two unrelated carriers, BM114 and GM06229b. Five PCR markers—three from chromosome 11q23, D11S1340, APOC3, and b1030-11, and two from chromosome 22q11, D22S941 and D22S264—were amplified from their respective normal chromosomes. A PCR product of the correct size was detected in normal human genomic DNA, and no PCR product was detected using a hamster DNA template (data not shown) or in the absence of any DNA template (H2O). A DNA ladder was used as a size standard.
Genomic Southern Hybridization Studies
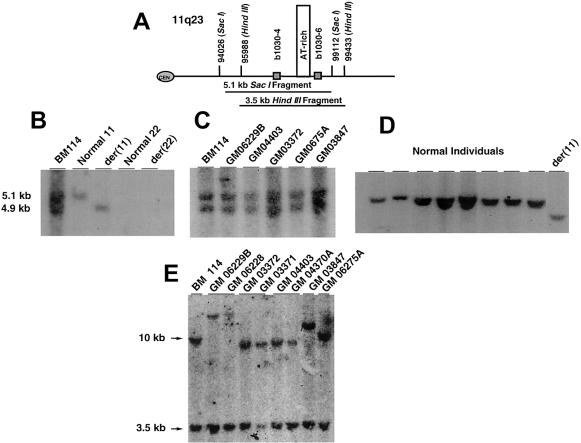
Previous genotyping studies of constitutional t(11;22) carriers demonstrated that the individuals examined are not related (Edelmann et al. 1999_c;_ Funke et al. 1999). To determine whether the chromosomal breakpoints in these unrelated carriers occurred in the same interval on both chromosomes 11 and 22, we performed genomic Southern hybridization studies to identify novel junction fragments in the derivative chromosomes. The probes used in these studies were generated from the genomic sequence on chromosome 11q23 adjacent to either side of the breakpoint intervals defined in figure 1 (Edelmann et al. 1999_c_). For the der(11) chromosome, genomic DNA was digested with _Sac_I and was probed with the b1030-4 PCR product, which should result in a 5.1-kb fragment on the normal chromosome 11, as indicated in the schematic in figure 2A. Initially, DNA from the hamster-human somatic hybrid cell lines, as well as the lymphoblastoid cell line generated from BM114, was probed with the b1030-4 probe. A novel 4.9-kb fragment was detected in the hamster-human somatic hybrid cell line carrying the der(11) chromosome, a 5.1-kb fragment was detected in the hamster-human somatic hybrid cell line carrying the normal 11 chromosome, and both the 4.9- and 5.1-kb bands were detected in the parental lymphoblastoid cell line (fig. 2_B_). In five additional carriers, both the 4.9- and 5.1-kb signals were present (fig. 2_C_); however, in eight additional noncarriers, only the signal from the 5.1-kb band was detected (fig. 2_D_). The finding that all of the junction fragments were detected with same probe and were the same size indicated a precise recombination event and also that the site of chromosome exchange in all six unrelated carriers may be the same.
Figure 2.
Human genomic Southern blot hybridization analysis. A, Partial restriction map of the 11q23 breakpoint interval highlighting the _Sac_I and _Hin_dIII restriction endonuclease sites and the two probes—b1030-4, to detect the der(11) chromosome, and b1030-6, to detect the der(22) chromosome. The AT-rich repeat is boxed. B, Genomic DNA from the BM1 14 lymphoblastoid cell line and resulting hamster-human somatic cell hybrid cell lines were probed with the radiolabeled b1030-4 PCR product. C, DNA was purified from the t(11;22) carriers shown and probed with b1030-4. D, DNA was purified from eight unaffected individuals and the der(11) chromosome from BM114 and probed with b1030-4. E, DNA was isolated from carriers and der(22) offspring and probed with b1030-6.
To identity novel junction fragments from the der(22) chromosomes, a _Hin_dIII digest was used, and genomic DNA was probed with the b1030-6 PCR product. The probe should hybridize to a 3.5-kb fragment in the normal 11 chromosome (fig. 2_A_). In all of the carriers and their der(22) offspring that were examined, signals from the novel junction fragments were detected from unrelated cases, ranging in size from 10 kb to 12 kb; however, carriers and their own der (22) offspring had the same-sized junction fragment (family members, carrier first, followed by der(22) offspring, underlined; fig. 2_E_). On the basis of the results obtained from the der(11) genomic Southern blot, it is evident that the chromosome breakpoints are similar; however, the breakpoints on the der(22) chromosome appear to be different. However, the size variation may represent a _Hin_dIII restriction fragment–length polymorphism between the unrelated individuals. Similar polymorphisms have been detected in the LCR22s (Tapia-Páez et al. 2000). The data presented in the next section rule out the possibility of varying chromosomal breakpoints and supports the likelihood of sequence polymorphisms causing the banding pattern observed on the autoradiogram.
