Bap, a Staphylococcus aureus Surface Protein Involved in Biofilm Formation (original) (raw)
Abstract
Identification of new genes involved in biofilm formation is needed to understand the molecular basis of strain variation and the pathogenic mechanisms implicated in chronic staphylococcal infections. A biofilm-producing Staphylococcus aureus isolate was used to generate biofilm-negative transposon (Tn917) insertion mutants. Two mutants were found with a significant decrease in attachment to inert surfaces (early adherence), intercellular adhesion, and biofilm formation. The transposon was inserted at the same locus in both mutants. This locus (bap [for biofilm associated protein]) encodes a novel cell wall associated protein of 2,276 amino acids (Bap), which shows global organizational similarities to surface proteins of gram-negative (Pseudomonas aeruginosa and Salmonella enterica serovar Typhi) and gram-positive (Enteroccocus faecalis) microorganisms. Bap's core region represents 52% of the protein and consists of 13 successive nearly identical repeats, each containing 86 amino acids. bap was present in a small fraction of bovine mastitis isolates (5% of the 350 S. aureus isolates tested), but it was absent from the 75 clinical human S. aureus isolates analyzed. All staphylococcal isolates harboring bap were highly adherent and strong biofilm producers. In a mouse infection model bap was involved in pathogenesis, causing a persistent infection.
Staphylococcus aureus is one of the most important pathogens in humans and animals. The pathogenesis of a particular S. aureus strain is attributed to the combined effect of extracellular factors and toxins, together with the invasive properties of the strain such as adherence, biofilm formation, and resistance to phagocytosis. Despite general agreement that biofilms are the basis for persistent or chronic bacterial infections (8), the understanding of the molecular mechanisms implicated in the biofilm formation process is still growing (39). Two steps appear to be involved in this process: (i) attachment of the bacterial cells to a surface (early adherence) and (ii) growth-dependent accumulation of bacteria in multilayered cell clusters (intercellular adhesion) (18). Different proteins (24, 37), including those of the family of microbial surface components that recognize adhesive matrix molecules (MSCRAMMs) (13), are involved in S. aureus adhesion. Specifically, MSCRAMMs may mediate S. aureus attachment to different cell types (11, 44) and abiotic surfaces once the adhesive host plasma constituents have covered the target surface (10, 29). However, the possible role of known MSCRAMMs or other molecules on the binding of S. aureus to inert surfaces in the absence of host constituents has not been thoroughly studied so far.
The icaABCD cluster, an operon present in Staphylococcus epidermidis and S. aureus (9, 20), participates in the intercellular adhesion step of biofilm formation by encoding proteins involved in the synthesis of the biofilm matrix polysaccharide poly-_N_-succinyl β-1-6 glucosamine (PIA-PNSG). The implication of ica in staphylococcal pathogenesis has been demonstrated in various animal models (30, 40). A relationship has been shown in vivo between the expression of ica in clinical S. epidermidis strains and infection (15). In addition, immunizations with PIA-PNSG efficiently protect against S. aureus infection (30).
Considerable effort has been made by different groups to associate S. aureus biofilm formation with different mechanisms of virulence and pathogenesis. In this context and using highly adherent strains, we have observed that S. aureus in vitro adherence and biofilm formation on inert or mammalian cell surfaces is associated with (i) exopolysaccharide production (6, 23); (ii) rough colony morphology phenotype in Congo red agar (CRA) (6); (iii) higher resistance to phagocytosis (32); (iv) lower susceptibility to antibiotics when forming biofilms (2); (v) higher capacity to attach to different surfaces and biomaterials used in orthopedic surgery, causing osteomyelitis (16); and (vi) higher capacity to colonize the ovine mammary gland, causing mastitis (6). In addition, active immunizations with exopolysaccharides extracted from a highly adherent S. aureus isolate have been shown to trigger protective immunity against mastitis (3). However, the genetic mechanisms underlying these observations have not been determined.
Transposon mutagenesis has been used to determine the genetics of biofilm formation in different bacterial species, including S. epidermidis (18, 33), Streptococcus gordonii (26), Escherichia coli (38), Pseudomonas fluorescens (36), and Pseudomonas aeruginosa (35), using polystyrene microtiter plates as substrate. With this methodological approach, we were able to identify in this study a new protein which is involved in S. aureus attachment to abiotic surfaces and biofilm formation in vitro and enhances infection in vivo.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
In order to find a strong biofilm forming strain, the ability of 350 S. aureus mastitis isolates to attach and form a biofilm on polystyrene microtiter plates was tested as previously described (9). Accordingly, a bovine subclinical mastitis isolate, S. aureus V329, was selected because of its strong biofilm production phenotype and antibiotic susceptibility profile, which facilitates genetic manipulations. This isolate was used to generate a genomic library and to obtain biofilm-negative transposon insertion mutants. S. aureus SA113, a restriction-negative and biofilm producer strain, and the biofilm-defective S. aureus SA113Δ_ica_, which contains a tetracycline resistance cassette that replaces the ica genes, were kindly provided by F. Götz (9). S. aureus RN4220, a restriction-negative strain, unable to form a biofilm, was kindly provided by J. C. Lee. Seventy-five human S. aureus strains and 50 coagulase-negative Staphylococcus strains from animals with bovine mastitis were analyzed for the presence of bap.
Staphylococcal strains were cultured in Trypticase soy agar (TSA) and in trypticase soy broth (TSB) supplemented with glucose (0.25%, wt/vol) when indicated. B2 broth (42) was used for biofilm formation on a glass surface. E. coli strains DH5α and BL21(DE3) were grown in Luria-Bertani medium. Media were supplemented when appropriate with erythromycin (20 μg/ml), ampicillin (50 μg/ml) and chloramphenicol (10 μg/ml for plasmid pBT2 [7] and 20 μg/ml for plasmid pID408 [31]). Plasmid pID408 contains the transposon Tn917, which includes the pBR322 amp or rop region that allows replication and selection of the plasmid in E. coli, and the temperature-sensitive replicon pE194ts and the Cmr gene of pTV32ts that allows replication and selection in S. aureus at 32°C. Plasmid pET-15b (Novagen) was used for protein expression in E. coli. The lambda vector EMBL-4 (Promega) was used to obtain a genomic library.
DNA manipulations.
