An Essential Role for the Substrate-Binding Region of Hsp40s in Saccharomyces cerevisiae (original) (raw)
Abstract
In addition to regulating the ATPase cycle of Hsp70, a second critical role of Hsp40s has been proposed based on in vitro studies: binding to denatured protein substrates, followed by their presentation to Hsp70 for folding. However, the biological importance of this model is challenged by the fact that deletion of the substrate-binding domain of either of the two major Hsp40s of the yeast cytosol, Ydj1 and Sis1, leads to no severe defects, as long as regions necessary for Hsp70 interaction are retained. As an in vivo test of this model, requirements for viability were examined in a strain having deletions of both Hsp40 genes. Despite limited sequence similarity, the substrate-binding domain of either Sis1 or Ydj1 allowed cell growth, indicating they share overlapping essential functions. Furthermore, the substrate-binding domain must function in cis with a functional Hsp70-interacting domain. We conclude that the ability of cytosolic Hsp40s to bind unfolded protein substrates is an essential function in vivo.
Keywords: Ydj1, Sis1, molecular chaperone, heat shock protein 70, yeast
Introduction
Hsp70s and Hsp40s cooperate in a wide range of cellular processes, including protein translation, translocation across membranes, and folding. Hsp70s bind unfolded or partially unfolded polypeptides in an ATP-regulated cycle (Hartl 1996; Bukau and Horwich 1998; Mayer et al. 2000). Hsp40s stimulate ATP hydrolysis, promoting the ADP-bound form of Hsp70, which has a higher affinity for unfolded protein substrates. In addition, many Hsp40s also bind unfolded polypeptide substrates and are able to prevent their aggregation independently of Hsp70 action. Based on the results of several in vitro studies, the current model of the cycle of Hsp70 and Hsp40 purports that Hsp40 first binds unfolded protein substrates and then transfers them to Hsp70 for folding (Langer et al. 1992; Liberek et al. 1995; Laufen et al. 1999).
Hsp40s are modular proteins. The signature J domain binds the ATPase domain of Hsp70 and is required for stimulating ATP hydrolysis. In class I and II Hsp40s, the J domain is followed by a region rich in glycine and phenylalanine residues (G/F) (Cheetham and Caplan 1998). Class I Hsp40s are distinguished by a cysteine-rich region that has been implicated in substrate binding (Banecki et al. 1996; Szabo et al. 1996). In addition, class I and II Hsp40s contain a poorly conserved COOH terminus that also has been implicated in binding unfolded polypeptides (Lu and Cyr 1998a,Lu and Cyr 1998b).
Multiple Hsp40s exist in both prokaryotic and eukaryotic cells. Here, we focus on two cytosolic Hsp40 chaperones of Saccharomyces cerevisiae, Ydj1 and Sis1 (see Fig. 1) (Caplan and Douglas 1991; Luke et al. 1991). Lack of Ydj1, a class I Hsp40, results in a severe growth phenotype, especially at higher temperatures. Sis1 is an essential type II Hsp40. Some functional overlap exists between these two Hsp40s, as overexpression of SIS1 can partially rescue the growth defect of a ydj1 disruption strain. However, overexpression of YDJ1 does not restore viability to an sis1 disruption strain. Each of these Hsp40s is capable of stimulating the ATPase activity of the abundant essential cytosolic Hsp70, Ssa, and can cooperate with Ssa in refolding denatured luciferase (Lu and Cyr 1998a,Lu and Cyr 1998b).
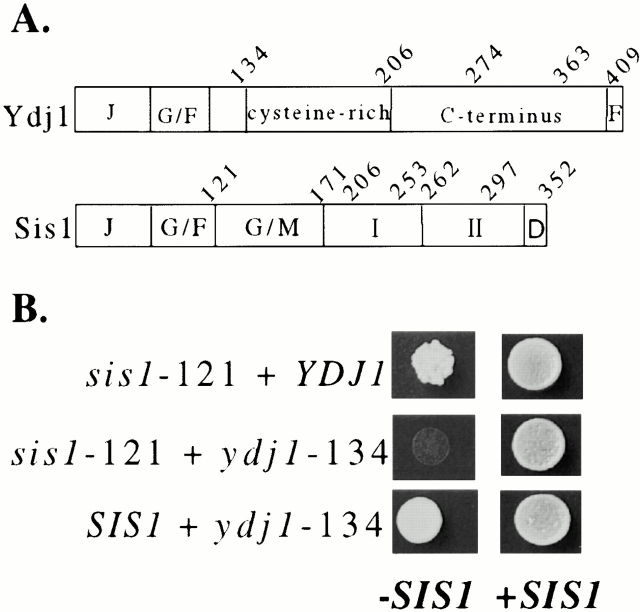
Figure 1.
