Local action of long-range repressors in the Drosophila embryo (original) (raw)
Abstract
Previous studies have identified two corepressors in the early Drosophila embryo: Groucho and dCtBP. Both proteins are recruited to the DNA template by interacting with short peptide motifs conserved in a variety of sequence-specific transcriptional repressors. Once bound to DNA, Groucho appears to mediate long-range repression, while dCtBP directs short-range repression. The short-range Krüppel repressor was converted into a long-range repressor by replacing the dCtBP interaction motif (PxDLSxH) with a Groucho motif (WRPW). The resulting chimeric repressor causes a different mutant phenotype from that of the native Krüppel protein when misexpressed in transgenic embryos. The different patterning activities can be explained on the basis of long-range silencing within the hairy 5′ regulatory region. The analysis of a variety of synthetic transgenes provides evidence that Groucho-dependent long-range repressors do not always cause the dominant silencing of linked enhancers within a complex _cis_-regulatory region. We suggest a ‘hot chromatin’ model, whereby repressors require activators to bind DNA.
Keywords: CtBP/Drosophila/embryo/Groucho/long-range repression
Introduction
Complex enhancers direct stripes and bands of gene expression in the early Drosophila embryo. These enhancers are typically 300 bp–1 kb in length and contain clustered binding sites for transcriptional activators and repressors (e.g. Hoch et al., 1992; Ip et al., 1992; Small et al., 1992). Different enhancers can work independently of one another within a common _cis_-regulatory region to direct composite patterns of gene expression. For example, the seven-stripe even-skipped (eve) expression pattern is activated by five separate enhancers located 5′ and 3′ of the transcription unit (Stanojevic et al., 1991; Small et al., 1992, 1996; Fujioka et al., 1999). The ability of these enhancers to function in an autonomous fashion depends on short-range transcriptional repressors that work over distances of <100 bp to inhibit, or quench, upstream activators (Gray et al., 1994). The binding of the Krüppel repressor to the stripe 2 enhancer does not interfere with the activity of the stripe 3 enhancer since Krüppel mediates repression only when positioned near upstream activators (Small et al., 1993; Gray and Levine, 1996). Consquently, Krüppel quenches Bicoid activators within the stripe 2 enhancer without interfering with the D-Stat activators bound to the stripe 3 enhancer (Small et al., 1996; Yan et al., 1996).
There are several short-range repressors in the early embryo, including Krüppel, Snail, Knirps and Giant (Gray et al., 1994; Arnosti et al., 1996; Gray and Levine, 1996; Hewitt et al., 1999). Most or all of these repressors interact with a common corepressor protein, dCtBP (Nibu et al., 1998a,b; Poortinga et al., 1998), which is the Drosophila homolog of a human protein that was found to attenuate the oncogenic activities of the adenovirus E1A protein (Sollerbrant et al., 1996; Schaeper et al., 1998). dCtBP is maternally expressed and ubiquitously distributed throughout early embryos. A variety of studies suggest that the dCtBP corepressor protein is recruited to the DNA template by interacting with a conserved sequence motif contained in most or all sequence-specific short-range repressors: PxDLSxK/R/H (Nibu et al., 1998a,b). There is emerging evidence that mammalian CtBP proteins also function as corepressors, although it is not known currently whether the mammalian repressors (e.g. bKLF, Ikaros and ZEB-1) only function over short distances (Turner et al., 1998; Koipally et al., 2000; Postigo et al., 2000).
A number of repressors can work when positioned far from upstream activators and the core promoter. For example, the binding of the Hairy repressor to a modified rhomboid lateral stripe enhancer (NEE) can cause the dominant silencing of a linked mesoderm-specific enhancer, even when the two enhancers are separated by >1 kb in the 5′ cis -regulatory region (Barolo and Levine, 1997). Hairy interacts with a second ubiquitous corepressor protein, Groucho (Paroush et al. 1994). Hairy– Groucho interactions depend on a conserved sequence motif at the Hairy C-terminus: WRPW (Fisher et al., 1996). These studies suggest that the dCtBP corepressor protein mediates short-range repression, while Groucho mediates long-range repression. The present study provides additional support for this possibility.
The long-range action of the Groucho corepressor poses a potential problem with regard to enhancer autonomy in complex promoter regions. In principle, the binding of a Groucho-dependent repressor could result in the dominant silencing of all enhancers located in the 5′ and 3′ regulatory regions of a target gene. This imposes a potentially severe constraint on the evolution of complex patterns of gene activity. To investigate this issue, we have examined the activities of chimeric repressor proteins that contain the DNA-binding domains of the short-range Krüppel or Snail repressors and the Groucho interaction sequences in the long-range Hairy repressor. These chimeric repressors were expressed in specific regions of transgenic embryos using defined, heterologous enhancers. The Krüppel–Hairy fusion protein causes altered patterns of segmentation gene expression that are consistent with the notion that Hairy–Groucho interactions convert Krüppel into a long-range repressor. However, the abnormal rhomboid expression pattern obtained with a similar Snail–Hairy fusion protein suggests that it does not function as a dominant silencer, but instead causes the local repression of a single enhancer. The subsequent analysis of a number of synthetic transgenes provides direct evidence that the long-range Hairy repressor does not always cause the dominant silencing of linked enhancers. We propose that repressors require linked activators to function and, consequently, long-range repressors can sometimes function in a short-range fashion.
