New Components of a System for Phosphate Accumulation and Polyphosphate Metabolism in Saccharomyces cerevisiae Revealed by Genomic Expression Analysis (original) (raw)
Abstract
The PHO regulatory pathway is involved in the acquisition of phosphate (Pi) in the yeast Saccharomyces cerevisiae. When extracellular Pi concentrations are low, several genes are transcriptionally induced by this pathway, which includes the Pho4 transcriptional activator, the Pho80-Pho85 cyclin-CDK pair, and the Pho81 CDK inhibitor. In an attempt to identify all the components regulated by this system, a whole-genome DNA microarray analysis was employed, and 22 PHO-regulated genes were identified. The promoter regions of 21 of these genes contained at least one copy of a sequence that matched the Pho4 recognition site. Eight of these genes, PHM1–PHM8, had no previously defined function in phosphate metabolism. The amino acid sequences of_PHM1 (YFL004w), PHM2 (YPL019c),PHM3 (YJL012c), and PHM4 (YER072w) are 32–56% identical. The phm3 and phm4 single mutants and the phm1 phm2 double mutant were each severely deficient in accumulation of inorganic polyphosphate (polyP) and Pi. The phenotype of the_phm5 mutant suggests that PHM5 (YDR452w) is essential for normal catabolism of polyP in the yeast vacuole. Taken together, the results reveal important new features of a genetic system that plays a critical role in Pi acquisition and polyP metabolism in yeast.
INTRODUCTION
Phosphate (Pi) is an essential nutrient for all organisms, used in the biosynthesis of diverse cellular components, including nucleic acids, proteins, lipids, and sugars. It is therefore essential for organisms to have evolved regulatory mechanisms for acquisition, storage, and release of this molecule (Torriani-Gorini_et al._, 1994).
In Saccharomyces cerevisiae, the PHO regulatory pathway regulates expression of the “PHO” genes, involved in the scavenging and specific uptake of Pi from extracellular sources (Johnston and Carlson, 1992; Oshima, 1997). The PHO regulatory system consists of at least five PHO-specific regulatory proteins, the Pho2 and Pho4 transcriptional activators, the Pho80-Pho85 cyclin-cyclin dependent protein kinase (CDK) complex, and the Pho81 CDK inhibitor (Figure 1). Pho84 is a high-affinity Pi transporter localized on plasma membrane, which has been shown to contribute to Pi uptake from culture medium (Bun-ya et al., 1991). PHO84 gene expression is activated by a Pi-starvation signal mediated by the PHO regulatory system. Additionally, the_PHO5_ gene encodes a repressible acid phosphatase which is localized to the periplasmic space.
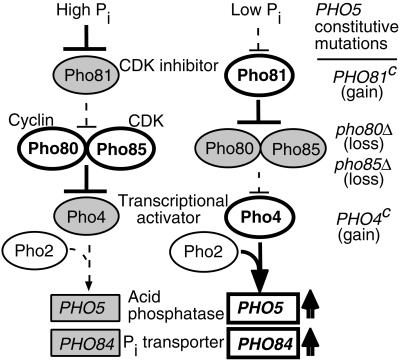
Figure 1.
PHO regulation system. A simplified schematic of the PHO regulation mechanism showing the five main regulator proteins is illustrated (Johnston and Carlson, 1992; Oshima, 1997). Ovals and boxes represent proteins and genes, respectively. Thick lines mean that the signals are transduced to the downstream component, while dotted lines indicate the absent of an interaction with the downstream component. Open ovals and boxes indicate active states, gray oval and boxes indicate inactive state. The _PHO5_-constitutive mutants used in this study are listed to the right of the corresponding mutated protein with the nature of the mutation indicated by “gain” or “loss” of function.
When the Pi concentration in the medium is low (∼ 0.2 mM Pi), the Pho81 protein inhibits the Pho80-Pho85 kinase activity, which in its active state catalyzes a hyperphosphorylation of Pho4 (Schneider et al., 1994; Ogawa_et al._, 1995). The hypophosphorylated form of Pho4 is preferentially localized to the nucleus, where together with Pho2, it activates target gene transcription (Kaffman et al., 1998;Komeili and O'Shea, 1999). Alternatively, when the Pi concentrations are high (∼ 10 mM Pi), the Pho80-Pho85 kinase phosphorylates Pho4. In addition to having a lower affinity for Pho2 and the nuclear import protein Pse1/Kap121, phosphorylated Pho4 is a preferred substrate of the nuclear export protein Msn5, resulting in extranuclear localization. Phosphorylated Pho4 is thus unable to activate target gene expression.
Besides PHO5 and PHO84, seven additional genes are known to be regulated by the PHO regulatory system; these include_PHO11_and PHO12 (homologs of PHO5),PHO8 (vacuolar alkaline phosphatase, Kaneko et al., 1987), PHO89 (Na/Pi cotransporter, Martinez and Persson, 1998), PHO86 (required for Pi uptake, Yompakdee et al., 1996), PHO81 and SPL2 (a homolog of_PHO81_, Flick and Thorner, 1998). The promoters of all nine previously recognized PHO-regulated genes have common motifs, CACGTG and/or CACGTT, as core sequences comprising the Pho4 binding site (Oshima, 1997). Both the regulating properties and the functions of the target genes point to the critical role played by the PHO regulatory system in Pi acquisition in yeast. Comprehensive identification and characterization of the PHO-regulated genes in the yeast genome is therefore likely to be an important step toward understanding the regulation and physiology of Pi metabolism.
DNA microarrays provide a systematic way to study the expression programs of the entire genome (DeRisi et al., 1997). Using DNA microarrays, we conducted an exhaustive search for yeast genes regulated by the PHO regulatory system. Several of the genes identified were of unknown function and were further characterized by gene disruption. Biochemical analysis of five of the novel PHO-regulated genes revealed them to be important in inorganic polyphosphate (polyP) metabolism.
PolyP, a linear polymer of up to hundreds of Pi residues linked by high-energy phosphoanhydride bonds, is ubiquitous in nature, having been found in all organisms examined (Kornberg, 1999).S. cerevisiae is known to accumulate large amounts of polyP in vacuoles, comprising 37% of the total cellular phosphate (Urech, 1978). The enzyme primarily responsible for polyP synthesis in_Escherichia coli_ is polyP kinase (PPK), and polyP is hydrolyzed to Pi by exopolyphosphatase (Kornberg, 1999). In S. cerevisiae, an exopolyphosphatase gene, PPX1, has been identified (Wurst et al., 1995), but a gene for PPK has not. Moreover, no gene homologous to the bacterial PPKs has been found in the genome databases of S. cerevisiae. Thus, the identify of the enzymes that mediate polyP metabolism in yeast and the other eukaryotes has been an important unsolved problem in metabolism of a centrally important nutrient.
MATERIALS AND METHODS
Strains and Media
S. cerevisiae strains used in this study are listed in Table 1. YPAD medium (Adams et al., 1997) was used as rich media. A Pi-depleted YPAD medium (YPAD-Pi) was prepared as described (Kaneko et al., 1982), and a high-Pi YPAD (YPAD+Pi) medium was prepared by addition of sodium phosphate (10 mM, pH 5.8) to the YPAD-Pi medium. High-Pi, low-Pi, and Pi-free synthetic media were prepared as described (Yoshida et al., 1989). YPAD medium buffered at pH 7.5 supplemented with 50 mM CaCl2 was prepared as described (Zhang et al., 1998).