PCR Strategy to Amplify the der(11) and der(22) Junction Fragments
Because the genomic Southern hybridization results indicated that the t(11;22) breakpoints on both 11q23 and 22q11 regions were likely to be similar, it was feasible to use a PCR-based strategy to amplify and examine the sequences at the breakpoint junctions. The region on chromosome 11q23 harboring the t(11;22) breakpoint between markers D11S1340 and APOC3 has been cloned and sequenced and did not contain low-copy repetitive sequences (GenBank accession number AC007707). It therefore would be possible to use a simple PCR strategy to amplify the chromosome breakpoint junctions by generating PCR primers from the 11q23 and 22q11 intervals. Unfortunately, the LCR22-3a disrupted by the constitutional t(11;22) translocation has been unclonable despite numerous attempts (Funke et al. 1999; Shaikh et al. 2000, Tapia-Páez et al. 2000). Because of the absence of sequence from the 22q11 breakpoint region, a simple PCR amplification strategy was ruled out. However, we examined the sequence on 22q11 in the derivative chromosomes from a balanced t(17;22) family affected with NF1 (Kehrer-Sawatski et al. 1997). The translocation breakpoint occurred in AT-rich repeats on chromosomes 17 and 22. All the mapped LCR22s have been cloned and sequenced except for LCR22-3a (Dunham et al. 1999). The 22q11 sequence present on the derivative chromosomes did not precisely match the reported sequence of these LCR22s, suggesting that the sequence might derive from the elusive LCR22-3a.
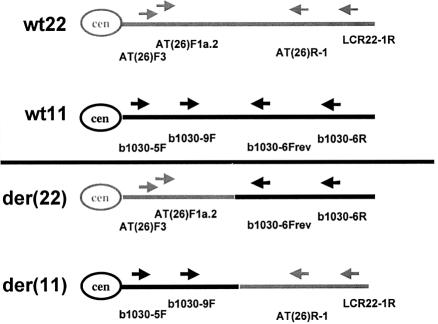
The similarity between the two translocation events led us to postulate a common mechanism for both events and allowed us to devise a PCR strategy to amplify the junction fragments from the t(11;22) carriers. To generate PCR primers to amplify across the derivative junction fragments, from the chromosome 22q11 interval, we designed a nested PCR approach as shown in figure 3, necessitated by the fact that the primary PCR products contained multiple species. The 22q11 PCR primer that we used, AT(26)R-1, was identical to the one used in the study of the t(17;22) translocation (Kehrer-Sawatski et al. 1997). The other three primers on 22q11, AT(26)F-3 and AT(26)F1a, were designed from the sequence on the der(17) and der(22) chromosomes (Kehrer-Sawatski et al. 1997). The primer, LCR22-1R, was designed from LCR22 sequences. The PCR primers b1030-5F, b1030-9F, b1030-6Frev, and b1030-6R were generated from the available sequence of chromosome 11q23 (GenBank accession number AC007707). The resulting 1,600-bp first-round der(11) PCR products and the 800-bp second-round products, as well as the 1,200-bp first-round der(22) PCR products and the 800-bp second-round PCR products from the carriers, were subjected to automated sequence analysis. A PCR product was not detected among unaffected individuals (data not shown).
Figure 3.
PCR strategy for cloning the der(11) and der(22) junction fragments. The positions of the outer and nested PCR primers in the sequence of the normal chromosome 22 (gray) and normal chromosome 11 (black) sequence are shown. The nested PCR strategy used to amplify the derivative chromosomes is depicted within the schematic of the derivative chromosomes. To amplify the der(22) junction fragment, outer primers AT(26)F3 and b1030-6R were used for the first-round PCR. The PCR product was diluted 50-fold and was used in a nested PCR reaction with primers AT(26)F1a.2 from chromosome 22 and b1030-6Frev from chromosome 11. For the (der)11 junction fragment, primers b1030-5F and LCR22-1R, were used for the first-round PCR, and primers b1030-9F and AT(26)R-1, were used for the second-round, or nested, PCR.