The COOH terminus of either Sis1 or Ydj1 is essential. (A) Domains of Ydj1 and Sis1. Ydj1: J domain (J), G/F region (G/F), cysteine-rich region (cysteine-rich), COOH terminus (C-terminus), and farnesylation signal (F). Sis1: J domain (J), G/F region (G/F), G/M region (G/M), domains I (I) and II (II) of the COOH terminus, and dimerization domain (D). (B) Strain JJ1146 (_sis1 ydj1/_YCp50:SIS1) was transformed with indicated combinations of plasmids containing wild-type YDJ1 (or SIS1) and truncation mutants _ydj1_-134 (or _sis1_-121). After overnight incubation at 23°C, equal amounts of cells were plated onto selective media to maintain wild-type SIS1 (+SIS1) or media containing 5-FOA to counterselect for _SIS1 (−_SIS1). Plates were incubated at 23°C for 5 d.
Both Ydj1 and Sis1 can bind unfolded polypeptides. A proteolytic fragment of Ydj1 encompassing amino acids (a.a.) 179–384, which contains half of the cysteine-rich region and most of the COOH terminus, has been shown to bind denatured substrates in vitro (Lu and Cyr 1998a). The region of Sis1 responsible for binding denatured substrates also maps to the COOH terminus (a.a. 171–352) (see Fig. 1 A), (Lu and Cyr 1998b). The recent crystal structure of this fragment revealed two similar domains, domain I and domain II, as well as a dimerization domain. Domain I contains a large hydrophobic depression and thus is proposed to contain the peptide-binding site (Sha et al. 2000).
Although it has been demonstrated that Ydj1 and Sis1 can bind substrates in vitro, the importance of this function in vivo is not clear. In a ydj1 disruption strain, expression of the first 134 a.a. of Ydj1, containing the J domain, G/F region, and a short spacer region, restores wild-type growth at the optimal growth temperature of 30°C (Johnson and Craig 2000). Likewise, in an sis1 disruption strain, a strain expressing only the J plus G/F region of Sis1 (Sis1-121) grows as well as a wild-type strain at 30°C and exhibits only a mild temperature–sensitive growth phenotype (Yan and Craig 1999). These results suggest that the ability of either Ydj1 or Sis1 to bind substrate is unnecessary. Therefore, we set out to critically test the requirements for Hsp40 function in vivo, using the cytosol of yeast as a test. Using a strain having chromosomal deletions of both YDJ1 and SIS1, we found that the COOH terminus of either Ydj1 or Sis1 must be present for viability and thus, despite limited sequence homology, share overlapping essential functions.
Materials and Methods
Yeast Strains
Strain JJ160 (ydj1::HIS3) (Johnson and Craig 2000) was crossed to the isogenic strain WY26 (sis1::LEU2) (Yan and Craig 1999) to yield JJ1146 (MAT a _trp1-1 ura3-1 leu2-3,112 his3-11,15 ade2-1 can1-100 met2-_Δ_1 lys2-Δ_2 ydj1::HIS3 sis1::LEU2)/pYW17(_SIS1_-YCp50). Strain JJ261 (MAT_α ade1-100 Δ_trp1 ura3-52 leu2-3,112 ydj1::HIS3 Δ hem1) was constructed by crossing the nonisogenic strains MH1 (MAT a, ura3-52 leu2-112 his4-519 ade1-100 Δ hem1) (Haldi and Guarente 1989) and the ydj1::HIS3 disruption strain JJ257 (Johnson and Craig 2000).
Plasmids and Additional Methods
Wild-type or mutant YDJ1 was supplied on a low copy LYS2 plasmid (Sikorski and Boeke 1991). Wild-type or mutant SIS1 was supplied on a low copy TRP1 plasmid (Yan and Craig 1999). After transformation of indicated plasmids into strain JJ1146, colonies were grown in media containing 5-fluroorotic acid (5-FOA) (USBiological) to counterselect for Ycp50-SIS1. YYS fusions (J plus G/F of Ydj1 fused to the G/M rich region and portions of the COOH terminus of Sis1) have been described (Yan and Craig 1999). The sis1-253 truncation mutant was selected in a genetic screen to isolate sis1 mutants that failed to grow in the absence of the COOH terminus of Ydj1. Sis1-253 levels expressed from a low copy vector were low (data not shown), thus this mutant was expressed on a multicopy vector in these studies. The COOH terminus of Sis1 (a.a. 170–352) was expressed under the control of the GPD promoter in p414GPD (Yan and Craig 1999) and moved to pRS317 as a Sac1-Sal1 fragment. Levels of Sis170-352 expressed from this plasmid were similar to wild-type Sis1 levels (data not shown). Plasmid Ycplac22-N134/ 121 was constructed to coexpress _ydj1_-N134 and _sis1_-121 from the same low copy plasmid (Gietz and Sugino 1988). Western blot analysis of Sis1 proteins using a polyclonal antibody raised against full-length Sis1 was performedas described (Yan and Craig 1999).