Table 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| NBW7 | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 | Ogawa and Oshima, (1990) |
| NBD4-1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 pho4Δ∷HIS3 | Ogawa and Oshima, (1990) |
| NBD82-1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 PHO4c-1 | Ogawa and Oshima, (1990) |
| NBD80-1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 pho80Δ∷HIS3 | Bun-ya et al., (1991) |
| NBD85A-1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 pho85Δ∷ADE2 | Our stock |
| NOF1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 PHO81c-1 | Our stock |
| NBD8184-1Ba | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 pho84Δ∷HIS3 | Our stock |
| NBM4Lf1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm1Δ∷CgLEU2 | This study |
| NBM19Hf1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm2Δ∷CgHIS3 | This study |
| NBM4L19H2a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm1Δ∷CgLEU2 phm2Δ∷CgHIS3 | This study |
| NBM12W2a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm3Δ∷CgTRP1 | This study |
| NBM72W7a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm4Δ∷CgTRP1 | This study |
| NBM452H1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm5Δ∷CgHIS3 | This study |
| NBM281H1a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 phm6Δ∷CgHIS3 | This study |
| NBM18H7a | _MAT_a ade2 his3 leu2 trp1 ura3 pho3-1 ctf19Δ∷CgHIS3 | This study |
| DBY7286 | _MAT_a ura3-52 | Our stock |
| CRY | _MAT_a ade2 his3 leu2 trp1 ura3 | Wurst et al., (1995) |
| CRXb | _MAT_a ade2 his3 leu2 trp1 ura3 ppx1Δ∷LEU2 | Wurst et al., (1995) |
| CB023b | _MAT_a ade2 his3 leu2 trp1 ura3 pep4Δ∷HIS3 prb1Δ∷hisG prc1Δ∷hisG | Wurst_et al._, (1995) |
| CRY-V4b | _MAT_a ade2 his3 leu2 trp1 ura3 vma4Δ∷URA3 | Wurst et al., (1995) |
| NB91-6B | _MAT_a his3 leu2 trp1 pep4 (=pho9-1) | Our stock |
Growth Conditions for RNA Preparation
For comparison of the gene expression pattern between the low- and high-Pi conditions, yeast strain NBW7 or DBY7286 was grown in 25 ml of the YPAD+Pi medium overnight, the cells were collected, washed with the YPAD-Pi medium two times, and inoculated into 500 ml of the YPAD+Pi and -Pi media to an OD600 of 0.05. The two cultures were shaken at 30°C to an OD600 of 0.5, the grown cells were harvested, and then frozen by immersion in liquid nitrogen. The cells were stored in − 80°C until RNA was prepared. For comparison of the gene expression pattern between the mutants and wild-type, yeast strains NBD82–1 (PHO4_c_-1), NBD80–1 (_pho80_Δ), NBD85A-1 (_pho85_Δ), NOF1 (PHO81_c_-1), and NBW7 (wild-type) were precultivated in 25 ml of YPAD overnight, the grown cells were collected washed with YPAD, and inoculated into 500 ml of YPAD to an OD600 of 0.1. The remainder of the procedure was the same as above.
DNA Microarray Analysis
The preparation of the yeast ORF DNA microarray, RNA preparation from yeast cells, probe preparation with fluorescence dye, hybridization, scanning of the hybridized array, and data processing were performed as described previously (Chu et al., 1998;Spellman et al., 1998). To compare gene expression patterns between the low- and high-Pi conditions, Cy3- or Cy5-labeled cDNA probes were prepared from the high- or low-Pi samples, respectively, by reverse transcription in the presence of Cy3- or Cy5-dUTP (Amersham Pharmacia Biotech., Piscataway, NJ), as previously described (DeRisi et al., 1997). To compare the gene expression patterns between mutant and wild-type cells, cDNA probes templated by mRNAs prepared from wild-type and mutant cells were labeled with Cy3-dUTP and Cy5-dUTP, respectively. The original array data are available on our web page on <http://cmgm.stanford.edu/pbrown/phosphate/>.
Construction of the S. cerevisiae Disruption Mutants
To make disruption mutants of PHM1, PHM2,PHM3, PHM4, PHM5, PHM6, and_CTF19_, PCR-mediated gene disruptions were performed as described (Sakumoto et al., 1999). pCgHIS3, pCgTRP1, pCgLEU2 harboring the Candida glabrata HIS3, TRP1, and_LEU2_ fragments on pUC19, respectively, were obtained from Dr. Y. Mukai (Osaka University), and used as templates to generate the PCR fragments for the gene disruptions. Deletion regions in the target genes were as follows: PHM1, +1 to +2387; PHM2, +1 to +2508; PHM3, +53 to +926; PHM4, +44 to +347; PHM5, +1 to +2025; CTF19, +1 to +1130;PHM6, +1 to +315 (nucleotide positions relative to A of initiation codon of ATG in each ORF). In each case, the deleted sequence was replaced with the marker genes indicated in Table 1. The disruption junctions were verified by colony-PCR (Adams et al., 1997).
Polyphosphate Overplus
Polyphosphate overplus (Harold, 1966) culture was performed as follows: The yeast strains indicated were grown in the YPAD-Pi media overnight, the grown cells were collected, washed with water, and resuspended in 10 ml of the synthetic Pi-free media to an OD600 of 0.5. The cultures were shaken for 2 h. at 30°C, then potassium phosphate (pH 5.8) was added (10 mM final concentration). After 2 more hours of cultivation, the cells were harvested, and then frozen by immersion in liquid nitrogen. For the polyP overplus at low pH, yeast strains were grown in the YPAD-Pi media overnight, the grown cells were collected, washed with water, and resuspended in 10 ml of the YPAD media supplemented with 10 mM (final) KH2PO4 and/or 10 mM (final) sodium acetate buffer (pH 4.0) to an OD600 of 0.5. The cultures were shaken for 2 h at 30°C, then the cells were harvested, and frozen by immersion in liquid nitrogen.
Cell Extract Preparation From Yeast
Cells in 150 μl of extraction buffer (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 1 mM EDTA) were mixed in a vortex mixer with 150 mg of acid-washed glass beads (0.5-mm diameter) for 2 min at 4°C, and microcentrifuged at 14,000 rpm for 10 min at 4°C. The aqueous phase was extracted by phenol:chloroform followed by chloroform and ether extractions. After removing ether in the samples by evaporation, their A260 values were measured to calculate their total RNA concentrations.
PolyP Detection by PAGE
PolyP analysis by PAGE was performed as described in Wurst_et al._ (1995) with the following modifications. Samples containing the indicated amounts of RNA were loaded with 7 μl of loading dye solution (1x TBE buffer [Sambrook et al., 1989], 10% sucrose, 0.05% bromophenol blue) on a 20% polyacrylamide gel (270-mm height x 165-mm wide x 0.5-mm thick) with 1x TBE buffer. Ten μg of sodium phosphate glass type 5, 15, or 35 (Sigma, St. Louis, MO) were loaded as a polyP size markers. Radioactive [γ-32P]ATP was also used as an additional size marker. The electrophoresis was run at 20 V/cm for ∼ 1.5 or 3 h, and the ATP-loaded gel piece was analyzed by a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). The nonradioactive piece of gel was soaked into 10% acetate, 10% methanol for 15 min, stained with the staining solution (0.5% Toluidine Blue O [Sigma], 25% methanol, 5% glycerol, 5% acetate) for 15 min, and destained with destaining solution (same as the staining without Toludine Blue O) for 10 min several times. ATP migrates at a position corresponding to polyP of between seven and eight Pi residues long. PolyP bands of sodium phosphate glass type P35 with 55 and 65 chain length on PAGE were extracted and used as markers to estimate longer chain lengths of polyP.
Enzymatic polyP Assays
PolyP was assayed by the nonradioactive method as described (Ault-Riché_et al._, 1998), without the Glassmilk purification steps. Since the major population of yeast polyP is < 60 Pi residues in length (Figure 4B), they are not able to be effectively trapped by Glassmilk. Concentrations of polyP in the samples were shown as mol of Pi residues per mg of total RNA.
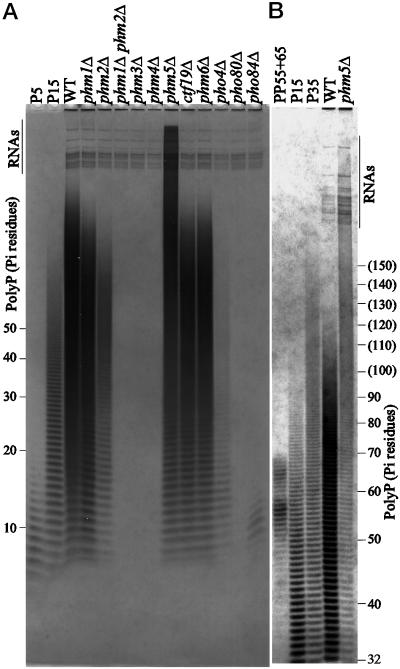
Figure 4.
