Pie1, a Protein Interacting with Mec1, Controls Cell Growth and Checkpoint Responses in Saccharomyces cerevisiae (original) (raw)
Abstract
In eukaryotes, the ATM and ATR family proteins play a critical role in the DNA damage and replication checkpoint controls. These proteins are characterized by a kinase domain related to the phosphatidylinositol 3-kinase, but they have the ability to phosphorylate proteins. In budding yeast, the ATR family protein Mec1/Esr1 is essential for checkpoint responses and cell growth. We have isolated the PIE1 gene in a two-hybrid screen for proteins that interact with Mec1, and we show that Pie1 interacts physically with Mec1 in vivo. Like MEC1, PIE1 is essential for cell growth, and deletion of the PIE1 gene causes defects in the DNA damage and replication block checkpoints similar to those observed in mec1Δ mutants. Rad53 hyperphosphorylation following DNA damage and replication block is also decreased in pie1Δ cells, as in mec1Δ cells. Pie1 has a limited homology to fission yeast Rad26, which forms a complex with the ATR family protein Rad3. Mutation of the region in Pie1 homologous to Rad26 results in a phenotype similar to that of the pie1Δ mutation. Mec1 protein kinase activity appears to be essential for checkpoint responses and cell growth. However, Mec1 kinase activity is unaffected by the pie1Δ mutation, suggesting that Pie1 regulates some essential function other than Mec1 kinase activity. Thus, Pie1 is structurally and functionally related to Rad26 and interacts with Mec1 to control checkpoints and cell proliferation.
When DNA replication is blocked and DNA damage occurs, checkpoints arrest the cell cycle, allowing DNA replication and repair to take place (13, 19). Loss of checkpoint control results in cell death or genetic instability that can lead to cancer. Checkpoint pathways are an evolutionarily conserved feature of eukaryotic cells. This conservation is exemplified by the family of genes encoding high-molecular-weight protein kinases, including ATM (mammals), ATR (mammals), MEC1 (Saccharomyces cerevisiae), TEL1 (S. cerevisiae), rad3+ (Schizosaccharomyces pombe), mei-41 (Drosophila melanogaster), and uvsB (Aspergillus nidulans) (6, 9, 18, 22, 23, 30, 37, 40, 48). Each of these genes falls into two family groups based on homology; ATM is related most closely to TEL1, while ATR is more related to MEC1, rad3+, mei-41, and uvsB (6, 40). This homology is not restricted to the kinase domain at the carboxyl terminus but extends over the length of the protein. The carboxyl-terminal kinase domain is structurally related to the catalytic domain of the phosphatidylinositol (PI) 3-kinases. Despite this similarity, none of these proteins has been shown to phosphorylate lipids. ATM, ATR, and Rad3 are all capable of phosphorylating protein substrates (5, 8, 28, 29). However, it remains to be determined how the kinase activity of these proteins is controlled in checkpoint responses. Moreover, little is known about whether these proteins form a complex with other proteins, although Rad3 has been recently shown to form a complex with Rad26 (12). The only Rad26 homolog identified so far is A. nidulans UVSD (40), but it has not been determined whether UVSD interacts with the ATR-related UVSB.
In budding yeast, Mec1 plays a critical role in the checkpoint controls. Three cell cycle-dependent DNA damage responses have been characterized in budding yeast, known as the G1-, S-, and G2/M-phase damage checkpoints. Mec1 is essential for all three DNA damage checkpoints (25), as well as the DNA replication block checkpoint (48). Tel1 is suggested to play an overlapping role with Mec1 in checkpoint responses (30). Besides MEC1 and TEL1, a number of genes have been identified that are involved in the DNA damage checkpoint and/or the DNA replication block checkpoint. These include CHK1, DDC1, MEC3, RAD9, RAD17, RAD24, and RAD53/MEC2 (2, 25–27, 35, 38, 46–48). RAD53 and CHK1, encoding protein kinases, function downstream of MEC1 in the checkpoint pathways. Rad53, like Mec1, plays a role in both replication block and all three DNA damage checkpoints (2, 48). Following DNA damage and replication block, Rad53 is hyperphosphorylated and activated by a mechanism dependent on Mec1 (36, 42). Thus, Mec1 and Rad53 comprise a central checkpoint pathway in budding yeast. Chk1 plays a role in the G2/M-phase DNA damage checkpoint control and is hyperphosphorylated following DNA damage in a Mec1-dependent manner (35). RAD9, RAD17, MEC3, DDC1, and RAD24 are required for all three DNA damage checkpoints (25). Rad9 is hyperphoshorylated after DNA damage, and the phosphorylated Rad9 protein binds to Rad53, possibly to modulate Rad53 activity (14, 43, 45). Genetic evidence has suggested that RAD17, RAD24, MEC3, and DDC1 operate in the same checkpoint pathway (25). Ddc1, Mec3, and Rad17 physically interact with each other but not with Rad24 (24). Instead, Rad24 forms a complex with Rfc2, Rfc3, Rfc4, and Rfc5 (16, 31, 38) and has been suggested to function upstream of the Rad17-Mec3-Ddc1 complex (24). Ddc1 is also hyperphosphorylated in a Mec1-dependent manner following DNA damage (33). Thus, Mec1 might regulate the Rad17-Rad24 checkpoint pathway by phosphorylating Ddc1. In addition to its role in checkpoint controls, MEC1 is essential for cell growth and the mec1Δ mutation is lethal. This lethality is suppressed by sml1 mutations or overexpression of RNR1. RNR1 encodes a large subunit of ribonucleotide reductase, while SML1 encodes a small protein that binds to Rnr1 (10, 50). Deletion of SML1 causes an increase in the deoxynucleoside triphosphate pool; thus, Mec1 may facilitate DNA replication by inhibiting Sml1 and thereby increasing the pool of available deoxynucleoside triphosphate (50). Although roles for Mec1 in checkpoint control and cell proliferation have been characterized, no protein has been identified that interacts with and/or regulates Mec1.
In this paper, we describe the isolation of PIE1 in a yeast two-hybrid screen searching for proteins that interact with Mec1. We show that Pie1 interacts physically with Mec1 in vivo. The pie1Δ mutation confers the same phenotype as the mec1Δ mutation with respect to cell growth and checkpoint responses. Thus, Pie1 plays a critical role in checkpoint control and cell proliferation by interacting with Mec1.
MATERIALS AND METHODS
Strains, media, and general methods.
The yeast strains used in this study are isogenic and are listed in Table 1. Standard genetic techniques were used for manipulating yeast strains (17, 21). Synthetic complete (SC) medium containing 0.5% Casamino Acids and the appropriate supplements was used to maintain selection of URA3 and TRP1 plasmids.
TABLE 1.