Routine DNA manipulations were performed using standard procedures (41). Plasmid DNA was isolated from S. aureus strains using a Qiagen plasmid miniprep kit according to the manufacturer's protocol, except that the bacterial cells were lysed by lysostaphin (12.5 μg/ml; Sigma) at 37°C for 2 h before plasmid purification. Plasmids were transformed into staphylococci by electroporation, using a previously described procedure (25) with the following modifications: 10% glycerol was replaced by 0.5 M sucrose, and staphylococcal transformations were enhanced by inducing chloramphenicol acetyl transferase translation with subinhibitory concentrations of chloramphenicol (0.2 μg/ml) after electroporation.
For Southern hybridization, chromosomal DNA was purified as previously described (28), digested with _Eco_RI, and analyzed by agarose gel electrophoresis. Gels were blotted onto nylon membranes (Hybond-N 0.45-mm-pore-size filters; Amersham Life Science) using standard methods (41). A 971-bp PCR fragment (oligonucleotides bap-6m, 5′-CCCTATATCGAAGGTGTAGAATTGCAC-3′ [1807], and bap-7c, 5′-GCTGTTGAAGTTAATACTGTACCTGC-3′ [2777]) of the bap region was used as a DNA probe (numbers in parentheses correspond to the position in the gene of the first nucleotide contained in the PCR fragment). Labeling of the probe and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labeling and chemiluminescence detection kit (Roche).
All the enzymes for DNA manipulation were supplied by MBI Fermentas, Roche, and Amersham Pharmacia Biotech; assays were performed as recommended by the manufacturer. Oligonucleotide primers were purchased from Life Technologies.
Transposon mutagenesis.
S. aureus strain V329 harboring pID408 (V329:pID408) was grown overnight in TSA-chloramphenicol at 30°C. A single colony of V329:pID408 was inoculated in 1 ml of TSB-chloramphenicol (5 μg/ml) and incubated for 1 h at 30°C. Subsequently, 100 μl of this culture were spread on TSA-erythromycin plates and incubated for 18 h at 44°C. Transposon insertion mutants were subcultured on TSA-erythromycin plates. To exclude the possibility of contamination, mutants were compared with the parental strain by coagulase DNA typing (21).
Adherence studies. (i) Early adherence to an inert surface.
Early adherence to a polystyrene surface was determined as previously described (14), with the following modifications. S. aureus strains were grown overnight in TSB supplemented with 0.25% glucose at 37°C. Overnight cultures were adjusted with TSB–0.25% glucose to an optical density at 578 nm (OD578) of 0.1. Ten milliliters of each suspension was added to two polystyrene petri dishes and incubated for 1 h at 37°C. Petri dishes were washed at least five times with phosphate-buffered saline (PBS). Cells were fixed with Bouin solution and Gram stained. Adherent bacterial cells were observed by oil immersion microscopy and counted (results represent the means of four different microscopic fields). Each experiment was repeated three times.
(ii) Biofilm assay.
A late adherence assay was carried out essentially as described elsewhere (18). Briefly, S. aureus strains were grown overnight in TSB at 37°C. The culture was diluted 1:40 in TSB–0.25% glucose, and 200 μl of this cell suspension was used to inoculate sterile 96-well polystyrene microtiter plates (lwaki). After 12 h, medium was replaced, and 12 h later, the wells were gently washed three times with 200 μl of sterile PBS, dried in an inverted position, and stained with 0.1% safranin for 30 s. Wells were rinsed again, and the absorbance was determined at 490 nm (Micro-ELISA Autoreader; Elx800 Bio-Tek instruments). Each assay was performed in triplicate and repeated five times.
Verification of the classification of strains as biofilm producers and nonproducers was carried out by different methods.
(a) Formation of cell aggregates in a cell suspension.
Cells were grown overnight in 5 ml of TSB–0.25% glucose at 37°C and examined macroscopically and microscopically for the presence or absence of aggregates (intercellular adhesion).
(b) Macroscopic observation of biofilm on glass.
Cells were grown in 50 ml of B2 at 37°C, using a glass container, without shaking, for 2 days, and the walls of the container were visually (macroscopically) examined for the presence or absence of a white biofilm layer.
(c) Colonization of other materials.
The capacity to form a biofilm layer was verified using polyvinylchloride (PVC) plastic as a target surface and a phase-contrast microscope (magnification, ×1,000; Nikon Optiphot-2 microscope), as previously described (38).
(d) Colony morphology in CRA.
Colony morphology was determined in CRA as previously described (6, 48), with rough colonies being indicative of biofilm formation (positive result) and smooth colonies being associated with a deficiency in biofilm formation.
PIA-PNSG detection.
PIA-PNSG production in S. aureus was detected as described by Cramton et al. (9). Briefly, cells were grown overnight in TSB supplemented with 0.25% glucose, the optical density was determined, and the same number of cells (2 to 4 ml) from each culture was resuspended in 50 μl of 0.5 M EDTA (pH 8.0). Cells were then incubated for 5 min at 100°C and centrifuged to pellet the cells, and 40 μl of the supernatant was incubated with 10 μl of proteinase K (20 mg/ml; Sigma) for 30 min at 37°C. After addition of 10 μl of Tris-buffered saline (20 mM Tris-HCl, 150 mM NaCl [pH 7.4]) containing 0.01% bromophenol blue, 10 μl was spotted on a nitrocellulose filter using a Bio-Dot Microfiltration apparatus (Bio-Rad), blocked overnight with 5% skim milk in PBS with 0.1% Tween 20, and incubated for 2 h with an anti-S. aureus PIA-PNSG antibody diluted 1:10,000 (30). Bound antibodies were detected with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories, Inc.) diluted 1:10,000, and the Amersham ECL (enhanced chemiluminescence) Western blotting system.
Gene identification.
A previously described method (31) was used to clone the chromosomal DNA flanking the transposon insertion points in mutants with attenuated biofilm production. Briefly, 2.5 μg of S. aureus chromosomal DNA from each mutant was restricted with _Eco_RI, resuspended in 200 μl of ligation buffer (Promega), and self-ligated for 12 h at 16°C. The ligated products were transformed into E. coli DH5α, plated onto Luria-Bertani agar containing ampicillin, and incubated at 37°C overnight. Plasmid DNA was extracted from a single ampicillin-resistant colony using a Qiagen plasmid miniprep kit. Chromosomal DNA sequences flanking the transposon were obtained using primer Tn_917_-3c (5′-AGAGAGATGTCACCGTCAAGT-3′), which corresponds to the inverted repeat region located 70 bp from the erm_-proximal end of Tn_917.