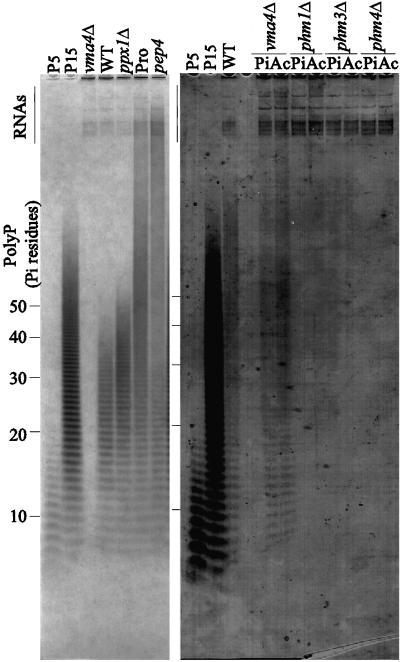
PolyP analysis by PAGE for various_phm_ mutants. Each polyP sample, containing 10-μg RNA, prepared from indicated strains after the polyP overplus culture, was loaded on 20% polyacrylamide gel, and subjected to electrophoresis at 20 V/cm for 1.5 h (panel A) or 3 h (panel B). The strains used were NBW7 (WT), NBM4Lf1 (_phm1_Δ), NBM19Hf1 (_phm2_Δ), NBM4L19H2 (_phm1_Δ_phm2_Δ), NBM12W2 (_phm3_Δ), NBM72W7 (_phm4_Δ), NBM452H1 (_phm5_Δ), NBM18H7 (_ctf19_Δ), NBM281H1 (_phm6_Δ), NBD4–1 (_pho4_Δ), NBD80–1 (_pho80_Δ), and NBD8184–1 (_pho84_Δ). Several RNA bands are visible in a region near the top of the both gels, indicated by “RNAs”. Marker samples containing 10 μg of sodium phosphate glass type 5, 15, and 35 were loaded on the P5, P15, and P35 lanes, respectively. Extracted polyP samples corresponding to 55 and 65 Pi residues were also loaded as size markers (PP55 +65). Chain lengths of the polyP ladders determined by the markers (Materials and Methods) are indicated to the left and right of panel A and B, respectively. The migration positions indicated with the chain length in the parenthesis on panel B are estimated from a standard curve.
PHM2-GFP Fusion Experiment
pPHM2-GFP was constructed by insertion of the PCR fragment corresponding to nucleotide positions from − 393 to + 2505 of_PHM2_ (relative to A of initiation codon of ATG) into an_Eco_RI-_Hin_dIII gap of pTS395, a_GFP_-expressing yeast vector provided by Dr. D. Botstein. Yeast transformant NBM-4L19H1[pPHM2-GFP] was grown in synthetic low-Pi medium lacking uracil for > 3 h at 30°C. Fluorescent dye, FM4–64 (Molecular Probes, Eugene, OR) , which has been reported to stain vacuolar membrane (Vida and Emr, 1995), was added to 30 μM, and incubated for an additional 15 min. Ten-microliter samples were examined by fluorescent microscope Axioplan2 (Carl Zeiss, Thomwood, NY) following the procedure of Vida and Emr (1995) and Adams et al. (1997).
Pi Uptake Assay
A Pi uptake assay was performed following the procedure of Bun-ya et al. (1991). Yeast strains were grown in the YPAD-Pi media overnight, the grown cells were collected, washed with water, and resuspended in 50 ml of synthetic Pi-free media to an OD660 of 0.1. The cultures were shaken for 2 h at 30°C, and their OD660 values were remeasured. Potassium phosphate (0.1 mM, pH 5.8) and radioactive phosphate (32PO4; 1 μCi/ml) were added to cultures, and shaken. Five-milliliter samples were taken at indicated intervals and immediately filtered though a HA filter (3.5-mm diameter, Millipore). The cells trapped on the filter were washed with 10 ml of synthetic Pi-free media. The radioactivity trapped on the membrane filters was quantitated by a liquid scintillation counting.
RESULTS
Whole Genome Survey for the PHO-Regulated Genes
In order to identify all of the genes regulated by the PHO regulatory pathway, we used DNA microarrays fabricated as described previously (DeRisi et al., 1997, Chu et al., 1998; DeRisi et al., 1997, Chu et al., 1998;Spellman et al., 1998). A cDNA probe prepared from poly(A)+ RNA isolated from a wild-type yeast strain, NBW7, cultivated in low-Pi media (YPAD-Pi), was labeled with Cy5 fluorescent dye, while a cDNA probe prepared from the same strain cultivated in high-Pi media (YPAD+Pi) was labeled with Cy3 fluorescent dye. The Cy5- and Cy3-labeled cDNA probes were mixed and hybridized to the microarray. As expected, the transcript levels of both PHO5 and PHO84 were elevated in low-Pi media in two independent experiments (Figure 2A). In a repeat experiment using strain DBY7286, derived from S288C, (the standard S. cerevisiae strain for the genome database), most of the highly derepressed genes we found in NBW7 were consistent with those identified in DBY7286 (Figure 2) suggesting similar PHO regulation in the two strains.
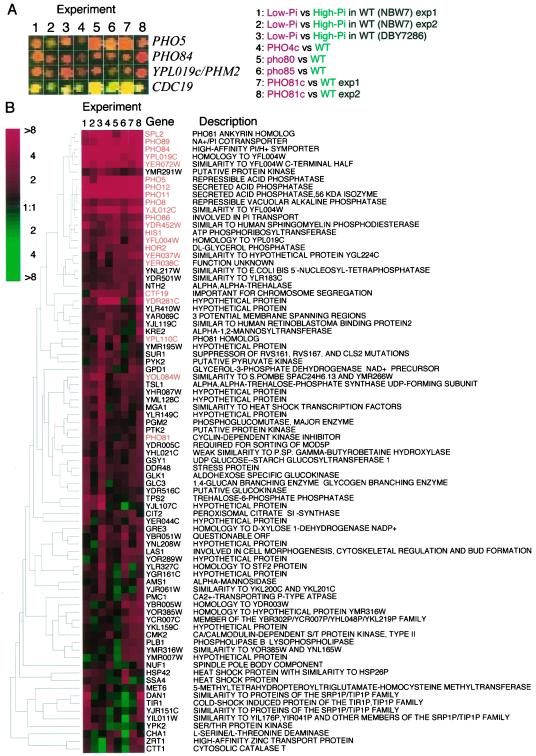
Figure 2.
DNA microarray analysis. (A) Fluorescent scanning images of the spots for the typical PHO-regulated genes,PHO5, PHO84, and_YPL019c/PHM2_, and a non PHO-regulated gene,CDC19. The column number indicates experiments performed as follows, 1 and 2: comparison between NBW7 (wild-type) cultivated in low- (YPAD-Pi, Cy5) and high-Pi media (YPAD+Pi, Cy3); 3: DBY7286 (wild-type) cultivated in low- (YPAD-Pi, Cy5) and high-Pi media (YPAD+Pi, Cy3); 4: comparison between NBD82–1 (PHO4_c_-1, Cy5) and NBW7 (Cy3); 5: comparison between NBD80–1 (_pho80_Δ, Cy5) and NBW7 (Cy3); 6: comparison between NBD85A-1 (_pho85_Δ, Cy5) and NBW7 (Cy3); 7 and 8: comparison between NOF1 (PHO81_c_-1, Cy5) and NBW7 (Cy3). All strains used in experiment 4, 5, 6, 7, and 8 were cultivated in YPAD media. (B) Cluster analysis of the DNA microarray data. Eight independent DNA microarray data were analyzed the Cluster program (Eisen et al., 1998). Genes showing more than twofold induction in any experiments correspond to the rows, and the experiments are the columns. Red represents a higher level of expression in the low-Pi conditioned wild-type cells compared with the high-Pi conditioned cells or in the mutant strains compared with the wild-type strains. The color saturation represents the magnitude of the expression ratio, as indicated by the scale at the top left of the figure. Black indicates no detectable difference in expression levels; gray denotes a missing observation. The rows, representing series of observation on individual genes, were ordered based on the similarity of their expression patterns in the eight experiments (Eisen et al., 1998). The dendrogram to the left of the figure represents the correlation between expression patterns, as described by Eisen et al., (1998). Names of the genes listed in Table 2 are emphasized by red color.
