Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing (original) (raw)
Abstract
Alternative pre-mRNA splicing is a major cellular process by which functionally diverse proteins can be generated from the primary transcript of a single gene, often in tissue-specific patterns. The current study investigates the hypothesis that splicing of tissue-specific alternative exons is regulated in part by control sequences in adjacent introns and that such elements may be recognized via computational analysis of exons sharing a highly specific expression pattern. We have identified 25 brain-specific alternative cassette exons, compiled a dataset of genomic sequences encompassing these exons and their adjacent introns and used word contrast algorithms to analyze key features of these nucleotide sequences. By comparison to a control group of constitutive exons, brain-specific exons were often found to possess the following: divergent 5′ splice sites; highly pyrimidine-rich upstream introns; a paucity of GGG motifs in the downstream intron; a highly statistically significant over-representation of the hexanucleotide UGCAUG in the proximal downstream intron. UGCAUG was also found at a high frequency downstream of a smaller group of muscle-specific exons. Intriguingly, UGCAUG has been identified previously in a few intron splicing enhancers. Our results indicate that this element plays a much wider role than previously appreciated in the regulated tissue-specific splicing of many alternative exons.
INTRODUCTION
Alternative pre-mRNA splicing is an important mechanism for regulating gene expression during development. As many as 30% of human genes utilize alternative RNA processing to generate, from a single gene, mature mRNAs with differences in exon composition at the 5′-end, within internal coding regions or at the 3′-end (1,2). Most importantly, the regulated inclusion or exclusion of selected exons facilitates synthesis of multiple protein isoforms with differences in structural/functional properties. Many examples are known in which the resulting protein isoforms can have different or even antagonistic activities with respect to transcriptional activation, ligand interactions at the cell surface, intracellular binding interactions among cytoskeletal components, subcellular localization or differences in enzymatic activity (see for example 3,4). In complex genes combinatorial alternative splicing of multiple alternative exons can generate dozens or even hundreds of distinct isoforms (5–7). Processes as fundamental as the sex determination pathway in Drosophila (8) or the life cycle of many viruses (9) are regulated in a large part via alternative pre-mRNA splicing events.
Given the size of the human genome and the abundance of alternatively spliced genes, it is likely that thousands of internal coding exons within the human genome are subject to alternative splicing. It is of great biological interest to understand the nature of the signals, encoded within the pre-mRNA, that are responsible for mediating these precisely regulated splicing events. Computational analysis of genomic DNA sequences has previously played an important role in defining the splice site signals located at 5′ and 3′ intron–exon boundaries of many constitutive exons (10) and in defining a consensus branch point sequence upstream of the 3′ splice acceptor site (11). These landmarks, which represent interaction sites for the nuclear machinery required for exon recognition and splicing, are also very useful for prediction of gene structure in computational analysis of human genome sequences (for reviews see 12–14). Similar studies have revealed non-random sequence composition of the proximal intron sequences, including an abundance of G-rich elements in the downstream region (15,16).
An increasing body of evidence indicates that RNA sequence elements important for regulation of pre-mRNA splicing can be located outside the traditional splice sites, either internally within the exon or in the flanking intron sequence. The concept of splicing ‘enhancers’ and ‘silencers’ that promote or inhibit splicing at neighboring splice sites, analogous to similarly named elements that participate in transcriptional regulation, is now well established. Numerous laboratories are actively pursuing classification of RNA sequences that function as splicing regulatory elements, as well as characterization of the relevant nuclear splicing factor proteins that interact at these sites. Important progress has been made recently with the finding that many candidate regulatory proteins are widely expressed members of two classes: hnRNP proteins (17,18) and SR (serine/arginine-rich) proteins (19,20). The RNA binding specificity for some of these factors has been characterized via biochemical binding assays, leading to the definition of consensus binding sites. In a few cases candidate tissue-specific splicing factors, such as nPTB (21) and NOVA-1 (22), have been identified as playing an important role in regulation of selected exons in the brain. However, biochemical studies of this nature are inherently limited to analysis of one or a few regulated splicing events. It is not known yet whether these candidate brain-specific splicing proteins play a role in only a limited repertoire of exons or a more general role in regulating many alternative exons. Thus, the critical question of how tissue-specific regulation of alternative splicing is controlled remains poorly understood.