der(11) and der(22) Junction Fragment PCR Products
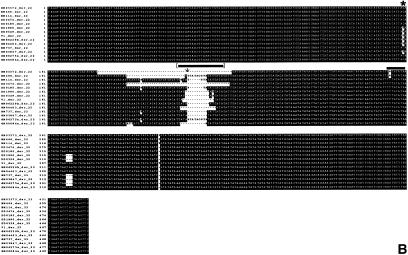
Approximately 500 bp of the der(11) and der(22) PCR products harboring the site of strand exchange from 13 unrelated carriers—including two de novo patients, DD0185 and BM737, as well as a single der(22) patient, GM00084a, for whom we did not have information about the parents—were aligned using the Clustal W program, as shown in figure 4. The sequence was compared across this interval and was found to be 98%–99% identical between samples for both derivative chromosomes. The sequence of the der(22) junction fragment in der(22) patients was identical to their carrier parents, so only the carriers are shown. The carrier GM06275a had the most extensive sequence on both derivative chromosomes in the interval shown in figure 4 and was used as a basis for comparison of the sequences from the other carriers and der(22) patients.
Figure 4.
Alignment of sequence from derivative 11 and 22 chromosomes. The PCR strategy outlined in figure 3 was used to amplify der(11) and der(22) junction fragments, ∼800 bp in size, from 13 unrelated carriers of the (11;22) translocation. Two of the carriers, BM737 and DD0185, represent de novo cases. Sequence from positions 1–533, for the der(11) chromosome (4A), and sequence from positions 1–487, for the der(22) chromosome (4B), in the carrier with the least sequence loss is shown for all individuals and was aligned using the Clustal W program. The der(22) offspring and their carrier patients had identical sequence, in both derivative chromosomes, and the results were not shown, except for GM00084a, because it was an isolated case and the parents were not available. Regions of nucleotide alterations are highlighted (white background) and regions of deletions are denoted by dashes. The bold bar at positions 225–232 in the der(11) chromosome and positions 252–259 in the der(22) chromosome depict the positions of the regions of strand exchange proposed elsewhere (Kurahashi et al. 2000_a_). The bracketed bold bar at positions 373–395 in the der(11) chromosome and positions 220–240 in the der(22) chromosome depict the intervals hypothesized to be the sites of chromosome breakage and strand exchange in the carriers. The hatched bar at positions 385–389 in the der(11) chromosome and the arrow at position 225 in the der(22) chromosome depict the positions of the balanced t(17;22) breakpoint junction (Kehrer-Sawatski et al. 1997). The stars at position 343, in the der(11) chromosome, and position 159, in the der(22) chromosome, represent sequence polymorphisms.
The alignment of the der(11) and der(22) junction PCR products had some mismatches and deletions, perhaps representing sequence polymorphisms. One example of this is an A→G change on the der(11) chromosome (fig. 4_A_), and a second example is a T→C change on the der(22) chromosome (fig. 4_B_). One striking finding was a staggered set of deletions in the interval at positions 369–429 in the der(11) fragment and positions 182–242 on the der(22) fragment in all carriers except GM06275a (fig. 4_A_ and4_B_). The site of the balanced t(17;22) translocation breakpoints on chromosome 22 occurred in the same interval (fig. 4_A_ and4_B_). This was determined by analysis of the available genomic sequence on chromosome 17 (GenBank accession number AC004526) and by deduction of the sequence of the normal chromosome 22 from the remaining sequence. This was necessary because the genomic sequence is not available for LCR22-3a, as previously mentioned.
The presence of deletions on the derivative chromosomes was reminiscent of similar larger deletions that were found on 11q23 by comparing the sequence of BAC 442E11 (AC007707), to the sequences of BACs 87O6, 666H20, and 788E7 (data not shown). The fact that deletions occurred in the BACs was verified further when the sequence was compared with that obtained by direct PCR amplification of human genomic DNA (Kurahashi et al. 2000_a_). Approximately 200 bp of additional sequence was ascertained. The position of the t(11;22) breakpoints on the derivative chromosomes that were reported elsewhere are indicated (bold bar; Kurahashi et al. 2000_a_). We have taken similar approaches and we were able to extend this sequence further by an additional 53 bp. The genomic sequence that has been ascertained is likely to still be incomplete because of difficulties in its cloning, PCR amplification, and sequencing, perhaps because of its AT-rich palindromic nature (Viswanathan et al. 1999). However, the additional genomic sequence enabled us to reexamine the site of chromosome exchange. The breakpoints we suggest are 135 bp distal on the der(11) chromosome and 59 bp proximal on the der(22) chromosome to those previously proposed (Kurahashi et al. 2000_a_).