Strains used in this studya
| Strain | Genotype |
|---|---|
| KSC006 | MATaade1 his2 trp1 ura3 leu2 |
| KSC783 | MATamec1-1 sml1-1 trp1 ura3 leu2 |
| KSC1178 | MATasml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1180 | MATapie1Δ∷TRP1 sml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1186 | MATamec1Δ∷LEU2 sml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1196 | MATamec1Δ∷LEU2 pie1Δ∷TRP1 sml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1212 | MATaMEC1-HA∷URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1213 | MATaPIE1-myc∷TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1214 | MATaMEC1-HA∷URA3 PIE1-myc∷TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1215 | MATaMEC1-HA∷URA3 sml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1234 | MATapie1Δ∷LEU2 sml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1286 | MATaMEC1-HA∷URA3 pie1Δ∷LEU2 sml1Δ∷LEU2 ade1 his2 trp1 ura3 leu2 |
Plasmids and gene disruptions.
To construct the amino-terminal HA-tagged version of MEC1, the 5′ noncoding and amino-terminal regions of MEC1 gene were amplified by PCR with the 5′ noncoding HA-MEC1 primers KS113 (5′-TACGCGTAATCTGGAACATCGTATGGATATGATTCCATGCAGTCTTGT-3′) and KS006 (5′-AATTAACCCTCACTAAAGGGAAC-3′) or the amino-terminal HA-MEC1 primers KS114 (5′-TACGCGTATCCTTATGACGTACCAGATTATGCGGAATCACACGTCAAATATC-3′) and KS007 (5′-TTTTACGACTCACTATAGGGCGA-3′). The YEp-MEC1-HA plasmid was constructed by a three-part ligation of the _Eco_RI-_Mlu_I-treated, PCR-amplified 5′ noncoding fragment and the _Hin_dIII-_Mlu_I-treated, PCR-amplified amino-terminal fragment with _Eco_RI-_Hin_dIII-linearized YEp-MEC1 (41). To construct the kinase-negative version of MEC1 (mec1-KN), in vitro mutagenesis was performed, with aspartic acid changed to serine at position 2224 and asparagine changed to serine at position 2229, by PCR using YEp-MEC1 with the oligonucleotide primers KS006, KS106 (5′-CACGTTTTGGCTAGATATTGCGGCC-3′), KS118 (5′-ATATTAGGTCTAGGATCCAGGCACTGTGAAAGCATATTACTA-3′), and KS119 (5′-ATCTAGTAATATGCTTTCACAGTGCCTGGATCCTAGACCTAATAT-3′). The _Kpn_I-_Bst_EII fragment of YEp-MEC1-HA was replaced by a 0.4-kb _Kpn_I-_Bst_EII fragment from the PCR product, generating YEp-MEC1-KN-HA. To create pBD-MEC1(2–2368), the _Mlu_I-_Sal_I fragment of YEp-MEC1-KN-HA was cloned into the _Mlu_I-_Sal_I sites of plasmid pGBDm. pGBDm is a modified version of pGBDU-C1 (20), in which the multicloning site contains the _Mlu_I restriction site (obtained from T. Naiki). To construct the pBD-MEC1(2–938) and pBD-MEC1(2–1399) plasmids, pBD-MEC1(2–2368) was treated with _Afl_II-_Sal_I or _Nhe_I-_Sal_I, respectively, blunted, and self-ligated. A fragment containing a central region of MEC1 was obtained by PCR using primers KS491 (5′-TGCGGATCCGAGAAAACTGGCAACCCTTTC-3′) and KS492 (5′-GATGTCGACTTAAGTCCTTAATGGATCCTCGTTTG-3′). The PCR fragment was digested with _Bam_HI and _Sal_I and then cloned into the _Bam_HI-_Sal_I sites of pGBDU-C1, creating pBD-MEC1(496–1590). The amino-terminal Mec1 fragment (from positions 2 to 500) was cloned into the _Mlu_I-_Sal_I sites of plasmid pGBDm after the corresponding fragment was amplified by PCR with primers KS488 (5′-TCGGTCGACTTAGTTGCCAGTTTTCTCAATATCGC-3′) and KS489 (5′-AAAACGCGTATCCTTATGACGTAC-3′). To construct pBD-MEC1(1586–2368), the _Bam_HI-_Sal_I fragment from pBD-MEC1(2–2368) was cloned into the _Bam_HI-_Sal_I sites of pGDBU-C3 (20). To construct YIp-MEC1-HA, the _Spe_I-_Xho_I fragment from YEp-MEC1-HA and a _Kpn_I-_Spe_I fragment of the 5′ noncoding sequence of MEC1 were cloned into the _Kpn_I-_Xho_I sites of pRS306. The PIE1 gene was cloned by PCR with primers KS418 (5′-CGAXGAATTCCAACACGAAATCCAGTTCTTCGACC-3′) and KS419 (5′-TGTGGTCGACGTTCTTTCCATGTTCAAAAGAAGAC-3′). After treatment with _Eco_RI-_Sal_I, the fragment was subcloned into YCplac22 and YCplac33 (15), creating YCpT-PIE1 and YCp-PIE1, respectively. To create pAD-PIE1, the PIE1 open reading frame was cloned into the _Bam_HI-_Sal_I sites of pGAD-C1 (20) by PCR using primers KS436 (5′-CTCGGATCCATGAGACGAGAAACGGTGGGT-3′) and KS437 (5′-CTCGTCGACCTTACAGTCCCATTGAGAT-3′). The myc epitope sequence was fused to the sequence encoding the amino- terminal Pie1 by PCR using primers KS427 (5′-CTCACGCGTGTTCAAATCTTCCTCAGATATCAGTTTCTGTTCTCGTCTCATCTA TAATAGAAATAT-3′) and KS428 (5′-CTCACGCGTGAGCAAAAGCTCATTTCTGAAGAGGACTTGAATGAAACGGTGGGTGAATTTTCT-3′). After treatment with _Mlu_I and self-ligation, YCpT-PIE1-myc was obtained. To create YIp-PIE1-myc, YCpT-PIE1-myc was digested with _Bgl_II and self-ligated. MEC1-HA and _PIE1_-myc strains were obtained after transforming YIp-MEC1-HA and YIp-PIE1-myc after treatment with _Psh_AI and _Pst_I, respectively. To create YCpT-PIE1-ΔC-myc, in vitro mutagenesis to create a termination codon was performed by PCR with primers KS461 (5′-CCAGAATATATCGAAGAATTGAAGATGCAATAACCGCGGAAA-3′) and KS462 (5′-CTTGCATTATTCTCCCCGTTCTTTTATTCCGCGGAAA-3′) and the _Ned_I-_Sal_I fragment of YCpT-PIE1-myc was replaced by the corresponding PCR product. To create YCpT-PIE1-KA-myc, the open reading frame was amplified by PCR with primers KS481 (5′-AATCCGCGGCACGTAAGATAAGTGATAATTTACTGAAAAAAAATATGGT-3′) and KS482 (5′-GTGCCGCGGATTGTGGTGATTGAGGTTTTGC-3′) and the _Mlu_I-_Xba_I fragment of YCpT-PIE1-myc was replaced by the corresponding PCR fragment. The replaced PCR fragments were completely sequenced. The _Sal_I fragment from YEp-Rad53 (41), containing the sequence encoding the carboxyl-terminal half of Rad53, was cloned into _Sal_I-treated pGEX-5X-2 (Amersham Pharmacia Biotech), creating pGST-Rad53. YCp-RAD53-HA was described previously (41). The PIE1 and SML1 genes were deleted by PCR-based deletion using the Candida glabrata TRP1 and LEU2 genes (obtained from K. Kitada). The disruption of MEC1/ESR1 was described previously (22). The disruption of each gene was confirmed by PCR. The heterozygous diploids were then sporulated, and the tetrads were dissected. The tagged constructs (MEC1-HA and PIE1-myc) expressed appropriate-sized proteins from their own promoter and complemented their respective null mutations with respect to cell growth and sensitivity to DNA-damaging agents.