Hap1 Activity in ydj1 Mutant Strains
To assay Hap1 activity, strain JJ261 expressing wild-type YDJ1, _ydj1_-104, or the indicated Ydj1–Sis1 fusion protein was transformed with a UAS1/_CYC1_-lacZ reporter plasmid (Guarente et al. 1984). After overnight incubation at 25°C in the presence of high concentrations of δ-aminolevulinic acid (ALA) (250 μg/ml), β-galactosidase assays were performed as described (Johnson and Craig 2000).
Results and Discussion
The COOH Terminus of either Ydj1 or Sis1 Is Essential for Viability
The nonessentiality of the substrate-binding regions of Sis1 and Ydj1 in vivo contrasts with the proposed critical role of Hsp40s in binding denatured substrates before presentation of those substrates to Hsp70. Two possibilities come to mind. Either binding of unfolded protein substrates is not critical for Hsp40 function in the cytosol, or there is significant functional overlap between Sis1 and Ydj1, and either protein can carry out this function.
To test these ideas, we created a strain that contains chromosomal disruptions of both YDJ1 and SIS1. Using combinations of plasmids expressing mutant and wild-type YDJ1 and SIS1 genes, we tested the importance of the COOH-terminal regions of these Hsp40s while maintaining the J plus G/F regions required for interaction with Hsp70. We constructed an sis1 ydj1 strain that carries a wild-type copy of SIS1 on a plasmid harboring URA3 to allow viability. To test for the ability of introduced genes to allow cell growth, transformants were spotted onto plates containing 5-fluoroorotic acid (5-FOA) to select for cells having lost the _URA3_-based plasmid containing SIS1. As expected from previous results (Yan and Craig 1999), a strain carrying a combination of wild-type YDJ1 and sis1-121, encoding the J plus G/F regions, was viable (Fig. 1 B). Similarly, the combination of wild-type SIS1 plus _ydj1_-134, encoding the J plus G/F plus spacer regions, was viable (Johnson and Craig 2000). However, the combination of _sis1_-121 plus _ydj1_-134 did not support viability, even though the regions necessary for Hsp70 interaction were retained. These results indicate that the COOH-terminal substrate-binding region of either Ydj1 or Sis1 is required in vivo, suggesting that Ydj1 and Sis1 may perform overlapping functions in the cell.
Localization of the Essential Region in the COOH Terminus of Sis1
Substrate binding is likely the essential function of the COOH terminus of Sis1, as a proteolytic fragment encompassing a.a. 171–352 is capable of binding denatured substrates in vitro (Lu and Cyr 1998b). Recently, the crystal structure of this fragment revealed three domains (Fig. 2 C). Domains I and II have similar folds. However, domain I contains a prominent hydrophobic depression and thus was proposed to contain the substrate-binding site. In addition, the extreme COOH terminus (a.a. 338–352) is a dimerization domain (Sha et al. 2000).
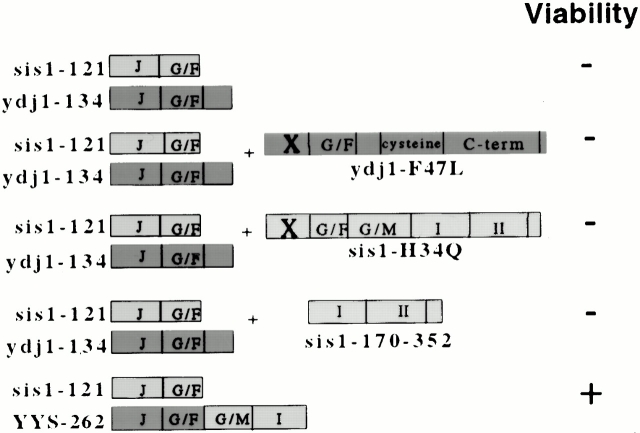
Figure 2.