To evaluate if these gene expression changes were dependent on the known PHO regulatory factors, further DNA microarray analyses were performed comparing the parental strain NBW7 to four strains carrying mutations in components of the PHO regulatory system (Figure 1): 1) a gain-of-function mutation of the PHO4 gene encoding transcriptional activator named_PHO4_c-1 (Ogawa and Oshima, 1990); 2) a pho80 deletion mutant; 3) a pho85 deletion mutant; and 4) _PHO81_c-1 a gain-of-function mutation in the PHO81 gene, encoding the CDK inhibitor for Pho80-Pho85 (Ogawa et al., 1995). In all cases PHO5 was expressed (Figure 2A) irrespective of Pi concentration (YPAD media). A total of eight DNA microarray experiments were performed, including two independent repeated experiments. Cluster analysis of the combined data is shown in Figure 2B. Transcript levels of > 80 genes changed significantly in response to one or more of the conditions we total.
All nine genes that have previously been reported to be PHO-regulated (PHO5, PHO11, PHO12, PHO8,PHO84, PHO89, PHO86 PHO81 and_SPL2_) were successfully identified (Figure 2B).PHO5, PHO11, PHO12, PHO84,PHO89, and SPL2 had high differential expression ratios (more than fivefold) whereas PHO8 and_PHO86_ had lower differential ratios (between two and fivefold), consistent with previous results from Northern analysis (Kaneko et al., 1987; Bun-ya et al., 1991;Yompakdee et al., 1996). The nine known PHO-regulated genes can be categorized into three groups according to their function (Table2). A third member of the_PHO81_ family, YPL110c, also seems to be regulated by the PHO regulatory pathway (Figure 2B) in a manner similar to_PHO81_. Twelve additional genes showed clear evidence of PHO regulation in our experiments (Figure 2B and Table 2). Three of these genes (YPL019c, YJL012c and YER072w) had levels of differential expression similar to PHO5 and_PHO84_, in nearly all of the eight experiments. Interestingly, the putative proteins encoded by these three genes share homology with YFL004w as described later (Figure3A). The expression of YFL004w was also regulated by the PHO pathway, with differential expression levels similar to PHO8 and PHO86 (Figure 2B). Because of their homology and high differential expression ratios, we speculated that these four genes, which we have named PHM1 through PHM4 (_ph_osphate _m_etabolism genes), might have important functions in Pi metabolism.
Table 2.
The PHO regulated genesa
| ORF | Gene | No. of Pho4 binding site on the promoter | Descriptionb | |
|---|---|---|---|---|
| CACGTG | CACGTT | |||
| Phosphatases | ||||
| YBR093c | PHO5 | 1 | 1 | Repressible acid phosphatase (p60) |
| YAR071w | PHO11 | 1 | 1 | Repressible acid phosphatase (p56) |
| YHR215w | PHO12 | 1 | 1 | Repressible acid phosphatase (p58) |
| YDR481c | PHO8 | 1 | 1 | Repressible vacuolar alkaline phosphatase |
| Pi transporter | ||||
| YML123c | PHO84 | 2 | 2 | High-affinity H+/Pi cotransporter |
| YBR296c | PHO89 | 1 | 1 | Na+/Pi cotransporter |
| YJL117w | PHO86 | 1 | 1 | Involved in Pi transport |
| PHO81 family | ||||
| YGR233c | PHO81 | 1 | 0 | Cyclin-dependent kinase inhibitor |
| YPL110c | 1 | 1 | PHO81 homolog | |
| YHR136c | SPL2 | 4 | 0 | PHO81 (ankyrin) homolog |
| YFL004w family | ||||
| YFL004w | PHM1c/VTC2e | 1 | 0 | Involved in polyP synthesisf |
| YPL019c | PHM2c/VTC3e | 2 | 1 | Homology to YFL004w |
| Involved in polyP synthesisf | ||||
| YJL012c | PHM3c/VTC4e | 2 | 0 | Homology to YFL004w |
| Involved in polyP synthesisf | ||||
| YER072w | PHM4c/VTC1e/NRF1e | 1 | 1d | Homology to YFL004w |
| Involved in polyP synthesisf | ||||
| Vacuolar transporter chaperone | ||||
| Others | ||||
| YER062c | HOR2/GPP2 | 1 | 0 | DL-glycerol phosphatase |
| YPL018w | CTF19 | 2 | 1 | Important for chromosome segregation |
| YER055c | HIS1 | 1 | 0 | ATP phosphoribosyltransferase |
| YDR452w | PHM5c | 1d | 0 | Similar to acid sphingomyelinase |
| Supposed vacuolar polyphosphatasef | ||||
| YDR281c | PHM6c | 1 | 2 | Hypothetical protein |
| YOL084w | PHM7c | 1d | 0 | Similarity to S. pombe SPAC24H6.13 and YMR266w |
| YER037w | PHM8c | 0 | 1 | Similarity to hypothetical protein YGL224c |
| YER038c | 0 | 0 | Function unknown |
Figure 3.
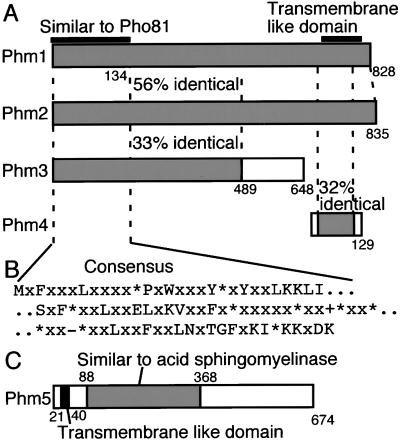
(A) Homology between Phm1, Phm2, Phm3, and Phm4. Shaded and white boxes represent homologous and nonhomologous regions, respectively, and their identities to the Phm1 sequence are indicated by percentage. The region homologous to Pho81 and the putative transmemembrane region are indicated above the Phm1 box. (B) Consensus sequences in the N-terminal regions of nine S. cerevisiae proteins, including Pho81. Hydrophobic, positive- and negative-charged amino acid residues are indicated by “*,” “+,” and “-,” respectively. (C) The putative Phm5 protein structure. The region of sequence similarity to acid sphingomyelinase in human and C. elegans is indicated by a shaded box. The putative transmembrane region is indicated by a filled box.
Among the other newly identified PHO-regulated genes listed in Table 2, the functions of three had been previously described:HOR2/GPP2 (YER062c), CTF19 (YPL018w), and HIS1 (YER055c). The_HOR2_/GPP2 gene encodes glycerol phosphatase (Norbeck et al., 1996) which hydrolyzes the phosphate bond of glycerolphosphate, releasing free Pi. The His1 enzyme (ATP phosphoribosyltransferase)-catalyzed reaction can produce pyrophosphate from ATP and phosphoribosylpyrophosphate. While both_HIS1_ and HOR2 encode enzymes that catalyze reactions that release Pi, the special significance of these reactions for the cell's Pi economy is not obvious. The Ctf19 protein has previously been identified as a subunit of the centromere-binding complex (Ortiz et al., 1999), and has no recognized role in Pi metabolism or regulation. A possible explanation for the observed differential expression could be that_CTF19_ shares promoter sequences with the “head-to-head” oriented adjacent gene YPL019c/PHM2, which was highly differentially expressed under PHO regulation. More plausibly, it may have an additional, unrecognized role in Pi metabolism.
Two “core” Pho4 binding sequences, CACGTG and CACGTT have previously been described. At least one of these sequences is found within 500 bp upstream of the coding sequence of 21 of the 22 putative PHO-regulated genes listed in Table 2, a fraction significantly higher than the 20% of all yeast ORFs that have one of these sequences within 500 bp upstream (1287 ORFs out of 6282). Of the 14 genes with more than two putative Pho4 binding sites, 9 had the high derepression ratios described above. Higher levels of differential expression showed some correlation to the presence of both of the two types Pho4 binding sites. Of the 22 putative PHO-regulated genes, only YER038c had no consensus Pho4-binding sites in its promoter region. The 3′ end of the predicted coding region of the adjacent gene,YER037w, is located only 4 bp from the YER038c ORF, which had a very similar observed expression pattern (Figure 2). It is therefore likely that the apparent PHO regulation of_YER038c_ reflects hybridization to the 3′ untranslated region of YER037w.