Two-hybrid screening.
Yeast two-hybrid screening of an S. cerevisiae expression library (a gift from P. James) was carried out as described previously (24) using pBD-MEC1(2–1399) as bait. After transformation with the library, approximately 100 colonies of PJ69-4A cells carrying pBD-MEC1(2–1399) grew on selective medium containing 10 mM 3-aminotriazole (AT). A transformation efficiency test indicated that 4 × 106 Ura+ Leu+ transformants were obtained in this screening. A total of 48 plasmids retested as positive, and 17 of these were chosen for further analysis. Restriction and sequence analyses followed by a DNA database search revealed that 12 of these plasmids contained YDR499/PIE1.
UV radiation and drug sensitivities.
Yeast cells were precultured in yeast extract-peptone-dextrose (YEPD) or SC medium appropriate to select for TRP1 and/or URA3 plasmids. The cells were then diluted in YEPD and allowed to grow at 30°C for 3 h before being subjected to UV irradiation and drug treatment. The UV radiation sensitivity assay was performed as described previously (41). Methyl methanesulfonate (MMS) sensitivity was determined as described previously (41). Cells were incubated with MMS at 30°C for 30 min. Incubation was terminated by addition of sodium thiosulfate to a final concentration of 5%. The hydroxyurea (HU) sensitivity assay was performed as described previously (38).
UV and MMS synchrony experiments.
To analyze cell cycle delay at the G2/M transition, log-phase cultures at 30°C were prearrested with 6 mg of α-factor per ml for 120 min, washed with water, and then released for 120 min into YEPD containing 15 mg of nocodazole per min to synchronize cells in G2/M. Cells arrested in G2/M were spread on YEPD plates and irradiated with a 254-nm UV lamp at 75 J/m2. The cells were then washed to remove nocodazole and released into fresh YEPD containing 1% dimethyl sulfoxide at 30°C. At timed intervals, cells were withdrawn and stained with 4′,6-diamidino-2-phenylindole (DAPI) for microscopic examination. An MMS synchrony experiment to monitor S-phase regulation and a UV synchrony experiment to analyze cell cycle delay at the G1/S transition were carried out as described previously (31).
Immunofluorescence microscopic analysis.
Yeast cells were grown in YEPD medium at 30°C. To examine spindle elongation at 30°C, the culture was synchronized in the G1 phase by addition of 6 mg of α-factor per ml at 30°C for 2 h. The cells were then washed to remove α-factor and released into YEPD containing 100 mM HU at 30°C. Aliquots of cells were removed and processed for DNA flow cytometry analysis, viability assessment, and indirect immunofluorescence microscopy as described previously (41).
Immunoblotting.
Protein extracts for immunoblotting were prepared and resolved sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (41). The proteins were then transferred to nylon membranes, subjected to immunoblot analysis with the monoclonal anti-HA (3F10, 12CA5, or 16B12), anti-myc (9E10), or anti-nuclear pore complex protein (MAb414) antibodies (4) or the polyclonal anti-Ssb1 antibodies (obtained from S. Nishikawa), and detected using an ECL kit (Amersham Pharmacia Biotech).
Nuclear and cytoplasmic fractionation.
The nucleus and cytoplasm were separated essentially as described previously (3). Yeast cells were precultured in SC medium appropriate to select for TRP1 plasmids. They were diluted in YEPD and allowed to grow at 30°C for 3 h. Cells at an optical density at 600 nm of 100 were collected by centrifugation and incubated in 50 mM Tris-H2SO4 (pH 9.0)–10 mM dithiothreitol at 30°C for 5 min. They were then converted into spheroplasts in 5 ml of sorbitol buffer (1.2 M sorbitol, 40 mM Tris-HCl [pH 8.0], 10 mM dithiothreitol) containing 500 μg of Zymolyase 100T after incubation at 30°C. Spheroplasts were loaded on 7.5% Ficoll-sorbitol buffer (7.5% Ficoll in 1.2 M sorbitol, 40 mM Tris-HCl [pH 8.0], and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) and harvested by centrifugation at 4000 × g for 5 min at 4°C. The spheroplasts were resuspended in 7 ml of 20% Ficoll in phosphate buffer (20 mM potassium phosphate [pH 6.5], 1 mM MgCl2, 1 mg each of leupeptin and pepstatin per ml, 0.5% aprotinin, 1 mM PMSF, 10 mM benzamidine-HCl), gently homogenized (25 strokes in a Dounce homogenizer), and centrifuged at 8,000 × g for 5 min. After recentrifugation at 8,000 × g for 10 min, the supernatant was loaded onto 30% Ficoll in phosphate buffer and centrifuged at 24,000 rpm for 60 min in a Beckman SW60Ti rotor. The 20% Ficoll layer was recovered as a cytoplasmic fraction, and the precipitates were suspended in lysis buffer as a nuclear fraction.
Immunoprecipitation and kinase assay.
Yeast cells were precultured in YEPD or SC medium appropriate to select for TRP1 and/or URA3 plasmids. The cells were then diluted in YEPD and allowed to grow at 30°C for 3 h. When treated with MMS, the culture was incubated in 0.1% MMS for 1 h. The cells (optical density at 600 nm = 40) were pelleted, washed, and resuspended in 150 ml of lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 1 mM sodium orthovanadate, 1 mM dithiothreitol, 0.1% Triton X-100, 40 mM β-glycerophosphate, 15 mM _p_-NO2-phenylphosphate, 1 mg of leupeptin per ml, 1 mg of pepstatin per ml, 0.5% aprotinin, 100 mg of APMSF per ml). An equal volume of glass beads was added, and the cells were lysed by vortexing. Extracts were clarified by 15 min of centrifugation at 4°C. The supernatant was diluted with lysis buffer and incubated at 4°C for 2 h with protein A-Sepharose beads bound with anti-HA or anti-myc antibodies. Protein concentration was determined by a protein assay (Bio-Rad). Immunoprecipitates were washed four times with lysis buffer and twice with kinase buffer (20 mM sodium HEPES [pH 7.5], 10 mM MgCl2, 4 mM MnCl2). For coimmunoprecipitation experiments, immunoprecipitates were boiled immediately in 1 × SDS-PAGE sample buffer. For the kinase assays, immunoprecipitates were separated into equal portions for immunoblotting and the kinase reaction. The kinase reaction was initiated in 50 μl of kinase buffer by the addition of 10 mCi of [γ-32P]ATP (3,000 Ci/mmol) (Amersham Pharmacia Biotech), 1 μg of glutathione _S_-transferase (GST)–Rad53, and ATP to 50 μM. Reactions were terminated by addition of 5× sample buffer and boiling for 5 min. The eluted proteins were separated by SDS-PAGE, and the gels were dried and autoradiographed.