Sequence on the Deduced Parental Chromosomes 11, 17, and 22 Is Palindromic
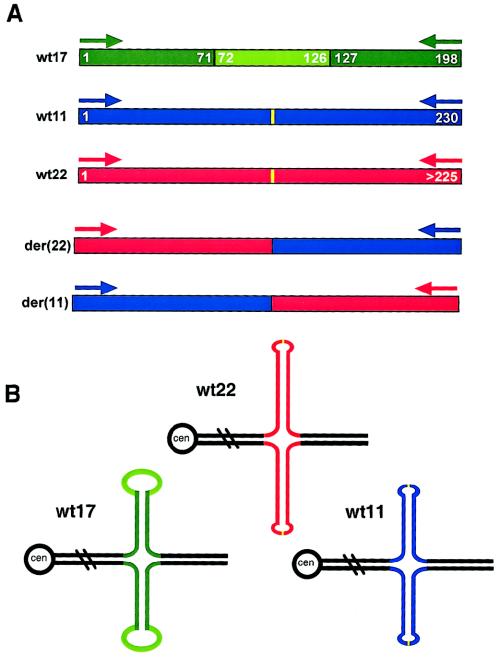
When the sequences of the deduced parental chromosomes 17 and 22 were analyzed, it was found that the AT-rich repeats were palindromic (fig. 5_A_). The t(17;22) breakpoint occurred in an interval on chromosome 11 consisting of a unique sequence that equally divided the two 71-bp blocks of inverted repeats (fig. 5_A_ and 5_B_). On chromosome 22, the site of strand exchange occurred at the center of symmetry of a nearly perfect, >225-bp palindrome (fig. 5_A_ and B), as demonstrated by the fact that the same primers could amplify both derivative chromosomes (Kehrer-Sawatski et al. 1997). This also occurred in the chromosome 11 and 22 intervals implicated in the t(11;22) translocation. A 230-bp palindromic sequence and a >225-bp, AT-rich palindromic sequence were present on the deduced parental chromosomes 11 and 22, respectively (fig. 5A). Although the sequences of both chromosomes 11 and 22 were related in their AT-rich nature and in the fact that they were palindromic, their sequences were not identical. Therefore, intrastrand base-pairing forming putative hairpin structures on the normal chromosomes 11 and 22 could not form in the derivatives (fig. 5_A_ and 5_B_).
Figure 5.
Schematic representation of the chromosome 17, 11, and 22 intervals. A, The relative orientation of palindromic sequence on the three chromosomes in the vicinity of the chromosome breakpoint junction is depicted by the arrows. The wild-type chromosome 17 has a palindromic sequence of 71 bp (dark green) on either side of a unique, nonpalindromic sequence of 55 bp (light green). The wild-type chromosomes 11 and 22 have a palindrome structure of 230 bp (blue) and ⩾225 base pairs (red), respectively. The full sequence of chromosome 22q11 in the vicinity of these breakpoint junctions is not known, indicating that the palindrome may extend well beyond the 225-bp interval shown. The yellow bar denotes the center of symmetry of the palindrome and the position of the deletions in the carriers (bracketed bold bar, fig. 4). The numbers shown here refer to the beginning and end of the palindromic blocks and not the numbers shown infigure 4. The derivative chromosomes represent the recombinant chromosomes, which are no longer palindromic. B, The palindromic sequences in the normal chromosomes 17, 11, and 22 can theoretically form hairpins or cruciform structures as indicated. The hairpin structures on chromosomes 11 and 22 can extend the entire length of the palindrome, and the loop is assumed because of steric hindrance. In contrast, there is a stem-loop structure hypothesized to form on chromosome 17, and the light green interval depicts the region that lacks base pairing.
Examination of the sequence in the deduced parental chromosomes showed a 20-bp region of potential sequence identity in the center of symmetry of the palindrome (bracketed bar,fig. 4). However, this is difficult to define precisely, because of the lack of complete parental genomic sequence of the 11q23 and 22q11 intervals. Interestingly, the center of the 20-bp region of putative identity between chromosomes 11 and 22 is also the interval where the staggered deletions mapped and is precisely where the t(17;22) breakpoint occurred (fig. 5_A_).