Construction of genomic library.
The lambda vector EMBL-4 was used to construct a genomic library of S. aureus V329 according to the manufacturer's instructions (Promega). Chromosomal DNA of S. aureus V329 was digested with _Eco_RI and ligated into vector EMBL-4 restricted with _Eco_RI. A 200-bp PCR fragment of the bap flanking region was used as a DNA probe (oligonucleotides 556-1m, 5′-CTGTCCATATTTGGACTGTG-3′, and 556-2c, 5′-CTTATAGATGTGCGTAGTC-3′). Labeling of the probe and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labeling and chemiluminescence detection kit (Roche).
DNA sequencing and computer analysis.
A genomic _Hin_dIII fragment including the bap gene was cloned in pBT2 plasmid (pBT2:Bap). The nucleotide sequence was determined by the dideoxy chain termination method, using an ABl 377 model automatic sequencer (PE Biosystems; Foster City, Calif.) at the DNA Sequencing Service of the IBMCP-UPV (Valencia, Spain). For C-repeat sequencing, a genomic DNA _Xba_I-_Eco_RI fragment containing this region was subcloned in pBT2. Nested deletions were generated (Erase-a-base system; Promega). Double DNA sequencing of this region was carried out using the primers pBT2-1c (5′-GGACGATATCCCGCAAGAGGCCCG-3′) and pBT2-2c (5′-GGTGCCGAGGATGACGATGAGCGC-3′), corresponding to the flanking region of the plasmid pBT2 cloning site.
Homology searches were carried out using the BLAST 2.0 program (1) at the NCBl server. The cloned sequence was compared against those in the GenBank database and the publicly available S. aureus genomes (The Institute for Genomic Research, University of Oklahoma, and Sanger Centre).
Complementation studies.
To prove that the biofilm-deficient phenotype of the mutants was due to the disruption of bap, mutants were complemented with plasmid pBT2 carrying a 9.2 kb Hin_dIII fragment from the wild-type strain, including the bap gene under the control of its own promoter (pBT2:Bap). Plasmid pBT2:Bap was transformed by electroporation into S. aureus strains m556, RN4220, SA113, and SA113Δ_ica. Stable expression of Bap was analyzed in total bacterial extracts by Coomassie staining of proteins run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels.
SDS-PAGE and Western blot analysis.
Bacteria were grown overnight in TSB (supplemented with 10 μg of chloramphenicol/ml in the case of complemented strains) at 30°C. Following centrifugation of 1 ml of culture, cells were harvested, washed, and finally resuspended in 75 μl of PBS buffer containing lysostaphin (12.5 μg/ml; Sigma). After 2 h of incubation at 37°C, cells were resuspended in 1 volume of Laemmli buffer and boiled for 10 min. After centrifugation, 20 μl of the supernatant was used for SDS-PAGE (10% separation gel, 4.5% stacking gel) and proteins were stained with Coomassie brilliant blue R250 (0.25%; Sigma).
For Western blot analysis, protein extracts were prepared and analyzed by SDS-PAGE as described above and blotted onto Immobilon P membrane (Millipore). Anti-Bap serum was diluted 1:2,500 with Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 7.5], 150 mM NaCl) and immuno-absorbed with 5% skim-milk. Alkaline phosphatase-conjugated goat anti-rabbit Immunoglobulin G (Sigma) diluted 1:15,000 in TBS–5% skim milk was used and the subsequent chemiluminescence reaction (with CSPD [Roche]) was recorded.
Expression of the N-terminal region of Bap in E. coli.
A 1,919-bp DNA sequence corresponding to nucleotides (nt) 1 through 1919 of the bap gene was amplified by PCR with primers bap-2m (5′-GGGGGGCATATGGGAAATAAACAAGGTTTTTTACC-3′) and bap-3c (5′-GGGGGGATCCCCAACCTCGTCAATGGTTAAGTCAGC-3′) (_Nde_I and _Bam_HI restriction sites are shown in boldface type). The amplified product was restricted with _Nde_I and _Bam_HI and cloned in frame downstream from the His tag sequence in the pET-15b vector. The nucleotide sequence of the cloned bap fragment was verified by sequence analysis. Purified plasmid DNA was used to transform the expression host BL21(DE3), and the fusion protein was produced as specified by the manufacturer (Novagen). After disrupting the cells by sonication, the recombinant protein was purified by immobilized metal affinity chromatography using a cobalt-based resin (Clontech).
Production of rabbit polyclonal antiserum.
Polyclonal antiserum to purified recombinant protein was raised as previously described (43). For the initial dose (day 1), 100 μg of antigen in complete Freund's adjuvant was injected subcutaneously. Booster doses (50 μg) were administered intramuscularly on days 14 and 42 in incomplete adjuvant. Blood was collected from the marginal ear vein at 2-week intervals after booster administration, and antibody titer in serum was determined.
Experimental infection.
A mouse foreign body infection model was used to determine the role of bap in the pathogenesis of S. aureus. A total of 53 adult male mice (Swiss-Albino, B&K Universal, Barcelona, Spain) were used. A 1-cm segment of intravenous catheter (22G1"; B. Braun) was aseptically implanted into the subcutaneous interscapular space. Each group of nine mice was inoculated with 1.5 × 105 CFU of either S. aureus V329 or the Bap-defective S. aureus strain m3591. Three animals were euthanatized by cervical dislocation on days 4, 7, or 10 postinfection. The catheter was aseptically removed, placed in a sterile microcentrifuge tube containing 1 ml of PBS, and vortexed at high speed for 3 min. The number of bacteria was determined by plate count. The experiment was repeated three times.
In each experiment, an extra group of animals was inoculated with vehicle (PBS) and served as a negative control. In addition, to exclude the possibility of contamination, bacteria recovered at the end of the experimental period were compared with the parental strains by coagulase DNA typing (21) and with regard to growth in TSA-erythromycin, CRA colony morphology phenotype, and biofilm formation capacity (see above).
Statistical analysis.
A two-tailed Student's t test was used to determine the differences between groups in biofilm formation. A nonparametric test (Mann-Whitney U test) was used to assess significant differences in bacterial recovery within groups in primary adherence and experimental infection. For analysis of the cure ratio, a two-by-two contingency table was produced and Fischer's exact test was applied. Differences were considered statistically significant when P was <0.05 in all cases.
Nucleotide sequence accession number.
The DNA sequence reported in this article has been deposited in the GenBank nucleotide sequence database under accession number AF288402.