RESULT
Pie1 interacts physically with Mec1.
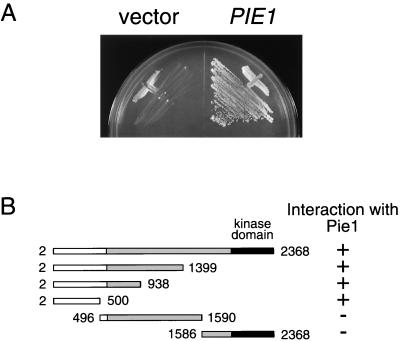
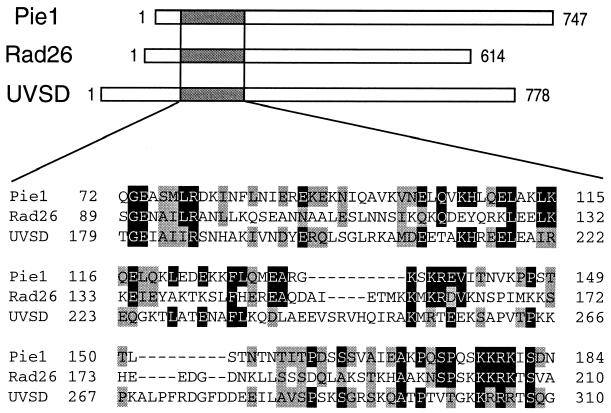
In an attempt to identify Mec1-interacting proteins, we performed a yeast two-hybrid screen of a budding yeast expression library. As bait we used a derivative of Mec1 lacking its kinase domain (amino acids 2 to 1399 of Mec1). We isolated 17 positive clones and found that 12 of these contained YDR499 fused to the transcriptional activation domain. We therefore chose YDR499 for further analysis. Hereafter, YDR499 is designated a PIE1, encoding a protein interacting with Mec1/Esr1. The full-length Mec1 protein was also found to interact with Pie1 in the two-hybrid assay (Fig. 1A). The predicted PIE1 gene product of 747 amino acids has a calculated molecular mass of 86 kD. A database search demonstrated that Pie1 has a region weakly homologous to the S. pombe Rad26 (1) and A. nidulans UVSD (40) proteins (Fig. 2). It has been shown that the Rad26 protein interacts physically with the ATR family protein Rad3 (12).
FIG. 1.
Interaction between Mec1 and Pie1 in the two-hybrid assay. (A) Pie1 interacts with Mec1 in the two-hybrid assay. Strain PJ69-4A carrying pBD-MEC1(2–2368) was transformed with pAD-PIE1 or the control vector. Transformants were streaked on an SC-Ura-Leu-His plate containing 10 mM AT. (B) Identification of the Mec1 region required for its interaction with Pie1. Strain PJ69-4A carrying pAD-PIE1 was transformed with pBD-MEC1(2–2368), pBD-MEC1(2–1399), pBD-MEC1 (2–938), pBD-MEC1(2–500), pBD-MEC1(496–1590), or pBD-MEC1(1586–2368). The kinase domain and central homologous region of Mec1 are indicated as black and gray bars, respectively. Transformants were streaked on an SC-Ura-Leu-His plate containing 10 mM AT. Interaction with Pie1 was assessed by measuring the growth of transformants.
FIG. 2.
Structure and alignment of Pie1, S. pombe Rad26, and A. nidulans UVSD. The Pie1, S. pombe Rad26, and A. nidulans UVSD proteins contain 747, 614, and 778 amino acids, respectively. Alignment of the conserved regions of these three proteins is shown below. Amino acids that are identical or conserved are indicated by black and gray boxes, respectively.
To examine whether Pie1 interacts physically with Mec1 in vivo, we performed immunoprecipitation experiments. For this purpose, we generated HA-tagged MEC1 and myc-tagged PIE1 constructs and replaced the corresponding genomic copies with the tagged constructs. Extracts were prepared from cells and subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitates were then probed with antibodies against the HA and myc epitopes. When probed with anti-HA antibodies, bands corresponding to Mec1 were detected in MEC1-HA cells. When immunoblotted with anti-myc antibodies, bands corresponding to Pie1-myc were detected only in cells expressing both Mec1-HA and Pie1-myc (Fig. 3). In the converse experiment, extracts were subjected to immunoprecipitation with anti-myc antibodies. The immunoprecipitates were then analyzed by immunoblotting with antibodies against the HA and myc epitopes. When probed with anti-HA antibodies, Pie1-myc was detected only in anti-HA immunoprecipitates from extracts of cells expressing both Mec1-HA and Pie1-myc (Fig. 3). These observations demonstrate that Pie1 and Mec1 physically interact in vivo.
FIG. 3.
Interaction between Mec1 and Pie1 in vivo. Extracts were prepared from MEC1-HA (KSC1212), PIE1-myc (KSC1213), and MEC1-HA PIE-myc (KSC1214) cells, and subjected to immunoprecipitation (IP) with anti-HA (left panel) or anti-myc (right panel) antibodies. The immunoprecipitates were separated by SDS-PAGE and subjected to immunoblot analysis with anti-HA or anti-myc antibodies.
The Mec1 protein is divided into three regions, similar to other ATR family proteins (6). The carboxyl-terminal kinase domains are highly conserved among the ATR family proteins, and the central regions also exhibit significant homology. In contrast, no apparent homology is observed among the amino-terminal regions. To delineate the region of Mec1 that interacts with Pie1, we constructed baits containing various fragments of MEC1 and tested their ability to interact with Pie1 in the yeast two-hybrid system (Fig. 1B). We found that Pie1 interacts with the amino-terminal region of Mec1, corresponding to amino acids 2 to 500, a region that is not conserved among the ATR family members.
PIE1 is essential for cell growth.
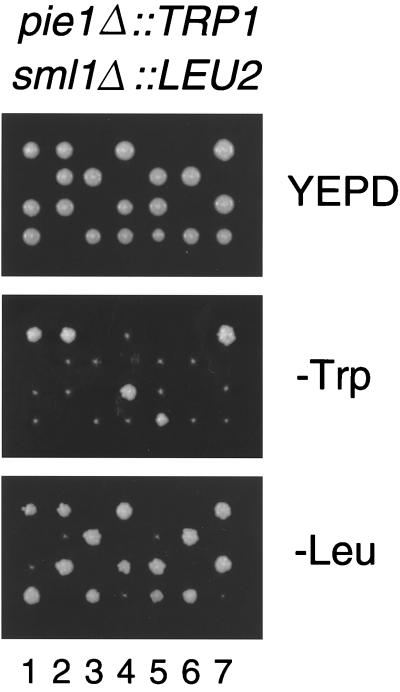
To examine the PIE1 function, we created a null strain by gene disruption (see Materials and Method). A heterozygous PIE1/pie1 Δ∷TRP1 diploid was sporulated, and only two spores were viable in each tetrad. All the viable spores were Trp−, containing the wild-type PIE1 gene. These results indicate that PIE1 is essential for cell growth. MEC1 is required for cell growth, in addition to checkpoint controls, and the lethality of the mec1Δ mutation is suppressed by sml1Δ mutations (50). To examine whether the pie1Δ lethality is also rescued by the sml1Δ mutation, the pie1Δ/PIE1 sml1Δ/SML1 diploids were sporulated. Trp+ cells were recovered, and all were found to be Leu+, indicating that pie1Δ sml1Δ cells are viable (Fig. 4). The pie1Δ sml1Δ double-mutant cells grew as well as the wild-type and sml1Δ mutant cells did. Furthermore, the sml1Δ mutation similarly suppressed the lethality of a mec1Δ pie1Δ double mutation (see below). Thus, the viability loss caused by the pie1 disruption is rescued by the sml1Δ mutation, suggesting that Pie1 and Mec1 have the same function in cell growth.