Discussion
Cruciform Structure Causes Genomic Instability
The data we present in this report definitively prove that the constitutional t(11;22) translocation in unrelated carriers disrupts AT-rich repeats, as described elsewhere (Edelmann et al. 1999_c_), and not Alu sequences, as was suggested recently (Hill et al. 2000). We hypothesize that the problem with the experiments leading to the incorrect interpretation of the site of chromosome breakage and exchange (junction fragment) could have occurred in the final stages of the study (Hill et al. 2000), when inverse PCR was performed. We suggest an altered PCR product was generated, perhaps by an intermolecular ligation or rearrangement instead of an intramolecular ligation, using the primers described (Hill et al. 2000). The primer sets for the nested PCR used to amplify the der(11) chromosome would have generated a correct junction-fragment PCR product from a correct intramolecular reaction, albeit of a larger size than reported. The primer set for the amplification of the der(22) junction fragment would not have generated a PCR product, as the primers from the chromosome 11 region would be missing. Hence, a PCR product from der(22) patients lacking a der(11) chromosome should not be detected.
We examined the sequence of the AT-rich repeats and found that they are inverted in orientation and are referred to as “palindromic.” The palindromic sequences present in normal chromosomes 11, 17, and 22 have the potential to form intrastrand base-pairing, resulting in hairpins on one strand or cruciform structures, as illustrated in figure 5B. We hypothesize that the center of the palindrome, the tip of hairpin, is susceptible to double-strand breaks, followed by illegitimate recombination events resulting in central asymmetric deletions. This hypothesis is based on previous studies of recombination from bacteria through mammals. In the bacteria Escherichia coli and the yeast Saccharomyces cerevisiae, palindromes are highly unstable, because such sequences can form hairpins that are difficult to replicate, leading to cleavage of the DNA and loss of the hairpin, resulting in stabilization of the locus (Gordenin et al. 1993; Henderson and Petes 1993; Ruskin and Fink 1993; Connelly et al. 1996; Cromie et al. 2000). Further support for the unstable nature of palindromic sequences in bacteria and yeast came from the analysis of the genomic intervals surrounding the translocation on chromosomes 11q23 and 22q11. The genomic interval on 22q11 harboring the t(11;22) translocation has not been cloned, despite numerous attempts (Funke et al. 1999; Kurahashi et al. 2000_a;_ Tapia-Páez et al. 2000). To determine the site of chromosome breakage on 11q23 in the derivative chromosomes, we attempted to generate a consensus genomic sequence by performing PCR amplification using surrounding primers with all available genomic sequences. As mentioned above, all BAC clones examined contained deletions of precisely the interval containing the AT-rich inverted repeat. A putative hairpin or cruciform structure in the normal 11q23 and 22q11 regions perhaps precludes typical molecular-biological analyses.
To ascertain the extent of instability in mammalian cells, studies delineating recombination events that involve palindromic transgenes have been performed in mice. In one study, two different palindromic transgenes, several kilobases in size, were tested for meiotic and mitotic instability (Collick et al. 1996). The long inverted repeats showed significant instability, and examination of the resulting mutations within the palindromic sequences suggested complex mechanisms, including central palindromic deletions (Collick et al. 1996). In a second study, of a 15.3-kb palindromic transgene, the most common rearrangement involved asymmetric deletions of 100–200 bp, near the central point of symmetry (Akgün et al. 1997). It was hypothesized that the small disruption of symmetry in the palindrome could subsequently stabilize the locus (Akgün et al. 1997). In the study, the majority of the rearrangements involved illegitimate or nonhomologous recombination events, as detected by Southern blot hybridization (Akgün et al. 1997). Akgün and colleagues (1997) hypothesized two different models to explain their results. In one model, replication slippage could occur, generating two-sided palindrome deletions spanning the central axis of symmetry or tip of the hairpin. This is analogous to the mechanism described for bacteria and yeast. In the second model, a single-stranded nick in the tip of the hairpin would result in a double strand break, and this might serve as an initiating event for illegitimate recombination. The evidence presented here, regarding the mechanism for the constitutional t(11;22) translocation and the observed products of recombination, is consistent with the second model.
Molecular Basis of Constitutional Translocations
De novo structural chromosome rearrangements are quite common, occurring in .67/1,000 amniocentesis specimens, and, of these, 1/2,000 is due to reciprocal translocations (Warburton 1991). The frequency of translocations in couples with recurrent spontaneous abortions is 20-fold higher than in individuals without recurrent spontaneous abortions, perhaps because of meiotic nondisjunction events leading to aneuploidy (Michels et al. 1982; Campana et al. 1986). To understand the molecular basis of de novo constitutional translocations, it is necessary to analyze the sequences of the sites of chromosome exchange and compare them with each other. With respect to the chromosome 22q11 region, the breakpoint junctions for a few distinct de novo balanced translocation cases have been cloned and sequenced, but no common mechanism has been identified.