Results
Krüppel–Hairy and Snail–Hairy chimeric repressors were expressed in transgenic embryos. The entire Krüppel coding sequence (composed of 502 codons) was fused in-frame with the hairy 3′ coding region (codons 255–337) in an effort to convert Krüppel into a long-range repressor (Figure 1). The Hairy sequence includes the Groucho interaction motif, WRPW, and a weak dCtBP motif, PLSLVIK (Poortinga et al., 1998). The strong dCtBP interaction motif in Krüppel, PEDLSMH, was mutagenized in order to circumvent potential competition between the dCtBP and Groucho corepressors (Zhang and Levine, 1999). The resulting Krüppel–Hairy fusion protein was expressed in the ventral mesoderm of transgenic embryos using a modified PE enhancer from the twist promoter region (Jiang et al., 1991; Jiang and Levine, 1993). A similar Snail–Hairy fusion protein was misexpressed using a modified eve stripe 2 enhancer (Kosman and Small, 1997; Figure 1). In both cases, an FRT–stop–FRT cassette was inserted between the promoter and coding region in order to block the expression of potentially deleterious proteins that might cause dominant lethality and prevent the selection of transgenic strains (Struhl and Basler, 1993). Once the lines were established, the stop cassette was removed by crossing the transgene into a strain that expresses the FLP recombinase under the control of a testis-specific tubulin promoter (Kosman and Small, 1997).
Fig. 1. Summary of expression vectors. Krüppel and snail coding sequences were placed under the control of heterologous twist and eve enhancers, respectively. The twist enhancer (‘twi’ in the diagrams) corresponds to two tandem copies of a modified 250 bp PE enhancer sequence that contains optimal Dorsal operator sites and Twist bHLH-binding sites. Two tandem copies of a modified eve stripe 2 enhancer (‘st2’ in D) were used to misexpress Snail. An FRT–stop–FRT cassette was inserted between the promoter and coding region to circumvent lethality resulting from the misexpression of these regulatory genes. (A) The wild-type Krüppel coding region was placed under the control of the modified twist enhancer. The encoded protein is composed of 502 amino acids and contains a dCtBP corepressor interaction motif, PEDLSMH, at position 464. (B) Same as (A) except that the Krüppel coding region was mutagenized to disrupt the dCtBP motif. The first three amino acids were converted into alanines. This protein is unable to bind dCtBP in vitro and has only limited repressor function in vivo (Nibu et al., 1998b). (C) Same as (B) except that a C-terminal portion of the Hairy repressor was attached in-frame to the 3′ end of the Krüppel coding region. The Hairy sequence contains the weak dCtBP-binding motif, PLSLVIK, and also contains the strong Groucho corepressor-binding motif, WRPW. (D) The snail–hairy fusion gene was placed under the control of the eve stripe 2 enhancer. Both dCtBP motifs in the snail coding sequence were left intact. The same portion of the Hairy protein as used in (C) was attached to the 3′ end of the snail coding region.
The Krüppel–Hairy fusion protein mediates long-range repression
The twi–Krüppel–hairy transgene causes the fusion of the fifth to eighth abdominal segments, whereas the twi–Krüppel transgene results in the fusion of the sixth and seventh abdominal segments (data not shown). A number of segmentation genes were examined to determine the basis of the distinct patterning activities of the chimeric and native repressors.
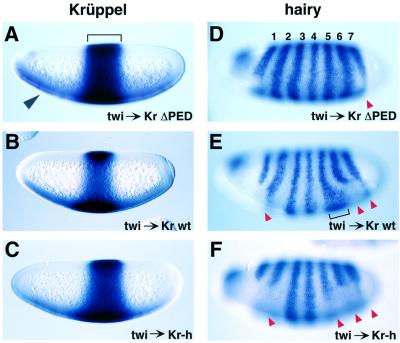
hairy is expressed in a series of seven transverse stripes along the length of the embryo (Howard et al., 1988). There is also a patch of staining near the anterior pole (e.g. Figure 2D). The first stripe is repressed in ventral regions prior to the onset of gastrulation. The twi–Krüppel transgene lacking the dCtBP interaction motif (PEDLSMH; see Figure 1) leads to a slight alteration in the hairy expression pattern. Stripes 5, 6 and 7 are sometimes shifted, and there may be a slight reduction in the expression of stripe 7 (arrowhead, Figure 2D). The twi–Krüppel transgene containing the wild-type Krüppel coding region causes a far more severe alteration in the hairy expression pattern (Figure 2E). Stripes 2 and 6 are consistently repressed in ventral regions (arrowheads). There is also an expansion of stripe 5 (bracket). These changes in stripes 2, 5 and 6 appear to require the dCtBP interaction motif since the wild-type and mutant transgenes exhibit similar levels of expression in ventral regions (Figure 2A–C).
Fig. 2. Misexpression of Krüppel alters the hairy expression pattern. Embryos were collected from the indicated transgenic strains and hybridized with either a digoxigenin-labeled Krüppel antisense RNA probe (A–C) or a hairy probe (D–F). The different Krüppel coding sequences exhibit similar levels of expression in ventral regions of transgenic embryos. Note that these ectopic patterns are considerably weaker than the endogenous Krüppel pattern, which is seen as a broad band in central regions. Activated forms of the transgenes were obtained from transgenic males carrying the FLP recombinase under the control of the testis-specific tubulin promoter. (A and D) Krüppel and hairy expression patterns, respectively, in transgenic embryos carrying the mutant Krüppel coding region lacking the dCtBP repression domain (Figure 1B). The hairy expression pattern is essentially normal, although the spacing between stripes is sometimes irregular, and there may be a slight reduction in the ventral expression of stripe 7 (red arrowhead in D). The repression of hairy stripe 1 in ventral regions is also observed in wild-type embryos (not shown). (B and E) Krüppel and hairy expression patterns, respectively, in transgenic embryos carrying the wild-type Krüppel coding sequence. There is consistent ventral repression of stripes 2, 6 and 7 (red arrowheads). Stripe 5 sometimes exhibits an expanded pattern, possibly due to the repression of the endogenous Giant repressor (not shown). (C and F) Krüppel and hairy expression patterns, respectively, in transgenic embryos carrying the Krüppel–hairy fusion gene. There is a consistent repression of stripe 5, in addition to stripes 2, 6 and 7, as seen with the wild-type Krüppel transgene (red arrowheads in F; compare with E). Stripe 5 is not repressed by the wild-type Krüppel transgene.