Among the candidate intronic regulatory sequences identified in biochemical studies of individual pre-mRNAs are the following: (i) the hexanucleotide UGCAUG, located in the intronic enhancer for alternative exon N1 in the c-src gene (23) and in intronic regulatory sequences near several other alternative exons (24–27); (ii) the motif (A/U)GGG, reported to enhance the splicing of an alternative exon in the chicken β-tropomyosin gene (28); (iii) CUG repeats, located within muscle-specific intron enhancer elements downstream of exon 5 of the chicken cardiac troponin T (cTNT) gene (29,30); (iv) UCAY, the core binding site for brain-specific splicing factor NOVA-1, which behaves as a splicing enhancer (31). These sites are operationally defined as splicing enhancer elements. Intron splicing silencers such as polypyrimidine tract-binding protein (PTB) may act through binding to inhibitory elements related to the sequences CUCUCU (32), UUCUCU (33) and UCUU (34). To test the generality of these candidate regulatory sequences and to search for potential new regulatory elements we have applied a computational approach to analyze the intron sequences of a larger sample of tightly controlled alternative exons. The most important finding was that the hexanucleotide UGCAUG was located with much higher frequency in the proximal intron sequences downstream of alternative exons than in the same region downstream of constitutive exons. This highly statistically significant result indicates that UGCAUG may play a more general role than previously recognized in regulation of tissue-specific alternative splicing.
MATERIALS AND METHODS
Collection of the alternative exon data set
The final sample collected for this study was collated from various literature and genetic databases and consisted predominantly of exons that are specifically included during pre-mRNA splicing in the brain but skipped during processing of the same transcripts in other tissues. A few of the brain-specific exons used in this study exhibited more selective expression patterns, being included in a subset of neurons but excluded in other neurons. cDNA sequences corresponding to each of the alternatively spliced RNAs were used to query the GenBank database for the corresponding genomic sequences, from which the flanking intron sequences could be retrieved. In a few cases the intron sequence information was already available in annotated sequence files, but in most cases the relevant sequences were identified in anonymous high throughput genomic sequence (htgs) entries. All genomic entries initially identified in this manner were required to satisfy two criteria in order to be verified as the appropriate sequences for analysis. First, candidate genomic clones were required to contain not only the tissue-specific alternative exon, but also one or more of the neighboring constitutive exons, all with a sequence essentially identical to the original cDNA. Secondly, each alternative exon was examined to confirm that it was appropriately flanked by consensus 5′ and 3′ splice sites that followed the GT..AG rule for sequences at the intron boundaries. Together these criteria ensured that the appropriate alternative exon in its proper genomic setting had been selected for analysis. These alternative exons, together with 1000 nt of upstream and downstream intron sequences, represented the data set for computational analysis. It should be noted that some of the original alternative splicing data were based on studies of mouse or rat, however, due to the greater availability of human genomic sequences we utilized predominantly human intron sequences for the subsequent analyses. Although it is known that some alternative splicing events are not conserved across species, we expect that most or all of the exquisitely regulated splicing events under analysis should be conserved. In any event, inadvertent inclusion of an unregulated exon would likely cause a slight underestimation of the statistical significance of our findings and would therefore not invalidate our main conclusions. All intron sequences collected in this fashion are available at http://www-gsd.lbl.gov/~dubchak/splicing-data. The alternative brain exons we studied are listed in Table 1.