FIG. 4.
_pie1_Δ lethality is suppressed by the sml1 Δ mutation. The pie1Δ∷TRP1/+ sml1Δ∷LEU2/+ diploid was sporulated and dissected on a YEPD plate. Seven tetrads are displayed vertically. Sporulated tetrads grown up on a YEPD plate were replica-plated to a SC-Trp or SC-Leu plate. Cells proliferating on SC-Trp are all Leu+, indicating that pie1Δ∷TRP1 sml1Δ∷LEU2 double mutants are viable.
Effects of the pie1Δ mutation on the DNA damage and replication block checkpoints.
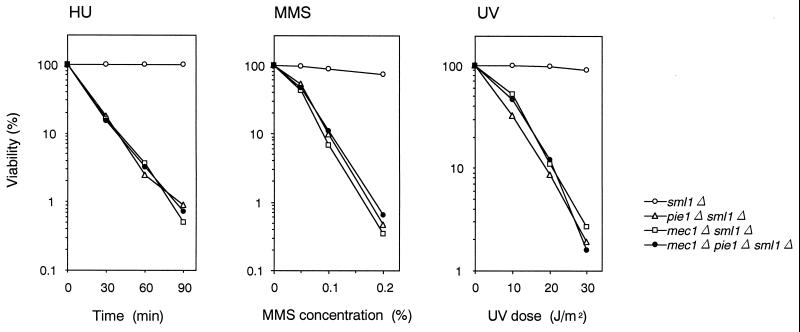
Suppression of pie1Δ lethality by the sml1Δ mutation allowed us to examine the phenotype associated with a complete loss of PIE1 function. We tested the sensitivity of pie1Δ mutants to HU, MMS, and UV light and found that pie1Δ sml1Δ strains are significantly more sensitive than sml1Δ strains are (Fig. 5). We then compared the sensitivity of pie1Δ, mec1Δ, and double-mutant mec1Δ pie1Δ strains in a sml1Δ background. In a sml1Δ background, the pie1Δ strain showed the same sensitivity to HU, MMS, and UV irradiation as the mec1Δ and mec1Δ pie1Δ strains did (Fig. 5), suggesting that Pie1 and Mec1 function in the same pathway following DNA damage and replication block.
FIG. 5.
Sensitivity of pie1Δ, mec1Δ, and mec1Δ pie1Δ mutants to HU, MMS, and UV. sml1Δ (KSC1178), pie1Δ sml1Δ (KSC1180), mec1Δ sml1Δ (KSC1186), and mec1Δ pie1Δ sml1Δ (KSC1196) cells were grown to log phase and treated with HU, MMS, or UV light. The viability of cells was estimated as described in Materials and Methods.
We next tested whether pie1Δ mutants are defective in their DNA damage and replication block checkpoints. We first examined the G2/M-phase DNA damage checkpoint by monitoring mitotic division following UV irradiation (Fig. 6). When cell cultures were released from nocodazole arrest after UV irradiation, wild-type cells exhibited delayed nuclear division while pie1Δ cells proceeded through mitosis at the same rate as mec1Δ cells did. Similarly, the pie1Δ strains progressed as fast as the mec1Δ strains through the G1/S transition and S phase following DNA damage (data not shown). We further examined the DNA replication block checkpoint in pie1Δ mutants. Cells were synchronized with α-factor and released into medium containing HU. At 120 min after the release into HU, wild-type cells were arrested as large budded cells with short spindles, while one-third of pie1Δ and mec1Δ mutants exhibited elongated spindles (Table 2). These results indicate that pie1Δ mutants are as defective as mec1Δ mutants in the G1-, S- and G2/M-phase DNA damage and the replication block checkpoints.
FIG. 6.
G2/M-phase DNA damage checkpoint in pie1Δ and mec1Δ mutants. sml1Δ (KSC1178), pie1Δ sml1Δ (KSC1180), and mec1Δ sml1Δ (KSC1186) cells were arrested with nocodazole and irradiated or not irradiated with UV. At the indicated times after release of UV-irradiated (+UV) and unirradiated (−UV) cultures from nocodazole, the percentage of uninucleate large budded cells was scored by DAPI staining.
TABLE 2.
DNA replication block checkpoint in _pie1_Δ and _mec1_Δ cellsa
| Genotype | % of cells with: | |
|---|---|---|
| Short spindle | Elongated spindle | |
| _sml1_Δ | 98 | 2 |
| _pie1Δ sml1_Δ | 63 | 37 |
| _mec1Δ sml1_Δ | 65 | 35 |
Rad53 is phosphorylated in response to DNA damage and replication block in a Mec1-dependent manner, and this phosphorylation correlates with activation of checkpoint pathways (36, 42). We therefore examined whether PIE1 is also required for the Rad53 phosphorylation. As observed in mec1Δ mutants, Rad53 phophorylation following HU and MMS treatment was significantly reduced in pie1Δ mutants (Fig. 7). These results demonstrate that PIE1 and MEC1 play similar roles in the DNA damage and replication block checkpoint controls.
FIG. 7.
HU- and MMS-induced Rad53 modification in pie1Δ and mec1Δ mutants. sml1Δ (KSC1178), pie1Δ sml1Δ (KSC1180), and mec1Δ sml1Δ (KSC1186) cells carrying YCp-RAD53-HA were left untreated or treated with 10 mg/of HU per ml for 120 min or 0.1% MMS for 30 min and then subjected to immunoblot analysis as described in Materials and Methods.
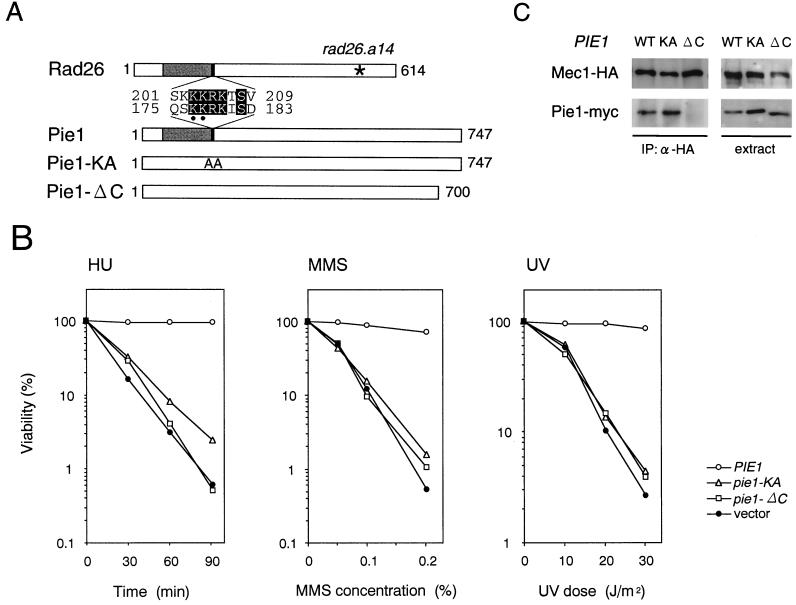
Analysis of the conserved and carboxyl-terminal regions of Pie1.