Previous studies have shown that low levels of Krüppel are sufficient to repress the stripe 6 enhancer, while higher concentrations are required to repress stripe 5 (Langeland et al., 1994). The twi–Krüppel transgene directs the expression of relatively low levels of Krüppel repressor in ventral regions (Figure 2A–C). These low levels may be sufficient to bind the stripe 6 enhancer, but are insufficient to bind the low-affinity operator sites within the stripe 5 enhancer. Once bound to the stripe 6 enhancer, Krüppel functions as a short-range repressor and does not influence other enhancers in the hairy 5′ regulatory region (see Discussion).
The twi–Krüppel–hairy transgene results in the consistent repression of stripe 5 in addition to stripes 2, 6 and 7 (arrowheads, Figure 2F). Unlike the native Krüppel protein, it would appear that the Krüppel–Hairy fusion protein does not work solely within the limits of the stripe 6 enhancer, but instead functions over long distances to repress the stripe 5 enhancer and/or core promoter. Repression of stripe 5 does not appear to depend on augmented levels of the Krüppel–Hairy fusion product since the transgene is expressed at about the same levels as the wild-type transgene (Figure 2C; compare with B).
A Snail–Hairy fusion protein does not mediate dominant repression
The rhomboid expression pattern was used to distinguish between short- and long-range repression along the dorsoventral axis of the early embryo (Figure 3). rhomboid is expressed in two different cell types: the neurogenic ectoderm and the amnioserosa (Bier et al., 1990). These two patterns are probably controlled by separate enhancers. Lateral stripes in the neurogenic ectoderm are regulated by a 300 bp enhancer, the NEE, located ∼1.7 kb upstream of the rhomboid transcription start site (Ip et al., 1992). Expression in the amnioserosa depends on _cis_-regulatory sequences located either 5′ of –2.2 kb or >2 kb downstream of the rhomboid transcription unit (Bier et al., 1990).
Fig. 3. A Snail–Hairy fusion protein mediates local repression. Wild-type and transgenic embryos were hybridized with either a digoxigenin-labeled rhomboid or snail RNA probe. Embryos are oriented with anterior to the left and dorsal up. (A) snail expression pattern in a wild-type embryo. Staining is restricted to ventral regions that will invaginate to form the mesoderm. (B) snail expression pattern in a transgenic embryo carrying the snail–hairy fusion gene under the control of the eve stripe 2 enhancer (see Figure 1). Staining is detected in both the ventral mesoderm and an ectopic stripe (arrowhead). (C) rhomboid expression pattern in a wild-type embryo. In this lateral view, staining is detected in both ventral regions (the lateral neurogenic stripes) and in the dorsal ectoderm. (D) rhomboid expression pattern in a transgenic embryo carrying the st.2–snail–hairy fusion gene. The ectopic stripe of snail–hairy expression creates a gap in the neurogenic stripes (arrowhead). However, staining in the dorsal ectoderm is not affected (arrow).
A Snail–Hairy fusion protein was misexpressed in transgenic embryos using the eve stripe 2 enhancer (Figure 3B; summarized in Figure 1). These embryos exhibit an abnormal rhomboid expression pattern, including a gap in the neurogenic pattern in the vicinity of stripe 2 (arrowhead, Figure 3D). This gap probably results from the binding of the ectopically expressed Snail–Hairy repressor to the rhomboid NEE.
Despite the fact that the Snail–Hairy fusion repressor contains the same region of Hairy that mediates long-range repression by the Krüppel–Hairy fusion protein, only the NEE appears to be repressed; the amnioserosa pattern is unaffected (arrow, Figure 3D, compare with C). A similar pattern of rhomboid repression was obtained with a stripe 2–snail transgene that expresses the wild-type, short-range Snail repressor (Nibu et al., 1998a). Thus, it would appear that the Snail–Hairy fusion protein does not cause the dominant silencing of amnioserosa regulatory sequences.
Hairy can permit enhancer autonomy
The failure of Snail–Hairy to mediate long-range repression within the _rhomboid cis_-regulatory region was unexpected since the Krüppel–Hairy fusion protein appears to work over long distances within the hairy locus (Figure 2 ). Perhaps the Snail–Hairy fusion protein is somehow defective due to competitive interactions between the strong dCtBP interaction motifs in the Snail moiety and the WRPW motif in Hairy (Zhang and Levine, 1999). Alternatively, it is possible that long-range repressors can sometimes permit enhancer autonomy. To investigate this issue, we examined the ability of the native Hairy repressor to work over long distances within synthetic _cis_-regulatory regions containing defined enhancers (Figure 4).