RESULTS
Production and characterization of Tn_917_ mutants.
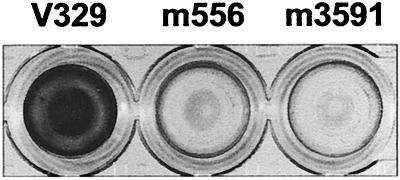
Upon transfer of plasmid pID408 into the adherent S. aureus strain V329, a collection of approximately 4,000 random Emr Cms transposon insertions was screened for their ability to form a biofilm. Two mutants, designated m556 and m3591, had lost the ability to form a biofilm (Fig. 1) and exhibited a growth rate indistinguishable from that of the wild type. Southern hybridization analysis of _Eco_RI-digested chromosomal DNA using a Tn_917_-specific probe revealed that each mutant had a single transposon insertion (data not shown).
FIG. 1.
Biofilm formation phenotype. Differences between the wild-type strain V329 and mutants m556 and m3591 in the capacity to form a 24-h biofilm on polystyrene microtiter plates after staining with safranin. The ELISA plate colors correspond to OD490s of 1.3, 0.17, and 0.18, respectively.
Sequencing of the DNA flanking the transposon insertion sites in both mutants revealed that the transposon insertions disrupted the same gene, but at different positions. When the DNA sequences were compared with those in the GenBank and the S. aureus databases using the BLAST program, no significant similarity with any sequence was found.
The putative gene involved in biofilm formation was cloned from a lamba genomic library as a 16-kb fragment detected by hybridization to the region flanking the transposon insertion site. The nucleotide sequence of this 16-kb fragment revealed that the transposon disrupted an unusually large gene, designated bap (coding for a biofilm-associated protein, Bap), consisting of 6,831 nt and harboring a potential ribosome-binding site (TGAGG) located 9 nt upstream of the ATG initiation codon. The presence of a putative promoter and transcription terminator sequence upstream and downstream of bap suggests that this gene is not part of an operon. Hence, polar effects of the transposon insertion are unlikely. The deduced amino acid sequence of Bap consists of 2,276 amino acids (aa), with a theoretical molecular mass of ≈239 kDa.
Bap expression allows and enhances biofilm formation.
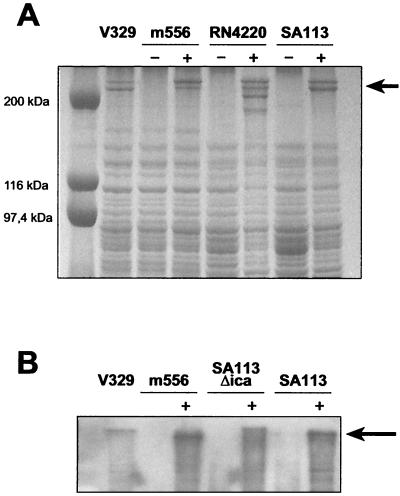
A double band that migrated at a position corresponding to 230 and 240 kDa was reproducibly detected by SDS-PAGE of the total protein extract from the wild-type bacteria but not from the m556 mutant (Fig. 2A). Bands of similar mobility were also detected in S. aureus strains which were complemented with the bap gene in pBT2:Bap. In addition, a band of the expected size was recognized by polyclonal antibodies raised against the first 640 aa of Bap (Fig. 2B), strongly suggesting that the band present in the wild type and complemented strains is the product of the bap gene.
FIG. 2.
(A) SDS-PAGE of protein extracts of wild-type strain V329, m556, RN4220, SA113, and the corresponding complemented strains: m556 pBT2:Bap (m556, lane +), RN4220 pBT2:Bap (RN4220, lane +), and SA113 pBT2:Bap (SA113, lane +). Note that a double protein band of 230 and 240 kDa is present in the wild-type strain as well as in all the strains harboring plasmid pBT2:Bap. (B) Study of the presence of Bap by Western blotting. An approximately 240-kDa band is recognized by polyclonal antibodies against the first 640 aa of Bap only in the wild-type strain V329 and complemented strains: m556 pBT2:Bap (m556, lane +), SA113Δ_ica_ pBT2:Bap (SA113Δica, lane +), and SA113 pBT2:Bap (SA113, lane +).
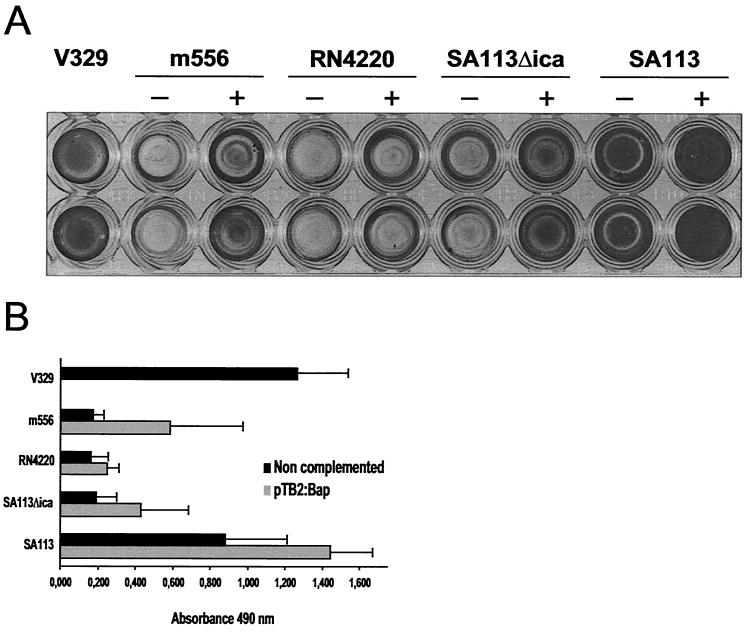
Analysis of the capacity to form a biofilm (on a polystyrene surface after 24 h) demonstrated that all strains expressing Bap, including S. aureus SA113, a strain whose biofilm formation has been related to the ica product (PIA-PNSG), showed an enhanced capacity to form a biofilm, implicating Bap in this process (Fig. 3). Significant differences in adherence were detected between the parental strain and its isogenic mutant, as well as between noncomplemented and complemented strains (P < 0.01). It is important to note that the capacity for biofilm formation was not completely restored in the complemented m556 mutant, although the expression level of the protein was similar to that in the wild type.