PHM1, PHM2, PHM3, and PHM4 Are Involved in polyP Accumulation
The predicted coding regions Phm1 and Phm2 are similar in length (828 and 835 amino acid residues for Phm1 and Phm2, respectively) and 58% identical in amino acid sequence. The predicted coding region of Phm3 is smaller (648 amino acids) and has 33% identity to the N-terminal region of Phm1 (Figure 3A). Phm4 is predicted to encode a protein of 129 amino acids, with 32% identity to the C-terminal region of Phm1, which contains three putative transmembrane spanning regions. Phm3 and Phm4 share no homology. Interestingly, the N-terminal regions (amino acids 1–135, Figure 3A) of Phm1, Phm2, and Phm3 have significant similarity to the N-terminal regions of Pho81, and its homolog Ypl110c (Figure 3B). Furthermore, this similarity is shared among a total of nine S. cerevisiae ORFs, including the putative phosphate transporter Pho87 (Bun-ya et al., 1996), two proteins homologous to Pho87, Yjl198w and Ynr013, and Syg1 (Spain_et al._, 1995), a multi-copy suppressor for a GPA1 deletion. The similarities were confined to the N-terminal region (< 300 amino acids) in all of the nine proteins. Homologous sequences can also be found in the databases of Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, mouse, and human (Battini et al., 1999).
To address the function of the PHM1, PHM2,PHM3, and PHM4 genes, strains carrying deletions of each of these genes were constructed (see MATERIAL AND METHODS). All five mutant strains, including a _phm1_Δ _phm2_Δ double mutant, were viable in rich media (YPAD), and no significant growth defects were observed in low-Pi synthetic media. All five were able to produce acid phosphatases in response to low Pi. When we analyzed polyP accumulation, however, the mutants showed striking phenotypes.
It is known that S. cerevisiae accumulates a large amounts of polyP in vacuoles under conditions of high Pi preceded by a period of Pi starvation. This is referred to as the “polyphosphate overplus” phenomenon (Harold, 1966). PolyP chains in extracts from yeast were analyzed by PAGE followed by staining with a Toluidine Blue dye, which stains polyP as well as nucleic acids. The extract from wild-type cells, NBW7, after the polyP overplus treatment (see MATERIAL AND METHODS) resulted in two distinct populations of stained molecules as is shown in Figure4A. The upper population represents RNA, whereas the lower “ladder-like” population is polyP, confirmed by pretreatment with either RNase A or exopolyphosphatase (our unpublished results). The chain lengths of the polyP bands were determined by comparing them with polyP marker ladders run in a nearby lane. The distribution of the polyP chain length in the NBW7 strain was broad (∼100 Pi residues) with a median length of ∼ 60 Pi residues (Figure 4B). The cellular concentration was measured at 19.4 μmol (1.55 mg) of polyP/mg total RNA by an enzymatic assay (Ault-Riché_et al._, 1998) (Table 3).
Table 3.
PolyP amounts in the various mutants
| Strain | Mutation | PolyP (μmol/mg of RNA) |
|---|---|---|
| NBW7 | wild type | 19.4 |
| NBM-4Lf1 | phm1Δ | 11.3 |
| NBM-19Hf1 | phm2Δ | 2.77 |
| NBM-4L19H2 | phm1Δ phm2Δ | <0.01 |
| NBM-12W2 | phm3Δ | <0.01 |
| NBM-72W7 | phm4Δ | <0.01 |
| NBM-452H1 | phm5Δ | 12.4 |
| NBM-18H7 | ctf19Δ | 9.73 |
| NBM-281H1 | phm6Δ | 8.93 |
| NBD4-1 | pho4Δ | 0.417 |
| NBD80-1 | pho80Δ | <0.01 |
| NBD8184-1B | pho84Δ | 0.015 |
The effects of _PHM1_-PHM4 on polyP accumulation following the polyP overplus treatment were analyzed by gel electrophoresis (Figure 4) and enzymatic assay (Table 3). Total polyP was slightly reduced in the _phm1_Δ mutant. The_phm2_Δ mutant had significantly reduced total polyP (14% of wild-type), and the median size of the polyP molecules was significantly smaller than that in the wild-type. PolyP was undetectable in the _phm1_Δ phm2_Δ double mutant. This clearly demonstrates that the PHM1 and_PHM2 genes have redundant functions in polyP accumulation. Both the _phm3_Δ and _phm4_Δ mutants lacked detectable polyP. These results suggest that Phm1, 2, 3 and 4 proteins are involved in the accumulation of polyP and that the products of Phm3, Phm4 and either Phm1 or Phm2 are required for polyP accumulation.
Phm2 is Localized to the Vacuole
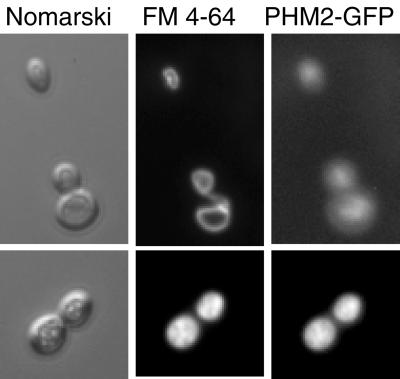
More than 90% of total polyP in yeast is localized to the vacuole (Urech, 1978). Since the Phm1–Phm4 proteins are involved in polyP synthesis, we speculated that the Phm1–Phm4 proteins were also localized to the vacuole. To address this question, a_PHM2_-GFP fusion gene, which encodes the full_PHM2_ ORF fused to the GFP ORF, was constructed and expressed in wild-type and _phm1_Δ _phm2_Δ double mutant strains. The fusion protein was active;_PHM2_-GFP was able to complement the polyP deficient phenotype of the _phm1_Δ _phm2_Δ mutant (our unpublished results). Phm2-GFP indeed appeared to be localized to the vacuoles (Figure 5) by colocalization with the vacuolar membrane marker FM4–64 (Vida and Emr, 1995).
Figure 5.
Localization of Phm2-GFP. The identical frames of the yeast transformant NBM4L19H1[pPHM2-GFP] were shown for Normarsky (left column), FM 4–64 fluorescence(center column), and GFP fluorescence (right column) images.
While this study was in progress, Cohen et al. (1999) reported on a gene family involved in a vacuolar transporter chaperon,VTC1, VTC2, VTC3, and VTC4 in S. cerevisiae. These four genes are identical to_PHM4_, PHM1, PHM2, and PHM3, respectively. In their report, the Phm4/Vtc1 protein was originally found in the same fraction as a Vma10 protein, a subunit of vacuolar H+-ATPase (v-ATPase) localized on the vacuolar membrane. Furthermore, Phm3/Vtc4, which does not have a putative transmembrane domain, was found in the membrane fraction only in the presence of Phm4/Vtc1 and either Phm1/Vtc2 or Phm2/Vtc3. These finding support our hypothesis that the Phm1 through 4 proteins form a complex on the vacuolar membrane. In addition, an S. pombe protein, Nrf1, with an amino acid sequence 78% identical to that of Phm4/Vtc1, was recently reported as a vacuolar membrane protein (Murray and Johnson, 2000).
PolyP Synthesis Is Influenced by v-ATPase Activity but It Is Not Essential
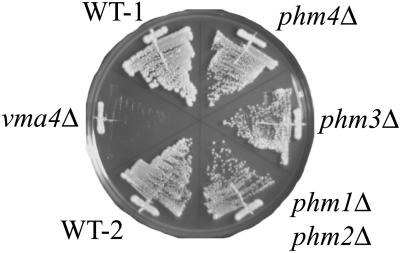
Previous work has shown that polyP accumulation is dependent upon v-ATPase activity (Wurst et al., 1996). A vma4 mutant strain, in which v-ATPase activity is completely deficient (Zhang et al., 1998), was reproducibly found to have no accumulation of polyP under the polyP overplus conditions (Figure6A). Cohen et al., (1999) reported that the v-ATPase activity in a_phm4_Δ/_vtc1_Δ mutant was 10–30% of that in wild-type cells. Cells with mutations in v-ATPase are characteristically deficient in respiration and sensitive to media containing 60 mM CaCl2 at pH 7.5 (Zhang et al., 1998). To address the possibility that the Phm1 through 4 proteins could be necessary for v-ATPase activity, and thus indirectly for the accumulation of polyP, the phm disruptants were tested for growth on nonfermentable carbon sources and in the presence of CaCl2. Growth of the _phm1_Δ_phm2_Δ, _phm3_Δ, and phm4_Δ mutants in 60 mM CaCl2 was similar to that of the wild-type strain (Figure 7), whereas the_vma4 mutant did not grow. Moreover, _phm1_Δ_phm2_Δ, _phm3_Δ, and _phm4_Δ mutants were able to grow on rich media with ethanol or glycerol as a sole carbon source, while the vma4 mutant was not (our unpublished results). These data suggest that the phm mutants retain v-ATPase activity.