One of these is the balanced t(2;22) translocation in a patient with VCFS/DGS (ADU) (Budarf et al. 1995; Levy et al. 1995; Sutherland et al. 1996). The translocation on 22q11 occurred within LCR22 sequences ∼250 kb distal to the common proximal breakpoint in patients with VCFS/DGS (Edelmann et al. 1999_a_). Examination of the sequences at the site of chromosome exchange and comparison with normal chromosomes 2 and 22 did not reveal extensive sequence homology or AT-rich repetitive sequences. Since only small (6-bp) stretches of sequence homology were present in the normal chromosomes, and, since 11 bp of sequences were deleted (Budarf et al. 1995), it is unlikely that the translocation occurred as a result of homologous recombination mechanisms. A similar situation was observed for a balanced t(11;22) translocation in infant twins with myeloid leukemia occurring as a result of the fusion of the MLL gene and the PNUTL (CDCREL-1) gene in the 22q11 region (Megonigal et al. 1998). Short homologous sequences (2–6 bp) mapped to the vicinity of the breakpoint junctions in the normal genomic intervals, but no extensive homologies or AT-repetitive sequences were present.
In the case of a constitutional pericentric inversion of chromosome 8, the site of chromosome breakage occurred in a small, nonrepetitive interval flanked by an interval containing highly repetitive elements such as Alu, LINE, and long terminal repeat (LTR) sequences (Graw et al. 2000). It was hypothesized that the repetitive elements might have contributed to chromosome instability (Graw et al. 2000). These three examples of balanced translocations are representative of a handful of other balanced translocations studied at high resolution (Krebs et al. 1997; Kurahashi et al. 1998; Millar et al. 2000).
As mentioned above, the results ascertained from these studies are not consistent with a single mechanism leading to the translocations. Precise definition of the t(17;22) and t(11;22) translocations described in this report may provide clues to only a subset of possible mechanisms leading to chromosome rearrangements. In those patients harboring interstitial deletions or duplications in the genome, there is evidence that more-straightforward, homologous recombination mechanisms between low- or high-copy repetitive elements occurred (reviewed in Lupski 1998). In some genomic disorders, homologous recombination events have been mediated by high-copy repetitive sequences, such as Alu or LINE elements (Calabretta et al. 1982). In other cases involving deletions or duplications >1 Mb in size, homologous recombination events between low-copy repetitive sequences, such as gene clusters, have been implicated, as in VCFS/DGS.
Mechanism of VCFS/DGS Deletions
More than 90% of patients with VCFS/DGS with hemizygous deletions have the same 3-Mb deletion (Carlson et al. 1997). Both inter- and intrachromosomal homologous recombination events between two LCR22s mediate the deletion (Baumer et al. 1998; Edelmann et al. 1999_a_). We have constructed high-resolution cosmid/PAC/BAC contig–based physical maps of the two LCR22s that harbor the common chromosomal breakpoints in patients with VCFS/DGS (Edelmann et al. 1999_a_), and they have been sequenced (Dunham et al. 1999). Within the two LCR22s, AT-rich repetitive sequences are interspersed. To determine whether inverted AT-rich repeats, such as those present in the derivative chromosomes, are present in these two 3-Mb LCR22s, we examined all of them and found that, although AT-rich sequences are present, none of the them comprise the inverted repeats that are present in LCR22-3a. On the basis of the lack of extensive homology between chromosomes 11 and 22, and of the deletion of sequences at the site of strand exchange, it is unlikely that homologous recombination mechanisms mediate the t(11;22) translocation. It is equally unlikely that AT-rich inverted repeats mediate the 3-Mb deletion in patients with VCFS/DGS, because of their absence from the implicated LCR22s. It should be noted, however, that resequencing the LCR22s in different individuals might reveal palindromic sequences, since the genomic sequence of chromosome 22 derives from only a few different individuals. It is also possible that the inverted sequences might have been deleted in the BAC clones, as has occurred in the 11q23 interval.