One region within Pie1 is homologous to Rad26 and UVSD (1, 40), and no other regions exhibit significant homology. It has been shown that the rad26.a14 mutation, which localizes to the carboxyl terminus, confers a defect in response to replication block but not to DNA damage (44) (Fig. 8A). We were therefore interested in examining a role for the carboxyl-terminal Pie1 in the response to DNA damage and replication block. Because there is no significant sequence homology within Pie1 to Rad26 and UVSD, we constructed a truncated mutation, pie1-ΔC, in which the carboxyl-terminal 47 amino acids are deleted (Fig. 8A). We transformed the plasmid carrying the pie1-ΔC mutation gene into pie1Δ sml1Δ cells and examined their sensitivity to DNA damage and HU treatment. The cell carrying the pie1-ΔC mutation showed the same sensitivity as the mutant cells carrying the control vector (Fig. 8B). Furthermore, the pie1-ΔC mutation gene failed to complement the pie1Δ lethality (data not shown). We next used coimmunoprecipitation to test whether the Pie1-ΔC protein interacts with Mec1. The pie1Δ sml1Δ cells expressing Mec1-HA cells were transformed with centromeric plasmids carrying PIE1-myc or pie1-ΔC–myc. Extracts were prepared from transformants and subjected to immunoprecipitation with anti-HA antibodies. Then the immunoprecipitates and extracts were probed with antibodies against the HA and myc epitopes. Although cells expressed the Pie1-myc and Pie1-ΔC–myc mutant proteins at a similar level, Pie1-ΔC–myc did not coprecipitate with Mec1-HA (Fig. 8C). These results suggest that the carboxyl terminus of Pie1 is required for interaction with Mec1.
FIG. 8.
Analysis of mutations of Pie1 at the conserved domain and the carboxyl terminus. (A) Mutations of Pie1 at the conserved and carboxyl-terminal regions. The structures and conserved regions of Pie1 and S. pombe Rad26 are shown. The pie1-KA mutation changes lysine to alanine at amino acid positions 177 and 178 (indicated by dots) within a conserved region. The pie1-ΔC gene product lacks the carboxyl-terminal 47 amino acids. An asterisk marks the location of the rad26.a14 mutation. (B) Sensitivity of the pie1-KA and pie1-ΔC mutants to HU, MMS, and UV. pie1Δ sml1Δ (KSC1234) cells were transformed with YCpT-PIE-myc, YCpT-PIE1-KA-myc, YCpT-PIE1-ΔC-myc, or the control vector. The transformants were grown to log phase and treated with HU, MMS, or UV light. The viability of cells was estimated as described in Materials and Methods. (C) Interaction of the Pie1-KA and Pie1-ΔC mutant proteins with Mec1. Extracts were prepared from MEC1-HA pie1Δ sml1Δ (KSC1286) cells carrying YCpT-PIE-myc (WT), YCpT-PIE1-KA-myc (KA), or YCpT-PIE1-ΔC-myc (ΔC) and subjected to immunoprecipitation (IP) with anti-HA antibodies. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-HA or anti-myc antibodies.
We next examined the significance of the region within Pie1 that is homologous to Rad26 and UVSD. We constructed a mutation (pie1-KA) in which the lysines at positions 177 and 178 were changed to alanines within a stretch of basic amino acids that is the most homologous region within Pie1 (Fig. 8A). We transformed the pie1-KA plasmid in pie1Δ sml1Δ mutants and found that these transformants were as sensitive to HU, MMS, and UV light as were the cells carrying the control vector (Fig. 8B). Moreover, the pie1-KA gene did not complement the pie1Δ lethality (data not shown). We also examined the interaction of the Pie1-KA–myc mutant proteins with Mec1-HA by co immunoprecipitation. Similar to the wild-type Pie1-myc protein, the Pie1-KA–myc mutant protein coprecipitated with Mec1-HA (Fig. 8C). Some short amino acid sequences rich in the basic amino acids lysine and arginine are known to function as a nuclear localization signal (11). It is possible that the pie1-KA mutation might cause mislocalization of Pie1 along with Mec1. We therefore examined the intracellular distribution of Mec1 and Pie1 in wild-type and pie1-KA mutant cells (Fig. 9). Whole-cell extracts were fractionated into two separate cytoplasmic and nuclear fractions. Equal volumes of these fractions and whole-cell extract were analyzed on immunoblots to detect Mec1-HA and Pie1-myc proteins. As control for the fractionation, we assessed each fraction for the presence of the cytoplasmic heat shock protein Ssb1 and a nuclear pore complex protein (4). Both Mec1-HA and Pie1-myc were found to exist in both cytoplasmic and nuclear fractions from wild-type cells. The pie1-KA mutation did not affect their intracellular distribution. Moreover, the intracellular distribution of Mec1-HA was not significantly altered in pie1Δ mutants (Fig. 9). These results indicate that the region within Pie1 that is homologous to Rad26 and UVSD is essential for Pie1 function but is not essential for its interaction with Mec1 or for the intracellular localization of Pie1 and Mec1.
FIG. 9.
Effect of the pie1-KA mutation on the intracellular distribution of Mec1 and Pie1. MEC1-HA pie1Δ sml1Δ (KSC1286) cells carrying YCpT-PIE-myc, YCpT-PIE1-KA-myc, or the control vector YCplac22 were harvested and spheroplasted. Spheroplasts were homogenized to prepare whole-cell extracts (W) and then separated into the cytoplasmic (C) and nuclear (N) fractions. Samples from each fraction were separated by SDS-PAGE and immunoblotted with anti-HA, anti-myc, anti-Ssb1, or anti-nuclear pore complex (NPC) antibodies.
Effect of the pie1Δ mutation on the Mec1 kinase activity.