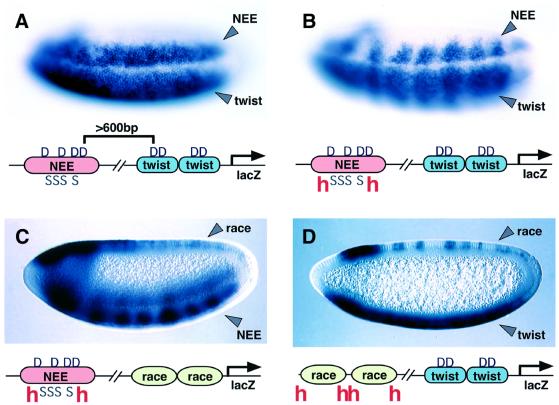
Fig. 4. Local action of the Hairy repressor. Transgenic embryos express the indicated lacZ reporter genes (see diagrams below embryos). Embryos were stained with a digoxigenin-labeled lacZ antisense RNA probe and are oriented with anterior to the left. (A) Lateral view of an embryo expressing the NEE-2×PE (twist) transgene. A composite lacZ staining pattern is observed, including lateral stripes in the neurogenic ectoderm (arrowhead ‘NEE’) and expression in the ventral mesoderm (arrowhead ‘twist’). The binding of the short-range Snail repressor to the NEE (indicated by ‘S’ in the diagram) does not interfere with the expression of the twist enhancer. (B) Same as (A) except that the 5′ NEE was modified to include two synthetic Hairy repressor sites (‘h‘, see diagram). The Hairy repressor mediates periodic repression of the NEE pattern (arrowhead ‘NEE’). The binding of Hairy to the modified NEE also causes periodic repression of the mesoderm pattern (arrowhead ‘twist’). (C) lacZ staining pattern obtained with a transgene that contains the modified h-NEE-h enhancer placed 5′ of the race enhancer. As seen in (B), Hairy represses the modified NEE (arrowhead ‘NEE’), but fails to repress the race staining pattern in the dorsal ectoderm (arrowhead ‘race’). (D) lacZ staining pattern obtained with a transgene that contains tandem copies of a modified race enhancer placed 5′ of the twist enhancer sequences. The h-race-h enhancer exhibits periodic stripes of repression, presumably due to the binding of the Hairy repressor (arrowhead ‘race’). However, the binding of Hairy to h-race-h does not influence the staining pattern directed by the linked twist enhancer (arrowhead ‘twist’).
The rhomboid NEE was placed 340 bp upstream of the 2×PE twist enhancer (Jiang et al., 1991; Pan et al., 1991; Jiang and Levine, 1993), and attached to a lacZ reporter gene (see diagram below Figure 4A). An additive lacZ staining pattern is observed in transgenic embryos (Figure 4A). There is staining in both the lateral neurogenic ectoderm (arrowhead ‘NEE’) and the ventral mesoderm (arrowhead ‘twist’). Snail repressor bound to the distal NEE (Figure 4A, ‘S’ in the diagram) does not interfere with the ventral expression mediated by the proximal 2×PE. As shown previously (Barolo and Levine, 1997), this staining pattern is altered when two synthetic Hairy repressor sites are placed within the distal NEE (Figure 4B; see diagram). The rhomboid lateral stripes exhibit periodic sites of repression that coincide with the hairy expression pattern (arrowhead NEE, Figure 4B). The ventral mesoderm staining pattern also exhibits periodic repression (arrowhead ‘twist’), which suggests that the binding of Hairy to the modified NEE represses both enhancers.
Additional assays were performed with a similar lacZ transgene except that the proximal 2×PE was replaced with two tandem copies of the minimal 533 bp race enhancer (see diagram below Figure 4C). The race enhancer directs weak, uniform expression in the presumptive amnioserosa in the dorsal-most regions of early gastrulating embryos (Rusch and Levine, 1997; e.g. Figure 4C). Both the race and the rhomboid amnioserosa patterns appear to be regulated by a Dpp–Smad signaling pathway that is activated only in the dorsal ectoderm (e.g. Rusch and Levine, 1997; Ashe and Levine, 1999; reviewed by Podos and Ferguson, 1999). The modified NEE containing the synthetic Hairy sites directs abnormal lateral stripes that exhibit periodic repression along the anteroposterior axis (Figure 4C, arrowhead ‘NEE’). However, the race staining pattern is uniform in the amnioserosa (Figure 4D, arrowhead ‘race’), thereby raising the possibility that Hairy does not always silence linked enhancers. Alternatively, the activators that bind the race enhancer may be insensitive to repression by Hairy. To distinguish these possibilities, Hairy-binding sites were inserted in the race enhancer.
The resulting transgene contains two copies of the modified race enhancer placed 5′ of tandem twist enhancer sequences (see diagram, Figure 4D). The lacZ staining pattern directed by race exhibits periodic sites of repression (arrowhead ‘race’). This altered pattern (compare with Figure 4C) suggests that Hairy binds to the modified race enhancer and represses the associated activators. The irregularities in the pattern of repression probably reflect the varying widths of the hairy expression stripes in dorsal versus ventral regions of early embryos (e.g. Figure 2D). The twist enhancers direct uniform expression of the lacZ reporter gene in ventral regions (Figure 4D, arrowhead ‘twist’). This suggests that the binding of Hairy to the race enhancer does not lead to the dominant silencing of the linked twist enhancers. In contrast, a comparable modified h-NEE-h enhancer mediates efficient repression of twist (Figure 4B, arrowhead ‘twist’).