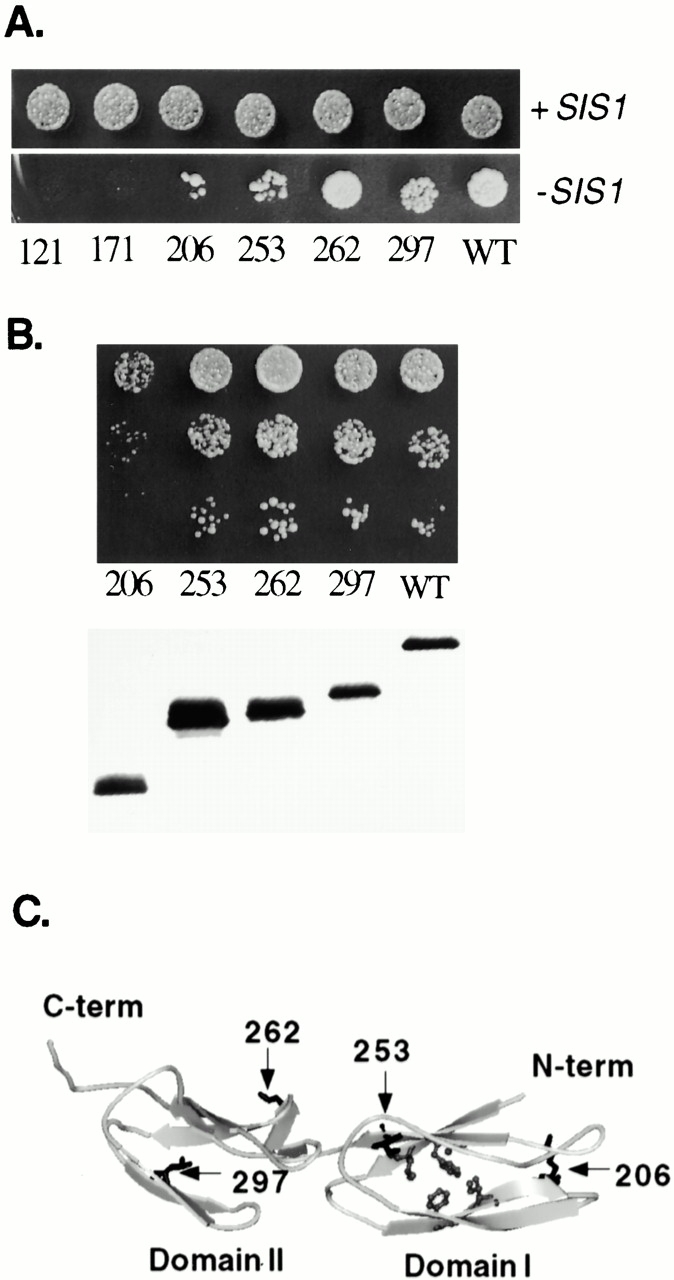
Essential region of the COOH terminus of Sis1. Strain JJ1146 (ydj1 sis1/YCp50:SIS1) expressing _ydj1_-134 was transformed with wild-type SIS1 or sis1 mutants expressing Sis1 fragments of increasing length. (A) After overnight incubation at 23°C, equal amounts of cells were plated onto selective media to maintain wild-type SIS1 (+SIS1) or media containing 5-FOA to counterselect for _SIS1 (−_SIS1). Plates were grown at 23°C for 5 d. (B) Colonies that appeared on 5-FOA plates were grown overnight, and serial dilutions were grown at 30°C for 2 d on selective media (top). Yeast extracts expressing wild-type or mutant Sis1 proteins were separated on a 15% SDS–acrylamide gel, transferred to nitrocellulose, and immunoblotted with an antibody against Sis1 (bottom). (C) Structure of substrate-binding region of Sis1. Sites of truncation mutants are labeled and marked with arrows. Shaded residues are postulated to interact with bound substrate (Sha et al. 2000). Image was generated using MSI WebLab™ ViewerPro. C-term, COOH terminus; N-term, NH2 terminus; WT, wild type.
To determine which sequences in the COOH-terminal fragment of Sis1 are essential in the absence of the COOH terminus of Ydj1, SIS1 truncation mutants were transformed into the sis1 ydj1 strain expressing _ydj1_-134. When transformants were spotted onto plates containing 5-FOA, colonies appeared in strains expressing _sis1_-206, -253, -262, -297, and wild type (Fig. 2 A). However, when cells picked from these colonies were tested more carefully (Fig. 2 B, upper panel), it became apparent that cells expressing Sis1-206 grew slowly at 30°C, even though this truncation is expressed at wild-type levels (Fig. 2 B, lower panel). Cells expressing Sis1-253, -262, or -297 exhibited wild-type growth rates.
These results clearly indicate that domain II (a.a. 260–336) and the dimerization region (a.a. 338–352) of Sis1 are not required for normal growth at 30°C even in the absence of the COOH terminus of Ydj1. However, domain I (a.a. 180–255) is required, as sis1-206 cells grow very slowly, and sis1-171 cells are inviable in this assay. Sha et al. 2000 proposed that, within domain I, residues V184 and L186 from β strand 1, F201, and I203 from β strand 2, and F251 from β strand 5 interact with bound substrate (Fig. 2 C), however, the role of these residues in substrate interaction has not yet been established. Sis1-262 and Sis1-253 contain these residues, whereas Sis1-206 truncates after β strand 2 and thus lacks F251 and β sheets that are probably important in stabilizing the domain. Our results suggest that Sis1-262 contains a functional substrate-binding domain. However, at this time, the only available in vitro assay of Sis1 chaperone function measures the ability to bind and refold denatured luciferase in cooperation with Hsp70 (Sha et al. 2000). Since activity in these in vitro assays is dependent on the presence of the dimerization domain of Sis1, we are unable to monitor the in vitro substrate-binding activity of Sis1-262 in the absence of domain II and the dimerization domain. However, our genetic results demonstrate a critical role of domain I in the absence of the COOH terminus of Ydj1 and are consistent with that role being substrate binding.