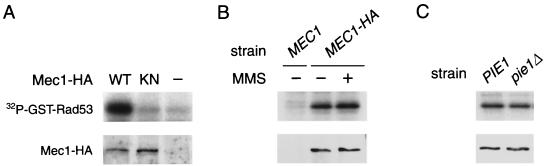
Members of the ATR family, for example, ATR and Rad3, have protein kinase activities. It is possible that Mec1, like the other family members, has a kinase activity and that the kinase activity is controlled by Pie1. Mec1 contains the motif DXXXXN at positions 2224 to 2229, which in conventional protein kinases plays a critical role in catalysis (49). We therefore constructed the kinase-negative mutation (mec1-KN) by changing amino acids in the conserved motif DXXXXN to SXXXXS. ATM and ATR phosphorylate the serine-glutamine/threonine-glutamine (SQ/TQ) cluster domain on Chk2, the mammalian homolog of Rad53, in vitro (29). Rad53 is considered to be a downstream target of Mec1 (36, 42) and has 16 SQ/TQ motifs; 7 of them are located in the amino-terminus, 1 is in the kinase domain, and 8 are in the carboxyl terminus. We therefore used a fusion protein consisting of GST and the carboxyl terminus of Rad53 (GST-Rad53) as a substrate. Extracts were prepared from cells carrying YEp-Mec1-HA, YEp-MEC1-KN-HA, or the control vector. The Mec1-HA or Mec1-KN-HA proteins were immunoprecipitated from these extracts with anti-HA antibodies and subjected to a kinase assay. GST-Rad53 was phosphorylated by Mec1-HA, whereas phosphorylation by Mec1-KN-HA was similar to the background level in the absence of HA-tagged proteins (Fig. 10A). GST alone was not efficiently phosphorylated by Mec1-HA (data not shown). These results show that Mec1 has an associated kinase activity to phosphorylate the carboxyl terminus of Rad53. We found that the mec1-KN mutation exhibits a phenotype very similar to the null mutation; this mutation does not complement mec1Δ sml1Δ cells with respect to sensitivity to HU, MMS, or UV light, nor does it complement the lethality of the mec1Δ mutation (data not shown). Together, these results indicate that Mec1 kinase activity is essential for checkpoint controls and cell growth. Because Rad53 is activated following DNA damage in a Mec1-dependent manner, we examined whether DNA damage increases the kinase activity of Mec1 from MEC1-HA cells in which the chromosomal MEC1 gene is replaced with the tagged construct. MMS treatment, however, did not affect Mec1-associated protein kinase activity (Fig. 10B). Since Pie1 functions in a complex with Mec1, we further asked whether Pie1 is required for Mec1 kinase activity. We examined the kinase activity associated with Mec1 prepared from MEC1-HA PIE1 sml1Δ and MEC1-HA pie1Δ sml1Δ cells. However, phosphorylation of GST-Rad53 was not decreased in extracts from pie1Δ sml1Δ cells compared with that in extracts from PIE1 sml1Δ cells (Fig. 10C). These results suggest that although Pie1 plays an essential role in checkpoint responses and cell growth, its role involves functions other than regulation of Mec1 kinase activity.
FIG. 10.
Protein kinase activity associated with Mec1. Cells grown to the mid-log phase were incubated with or without 0.1% MMS for 1 h and harvested for preparation of crude extracts. Extracts were subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitated HA-tagged Mec1 proteins were assayed for kinase activity using GST-Rad53 as a substrate, as described in Materials and Methods. In the top panel, 32P incorporation into GST-Rad53 was detected by autoradiography. In the bottom panel, the amount of the Mec1 protein used for the kinase assay was examined by immunoblotting. (A) mec1-1 sml1-1 (KSC783) cells carrying YEp-MEC1-HA (WT), YEp-MEC1-KN-HA (KN), or the control vector pRS426 (−), (B) MMS-treated (+) or untreated (−) sml1Δ (KSC1178) and MEC1-HA sml1Δ (KSC1215) cells; (C) MEC1-HA sml1Δ (KSC1215) and MEC1-HA pie1Δ sml1Δ (KSC1286) cells.
DISCUSSION
Genetic studies have demonstrated that Mec1 plays a critical role in checkpoint responses and cell growth in budding yeast. However, it is not clear how Mec1 is regulated by other proteins in the checkpoint responses and cell growth. In an attempt to address this question, we screened for proteins that associate with Mec1 in a two-hybrid system and identified PIE1/YDR499. A subsequent coimmunoprecipitation experiment revealed that Pie1 interacts physically with Mec1 in vivo. In this paper, we provide evidence demonstrating that Pie1 plays a critical role in checkpoint responses and cell growth by interacting with Mec1. First, similar to mec1Δ, the pie1Δ mutation is lethal and its lethality is suppressed by sml1Δ mutations. The lethality of mec1Δ pie1Δ double mutants is also suppressed by sml1Δ mutations. Tel1 plays an overlapping role with Mec1, and a high dosage of TEL1 can suppress lethality in mec1Δ and pie1Δ mutants (reference 30 and data not shown). Second, pie1Δ sml1Δ cells show the same sensitivity as mec1Δ sml1Δ cells following DNA damage and HU treatment. Furthermore, the mec1 and pie1 mutations are not additive with respective to sensitivity to DNA damage and HU treatment. Third, the pie1Δ and mec1Δ cells are equally defective in all the G1, S, and G2/M damage and replication block checkpoints. Finally, Rad53 is hyperphosphorylated following DNA damage and replication block, and this phosphorylation is dependent on Pie1 as well as Mec1. Thus, mec1Δ and pie1Δ mutants have identical phenotypes, indicating that Mec1 and Pie1 function by forming a complex in checkpoint responses and cell growth. Consistently, Paciotti et al. (32) and Rouse and Jackson (34) have characterized YDR499, designated DDC2 and LCD1, respectively, and shown that Pie1/Ddc2/Lcd1 interacts with Mec1 and plays a key regulatory role in checkpoint responses and cell growth.
To understand the regulatory role of Pie1, we examined whether Mec1 has an associated kinase activity and whether this kinase activity is regulated by Pie1. We first constructed a kinase-negative derivative of MEC1 (mec1-KN). The mec1-KN mutation was found to resemble the null mutation, because it failed to complement the lethality of the _mec1_Δ mutation and the sensitivity of _mec1_Δ mutants to HU, MMS, and UV light. Consistent with the hypothesis that Rad53 functions downstream of Mec1, there is a kinase activity associated with wild-type Mec1 that can phosphorylate the Rad53 protein. The kinase-negative derivative of Mec1 (Mec1-KN) cannot phosphorylate the Rad53 protein. These results suggest that Mec1 kinase activity is essential for the role of Mec1 in checkpoint responses and cell growth. However, Mec1 activity to phosphorylate the Rad53 protein in vitro is unaffected by DNA damage, although Rad53 phosphorylation following DNA damage requires Mec1 in vivo. Moreover, Mec1 kinase activity in vitro is not altered by deletion of PIE1, although _mec1_Δ and _pie1_Δ mutants show identical phenotypes. Mec1 exists in both the nucleus and cytoplasm, but the _pie1_Δ mutation does not significantly affect the intracellular localization of Mec1. So far, we have not obtained results demonstrating a regulatory role of Pie1. The catalytic submit of DNA-dependent protein kinase (DNA-PKcs) also contains a kinase domain structurally related to that of the phosphatidylinositol 3-kinase, as found in the ATR family proteins (39). Although not involved in checkpoint controls, DNA-PKcs may serve a model for understanding the regulation of the ATR family proteins. In vitro, DNA-PKcs is activated by binding double-stranded DNA ends in the presence of a heterodimer composed of the Ku70 and Ku86 subunits. In this manner, the activation of DNA-PKcs is dependent on both specific DNA structures and regulatory cofactors. It is possible that Pie1 targets Mec1 to specific DNA structures and/or other DNA binding proteins to activate the Mec1 kinase activity. Such DNA and/or proteins might be missing in our in vitro assay, so that Mec1 kinase activity could not be affected by DNA damage or deletion of PIE1. Alternatively, it is possible that Pie1 facilitates the recognition by Mec1 of specific substrates involved in checkpoint responses and cell proliferation. In this model, substrates for Mec1 might be modulated for recognition by Pie1 and then efficiently phosphorylated by Mec1. For example, Rad9 is hyperphosphorylated and is bound to Rad53 in response to DNA damage, and this Rad9-Rad53 interaction has been implicated in Rad53 activation (14, 43, 45). Mec1 might efficiently phosphorylate Rad53 only when it is associated with Rad9. Finally, it remains possible that Mec1 phosphorylates Pie1 and that phosphorylated Pie1 in turn, transduces a signal downstream in the cell growth and checkpoint responses. Paciotti et al. (32) have shown that Pie1/Ddc2 is phosphorylated by Mec1 following DNA damage and replication block, although the significance of the phosphorylation has not been demonstrated.