FIG. 3.
pBT2:Bap complementation studies involving biofilm formation in biofilm-defective mutant m556, non-biofilm-forming strain RN4220, and biofilm-forming strain SA113 and its ica_-defective mutant SA113Δ_ica. Biofilm formation capacity differences correspond to 24-h biofilm formed on polystyrene microtiter plates after staining with 0.1% safranin. The ELISA plates and mean OD490s obtained are shown. (A) ELISA wells corresponding to wild type V329, m556 (−) and m556 pBT2:Bap (+), RN4220 (−) and RN4220 pBT2:Bap (+), SA113Δ_ica_ (−) and SA113Δ_ica_ pBT2:Bap (+), and SA113 (−) and SA113 pBT2:Bap (+). (B) Mean optical density. Bars represent the mean values, and error bars represent the standard errors of the means (n = 5). Significant differences in adherence (P < 0.01) were noted between complemented and noncomplemented strains, as well as between the wild type and isogenic mutants.
Bap is involved in primary adherence and intercellular adhesion.
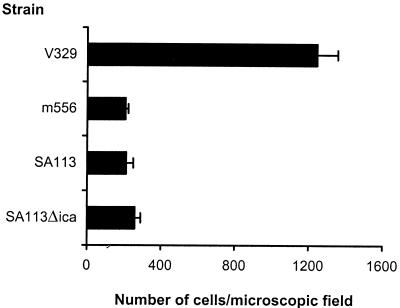
Primary adherence of S. aureus strains V329, m556, SA113, and SA113Δ_ica_ is illustrated in Fig. 4. S. aureus V329 adhered to polystyrene much more efficiently than the isogenic bap mutant m556 and strains SA113 and SA113Δ_ica_. In addition, the presence of cell-to-cell clusters was observed in V329 and SA113 cultures (incubated overnight in TSB–0.25% glucose) by phase-contrast microscopy (data not shown), but not in the mutant m556 and SA113Δ_ica_ cultures. Accumulation of cell aggregates at the bottom of the tube was macroscopically observed only in the case of wild type strains. These results strongly suggest that Bap is not only involved in intercellular adhesion and accumulation in multilayered cell clusters, as the product of the ica operon does, but also in primary attachment to an abiotic surface.
FIG. 4.
Primary attachment assay. Significant (P < 0.01) differences were detected between wild-type strain V329 and isogenic mutant m556. No differences were found between S. aureus SA113 and isogenic mutant SA113Δ_ica_. Bars represent the mean values, and error bars represent the standard errors of the means (n = 3).
Relationship between Bap and PIA-PNSG.
To determine Bap–PIA-PNSG interaction we tested PIA-PNSG production in Bap+ and Bap− strains. The strain V329 showed a low level of in vitro PIA-PNSG production. Similar results were obtained whith other natural Bap+ strains (data not shown). Inactivation of the bap gene in the mutant strain m556 reduced the levels of PIA-PNSG (Fig. 5). Complementation of the PIA-PNSG-producing strain SA113 with the bap gene strongly increased PIA-PNSG accumulation (Fig. 5). As expected, PIA-PNSG was no longer produced in the ica knockout strain SA113Δ_ica_ (Fig. 5).
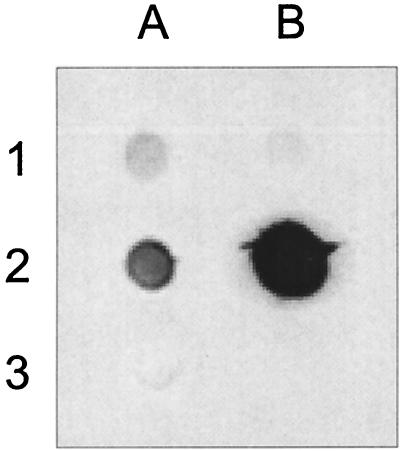
FIG. 5.
Dot blot analysis of S. aureus PIA-PNSG acummulation induced by Bap expression. S. aureus V329 (blot A1) and m556 (blot B1) produced low levels of PIA-PNSG. Complementation of S. aureus SA113 with pBT2-Bap plasmid strongly increased the levels of PIA-PNSG (blot B2) with respect to the wild-type strain (blot A2). PIA-PNSG was not detected in S. aureus SA113Δ_ica_ (blot A3).
Further phenotypic characterization of biofilm-defective mutants.
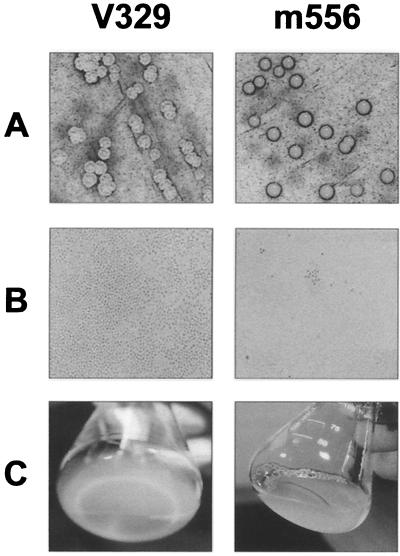
Consistent with the results on polystyrene microtiter plates, the wild-type strain (V329) grown on CRA exhibited a rough colony morphology typically associated with biofilm producers, whereas mutants m556 and m3591 produced a smooth colony morphology commonly found in non-biofilm-producing strains (Fig. 6A). Phase-contrast microscopic observation of late adherence to PVC plastic discs showed that the wild-type strain produced multiple layers of cells almost completely covering the PVC surface (Fig. 6B). In contrast, very few cells of the biofilm-defective mutant were attached to PVC plastic. Macroscopic examination of biofilms in a glass container revealed that upon 2 days of culture, the wild-type strain formed an obvious biofilm on the glass surface, but the mutants did not (Fig. 6C).
FIG. 6.
Phenotypic differences between the wild-type strain V329 and the defective mutant m556. (A) CRA colony morphology; (B) capacity to form a 24-h biofilm on PVC plastic, as observed by phase-contrast microscopy (magnification, ×1,000); (C) capacity to form a 48-h biofilm on the surface of a glass container (visual observation).
Structural features of Bap protein.