Figure 6.
(A) PolyP PAGE analysis of_vma4_ and pep4 mutants. PolyP samples were prepared from the strains, CRY-V4 (_vma4_Δ), CRY (WT), CRX (_ppx1_Δ), CB024 (Pro), and NB91–6A (pep4) cultivated by the polyP overplus method. The other conditions were as described in Figure 4A. (B) PolyP PAGE analysis of vma4 and phm mutants cultivated at low pH. PolyP samples were prepared from the strains, CRY (WT), CRY-V4 (_vma4_Δ), NBM4L19H2 (_phm1_Δ _phm2_Δ), NBM12W2 (_phm3_Δ), and NBM72W7 (_phm4_Δ) cultivated in YPAD-Pi media, followed by cultivation in YPAD supplemented with either 10 mM Pi (Pi), or 10 mM Pi and 10 mM sodium acetate buffer (Ac), for 2 h. The other conditions were as described in Figure 4A.
Figure 7.
Ca2+ sensitivities of_phm_ mutants at neutral pH. CRY (WT-1), CRY-vma4 (_vma4_Δ), NBW7 (WT-2), NBM4L19H2 (_phm1_Δ _phm2_Δ), NBM12W2 (phm3Δ), and NBM72W7 (_phm4_Δ) were streaked on the YPAD plate (pH 7.5) supplemented with 60 mM CaCl2, and incubated at 30°C for 3 days.
It is known that incubating yeast in low pH media can, in part, reverse the phenotype seen with v-ATPase mutants by lowering the intravacuolar pH via fluid-phase endocytosis (Nelson and Nelson, 1990). A low but significant level of polyP accumulation was detected when the_vma4_ mutant was incubated at low pH (Figure 6B). In contrast, the _phm1_Δ _phm2_Δ, _phm3_Δ and _phm4_Δ mutants failed to accumulate detectable polyP with the low pH incubation (Figure 6B). These results strongly suggest that the _PHM1_-PHM4 gene products are directly involved in polyP accumulation and that v-ATPase activity is not strictly essential for polyP synthesis.
PolyP Formation Prevents Short-term Saturation of Cellular Pi Accumulation
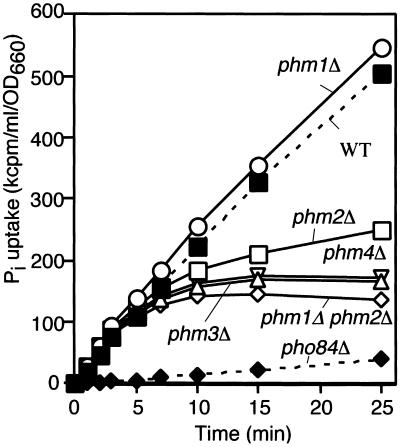
Phm1 through 4 proteins clearly play a role in polyP synthesis. The apparently paradoxical increase in the cell's ability to convert Pi into polyP in response to Pi starvation might represent a strategy for accumulating and holding precious Pi. We therefore measured Pi uptake in the_phm1_-phm4 mutants (Figure8). After cultivation in Pi-free synthetic media for 2h, NBW7 wild-type cells displayed a linear (nonsaturable) Pi uptake for up to 25 min when incubated in 0.1 mM Pi (twofold less concentration of synthetic low-Pi medium). In contrast, the _pho84_Δ mutant, which is deficient in H+/Pi cotransporter activity, showed very low uptake, as reported previously (Bun-ya et al., 1991). The phm1, 2,3, and 4 mutants each had unique saturable Pi-uptake profiles. The _phm1_Δ single mutant had an uptake profile similar to wild-type. The_phm2_Δ single mutant showed a rate of uptake similar to wild-type from 0 to 8 min, but after 8 min uptake was minimal. The Pi-uptake profiles of the _phm1_Δ_phm2_Δ double mutant, the _phm3_Δ, and the_phm4_Δ mutants were similar to wild-type for the first 5 min, after which uptake appeared to cease. This cessation of uptake in the phm mutants correlated well with the overall ability of each mutant to accumulate polyP. These data suggest that polyP accumulation is required, presumably as a sink, to sustain a high rate of Pi uptake.
Figure 8.
Pi uptake in phm mutants. Pi uptake activities in the strains NBW7 (WT), NBM4Lf1 (_phm1_Δ), NBM19Hf1 (_phm2_Δ), NBM4L19H2 (_phm1_Δ _phm2_Δ), NBM12W2 (_phm3_Δ), NBM72W7 (_phm4_Δ), and NBD8184–1B (_pho84_Δ) were tested as described in Materials and Methods.
PHM5 May Encode a Polyphosphatase in the Vacuole
The previously uncharacterized ORFs YDR452w,YDR281c, YOL084w, and YER037w were also found to be PHO-regulated. They are therefore named_PHM5_ through PHM8, respectively.YDR281c/PHM6 and YER037w/PHM8 were previously noted by Gray et al. (1998) to be induced by treatment with drugs that inhibit Cdc28 and Pho85 kinases. Among those four genes, Phm5 has significant homology to human and C. elegans acid sphingomyelinases (Figure 3C) which function in hydrolyzing phosphodiester bonds in sphingomyelin. When _phm5_Δ,_ctf19_Δ, and _phm6_Δ single disruptants were constructed and tested for polyP accumulation levels, we observed that the _phm5_Δ mutant had a unique distribution of longer chain length polyP when compared with wild-type (Figure 4A). PolyP levels and size in the _ctf19_Δ and _phm6_Δ mutants were indistinguishable from wild-type. The average polyP chain length in the_phm5_Δ mutant was over 150 Pi residues (Figure 4B). By enzymatic measurement, the total amount of polyP in the _phm5_Δ mutant was similar to that of wild-type (Table 3), indicating that polyP synthesis activity in_phm5_Δ was unaffected. These data suggest that the Phm5 protein is associated with, or is a polyphosphatase.
The deduced amino acid sequence of Phm5 (Figure 3C) includes a putative N-terminal membrane spanning domain, similar in size (27 amino acids), position and flanking amino acid sequences (rich in serine and positive charged residues) to that of the Pho8 protein, a PHO-regulated vacuolar alkaline phosphatase, which is anchored to the vacuolar membrane by a transmembrane domain at its N-terminus (Klionsky and Emr, 1989).
Wurst et al. (1995) reported that a mutant deficient in three vacuolar proteinases: proteinase A, proteinase B, and carboxypeptidase Y, has a longer polyP chain length distribution than the wild-type. This phenotype was reproducibly observed in our PAGE assay (Figure 6A). Furthermore, the phenotype was observed in a_pep4_ single mutant, which has a defect only in Proteinase A, which is required for maturation of several vacuolar enzymes, including Pho8. Together with Phm5′s similarity to sphingomyelinases, these data suggest that PHM5 encodes a polyphosphatase, which is matured by the vacuolar proteinase. Kumble and Kornberg (1996) purified a processed endopolyphosphatase of 35 kDa from yeast. Recently, it has been found that the 35-kDa protein contains amino acid sequences identical to the deduced Phm5 sequence (Sethuraman and Kornberg, personal communication).
Mutations in the PHO Regulator Genes Affect polyP Accumulation
Since the genes for polyP processing are regulated by the PHO regulatory system, polyP accumulation should be affected by mutations which affect this system. To investigate this, polyP levels under polyP overplus conditions were measured in _pho4_Δ,_pho80_Δ, and pho84_Δ mutants (Figure 4A). The_pho4 mutant, which is incapable of derepression of its target genes (Figure 1) accumulates a lower level of polyP than wild-type. The _pho84_Δ mutant, lacking a high-affinity Pi transporter, accumulates a very low but detectable level of polyP. The pho80_Δ mutant, surprisingly, did not accumulate detectable levels of polyP, despite the fact that PHO84, and PHM1 through_4 are highly expressed in this mutant. This paradoxical phenotype may be a consequence of an abnormal vacuole in this mutant (Nicolson et al., 1995).