Localization of the Essential Regions in the COOH Terminus of Ydj1
Having found the region of Sis1 that is critical in the absence of the COOH terminus of Ydj1, we asked the same question of Ydj1: what portion of Ydj1 is essential in the absence of the COOH terminus of Sis1? The substrate-binding region of Ydj1 has been localized between a.a. 179 and 384 of Ydj1 (Lu and Cyr 1998a), a fragment that contains part of the cysteine-rich region and most of the COOH terminus (Fig. 1 A). The extreme COOH terminus of Ydj1 contains the CAAX box, the site of a farnesylation modification thought to be involved in localization of Ydj1 to membranes (Caplan et al. 1992b). Truncation and point mutants of YDJ1, picked because they disrupt function of these regions, were transformed into the sis1 ydj1 strain that carried wild-type SIS1 on a _URA3_-based plasmid and also expressed sis1-121. The shortest truncation that allowed growth in the absence of full-length Sis1 was _ydj1_-274, which contains not only the J plus G/F region, but also the cysteine-rich region and 68 a.a. of the COOH terminus (Fig. 3 A). When retested for growth at 30°C, it was clear that _ydj1_-274 allowed cells to grow nearly as well as wild-type YDJ1.
Figure 3.
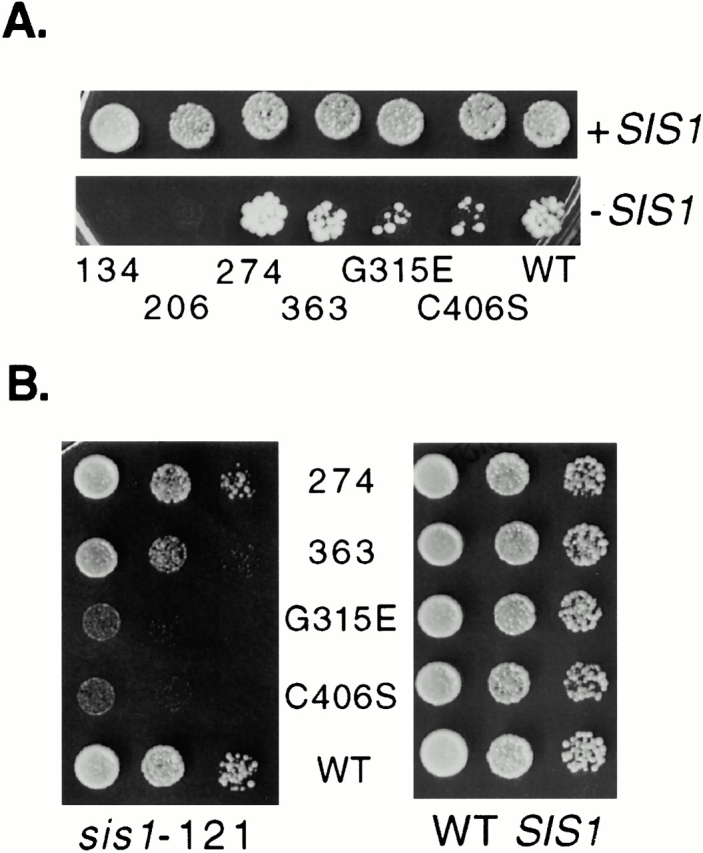
Essential region of the COOH terminus of Ydj1. Strain JJ1146 (ydj1 sis1/YCp50:SIS1) expressing _sis1_-121 was transformed with wild-type YDJ1 or the indicated ydj1 mutants. (A) After overnight incubation at 23°C, equal amounts of cells were plated onto selective media to maintain wild-type SIS1 (+SIS1) or media containing 5-FOA to counterselect for _SIS1 (−_SIS1). Plates were grown at 23°C for 5 d. (B) Colonies that appeared on 5-FOA plate (sis1-121) were grown overnight and serial dilutions were grown at 30°C for 2 d on selective media (left). Growth of ydj1 mutants on selective media after 2 d at 30°C in a ydj1 disruption strain (WT SIS1) (right). WT, wild type.
These results are consistent with substrate binding being a critical role of the region distal to the J plus G/F. Mutant DnaJ proteins having alterations in the cysteine-rich region have shown defects in suppression of aggregation of rhodanese and binding to some, but not all, protein substrates (Banecki et al. 1996; Szabo et al. 1996). Therefore, the cysteine-rich region is implicated in binding of at least some protein substrates. However, a fragment containing only the J plus G/F plus cysteine-rich region of Ydj1 (ydj1-206) did not support viability when sis1-121 was the only copy of SIS1 present. This result suggests that, in addition to the cysteine-rich region, the first 68 a.a. of the COOH terminus are required for the ability to bind substrates in vivo. Thus, our results are in agreement with studies in which a fragment of Ydj1 encompassing a.a. 179–384 bound denatured substrate (Lu and Cyr 1998a).