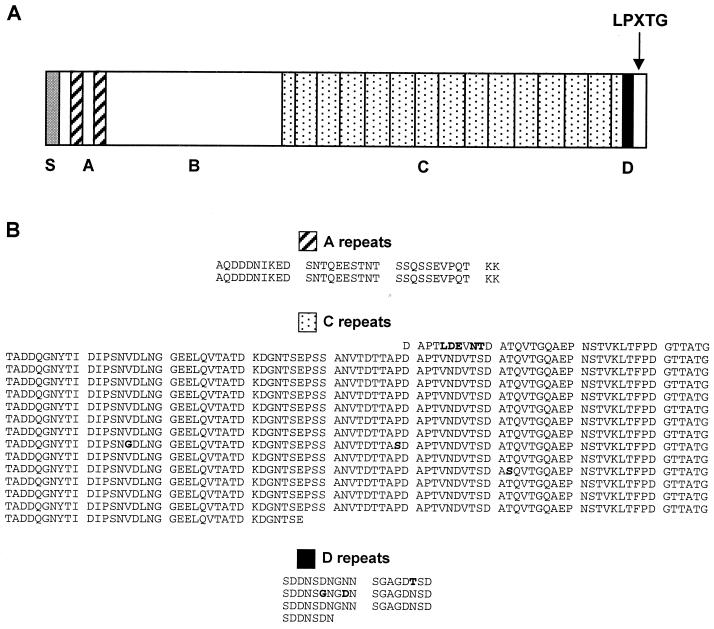
Analysis of the Bap primary amino acid sequence revealed the presence of a typical gram-positive amino-terminal signal sequence for extracellular secretion (first 44 aa) and a putative carboxy-terminal segment containing an LPXTG motif and a hydrophobic membrane-spanning domain followed by a series of positively charged residues typical of cell-wall-anchored surface proteins of gram-positive bacteria (34) (Fig. 7A). Following the putative signal peptide, the N-terminal region of Bap can be divided into two regions. Region A (aa 45 to 360) contains two short repeats of 32 aa (repeats A1 and A2) separated by 26 aa (Fig. 7B). BlastP searches of this region revealed no significant similarity scores among GenBank sequences. Region B, the remaining part of the N-terminal domain (aa 361 to 818), exhibited a significant similarity with an Enterococcus faecalis surface protein (Esp) (43), which is mostly found in infection-derived isolates.
FIG. 7.
(A) Structural analysis of Bap. S represents the signal sequence. The positions of the LPXTG motif and the A, B, C, and D domains are shown. Region A (aa 45 to 360) contains two short repeats of 32 aa (A1 and A2) separated by 26 aa. Region B (aa 361 to 818) represents the remaining part of the N-terminal domain. A spacer region (aa 819 to 947) followed by region C (aa 948 to 2139) consisting of 258-nt tandem repeats constitutes the central domain of Bap. The carboxy-terminal region of Bap consists of region D (aa 2148 to 2208), which contains three short repeats of 18 aa followed by an incomplete D repeat, and the LPXTG motif. (B) Alignment of the amino acid residues within repeat blocks A, C, and D of the Bap protein. Substitutions are shown in boldface type.
The central region of Bap (aa 819 to 2147) begins with a spacer region (aa 819 to 947) followed by 13 nearly identical 258-nt tandem repeat units encoding reiterations of an 86-aa sequence (C repeats, aa 948 to 2139) (Fig. 7A). The C-repeat region begins with a partial C-repeat sequence corresponding to the last 37 aa of a C repeat and ends with a partial C-repeat sequence corresponding to the 37 first aa of a C repeat (Fig. 7B). The C-repeat region accounts for 52% of the Bap protein, and each repeat shows high sequence identity. BlastN searches of the 258 nt encoding a C repeat reveals a strong similarity with a partial GenBank sequence (_Pst_I S. simulans, G. Thumm and F. Götz, accession number U66881), while BlastP searches of the C repeats exhibited high similarity with extracellular proteins from different species, including P. aeruginosa (E value: e-120; accession number AAG05263), Pseudomonas putida (E value: 3e-94; accession number AF182515), Synechocystis sp. (E value: 6e-94; accession number D63999), Salmonella enterica serovar Typhi (E value: 1e-82; accession number AF139831), and Enterococcus faecalis (E value: 5e-81; accession number AF034779).
The carboxy-terminal region of Bap comprises the D region and the LPXTG motif (Fig. 7A). The D region (aa 2148 to 2208) consists of three short repeats of 18 aa followed by an incomplete repeat comprising the first 7 aa of the D repeats (Fig. 7B). LPXTG is a consensus cell wall anchor motif found in most wall-associated surface proteins of gram-positive bacteria (34). BlastP searches of the D region revealed no significant similarity scores among GenBank sequences.
When the Bap protein was compared with products of S. aureus genomes using the BLAST program, no significant similarity with any sequence was found.
Distribution of the bap gene among staphylococcal species.
PCR amplification and Southern blot analysis using specific probes for the bap gene revealed that bap is present in only 5% of the 350 S. aureus bovine mastitis isolates tested. The presence of bap could not be detected in any of the 75 human S. aureus isolates studied. All the strains harboring bap were strong biofilm producers. Sequences that hybridized with the bap probe were present in 10% of the 50 coagulase-negative Staphylococcus isolates tested from animals with bovine mastitis (data not shown).
Experimental infection.
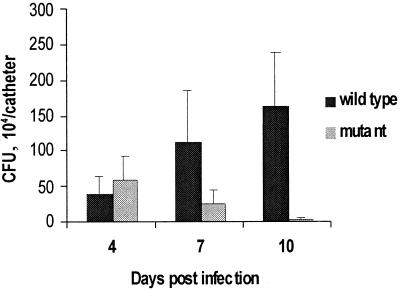
Bap contributed to the pathogenesis of S. aureus in the murine catheter-induced infection model (Fig. 8). Although at day 4 postinoculation differences between wild-type and mutant strains in the number of recovered bacteria per catheter were non significant, at day 7 postinoculation, the number of recovered bacteria (CFU) was 1.2 × 106 and 2.8 × 105 for the wild-type and mutant strains, respectively (P > 0.05). This difference increased by day 10, when the values were 1.7 × 106 and 3 × 104 CFU, respectively (P < 0.05).
FIG. 8.
Recovery of S. aureus V329 and its isogenic Bap mutant m3591 from implanted subcutaneous catheter in a mouse foreign body infection model. Bars represent the mean of CFU collected from catheters, and the error bars represent the standard errors of the means (n = 9). At day 10, differences between wild type and mutant were detected (P < 0.05). Bacteria were not detectable in control animals at the end of the experimental period.
The analysis of the cure ratio (number of animals infected/number of animals inoculated) (Table 1) showed that animals infected with the wild-type strain developed a persistent infection (six of nine animals infected at day 10). However, animals inoculated with the mutant strain were more capable of eliminating the infection (two of nine animals infected at day 10). These results indicate that Bap is an important factor determining the persistence of infection.