Discussion
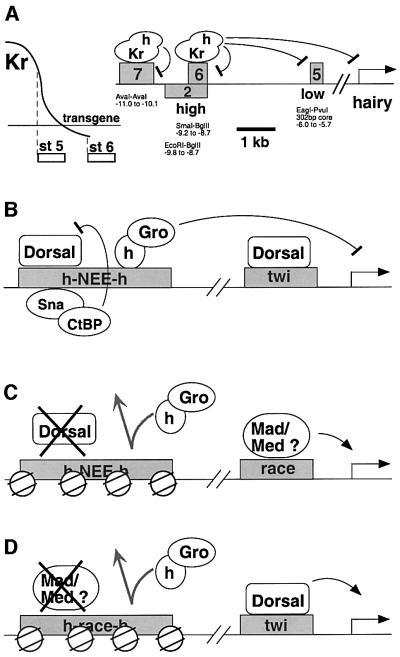
A comparison of the altered patterns of hairy expression obtained with the twi–Krüppel and twi–Krüppel–hairy transgenes provides evidence that dCtBP and Groucho mediate short- and long-range repression, respectively. The twi–Krüppel transgene causes the repression of hairy stripe 6, but not stripe 5. Previous studies have shown that the stripe 6 enhancer contains optimal, high-affinity Krüppel operator sites that can be occupied by the low levels of Krüppel produced in ventral regions by the twi–Krüppel transgene (Langeland et al., 1994). These low levels appear to be insufficient to bind the low-affinity sites within the hairy stripe 5 enhancer and, consequently, the native Krüppel protein works as a short-range repressor to inhibit stripe 6 expression without affecting stripe 5 expression (Figure 5A). In contrast, the twi–Krüppel–hairy transgene leads to the repression of both stripes 5 and 6. The binding of the Krüppel–Hairy fusion repressor to the stripe 6 enhancer appears to cause the dominant silencing of the neighboring stripe 5 enhancer over a distance of ∼2.5 kb in the hairy 5′ regulatory region (Figure 5A). An implication of these observations is that different repression domains exert distinct influences on embryonic patterning. Replacing the PxDLSxH motif (native Krüppel) with WRPW (Krüppel–Hairy) changes the regulatory activity of the Krüppel repressor.
Fig. 5. Summary of the hot chromatin model. The diagrams in (B–D) present the hypothetical on/off states of several enhancers located in different positions along the dorsoventral axis of the early embryo. (A) Summary of the hairy 5′ _cis_-regulatory region. hairy stripes 2, 5, 6 and 7 are regulated by four different enhancers in the 5′-flanking region. The stripe 5 and stripe 6 enhancers are separated by >2 kb. Previous studies (Langeland et al., 1994) have shown that the stripe 6 enhancer contains a series of high-affinity Krüppel-binding sites. Consequently, low levels of the Krüppel repressor gradient (see diagram on left) are sufficient to bind these operator sites and repress stripe 6 expression. The stripe 5 enhancer is active in regions containing low and intermediate levels of Krüppel since the enhancer contains low-affinity sites that are occupied only by high con centrations of the Krüppel protein. The repression of hairy stripe 5by the Krüppel–Hairy fusion protein (Figure 2) suggests that the binding of this repressor to the stripe 6 enhancer can work over a long range to repress the stripe 5 enhancer. (B) Summary of the synthetic h-NEE-h–2×PE (twi) transgene. The Snail repressor normally binds to the NEE and represses its activity in the ventral mesoderm. Snail recruits the dCtBP corepressor and works locally, within the limits of the NEE, and does not interfere with the activity of the linked twi (2×PE) enhancer. In contrast, the Hairy repressor recruits Groucho, and the Hairy–Groucho complex not only represses the modified NEE, but can also work over a distance to repress the linked twi enhancer. (C) Summary of the h-NEE-h–2×race transgene. Hairy fails to repress the race enhancer in dorsal regions of transgenic embryos. We propose that the h-NEE-h enhancer might be inaccessible for the binding of Hairy in dorsal regions where the race enhancer is active. The NEE is activated by the maternal Dorsal gradient. There is no Dorsal activator present in the dorsal ectoderm and, consequently, the h-NEE-h and NEE enhancers might not be available for binding Hairy (or Snail–Hairy) in these regions where the race and rhomboid amnioserosa enhancers are active. (D) Summary of the h-race-h–twist transgene. The race enhancer is probably activated by Mad and Medea, and other transcription factors that are restricted to the dorsal ectoderm (reviewed by Podos and Ferguson, 1999). The h-race-h enhancer might be condensed in ventral regions due to the absence of these activators. As a result, Hairy is unable to bind the modified race enhancer in ventral regions where the twist enhancer is expressed.
The Snail–Hairy fusion protein represses the rhomboid lateral stripes, but fails to repress the amnioserosa pattern. In contrast, the same Hairy repression domain permits Krüppel to function as a dominant silencer within the hairy 5′ regulatory region. There are several possible explanations for the failure of the Snail–Hairy repressor to silence rhomboid expression in the amnioserosa. Perhaps there is competition between dCtBP bound to the Snail moiety and Groucho bound to the Hairy moiety within the fusion protein (see Figure 1). The Krüppel–Hairy fusion protein was mutagenized to eliminate the dCtBP motif (PEDLSMH), whereas the Snail–Hairy fusion protein retains both dCtBP sequences. Previous studies suggest that the conversion of the weak dCtBP interaction motif near the Hairy C-terminus, PLSLVIK, into an optimal motif, PLDLSIK, disrupts the repressor function of an otherwise normal Hairy protein (Zhang and Levine, 1999). This result was taken as evidence that the dCtBP and Groucho corepressors interfere with one another when bound to closely linked motifs within the Hairy C-terminus. An argument against this explanation for the behavior of the Snail–Hairy fusion protein stems from the observation that the binding of Hairy to a modified NEE is sufficient to repress a linked mesoderm enhancer (twist PE), but not a similarly spaced race enhancer (Figure 4). Similarly, the binding of Hairy to a modified race enhancer fails to silence the mesoderm enhancer.