Table 1. Brain-specific alternative exons analyzed in this study.
| Gene name | Ref. | Source | Exon size (nt) | 5′ splice site/score | 3′ splice site/score | No. of ugcaug 0–100 |
|---|---|---|---|---|---|---|
| Ankyrin B, large insert | (46) | AC073240.2 | 6255 | AGgtattt = 74.3 | cattcacatcaaagA = 74.6 | 0 |
| FHL1B | (47) | AL078638.9 | 200 | CGgtaagt = 91.8 | tttgccatcctcagG = 93.0 | 0 |
| PMCA4 calcium pump | (48) | AL356980.4 | 178 | CAgtgagt = 76.1 | ctgattctttgcagA = 91.8 | 1 |
| SCN8 sodium channel | (49) | AF050730 | 123 | GGgtaaaa = 72.1 | ctgtttctgtgtagG = 87.3 | 1 |
| Amphiphysin II (region I) | (50) | AC012508 | 93 | AGgtgaca = 76.8 | cctccccaccccagC = 83.9 | 0 |
| N-type Ca channel | (51) | AC020707.4 | 63 | CCgtgagt = 75.0 | ttttgcatgtgcagT = 85.2 | 0 |
| NMDA-R1 exon 5 | (33,52) | Z32773 | 63 | AGgtatat = 74.3 | acattattcatcagA = 80.4 | 1 |
| CLCB | (53) | AC010297.3 | 54 | GTgtacgt = 67.9 | acctttccctcaagG = 83.5 | 0 |
| Myelin-associated glycoprotein, exon 12 | (54) | AC002132.1 | 45 | AGgtaggt = 89.2 | tccttctccaatagT = 78.6 | 1 |
| 4.1R exon 15 | (55) | AL357500.6 | 42 | AGgttagc = 84.1 | ttatggcaaaacagA = 71.8 | 0 |
| B-KSR1 | (56) | AC015688.3 | 42 | AGgtgagt = 96.7 | tcttctgtttaaagC = 80.5 | 1 |
| 4.1N | (57) | AL121895.21 | 36 | AGgtactg = 69.5 | ccacatcccactagC = 70.1 | 1 |
| 4.1B exon 15 | (58) | AC007445 | 36 | AGgtagaa = 69.2 | cttgatgctggcagT = 78.3 | 0 |
| HDlg | (59) | AC011322.3 | 36 | AGgtccat = 64.1 | gtctaatatgaaagT = 62.8 | 0 |
| KOR-3aa | (60) | U32929.1 | 34 | AGgtgagg = 92.2 | ctgtttttttccagC = 92.4 | 0 |
| Agrin exon 33a | (61) | M92657.1 | 33 | AGgtaagc = 94.2 | ctctcgtctcaaagC = 76.0 | 1 |
| MHC-B | (26) | AC011061.4 | 30 | AGgcaagt = 81.8 | tttgtaatgaacagC = 75.0 | 0 |
| NF1 exon 9a | (62) | AC004526.1 | 30 | CTgtaagt = 79.0 | aactgactacatagA = 66.1 | 1 |
| LAR tyrosine phosphatase | (63) | AL158083.3 | 27 | ATgtaagt = 87.2 | tctcccgcggtcagT = 79.1 | 0 |
| agrin exon 32a | (61) | M92657.1 | 24 | GCgtaagt = 78.7 | tctttgtttacaagC = 80.0 | 0 |
| Type II activin receptor | (64) | AC009480 | 24 | AGgtaaga = 94.4 | tttttcttcacaagC = 80.5 | 0 |
| GABAγ2 | (33,65) | AF165124.1 | 24 | AGgtataa = 68.6 | ctacaaacccaaagC = 63.2 | 1 |
| c-src, exon N | (66) | AL133293 | 18 | AGgtgtgt = 85.4 | tcgctggcccttagG = 78.2 | 1 |
| agrin exon 28 | (67) | AL390719.3 | 12 | AGgtactg = 69.5 | tcttcggaggccagA = 75.7 | 0 |
| FE65 | (68) | AF029234 | 6 | AGgtacta = 68.4 | gctgctggacccagA = 74.1 | 0 |
For control purposes we also analyzed a large set of complete gene sequences collected previously to test various gene prediction algorithms (13). This control set contained 570 gene sequences with a total of 1509 internal exons, most of which are presumably constitutuve exons whose exon–intron boundaries are already defined. Because our studies here focus on signals that regulate splicing of internal exons we excluded from analysis the first and last exons of every gene in the control sample. As an additional control in some experiments we analyzed a smaller sample of muscle-specific alternative exons that were collected exactly as described above for the brain-specific exons.