TABLE 1.
Analysis of cure ratio at various days postinoculation
| Bacteria | Cure ratioa | |||
|---|---|---|---|---|
| Day 4 | Day 7 | Day 10 | Totalb | |
| Wild type | 6/9 | 6/8 | 6/9 | 18/26 |
| Bap-deficient mutant | 4/9 | 5/9 | 2/9 | 11/27 |
DISCUSSION
In S. epidermidis, several surface proteins involved in biofilm formation have been described, three of which (SSP1, SSP2, and the AtlE autolysin [19, 45]) contribute to the primary attachment, and the other, a 140-kDa protein mediating intercellular adhesion, has been proposed as essential for accumulation of sessile bacteria on glass or polystyrene surfaces (22). The absence of homology among these proteins strongly suggests that staphylococci may use different approaches to form a biofilm. To our knowledge, data presented in this report describe the first S. aureus surface protein (Bap) directly involved in biofilm formation on abiotic surfaces in the absence of host plasma constituents.
To identify Bap, we have used the standard biofilm assay on microtiter plates for the screening of transposon mutants unable to adhere to the polystyrene surface. Surprisingly, in contrast to similar studies with other microorganisms like P. aeruginosa (35), E. coli (38), and Vibrio cholerae (47), where several genes were found to be involved in the biofilm formation process, this assay allowed identification of only two mutants in which the transposon affected the same 6,831-bp open reading frame designated bap.
The bap gene displayed little sequence similarity with known genes. However, the 2,276-aa bap product displayed an organizational similarity with an outer membrane protein of P. putida involved in adhesion to seeds (12) and with a surface protein of E. faecalis (Esp) of unknown function (43), but whose presence is highly correlated with the ability of this bacteria to produce a biofilm on abiotic surfaces (our unpublished results). The most remarkable feature of Bap is the presence of an extensive repeat region, in which the repeats are identical even at the nucleotide level. Although the biological function of the repeats has not been established, it is tempting to speculate that this region could serve to project the amino-terminal part of the protein from the cell surface and to promote interaction with abiotic surfaces, host cell components, or other bacteria. Analogous long repeats have been described in the alpha C protein of group B streptococci (46), where addition or deletion of repeat units leads to the expression of variant proteins which allow bacteria to escape the host immune response (27). Similar pathogenic mechanisms might occur in different Bap+ strains, in which a variable number of C repeats can be found in the structure of the bap gene.
Primary attachment, intercellular aggregation, and biofilm formation studies showed that Bap promotes primary attachment as well as the second step of biofilm formation, where intercellular adhesion plays an important role. In this context, since complementation of S. aureus SA113 with bap resulted in a significantly increased ability to form a biofilm and PIA-PNSG accumulation, it is tempting to speculate that PIA-PNSG and Bap may cooperatively affect cell-to-cell aggregation during the biofilm formation process. A possible explanation of the observation that biofilm formation capacity was not completely restored in the complemented m556 mutant is that the insertion of the transposon in mutant m556 resulted in the production of a truncated protein lacking the last 15 aa, and therefore the protein would be unable to anchor to the cell wall. The soluble truncated Bap protein may cover the abiotic surface and compete with the cell-associated Bap for the interaction with the abiotic surface.
CRA has been used to discriminate by colony morphology between biofilm and non-biofilm-forming strains of S. aureus (2, 6) and S. epidermidis (48), since they produce rough and smooth colonies, respectively. Congo red interacts with several proteins and proteinaceous fibrillar structures, such as curli fimbriae from E. coli (17) and the type III secretion machinery of Shigella flexneri (4). In the case of bap_-harboring S. aureus, loss of the Bap protein resulted in transformation of the rough colony morphology (of the wild-type V329 strain) into the smooth colony morphology (of the m556 mutant). A similar phenotypic variation has been described in S. epidermidis strains deficient in the production of PIA-PNSG (the ica product) (48) and S. aureus strain SA113Δ_ica (our unpublished results). When the expression of bap was restored in complemented strains, the rough phenotype appeared, suggesting the implication of Bap in this phenotypic variation. Why the deficiency in either the polysaccharide intercellular adhesin (PIA-PNSG) or a surface protein (Bap) results in the same phenotypic change is unknown.
Animal models have been useful to study the importance of different genes in the pathogenesis and virulence of S. aureus (30, 31). In this report, we used a mouse foreign body infection model to evaluate the role of Bap and the resulting biofilm formation process in the pathogenesis of S. aureus. Differences between the Bap-deficient mutant S. aureus m3591 and the parental strain in the capacity to colonize the catheter became more obvious at late stages of infection (by day 10), when the mutant strain showed a decreased persistence relative to the wild type. In the closely related species S. epidermidis, the expression of specific bacterial cell surface components appears to hinder the interaction of particular bacterial cell receptors with host proteins (5). In our model, the catheter may have become rapidly coated in vivo by host proteins after implantation and Bap might have hindered the interaction between bacterial receptors and the host proteins on the catheter. This may explain why a Bap-deficient mutant may be more prevalent than the wild-type strain at the initial stages of infection (up to day 4). Later on (days 7 to 10), biofilm formation would be strongly enhanced by Bap as infection proceeds, by promoting cell-to-cell interactions, bacterial accumulation, and persistence of infection.
In conclusion, this study describes a novel protein of S. aureus involved in biofilm formation on abiotic surfaces. Attachment to abiotic surfaces might not be necessarily related to attachment to biotic surfaces. In fact, there are examples that support the idea that bacterial biofilm formation can proceed through divergent pathways, depending on whether or not bacteria settle on a surface or in an environment that can provide nutrients (47). Other results demonstrate that the same factors may be involved in attachment to both types of surfaces (12). Probably, during the development of infections such as subclinical mastitis, biofilm formation could be an efficient way of persisting in the microenvironment of the udder, where shear forces arise during milking (6). The presence of Bap in S. aureus strains responsible for subclinical mastitis suggests that Bap may enhance intramammary adherence and biofilm formation, leading to the inefficacy of antibiotic treatment against biofilm bacteria and chronicity. Further studies to demonstrate this hypothesis are warranted.