We propose that Hairy can only bind active or ‘open’ enhancers (summarized in Figure 5). The NEE is activated by the maternal Dorsal nuclear gradient (Ip et al., 1992) and, consequently, it might contain activator proteins in both ventral and lateral regions of early embryos (Figure 5B). As a result, the binding of Hairy to the modified h-NEE-h enhancer can lead to the dominant silencing of a linked mesoderm enhancer (twist PE). In contrast, there is no Dorsal activator in dorsal regions of the early embryo, thereby rendering the h-NEE-h enhancer in a closed or condensed state (Figure 5C). This absence of activator might preclude the binding of Hairy so that the race enhancer is not silenced. Similarly, the race enhancer is probably activated by transcription factors that are restricted to dorsal regions, such as Zen and Smads (Figure 5D; Rusch and Levine, 1997). These activators are absent in ventral regions and, consequently, Hairy may be unable to bind the h-race-h enhancer and silence linked enhancers such as the twist PE.
The altered pattern of hairy expression caused by the Krüppel–Hairy fusion protein can be interpreted in the context of this ‘hot chromatin’ model (Figure 5A). There is evidence that hairy stripes 5, 6 and 7 are activated by a posterior gradient of the Caudal activator (Hader et al., 1998). The binding of the Krüppel–Hairy fusion protein to the optimal Krüppel operator sites in the stripe 6 enhancer would be expected to silence the neighboring stripe 5 enhancer due to the open conformation of the stripe 6 enhancer in those regions of the embryo where stripe 5 is expressed. Thus, the Caudal activator might bind to both enhancers in the position of stripe 5, thereby rendering the stripe 6 enhancer accessible to the Krüppel–Hairy fusion protein.
The dependence of repressors on activators might restrain long-range repressors and permit enhancer autonomy. This dependence might reflect the inherent properties of activators and repressors. Some activators recruit enzymes that decondense chromatin, and this may be essential for the binding of repressors in vivo (e.g. Mannervik et al., 1999). Short-range repression has been put forward as an important mechanism for enhancer autonomy (e.g. Small et al., 1993; Gray et al., 1994). We suggest that a second mechanism involves the reliance of repressors on activators for binding to target enhancers.
Materials and methods
In situ hybridization assays and transgenic embryos
Embryos were collected from transgenic adults, fixed and then hybridized with digoxigenin-labeled antisense RNA probes as described by Jiang et al. (1991). Transgenic strains were obtained by injecting _yw_67 embryos with various P-element transformation vectors as described by Rubin and Spradling (1982).
Preparation of expression vectors
Many of the experiments described in this study involved the misexpression of different Krüppel-coding sequences in ventral regions of pre-cellular transgenic embryos using a modified version of the PE enhancer from the twist 5′ regulatory region (Jiang and Levine, 1993). An FRT–stop–FRT cassette was inserted between the promoter and the Krüppel-coding sequence in order to circumvent dominant lethality and permit the isolation of transgenic lines (Struhl and Basler, 1993). A 1 kb _Not_I–_Not_I DNA fragment containing two tandem copies of the eve stripe 2 enhancer was removed from a previously described pCaSpeR transformation vector (Kosman and Small, 1997). This vector contains the eve stripe 2 enhancers upstream of an FRT–stop–FRT cassette. The 1 kb _Not_I–_Not_I fragment was replaced with a 0.5 kb _Not_I–_Not_I fragment containing two tandem copies of the PEeEt enhancer, which contains nucleotide substitutions that create optimal Dorsal operator sites and Twist basic helix–loop–helix (bHLH) E boxes (Jiang and Levine, 1993). This enhancer directs expression in the ventral-most 22–26 cells, which include the entire presumptive mesoderm and ventral regions of the neurogenic ectoderm.
A wild-type Krüppel cDNA encoding the full-length protein was isolated on a 2.1 kb _Pst_I–_Eco_RI fragment. This DNA also contains ∼60 bp of the 5′-untranslated region (5′-UTR) and ∼460 bp from the 3′-UTR. This fragment was cloned into a modified Bluescript SK+ plasmid, pBSK+/Asc2, which contains two _Asc_I sites in place of unique _Hin_cII and _Sac_I sites in the polylinker.
A mutant form of the Krüppel coding sequence, Krüppel Δ PED, lacks the dCtBP interaction motif. The PEDLSMH motif was converted into AAALSMH, which fails to mediate binding to dCtBP (described by Nibu et al., 1998b). The mutant Krüppel Δ PED coding region was also isolated on a 2.1 kb _Pst_I–_Eco_RI DNA fragment and cloned into the modified Bluescript vector (pBSK+/Asc2).
Unique _Nde_I and _Bgl_II sites were inserted between codon 502 and the stop codon within the Krüppel Δ PED cDNA. The following mutagenic oligonucleotide was used: 5′-CGGTGTGGTACTGGCctaAGATCTTGCCATATGTTGTTGATGGCC-3′ (cta, stop codon; ATG, codon 502; underlined sequence corresponds to the synthetic _Nde_I restriction site; double underlines correspond to a _Bgl_II site). A 250 bp _Nde_I–_Bgl_II fragment containing hairy codons 255–337 was prepared by PCR using appropriate primers. The 250 bp Nde_I–_Bgl II fagment was inserted into the _Nde_I and _Bgl_II sites of the mutagenized Krüppel Δ PED cDNA.