In summary, we have demonstrated that the recurrent constitutional t(11;22) translocation occurs in the same AT-rich palindromic intervals among unrelated carriers. We hypothesize that intrastrand base-pairing leads to hairpins or cruciform structures that are susceptible to double-strand breaks resulting in illegitimate recombination events. The fact that similar cruciform structures may form on chromosome 17 as well, in the region involved in the balanced t(17;22) translocation (Kehrer-Sawatski et al. 1997), is consistent with the possibility that this interval may be prone to breakage and may act as a recombination hotspot.
Acknowledgments
We would like to acknowledge the participating families for their donation of blood samples for the study and clinicians for their ascertainment. B.E.M. is supported by National Institutes of Health (NIH) grant PO-1 HD34980-02, American Heart Association and March of Dimes grant FY98-0414. L.E. is supported by NIH grant T32 CA09060. E.S. is supported by NIH training grant T32 GM07491-25.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for available genomic sequence on chromosomes 11q23 [AC007707] and 17 [AC004526])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for VCFS/DGS [MIM 192430/MIM 188400] and CES [MIM 115470])
References
- Akgün E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M (1997) Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17:5559–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA (1998) High level of unequal meiotic crossovers at the origin of the 22q11. 2 and 7q11.23 deletions. Hum Mol Genet 7:887–894 [DOI] [PubMed] [Google Scholar]
- Budarf ML, Collins J, Gong W, Roe B, Wang Z, Bailey LC, Sellinger B, Michaud D, Driscoll DA, Emanuel BS (1995) Cloning a balanced translocation associated with DiGeorge syndrome and identification of a disrupted candidate gene. Nat Genet 10:269–278 [DOI] [PubMed] [Google Scholar]
- Calabretta B, Robberson DL, Barrera-Saldana HA, Lambrou TP, Saunders GF (1982) Genome instability in a region of human DNA enriched in Alu repeat sequences. Nature 296:219–225 [DOI] [PubMed] [Google Scholar]
- Campana M, Serra A, Neri G (1986) Role of chromosome aberrations in recurrent abortion: a study of 269 balanced translocations. Am J Med Genet 24:341–356 [DOI] [PubMed] [Google Scholar]
- Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, Scambler P, Shprintzen R, Kucherlapati R, Morrow BE (1997) Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet 61:620–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W (1996) Instability of long inverted repeats within mouse transgenes. EMBO J 15:1163–1171 [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Leach DR (1996) The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells 1:285–291 [DOI] [PubMed] [Google Scholar]
- Cromie GA, Millar CB, Schmidt KH, Leach DR (2000) Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics 154:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGeorge A (1965) A new concept of the cellular basis of immunity. J Pediatr 67:907 [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE (1999_a_) Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 64:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999_b_) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, McCain N, Goldberg R, Pandita RK, Duong S, Fox J, Blumenthal D, Lalani SR, Shaffer LG, Morrow BE (1999_c_) A common breakpoint on 11q23 in carriers of the constitutional t(11;22) translocation. Am J Hum Genet 65:1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraccaro M, Lindsten J, Ford CE, Iselius L (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet 56:21–51 [DOI] [PubMed] [Google Scholar]
- Funke B, Edelmann L, McCain N, Pandita R, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, and Morrow BE (1999) Der(22) syndrome and velo-cardio-facial syndrome/DiGeorge syndrome share a 1.5 Mb region of overlap on chromosome 22q11. Am J Hum Genet 64:747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin DA, Lovachey KS, Degtyareya NP, Malkova AL, Perkins E, Resnick MA (1993) Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol 13:5315–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw SL, Sample T, Bleskan J, Sujansky E, Patterson D (2000) Cloning, sequencing, and analysis of inv8 chromosome breakpoints associated with recombinant 8 syndrome. Am J Hum Genet 66:1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guanti G (1981) The aetiology of the cat eye syndrome reconsidered. J Med Genet 18:108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Petes TD (1993) Instability of a plasmid-borne inverted repeat in Saccharomyces cerevisiae. Genetics 134:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AS, Foot NJ, Chaplin TL, Young BD (2000) The most frequent constitutional translocation in humans, the t(11;22)(q23;q11) is due to a highly specific Alu-mediated recombination. Hum Mol Genet 9:1525–1532 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G (1997) The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet 99:237–247 [DOI] [PubMed] [Google Scholar]