DISCUSSION
The Proteins Involved in the Pi Acquisition and Storage System in Yeast
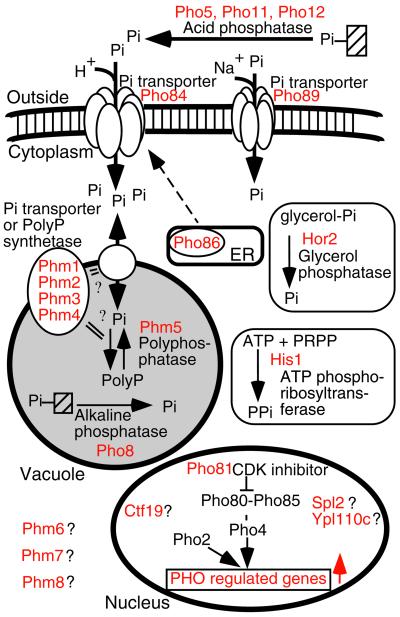
Using DNA microarray technology, we have identified at least 22 genes, including 13 novel genes, whose expression is regulated by the PHO pathway. Based on the premise that PHO-regulated genes are integral components of Pi metabolism, further molecular genetic approaches were undertaken to search for the function of the previously-uncharacterized genes. Our work has identified five of these genes as being involved in polyP metabolism. Thus, a total of 17 genes have now been found to function in Pi acquisition in yeast (Figure 9).
Figure 9.
Pi acquisition and storage system in_S. cerevisiae_. The known of predicted roles of proteins encoded by the PHO-regulated genes (names indicated in red) are schematized. See text for details.
It is interesting to consider how the products of those 17 genes work together as a physiological system for Pi acquisition and storage. When yeast encounter conditions of Pi starvation, the low Pi signal initiates Pho81 activity, which suppresses Pho80-Pho85 kinase activity. The ankyrin domain of the Pho81 protein is sufficient to inhibit Pho80-Pho85 activity (Schneider et al., 1994; Ogawa_et al._, 1995). The PHO-regulated Pho81 homologs_YPL110c_ and SPL2 share this domain, but their involvement in Pho80-Pho85 inhibition remains obscure (Flick and Thorner, 1998). Inhibition of the Pho80-Pho85 kinase results in an active Pho4 protein (hypophosphorylated form), which is localized to the nucleus where it acts as a specific transcriptional activator of PHO-regulated genes. Transcribed genes include PHO81, providing a positive feedback loop, which acts to keep Pho4 in its active form (Ogawa et al., 1995), resulting in high, continued expression of all PHO-regulated genes.
The Pi starvation signal triggers increased production of at least four types of phosphatases; 1) the acid phosphatases Pho5, Pho11, and Pho12, which are localized in periplasmic space; 2) the alkaline phosphatase Pho8, which is localized to the vacuole, 3) the glycerol phosphatase Hor2; 4) the putative polyphosphatase Phm5, localized in the vacuole. The acid and alkaline phosphatases are nonspecific, and hydrolyze a variety phosphorylated substrates, including nucleic acids, phosphosugars, phospholipids, and phosphoproteins. The glycerol phosphatase hydrolyzes the phosphate ester bond of glycerolphosphate, which is found in many sugar and lipid metabolites. All of these enzymes can contribute to increased levels of free Pi.
Under conditions of Pi starvation, expression of genes encoding the phosphate transporters, Pho84 and Pho89, are induced. Their optimal conditions are quite different: Pho84 transports Pi optimally at pH 4 and cotransports H+. Pho89, on the other hand, has optimal activity at pH 9.0 and cotransports Na+. This pair of Pi transporters work well in a wide range of environmental conditions in which yeast live. Expression of_PHO86_, a Pi transporter-related gene, is also increased in Pi starvation. The Pho86 protein was originally thought to form a complex with Pho84 and Pho87 (Bun-ya et al., 1996), however Lau et al. (2000) recently found that Pho86 is localized to the endoplasmic reticulum, where it functions in the proper localization of the Pho84 protein to the plasma membrane. Thus, Pho86 is now thought to act indirectly in Pi uptake.
The PHM1 through 4 genes, which we have shown to be involved in polyP synthesis, contribute to the Pi accumulation by a unique mechanism. Our results suggest that polyP synthesis is required for proper Pi accumulation. When polyP synthesis is critically slow, it can control the rate at which Pi is taken up by Pho84 membrane transporter. When polyP synthesis is slow, intracellular free Pi levels become high, which in turn acts as a direct negative feed back on the Pho84 membrane transporter. This critical intracellular Pi level is achieved after approximately 5 min of incubation in media containing 0.1 mM Pi. The phm mutants that lacked detectable polyP synthesis activity (_phm1_Δ_phm2_Δ double, _phm3_Δ and _phm4_Δ single disruptants) showed rapid initial uptake of Pi (like wild-type) but were incapable of further Pi uptake after ∼ 5 min. Interestingly, the phm2 mutant, which had some residual polyP synthesis activity (∼ 10% of wild-type) could continue to accumulate Pi after the initial 5-min period, but did so at a very reduced rate. In this mutant, it appears that the rate-determining step in Pi uptake in the first 5 min was controlled by the Pho84 membrane transporter, and after this time the rate was controlled by Pi to polyP conversion.
In this study, we have shown that polyP plays an important role in Pi accumulation and metabolism in yeast. The evidence for this involvement is not only metabolic but also genetic. Similar genetic interactions between polyP and Pi metabolism have been observed in E. coli. The promoter of the ppk-ppx operon, containing the genes for polyP kinase and exopolyphosphatase, includes a pho box, the response element for the Pi starvation signal mediated by the phoB-phoR two-component regulator (Kato et al., 1993). This suggests that the corresponding bacterial genes are regulated in a manner analogous to PHM1 through_4_ and PHM5, respectively. Vibrio cholerae, a Gram-negative bacterium, has a similar_ppk-ppx_ operon, with a pho box in its promoter, and its ppk mutant was unable to sustain a high rate of Pi accumulation (Ogawa et al., 2000). Thus, regulation by Pi appears to be a physiologically conserved feature of the genes for polyP metabolism, in both bacteria and yeast. Moreover, these results suggest that a major physiological role of polyP may be to promote long-term uptake and accumulation of Pi.
PHM5 Encodes a Vacuolar Polyphosphatase
Prior to this study, the PPX1 gene, encoding an exopolyphosphatase, was the only yeast gene implicated in polyP processing (Wurst et al., 1995). The expression of the_PPX1_ gene in this study showed no detectable response to Pi levels or perturbation of PHO regulation. Ppx1 protein is believed to be localized to the cytoplasm, which contains negligible amounts of polyP. Since > 90% of total cellular polyP is accumulated in vacuoles (Urech et al., 1978), the principal physiological polyphosphatase is likely to be vacuolar. The predicted vacuolar localization and PHO-regulation of Phm5 suggest that it is likely to contribute significantly to polyP degradation in vivo. Indeed, the phm5 mutation resulted in a marked increase in polyP chain lengths, whereas the ppx1 mutation resulted in a much smaller change (Figure 6A).
Phm1 through 4 Proteins Represent a New Type of polyP Synthesis System
Every Gram-negative bacterium for which the genome has been sequenced, has genes homologous to E. coli PPK (Tzeng and Kornberg, 1998). No genes homologous to PPK have been found in the genomes of Gram-positive bacteria, archea, or eukaryotes. This PPK is the only reported enzyme capable of synthesizing polyP, despite the fact that polyP has been found in every organism in which it has been sought, including Gram-positive bacteria, eukaryotes, and archea (Kornberg, 1999). Kornberg (1999) has speculated that mammalian cells synthesize polyP directly from the incorporated Pi without an ATP intermediate. Our results suggest that eukaryotic cells have an enzyme system for polyP synthesis completely different from the PPK of Gram-negative bacteria. PolyP synthesis in yeast requires v-ATPase activity, which produces proton motive force across the vacuolar membrane. We therefore speculate that the high-energy phosphoanhydride bonds in polyP are directly synthesized from Pi, using the proton motive force as a source of energy, by a vacuolar membrane-bound enzyme(s), analogous to the F-type ATPase in mitochondria. Based on this hypothesis, the Phm1 through 4 protein complex is the best candidate for this polyP synthesis enzyme in yeast.