Pie1 shares several functional properties with fission yeast Rad26. The rad26 deletion mutation confers the same checkpoint defect as the rad3 deletion mutation. Rad26 interacts physically with the ATR family protein Rad3 and undergoes Rad3-dependent DNA damage-induced phosphorylation (12). In A. nidulans, mutations in uvsD confer phenotypes similar to mutations in the ATR family member uvsB (40). The biochemical properties of the UVSB and UVSD proteins have not been examined yet. In addition to these functional similarities, both the Rad26 and UVSD proteins have limitted homology to Pie1 (1, 40). To examine the significance of the region conserved among these proteins, we replaced two conserved basic amino acid residues at this region of Pie1 with alanines. The resulting mutation, pie1-KA, was very similar to the pie1 null mutation with respect to cell growth and sensitivity to HU, MMS, and UV light. These results indicate that this homologous region is essential for the Pie1 function and suggest that the Pie1, Rad26, and UVSD proteins are functionally and structurally related to each other. We therefore expect that homologs of Pie1, Rad26, and UVSD will be found in higher eukaryotes. In D. melanogaster, mutations in mus304 were identified to confer the same checkpoint defects as mutations in the ATR family member mei-41 (7). Furthermore, mus304 and mei-41 were shown to act in the same genetic pathway during DNA repair and development (7). Although searches revealed no specific sequence homology, mus304 encodes a protein with a size similar to Pie1 and Rad26 and with a stretch of 4 basic amino acid residues in the corresponding region. Mus304 might be a Pie1-related protein.
The ATR family members are structurally and functionally related proteins found in a diverse range of organisms (6). These large proteins all contain a highly conserved kinase domain at the carboxyl terminus and a central region that displays significant homology. In Mec1, neither of these homologous regions is required for interaction with Pie1. However, the amino-terminal Mec1, which shows no apparent homology to the corresponding region of other ATR family members, was found to be required for interaction with Pie1. Not surprisingly, we also found that the carboxyl terminus of Pie1, which likewise shows no apparent homology to Rad26 and UVSD, is required for interaction with Mec1. It is possible that Rad26 and UVSD interact with the amino terminus of Rad3 and UVSB, respectively. To understand the role in the conserved region of Pie1, we examined whether the pie1-KA mutation has an effect on its interaction with Mec1 or the intracelluar localization of Mec1 and Pie1. However, neither was affected by the pie-KA mutation. These results indicate that this homologous region is essential for functions of Pie1 other than its interaction with Mec1 or intracellular localization. It will therefore be interesting to address other aspects, for example, whether Pie1 might interact with specific DNA structures and/or other checkpoint proteins through this region.
In summary, we identified Pie1 as a protein that interacts physically with Mec1 and showed that Pie1 and Mec1 function as a complex that is required for checkpoint responses and cell growth. Our result also suggests that Pie1 is a homologue of Rad26 and UVSD and that the region conserved among these three proteins is essential for Pie1 function. However, it remains to be determined exactly how Pie1 regulates Mec1 activity. Future work will focus on the interaction of the Mec1-Pie1 complex with specific DNA structures and/or other checkpoint proteins in checkpoint responses.
ACKNOWLEDGMENTS
T.W. and T.K. contributed equally to this work.
We thank T. Naiki for an initial two-hybrid screening; H. Ogawa, P. James, and S. Nishikawa for materials; T. Yoshihisa for technical suggestions and M. Lamphier for critical readings of the manuscript. We also thank John Rouse and Steve Jackson for communicating results prior to publication.
T.K. is a recipient of a JSPS predoctoral fellowship. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and General Research from the Ministry of Education, Science, Sports and Culture of Japan (K.M. and K.S.).
REFERENCES
- 1.AI-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Lehmann A R, Carr A M. Identification of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;4:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Aris J P, Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- 4.Aris J P, Blobel G. Yeast nuclear envelope proteins cross-react with an antibody against mammalian pore complex proteins. J Cell Biol. 1989;108:2059–2067. doi: 10.1083/jcb.108.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 6.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, Demaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky M H, Sekelsky J J, Tsang G, Hawley R S, Rubin G M. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000;14:668–678. [PMC free article] [PubMed] [Google Scholar]
- 8.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 9.Carr A M. Control of cell cycle arrest by Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- 10.Desany B D, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 12.Edwards R J, Bentley N J, Carr A M. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 13.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 14.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 15.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 16.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 18.Hari K L, Santerre A, Sekelsky J J, McKim K S, Boyd J B, Hawley R S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 19.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:229–234. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 20.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 22.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair, and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keegan K S, Holtzman D A, Plug A W, Christenson E R, Brainerd E E, Flaggs G, Bentley N J, Taylor E M, Meyn M S, Moss S B, Carr A M, Ashley T, Hoekstra M F. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 24.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longhese M P, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;17:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 28.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N J. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge S J. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 31.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paciotti V, Clerici M, Lucchini G, Longhese M P. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 33.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouse J, Jackson S P. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5793–5800. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge S J. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 37.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanajali S R, Simmons A, Clines G A, Sartiel A, Gatti R A, Chessa L, Sanal O, Lavin M F, Jaspers N G J, Taylor A M R, Arlett C F, Miki T, Weissman S M, Lovett M, Collins F S, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 38.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith G C M, Jackson S P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 40.Souza C P C D, Ye X S, Osmani S A. Checkpoint defects leading to premature mitosis also cause endoreplication of DNA in Aspergillus nidulans. Mol Biol Cell. 1999;10:3661–3674. doi: 10.1091/mbc.10.11.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 43.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 44.Uchiyama M, Galli I, Griffiths D J F, Wang T S-F. A novel mutant allele of Schizosaccharomyces pombe rad26 defective in monotoring S-phase progression to prevent premature mitosis. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinert T A, Hartwell L H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 48.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 49.Zakian V A. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 50.Zhao X, Muller E G D, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pool. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]