The wild-type, ΔPED and Krüppel–Hairy fusion sequences were isolated as _Asc_I–_Asc_I DNA fragments and inserted into the unique _Asc_I site within the 2×PEeEt expression vector. The _Asc_I site is located between the FRT–stop–FRT cassette and the 3′-UTR from the eve gene.
An _Eco_RI–_Xba_I DNA fragment containing codons 255–337 of the hairy coding region was inserted into synthetic _Eco_RI and _Xba_I sites created at the 3′ end of the snail coding sequence (residue 390) within a modified pBluescript vector. The encoded fusion protein contains the entire Snail-coding sequence and amino acids residues 255–337 of the Hairy C-terminus. An _Asc_I DNA fragment containing the snail–hairy coding sequence was inserted into the stripe 2–FRT–stop–FRT expression vector described by Kosman and Small (1997).
Synthetic reporter genes
The embryos shown in Figure 4A and B were obtained from transgenic strains described by Barolo and Levine (1997). The lacZ reporter genes were placed under the control of synthetic modular promoters containing the 300 bp rhomboid lateral stripe enhancer (NEE) placed upstream of the two tandem copies of the 250 bp PE enhancer from the twist promoter (Jiang and Levine, 1993). A synthetic Hairy repressor site was placed at each of the NEEs (Figure 4B).
The modified h-NEE-h enhancer described by Barolo and Levine (1997) was isolated as a 600 bp Bam HI–_Bst_XI DNA fragment. Synthetic _Bst_XI–_Bgl_II restriction sites were created within the _Bam_HI–_Bgl_II sites of a modified pBluescript II KS+ plasmid, pBlueG, which contains a unique _Bgl_II site in place of _Sma_I. A 340 bp _Pst_I–_Bam_HI (originally _Dra_I–_Dra_I) DNA fragment containing the CAT coding sequence was inserted into the Bgl II–_Pst_I sites of the h-NEE-h of the pBlueG plasmid. Two tandem copies of the minimal race enhancer (Rusch and Levine, 1997) were isolated on a 1 kb Not I–_Not_I DNA fragment. The _Not_I sites were blunted and inserted into the unique _Eco_RV site of the Bluescript plasmid containing the h-NEE-h enhancer. The _Hin_cII site in the polylinker was replaced with a _Not_I site. A 2.1 kb _Not_I–_Not_I fragment containing h-NEE-h, the CAT spacer and two copies of the race enhancer was inserted into the _Not_I site of a CaSpeR-AUG-βgal transformation vector (Thummel et al., 1988) containing the eve basal promoter, starting at –42 bp and continuing through codon 22 fused in-frame with lacZ (Small et al., 1992).
Hairy-binding sites were introduced into the minimal, 533 bp race enhancer via PCR amplification using two primers containing synthetic Hairy sites: 5′-TTAAGATCTGCGGCACGCGACATATCGATTGTTGTCTCATCGGCGGG-3′ and 5′-ATTGGATCCATGTCGCGTGCCGCCGCGAGCATTATTTATTTTTAATGCGAG-3′ (underlined sequences correspond to the Hairy-binding site).
The PCR product was digested with _Bgl_II and _Bam_HI, and cloned into pBlueG. Head to tail two tandem copies of h-race-h were prepared by digestion of the recombinant plasmid with either _Sca_I–_Bam_HI or _Sca_I–_Bgl_II; the two DNA fragments were ligated to each other and isolated on a 1.1 kb _Hin_dIII–_Xba_I DNA fragment. This fragment was inserted into the _Hin_dIII– _Not_I sites of the pBSK+/Asc2 vector along with a 340 bp _Xba_I–_Not_I DNA fragment containing the CAT coding sequence (used as a spacer). A 0.5 kb _Not_I–_Not_I DNA fragment containing two tandem copies of the 250 bp PE enhancer was inserted into the _Not_I site. A 1.9 kb _Asc_I–_Asc_I fragment containing two tandem copies of the h-race-h enhancer, the CAT spacer sequence and two copies of the twist PE enhancer was inserted into the _Asc_I site of the CaSpeR-AUG-βgal transformation vector (Thummel et al., 1988) containing the eve promoter fused in-frame with lacZ (Small et al., 1992).
Acknowledgments
Acknowledgements
We thank Drs Satoru Kobayashi and Akira Nakamura for providing the yw strain and advice on the injection of P-element transformation vectors. Y.N. is a fellow of the Japan Society for the Promotion of Science (JSPS). H.Z. is a fellow of the NIH. This work was supported by an NIH grant (GM 34431).