- Krebs I, Weis I, Hudler M, Rommens JM, Roth H, Scherer SW, Tsui LC, Fuchtbauer EM, Grzeschik KH, Tsuji, K, Kunz J (1997) Translocation breakpoint maps 5 kb 3′ from TWIST in a patient affected with Saethre-Chotzen syndrome. Hum Mol Genet 6:1079–1086 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Sakamoto M, Ono J, Honda A, Okada S, Nakamura Y (1998) Molecular cloning of the chromosomal breakpoint in the LIS1 gene of a patient with isolated lissencephaly and balanced t(8;17). Hum Genet 103:189–192 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML (2000_a_) Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22). Hum Mol Genet 9:1665–1670 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS (2000_b_) Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22). Am J Hum Genet 67:763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Demczuk S, Aurias A, Depetris D, Mattei MG, Philip N (1995) Interstitial 22q11 microdeletion excluding the ADU breakpoint in a patient with DiGeorge syndrome. Hum Mol Genet 4:2417–2419 [DOI] [PubMed] [Google Scholar]
- Lin AE, Bernar J, Chin AJ, Sparkes RS, Emanuel BS, Zackai EH (1986) Congenital heart disease in supernumerary der(22),t(11;22) syndrome. Clin Genet 29:269–275 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Goldberg R, Jurecic V, Morrow B, Carlson C, Kucherlapati RS, Shprintzen RJ, Baldini A (1995) Velo-cardio-facial syndrome: frequency and extent of 22q11 deletions. Am J Med Genet 57:514–522 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed] [Google Scholar]
- McTaggart KE, Budarf ML, Driscoll DA, Emanuel BS, Ferreira P, McDermid HE (1998) Cat eye syndrome chromosome breakpoint clustering: identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet Cell Genet 81:222–228 [DOI] [PubMed] [Google Scholar]
- Megonigal MD, Rappaport EF, Jones DH, Williams TM, Lovett BD, Kelly KM, Lerou PH, Moulton t, Budarf ML, Felix, CA (1998) t(11;22)(q23;q11.2) in acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci USA 95:6413–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels VV, Medrano C, Venne VL, Riccardi VM (1982) Chromosome translocations in couples with multiple spontaneous abortions. Am J Hum Genet 34:507–513 [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Gen 9:1415–1423 [DOI] [PubMed] [Google Scholar]
- Morrow B, Goldberg R, Carlson C, Das Gupta R, Sirotkin H, Collins J, Dunham I, O'Donnell H, Scambler P, Shprintzen R, Kucherlapati R (1995) Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am J Hum Genet 56:1391–1403 [PMC free article] [PubMed] [Google Scholar]
- Reiss JA, Weleber RG, Brown MG, Bangs CD, Lovrien EW, Magenis RE (1985) Tandem duplication of proximal 22q: a cause of cat-eye syndrome. Am J Med Genet 20:165–171 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Fink GR (1993) Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics 134:43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel A, Schmid W, Auf der Maur P, Moser H, Degenhardt KH, Geisler M, Grubisic A (1981) Incomplete trisomy 22. I. Familial 11/22 translocation with 3:1 meiotic disjunction: delineation of a common clinical picture and report of nine new cases from six families. Hum Genet 56:249–262 [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS (1999) Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet 65:1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS (2000) Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet 9:489–501 [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, Argamaso RV, Young D (1978) A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J 15:56–62 [PubMed] [Google Scholar]
- Sutherland HF, Wadey R, McKie JM, Taylor C, Atif U, Johnstone KA, Halford S, Kim UJ, Goodship J, Baldini A, Scambler PJ (1996) Identification of a novel transcript disrupted by a balanced translocation associated with DiGeorge syndrome. Am J Hum Genet 59:23–31 [PMC free article] [PubMed] [Google Scholar]
- Tapia-Páez I, O’Brien KP, Kost-Alimova M, Sahlen S, Kedra D, Bruder CE, Andersson B, Row BA, Hu P, Imreh S, Blennow E, Dumanski JP (2000) Fine mapping of the constitutional translocation t(11;22)(q23;q11). Hum Genet 106:506–516 [DOI] [PubMed] [Google Scholar]
- Viswanathan VK, Krcmarik K, Cianciotto NP (1999) Template secondary structure promotes polymerase jumping during PCR amplification. Biotechniques 27:508–511 [DOI] [PubMed] [Google Scholar]
- Warburton D (1991) De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 49:995–1013 [PMC free article] [PubMed] [Google Scholar]
- Zackai EH, Emanuel BS (1980) Site-specific reciprocal translocation t(11;22)(q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet 7:507–521 [DOI] [PubMed] [Google Scholar]