ACKNOWLEDGMENTS
We express our gratitude to F. Götz for providing us strains S. aureus SA113 and S. aureus SA113Δ_ica_, D. McKenney for the anti-S. aureus PIA-PNSG antibody, J. C. Lee for strain RN4220, R. Brückner for plasmid pBT2, D. W. Holden for plasmid pID408, L. Baldassarri and José Leiva for human S. aureus strains, and C. Peris for bovine Staphylococcus sp. strains. We also thank E. Grau for sequencing assistance and J. Saus for his support and helpful discussions.
This work was supported by grant BIO99-0285 from the Comisión Interministerial de Ciencia y Tecnología and grants from the Cardenal Herrera-CEU University and from the Departamento de Educación y Cultura del Gobierno de Navarra. Fellowship support for Carme Cucarella and Cristina Solano from the Cardenal Herrera-CEU University and from the Departamento de Educación y Cultura del Gobierno de Navarra, respectively, is gratefully acknowledged.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amorena B, Gracia E, Monzón M, Leiva J, Oteiza C, Pérez M, Alabart J L, Hernández-Yago J. Antibiotic susceptibility assay for Staphylococcus aureus in biofilms developed in vitro. J Antimicrob Chemother. 1999;44:43–55. doi: 10.1093/jac/44.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Amorena B, Albizu I, Baselga R. Use of liposome-immunopotentiated exopolysaccharide as a component of an ovine mastitis staphylococcal vaccine. Vaccine. 1994;12:243–249. doi: 10.1016/0264-410x(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani F K, Sansonetti P J, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun. 1997;65:4005–4010. doi: 10.1128/iai.65.10.4005-4010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldassarri L, Donelli G, Gelosia A, Simpson A W, Christensen G D. Expression of slime interferes with in vitro detection of host protein receptors of Staphylococcus epidermidis. Infect Immun. 1997;65:1522–1526. doi: 10.1128/iai.65.4.1522-1526.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baselga R, Albizu I, de la Cruz M, del Cacho E, Barberán M, Amorena B. Phase variation of slime production in Staphylococcus aureus: implications in colonization and virulence. Infect Immun. 1993;61:4857–4862. doi: 10.1128/iai.61.11.4857-4862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 8.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Cramton S E, Gerke C, Schnell N F, Nichols W W, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmi M, Vaudaux P, Lew D P, Vasey H. Role of fibronectin in staphylococcal adhesion to metallic surfaces used as models of orthopaedic devices. J Orthopaed Res. 1994;12:432–438. doi: 10.1002/jor.1100120316. [DOI] [PubMed] [Google Scholar]
- 11.Dziewanowska K, Patti J M, Deobald C F, Bayles K W, Trumble W R, Bohach G A. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect Immun. 1999;67:4673–4678. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinosa-Urgel M, Salido A, Ramos J L. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 2000;182:2363–2369. doi: 10.1128/jb.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster T J, Höök M. Surface proteins adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 14.Fournier B, Hooper D C. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182:3955–3964. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frebourg N B, Lefebvre S, Baert S, Lemeland J F. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J Clin Microbiol. 2000;38:877–880. doi: 10.1128/jcm.38.2.877-880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gracia E, Fernández A, Conchello P, Laclériga A, Paniagua L, Seral F, Amorena B. Adherence of Staphylococcus aureus slime-producing strain variants to biomaterials used in orthopaedic surgery. Int Orthopaed. 1997;21:46–51. doi: 10.1007/s002640050116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 18.Heilmann C, Gerke C, Perdreau-Remington F, Götz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 21.Hookey J V, Richardson J F, Cookson B D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iturralde M, Aguilar B, Baselga R, Amorena B. Adherence of ruminant mastitis Staphylococcus aureus strains to epithelial cells from ovine mammary gland primary culture and from a rat intestinal cell line. Vet Microbiol. 1993;38:115–127. doi: 10.1016/0378-1135(93)90079-m. [DOI] [PubMed] [Google Scholar]
- 24.Jönsonn K, McDevitt D, McGavin M H, Patti J M, Höök M. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J Biol Chem. 1995;270:21457–21460. doi: 10.1074/jbc.270.37.21457. [DOI] [PubMed] [Google Scholar]
- 25.Lee J C. Electrotransformation of staphylococci. Methods Mol Biol. 1995;47:209–216. doi: 10.1385/0-89603-310-4:209. [DOI] [PubMed] [Google Scholar]
- 26.Loo C Y, Corliss D A, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madoff L C, Michel J L, Gong E W, Kling D E, Kasper D L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci USA. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmur J. A procedure for isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 29.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 30.McKenney D, Pouliot K L, Wang Y, Murthy V, Ulrich M, Döring G, Lee J C, Goldmann D A, Pier G B. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 31.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 32.Monleón E, Pacheco M C, Luján L, Bolea R, Luco D F, Vargas M A, Alabart J L, Badiola J J, Amorena B. Effect of in vitro Maedi-Visna virus infection on adherence and phagocytosis of staphylococci by ovine cells. Vet Microbiol. 1997;57:13–28. doi: 10.1016/s0378-1135(97)00080-1. [DOI] [PubMed] [Google Scholar]
- 33.Muller E, Hübner J, Gutierrez N, Takeda S, Goldmann D A, Pier G B. Isolation and characterization of transposon mutants of Staphylococcus epidermidis deficient in capsular polysaccharide/adhesin and slime. Infect Immun. 1993;61:551–558. doi: 10.1128/iai.61.2.551-558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 37.Palma M, Haggar A, Flock J I. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J Bacteriol. 1999;181:2840–2845. doi: 10.1128/jb.181.9.2840-2845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 39.Pratt L A, Kolter R. Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol. 1999;2:598–603. doi: 10.1016/s1369-5274(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 40.Rupp M E, Ulphani J S, Fey P D, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Schenk S, Laddaga R A. Improved method for electroporation of Staphyloccocus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 43.Shankar V, Baghdayan A S, Huycke M M, Lindahl G, Gilmore M S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha B, François P P, Nüβe O, Foti M, Hartfort O M, Vaudaux P, Foster T J, Lew D P, Herrmann M, Krause K. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol. 1999;1:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 45.Veenstra G J, Cremers F F, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wastfelt M, Stalhammar-Carlemalm M, Delisse A M, Cabezon T, Lindahl G. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem. 1996;271:18892–18897. doi: 10.1074/jbc.271.31.18892. [DOI] [PubMed] [Google Scholar]
- 47.Watnick P I, Fullner K J, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziebuhr W, Krimmer V, Rachid S, Lößner I, Götz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]