References
- Arnosti D.N., Barolo,S., Levine,M. and Small,S. (1996) The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development, 122, 205–214. [DOI] [PubMed] [Google Scholar]
- Ashe H. and Levine,M. (1999) Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature, 398, 427–431. [DOI] [PubMed] [Google Scholar]
- Barolo S. and Levine,M. (1997) hairy mediates dominant repression in the Drosophila embryo. EMBO J., 16, 2883–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Jan,L.Y. and Jan,Y.N. (1990) rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev., 4, 190–203. [DOI] [PubMed] [Google Scholar]
- Fisher A.L., Ohsako,S. and Caudy,M. (1996) The WRPW motif of the hairy-related basic helix–loop–helix repressor proteins acts as a 4-amino-acid transcription repression and protein–protein interaction domain. Mol. Cell. Biol., 16, 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Miskiewicz,P., Raj,L., Gulledge,A.A., Weir,M. and Goto,T. (1999) Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers and multi-stripe positioning by gap gene repressor gradients. Development, 126, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S. and Levine,M. (1996) Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev., 10, 700–710. [DOI] [PubMed] [Google Scholar]
- Gray S., Szymanski,P. and Levine,M. (1994) Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev., 8, 1829–1838. [DOI] [PubMed] [Google Scholar]
- Hader T., La Rosee,A., Ziebold,U., Busch,M., Taubert,H., Jäckle,H. and Rivera-Pomar,R. (1998) Activation of posterior pair-rule stripe expression in response to maternal caudal and zygotic knirps activities. Mech. Dev., 71, 177–186. [DOI] [PubMed] [Google Scholar]
- Hewitt G.F. et al. (1999) Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development, 126, 1201–1210. [DOI] [PubMed] [Google Scholar]
- Hoch M., Gerwin,N., Taubert,H. and Jäckle,H. (1992) Competition for overlapping sites in the regulatory region of the Drosophila gene Krüppel. Science, 256, 94–97. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham,P. and Rushlow,C. (1988) Region-specific alleles of the Drosophila segmentation gene hairy. Genes Dev., 2, 1037–1046. [DOI] [PubMed] [Google Scholar]
- Ip Y.T., Park,R., Kosman,D., Bier,E. and Levine,M. (1992) The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev., 6, 1728–1739. [DOI] [PubMed] [Google Scholar]
- Jiang J. and Levine,M. (1993) Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell, 72, 741–752. [DOI] [PubMed] [Google Scholar]
- Jiang J., Kosman,D., Ip,Y.T. and Levine,M. (1991) The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev., 5, 1881–1891. [DOI] [PubMed] [Google Scholar]
- Koipally J. and Georgopoulos,K. (2000) Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem., 275, 19594–19602. [DOI] [PubMed] [Google Scholar]
- Kosman D. and Small,S. (1997) Concentration-dependent patterning by an ectopic expression domain of the Drosophila gap gene knirps. Development, 124, 1343–1354. [DOI] [PubMed] [Google Scholar]
- Langeland J.A., Attai,S.F., Vorwerk,K. and Carroll,S.B. (1994) Positioning adjacent pair-rule stripes in the posterior Drosophila embryo. Development, 120, 2945–2955. [DOI] [PubMed] [Google Scholar]
- Mannervik M., Nibu,Y., Zhang,H. and Levine,M. (1999) Transcriptional coregulators in development. Science, 284, 606–609. [DOI] [PubMed] [Google Scholar]
- Nibu Y., Zhang,H. and Levine,M. (1998a) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science, 280, 101–104. [DOI] [PubMed] [Google Scholar]
- Nibu Y., Zhang,H., Bajor,E., Barolo,S., Small,S. and Levine,M. (1998b) dCtBP mediates transcriptional repression by Knirps, Krüppel and Snail in the Drosophila embryo. EMBO J., 17, 7009–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.J., Huang,J.D. and Courey,A.J. (1991) Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev., 5, 1892–1901. [DOI] [PubMed] [Google Scholar]
- Paroush Z., Finley,R.L.,Jr, Kidd,T., Wainwright,S.M., Ingham,P.W., Brent,R. and Ish-Horowicz,D. (1994) Groucho is required for Drosophila neurogenesis, segmentation and sex determination and interacts directly with hairy-related bHLH proteins. Cell, 79, 805–815. [DOI] [PubMed] [Google Scholar]
- Podos S.D. and Ferguson,E.L. (1999) Morphogen gradients—new insights from Dpp. Trends Genet., 15, 396–402. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A. and Dean,D.C. (2000) Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc. Natl Acad. Sci. USA, 97, 6391–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Rusch J. and Levine,M. (1997) Regulation of a dpp target gene in the Drosophila embryo. Development, 124, 303–311. [DOI] [PubMed] [Google Scholar]
- Schaeper U., Subramanian,T., Lim,L., Boyd,J.M. and Chinnadurai,G. (1998) Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem., 273, 8549–8552. [DOI] [PubMed] [Google Scholar]
- Small S., Blair,A. and Levine,M. (1992) Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J., 11, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Arnosti,D. and Levine,M. (1993) Spacing ensures the autonomous expression of different stripe enhancers in the even-skipped promoter. Development, 119, 762–772. [PubMed] [Google Scholar]
- Small S., Blair,A. and Levine,M. (1996) Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol., 175, 314–324. [DOI] [PubMed] [Google Scholar]
- Sollerbrant K., Chinnadurai,G. and Svensson,C. (1996) The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res., 24, 2578–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D., Small,S. and Levine,M. (1991) Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science, 254, 1385–1387. [DOI] [PubMed] [Google Scholar]
- Struhl G. and Basler,K. (1993) Organizing activity of wingless protein in Drosophila. Cell, 72, 527–540. [DOI] [PubMed] [Google Scholar]
- Thummel C.S., Boulet,A.M. and Lipshitz,H.D. (1988) Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene, 74, 445–456. [DOI] [PubMed] [Google Scholar]
- Turner J. and Crossley,M. (1998) Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J., 17, 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Small,S., Desplan,C., Dearolf,C.R. and Darnell,J.E.,Jr (1996) Identification of a Stat gene that functions in Drosophila development. Cell, 84, 421–430. [DOI] [PubMed] [Google Scholar]
- Zhang H. and Levine,M. (1999) Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]