Other mutants that could be recovered on 5-FOA did not grow as well as ydj1-274 (Fig. 3 B). Although sequences required for substrate binding or other sequences important in allowing proper transient interactions with substrates have not been clearly established, the amino acid substitution G315D led to in vivo defects in binding of glucocorticoid receptor (Kimura et al. 1995). In vitro, G315D was found to be defective both in suppressing rhodanase aggregation and refolding denatured luciferase (Lu and Cyr 1998a). Consistent with a role in substrate interaction, an amino acid substitution at the same residue, ydj1-G315E, exhibited severe growth defects when sis1-121 was the only SIS1 gene present (Fig. 3 B). Although the most straightforward interpretation of these results is that G315 and surrounding residues are important for substrate binding, it is also possible that, in light of the robust growth of ydj1-274, COOH-terminal sequences can act negatively on more proximal residues, inhibiting their ability to function. Consistent with this idea, the truncation mutant ydj1-363, which results in removal of fewer residues from the COOH terminus than ydj1-274, and ydj1-C406S, which alters the farnesylation signal, grow poorly in the presence of sis1-121 compared with the truncation ydj1-274. The C406S mutation has been shown to result in a decrease in the membrane association of Ydj1 (Caplan et al. 1992b). The associated temperature-sensitive growth defect was attributed to this alteration in cellular localization. However, in other proteins, the farnesylation modification has been shown to not only affect cellular localization but also protein–protein interactions (Sinensky 2000); so it is possible that farnesylation has additional effects on Ydj1 function. Further studies will be required to determine how the farnesylation modification of Ydj1 affects in vivo chaperone activity.
The COOH Terminus of Sis1 Can Substitute for the COOH Terminus of Ydj1 In Vivo, but Only in Cis
The results presented above indicate that substrate binding is critical for Hsp40 function. In the current model of interaction between substrates, Hsp40 and Hsp70, substrate first bind the COOH terminus of Hsp40 and then are targeted to Hsp70 via interaction of the J domain with the ATPase domain of Hsp70 (Langer et al. 1992; Liberek et al. 1995; Laufen et al. 1999). The J domain of both Ydj1 and Sis1 have been shown to be important in vivo, as a single amino acid change in either J domain has severe effects on function. In a ydj1 disruption strain, an F47L alteration of Ydj1 causes both severe growth defects and defects in the maturation of Hsp90 substrates (Johnson and Craig 2000). The H34Q mutation of Sis1 disrupts the HPD motif in the J domain and causes a null phenotype in an sis1 disruption strain (Yan and Craig 1999).
We conducted in vivo tests to determine whether a functional J domain and COOH-terminal substrate-binding domain must be part of the same protein (Fig. 4). Growth of an sis1 ydj1 strain expressing _sis1_-121 and _ydj1_-134 could not be rescued by either H34Q Sis1 or F47L Ydj1. Nor did the COOH-terminal fragment of Sis1 (a.a. 170–352) expressed in trans with J plus G/F allow viability. These results indicate that the COOH terminus of Sis1 must be present in the context of a wild-type J domain to function, indicating that close cooperation between the two domains is essential for function in vivo. However, in support of the idea that the COOH-terminal regions can substitute for one another, a fusion protein (YYS-262) that joins the G/M region and domain I of Sis1 to the J plus G/F of Ydj1 (Yan and Craig 1999) allows viability (Fig. 4).
Figure 4.
The COOH terminus must cooperate with a functional J domain in cis. Strain JJ1146 (ydj1 sis1/YCp50:SIS1) expressing sis1-121 and ydj1-134 from Ycplac22-134/121 was transformed with plasmids expressing the indicated sis1 or ydj1 mutant. Viability was assayed by plating cells in the presence of 5-FOA to counterselect for wild-type SIS1.
YYS Fusion Proteins Partially Rescue Hap1 Defect In Vivo
Sis1 and Ydj1 have important functions in the cell, but few of their specific in vivo roles are known. Little is known about Sis1 function, although it has been reported to be required for the initiation of translation (Zhong and Arndt 1993). Ydj1 cooperates with the Hsp70 Ssa in the translocation of preproteins into the ER and mitochondria (Caplan et al. 1992a; Becker et al. 1996). In addition, Ydj1 has been shown to be required for the in vivo maturation of the heterologous proteins v-src and steroid receptors via the Hsp90 pathway (Kimura et al. 1995; Fliss et al. 1999; Johnson and Craig 2000), but native substrates have been elusive. However, recent evidence suggests that the transcription factor Hap1, which regulates the expression of CYC1 and other genes encoding respiratory functions in a heme-dependent manner (Zitomer and Lowry 1992), is such a substrate. When heme levels are low, Ydj1 and the yeast Hsp90, Hsp82, bind Hap1, and Hap1 activity is low. When heme levels increase, Ydj1 and Hsp82 dissociate from Hap1, and its transcriptional activity increases. Hsp82 is believed to play a role in the activation of Hap1, as decreased levels of Hsp8_2_ led to a large decrease in the overall activity of a Hap1 reporter gene construct without affecting Hap1 levels (Zhang et al. 1998).