Does a Pi acquisition system similar to the yeast system exist in higher eukaryotic cells? Kido et al. (1999) reported that expression of type II Na+/Pi cotransporter gene,NPT2, in rat kidney is derepressed by a dietary Pi starvation, and that the Pi signal-responsive element in its promoter contains the sequence, CACGTG, which is identical to the Pho4-binding site in yeast. Moreover, the N-terminal regions of Phm1, Phm2, and Phm3 contain conserved domains shared with six additional S. cerevisiae proteins, and similar protein structures can be found in genomes of many eukaryotes. Eight of the nine yeast proteins with this domain are related to Pi metabolism. We therefore refer to this domain as the “phosphate (Pi) domain” (Figure 3B). Since most of the homologous genes in other species have little or no functional characterization (except for human XPR1, gene encoding xenotropic and polytropic retrovirus receptor [Battini et al., 1999]), the function of the Pi domain may provide a useful lead for further functional investigations. Thus, further studies of the yeast system for Pi metabolism are likely to provide fundamental insights into Pi metabolism in all eukaryotes.
ACKNOWLEDGMENTS
We thank Dr. Y. Mukai in Osaka University for generously providing plasmids, Dr. A. Kornberg in Stanford University for his fruitful advice and discussion, Dr. D. Botstein in Stanford University for generously providing plasmids and his helpful discussion, and Dr. J. Krise for help with manuscript and discussion in course of this study. This work was supported by grant HG00983 from the NHGRI and by the Howard Hughes Medical Institute. P.O.B. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearn T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. [Google Scholar]
- Ault-Riché D, Fraley CD, Tzeng CM, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battini JL, Rasko JE, Miller AD. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bun-ya M, Shikata K, Nakade S, Yompakdee C, Harashima S, Oshima Y. Two new genes, PHO86 and PHO87, involved in inorganic phosphate uptake in Saccharomyces cerevisiae. Curr Genet. 1996;29:344–351. [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Cohen A, Perzov N, Nelson H, Nelson N. A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J Biol Chem. 1999;274:26885–26893. doi: 10.1074/jbc.274.38.26885. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick JS, Thorner J. An essential function of a phosphoinositide-specific phospholipase C is relieved by inhibition of a cyclin-dependent protein kinase in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:33–47. doi: 10.1093/genetics/148.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- Harold FM. Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev. 1966;30:772–794. doi: 10.1128/br.30.4.772-794.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Saccharomyces Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1992. pp. 193–281. [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Toh-e A, Oshima Y. Identification of the genetic locus for the structural gene and a new regulatory gene for the synthesis of repressible alkaline phosphatase in Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:127–137. doi: 10.1128/mcb.2.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Hayashi M, Toh-e A, Banno I, Oshima Y. Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. Gene. 1987;58:137–148. doi: 10.1016/0378-1119(87)90036-9. [DOI] [PubMed] [Google Scholar]
- Kato J, Yamamoto T, Yamaka K, Ohtake H. Cloning, sequence and characterization of the polyphosphate kinase-encoding gene (ppk) of Klebsiella aerogenes. Gene. 1993;137:237–242. doi: 10.1016/0378-1119(93)90013-s. [DOI] [PubMed] [Google Scholar]
- Kido S, Miyamoto K, Mizobuchi H, Taketani Y, Ohkido I, Ogawa N, Kaneko Y, Harashima S, Takeda E. Identification of regulatory sequences and binding proteins in the type II Sodium/Phosphate cotransporter NPT2 gene responsive to dietary phosphate. J Biol Chem. 1999;274:28256–28263. doi: 10.1074/jbc.274.40.28256. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- Kornberg A. Inorganic polyphosphate: a molecule of many functions. Prog Mol Subcell Biol. 1999;23:1–18. doi: 10.1007/978-3-642-58444-2_1. [DOI] [PubMed] [Google Scholar]
- Kumble KD, Kornberg A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J Biol Chem. 1996;271:27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- Lau W-TW, Howson RW, Malkus P, Schekman R, O'Shea EK. Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proc Natl Acad Sci USA. 2000;97:1107–1112. doi: 10.1073/pnas.97.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Persson BL. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:628–638. doi: 10.1007/s004380050776. [DOI] [PubMed] [Google Scholar]
- Murray JM, Johnson DI. Isolation and characterization of Nrf1p, a novel negative regulator of the Cdc42p GTPase in Schizosaccharomyces pombe. Genetics. 2000;154:155–165. doi: 10.1093/genetics/154.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H, Nelson N. Disruption of genes encoding subunits of yeast vacuolar H(+)-ATPase causes conditional lethality. Proc Natl Acad Sci USA. 1990;87:3503–3507. doi: 10.1073/pnas.87.9.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson TA, Weisman LS, Payne GS, Wickner WT. A truncated form of the Pho80 cyclin redirects the Pho85 kinase to disrupt vacuole inheritance in S. cerevisiae. J Cell Biol. 1995;130:835–845. doi: 10.1083/jcb.130.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbeck J, Pâhlman AK, Akhtar N, Blomberg A, Adler L. Purification and characterization of two isoenzymes of DL-glycerol-3-phosphatase from Saccharomyces cerevisiae: identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1996;271:13875–13881. doi: 10.1074/jbc.271.23.13875. [DOI] [PubMed] [Google Scholar]
- Ogawa N, Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, Noguchi K, Sawai H, Yamashita Y, Yompakudee C, Oshima Y. Functional domains of Pho81p, an inhibitor of the Pho85p protein kinase, in the transduction pathway for Pi signals in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, N., Tzeng, C.-M., Fraley, C.D., and Kornberg, A. (2000). Inorganic Polyphosphate in Vibrio. cholerae: genetic, biochemical, and physiologic features. J Bacteriol (in press). [DOI] [PMC free article] [PubMed]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- Sakumoto N, Mukai Y, Uchida K, Kouchi T, Kuwajima J, Nakagawa Y, Sugioka S, Yamamoto E, Furuyama T, Mizubuchi H, Ohsugi N, Sakuno T, Kikuchi K, Matsuoka I, Ogawa N, Kaneko Y, Harashima S. A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast. 1999;15:1669–1679. doi: 10.1002/(SICI)1097-0061(199911)15:15<1669::AID-YEA480>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schneider KR, Smith RL, O'Shea EK. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- Spain BH, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J. Truncated forms of a novel yeast protein suppress the lethality of a G protein a subunit deficiency by interacting with the b subunit. J Biol Chem. 1995;270:25435–25444. doi: 10.1074/jbc.270.43.25435. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani-Gorini A, Silver S, Yagil E. Phosphate in Microorganisms: Cellular and Molecular Biology. Washington D.C.: American Society for Microbiology; 1994. [Google Scholar]
- Tzeng CM, Kornberg A. Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol Microbiol. 1998;29:381–382. doi: 10.1046/j.1365-2958.1998.00887.x. [DOI] [PubMed] [Google Scholar]
- Urech K, Duerr M, Boller TH, Wiemken A. Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch Microbiol. 1978;116:275–278. doi: 10.1007/BF00417851. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst H, Shiba T, Kornberg A. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J Bacteriol. 1995;177:898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yompakdee C, Bun-ya M, Shikata K, Ogawa N, Harashima S, Oshima Y. A putative new membrane protein, Pho86p, in the inorganic phosphate uptake system of Saccharomyces cerevisiae. Gene. 1996;171:41–47. doi: 10.1016/0378-1119(96)00079-0. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kuromitsu Z, Ogawa N, Oshima Y. Mode of expression of the positive regulatory genes PHO2 and PHO4 of the phosphatase regulon in Saccharomyces cerevisiae. Mol Gen Genet. 1989;217:31–39. doi: 10.1007/BF00330939. [DOI] [PubMed] [Google Scholar]
- Zhang JW, Parra KJ, Liu J, Kane PM. Characterization of a temperature-sensitive yeast vacuolar ATPase mutant with defects in actin distribution and bud morphology. J Biol Chem. 1998;273:18470–18480. doi: 10.1074/jbc.273.29.18470. [DOI] [PubMed] [Google Scholar]