To determine whether Ydj1 also plays a role in the activation of Hap1, we examined Hap1 activity in ydj1 mutant strains containing a hem1 mutation, which allowed manipulation of heme levels. Varying amounts of the heme precursor, ALA, were added to the media to maintain high or low levels of heme. Hap1 activity was monitored by measuring transcription from the UAS1 of the CYC1 promoter linked to the lacZ gene (Guarente et al. 1984). In low levels of ALA (4 μg/ml), strains expressing mutant or wild-type YDJ1 had similarly low levels of β-galactosidase activity, 1–4 U (data not shown). In the presence of high concentrations of ALA (250 μg/ml), cells expressing wild-type YDJ1 had dramatically higher activity, 343 U. In contrast, cells expressing the J plus G/F region of Ydj1 (_ydj1_-104) had only 30 U of activity (Fig. 5 A), indicating that deletion of the substrate-binding region of Ydj1 causes an activation defect of Hap1, resulting in >10% of the activity observed in wild-type cells.
Figure 5.
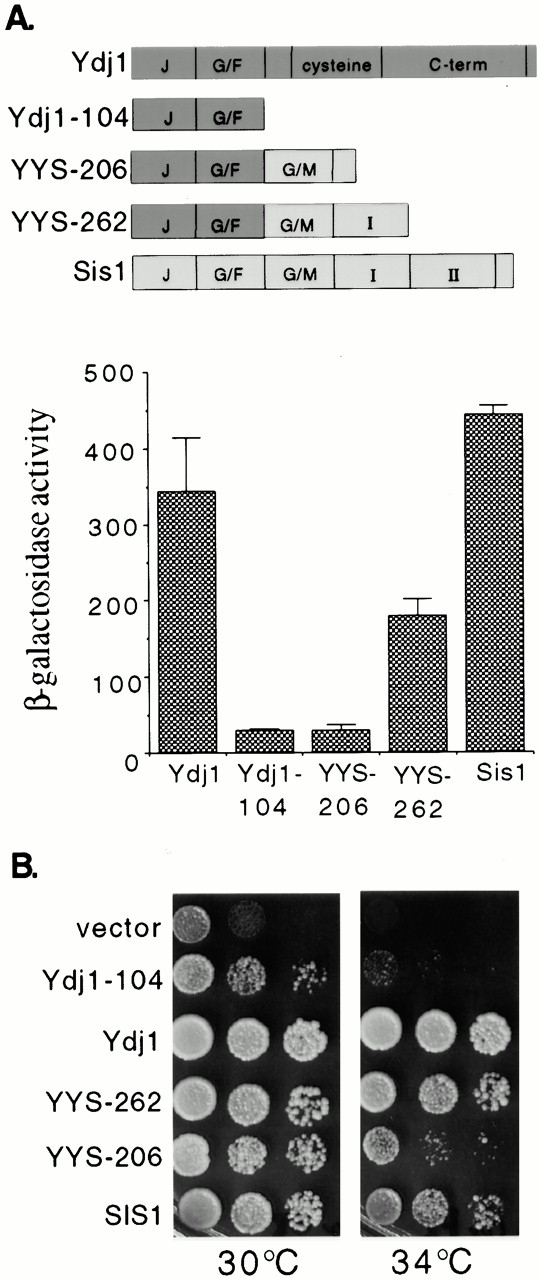
Partial rescue of ydj1 defects by YYS-262. (A) Hap1 was assayed in the ydj1 hem1 disruption strain JJ261 expressing full-length Ydj1, Ydj1-104, YYS-206, YYS-262, or overexpressing wild-type SIS1. β-Galactosidase activity of the UAS1/CYC1-lacZ reporter construct was measured in cells after overnight incubation at 25°C in the presence of high concentrations of ALA (250 μg/ml). (B) The ydj1 disruption strain JJ160 was transformed with plasmids expressing the indicated proteins. After overnight incubation at 25°C, 10-fold serial dilutions of cells were plated on rich media. Plates were grown for 2 d at the indicated temperatures.
Since the substrate-binding region of Ydj1 is required for activity of Hap1, we were able to directly test whether the substrate-binding region of Sis1 is able to replace Ydj1 in this function. For this, we used fusion proteins that join the J plus G/F of Ydj1 to the G/M region and domain I of Sis1 (Fig. 5 A). In the absence of ydj1, expression of SIS1 from a high copy plasmid restored wild-type levels of Hap1 activity. In addition, expression of YYS-262 partially restored Hap1 activity (180 β-galactosidase U), whereas cells expressing YYS-206 exhibited the same level of Hap1 activity as _ydj1_-104. Thus, YYS-262 is able to substantially rescue a specific defect, that is loss of Hap1 activity that arises in the absence of the substrate-binding region of Ydj1.
To determine if YYS-262 could more generally carry out Ydj1 functions, we tested its effect on growth of a ydj1 strain at 30°C and 34°C. As previously reported, the _ydj1_-104 construct partially rescues growth at 30°C but exhibits a severe temperature-sensitive phenotype (Johnson and Craig 2000). YYS-262 allowed near wild-type levels of growth at 30°C and 34°C (Fig. 5 A), but not 37°C (data not shown). YYS-206 could not support growth at 34°C, supporting evidence from Fig. 2 that this truncation of domain I is not fully functional. Thus, in addition to rescuing the Hap1 defect, domain I of the Sis1 COOH terminus can rescue the growth defect that arises in the absence of the substrate-binding region of Ydj1.
Summary
The COOH termini of Sis1 and Ydj1 share overlapping essential roles. In addition, the J domain of Ydj1 may substitute for the J domain of Sis1 (Yan and Craig 1999). In contrast, the essential function of the G/F region of Sis1 cannot be replaced by the G/F region of Ydj1 (Yan and Craig 1999), indicating a unique role of the Sis1 G/F region. Although the substrate binding domains of Sis1 and Ydj1 can substitute for one another under some conditions, they share only 29% sequence identity, compared with 66% sequence identity between the J domains. This difference likely results in some differences in substrate specificity, which may be responsible for the temperature-sensitive phenotype observed in the absence of the substrate-binding region of either Ydj1 or Sis1. Further studies will be needed to understand the specificity of Hsp40 interactions with Hsp70.
Acknowledgments
We thank L. Guarente for strain MH1 and the UAS1/_CYC1_-lacZ plasmid, and Avrom Caplan (Mount Sinai School of Medicine, New York, NY) for the _ydj1_-C406S mutant. In addition we thank Chris Pfund and Nelson Lopez for critical reading of this manuscript and helpful advice.
This work was supported by National Research Service Award 5F32 GM17139 (to J.L. Johnson) and National Institutes of Health grant 5RO1 GM31107 (to E.A. Craig).
Footnotes
Abbreviations used in this paper: 5-FOA, 5-fluroorotic acid; a.a., amino acids; ALA, δ-aminolevulinic acid; G/F, glycine and phenylalanine residues.
References
- Banecki B., Liberek K., Wall D., Wawrzynow A., Georgopoulos C., Bertoli E., Tanfani F., Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E.A. Functional interaction of cytosolic Hsp70 and DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Caplan A.J., Douglas M.G. Characterization of YDJ1a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A.J., Cyr D.M., Douglas M.G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism Cell. 71 1992. 1143 1155a [DOI] [PubMed] [Google Scholar]
- Caplan A.J., Tsai J., Casey P.J., Douglas M.G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in _S. cerevisiae_J. Biol. Chem. 267 1992. 18890 18895b [PubMed] [Google Scholar]
- Cheetham M.E., Caplan A.J. Structure, function and evolution of DnaJconservation and adaption of chaperone function. Cell Stress Chapererones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss A.E., Rao J., Melville M., Cheetham M.E., Caplan A.J. Domain requirements of DnaJ-like (Hsp40) molecular chaperones in the activation of a steroid hormone receptor. J. Biol. Chem. 1999;274:34045–34052. doi: 10.1074/jbc.274.48.34045. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae . Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Haldi M., Guarente L. N-terminal deletions of a mitochondrial signal sequence in yeast. Targeting information of delta-aminolevulinate synthase is encoded in non-overlapping regions. J. Biol. Chem. 1989;264:17107–17112. [PubMed] [Google Scholar]
- Hartl F.U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Johnson J.L., Craig E.A. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 2000;20:3027–3036. doi: 10.1128/mcb.20.9.3027-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Yahara I., Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M.K., Hartl F.U. Successive action of DnaK, DnaJ, and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Laufen T., Mayer M., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. Mechanism of regulation of Hsp70 chaperones by DnaJ co-chaperones. Proc. Natl. Acad. Sci. USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K., Wall D., Georgopoulos C. The DnaJ chaperone catalytically activates the DnaK chaperone to preferentially bind the sigma 32 heat shock transcriptional regulator. Proc. Natl. Acad. Sci. USA. 1995;92:6224–6228. doi: 10.1073/pnas.92.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Cyr D.M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding J. Biol. Chem. 273 1998. 5970 5978a [DOI] [PubMed] [Google Scholar]
- Lu Z., Cyr D.M. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1 J. Biol. Chem. 273 1998. 27824 27830b [DOI] [PubMed] [Google Scholar]
- Luke M., Suttin A., Arndt K. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J. Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P., Schroder H., Rudiger S., Paal K., Laufen T., Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- Sha B., Lee S., Cyr D.M. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Struct. Fold. Des. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Boeke J.D. In vitro mutagenesis and plasmid shufflingfrom cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sinensky M. Recent advances in the study of prenylated proteins. Biochim. Biophys. Acta. 2000;1484:93–106. doi: 10.1016/s1388-1981(00)00009-3. [DOI] [PubMed] [Google Scholar]
- Szabo A., Korszun R., Hartl F.U., Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Yan W., Craig E.A. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol. Cell. Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hach A., Wang C. Molecular mechanism governing heme signaling in yeasta higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol. Cell. Biol. 1998;18:3819–3828. doi: 10.1128/mcb.18.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong T., Arndt K.T. The yeast SIS1 protein, a DnaJ homolog, is required for initiation of translation. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- Zitomer R.S., Lowry C.V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae . Microbiol. Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]