Analysis of intron sequences
In order to evaluate the over- or under-representation of particular oligonucleotide sequences (or words) in the data set we first calculated the frequencies of each word w in our sample and control sets. The frequency of any word, ϕ(w), is calculated as follows:
ϕ(w) = Nw/(L – |w| + 1)
where L is the length of the sequence, Nw is the number of times w occurs in that particular sequence and |w| is the number of bases in the oligonucleotide. For each word the contrast score, c, is then the difference between its frequency in the brain data set and its frequency in the control data set. In order not to overestimate the effect of long runs of a single letter or a simple repeating sequence a word is not counted if it partially overlaps with the same word (a run of 10 consecutive A residues counts as only two AAAAA pentamers).
Since derivation of estimates for statistical significance of the observation is severely complicated by the biased and non-homogeneous composition of intronic sequences in the vicinity of intron–exon boundaries, making it impossible to apply the standard Bernoulli or Markov models, we have applied resampling statistics. The statistical significance of contrast value c for each candidate regulatory element in the brain sample was estimated by calculating the probability that c could be obtained by chance from a randomly selected subset of the control sample (equal in size to the brain sample). Contrast values were calculated similarly to above, to represent the difference in frequency of word w in each control subset versus its frequency in the entire control sample. The P value of contrast score c was defined as the fraction of random subsets that have any word with a higher contrast score then c. This is a stringent measure that is equivalent to the significance values in other programs.
The algorithm to calculate the scores of donor and acceptor sequences is based on Shapiro and Senapathy (35), available at http://www.genet.sickkids.on.ca/~ali/splicesitescore.html.
RESULTS
We have focused the current analysis on a specialized class of brain-specific alternative splicing events involving internal cassette exons that are expressed selectively in brain but not in other tissues. Excluded from this analysis were tissue-specific splicing events involving alternative promoters/first exons, utilization of alternative splice donor or acceptor sites and more complex cases involving co-regulation of multiple exons or mutually exclusive exon pairs. For this reason a number of the brain-specific splicing events described in an earlier study (36) are not included in our sample. Literature searches governed by these specific criteria ultimately revealed a total of 25 exons that were brain specific. Table 1 shows the list of exons collected for this study and literature citations to the original tissue-specific splicing data. Also provided are GenBank accession numbers for genomic clones containing these exons together with flanking intron sequences, as well as parameters such as exon size and splice site strength that are discussed in detail below.
As a control group with which to compare these brain-specific introns we utilized a large set of genes with defined exon–intron boundaries previously employed for gene prediction studies (13). Analysis of the intron sequences flanking exons of this control sample is important to screen out sequence elements that may be part of the constitutive exon recognition process and allow identification of elements critically associated with regulated brain-specific exons.
General properties of the flanking intron sequences
The nucleotide composition of the collected intron sequences was determined for both brain-specific and control groups, in order to reveal any potential bias in overall composition. As shown in Table 2, the regions upstream and downstream of the tissue-specific exons were not significantly different from the control introns when compared over a 1 kb region of flanking sequence. Over this span all of the intronic regions in both samples exhibited nucleotide compositions close to 25% each for A, C, G and T (range 23.4–27.8%). However, in the exon-proximal 100 nt a significant divergence from these ratios was observed. Specifically, guanine residues were moderately deficient in this region of the downstream intron (26.9% in controls, 23.0% in brain-specific sets); uridine residues were over-represented modestly in the downstream intron (26.6% in controls, 30.6% in brain-specific sets) and to a much greater extent in the upstream intron (28.8% in control, 35.7% in brain-specific sets). The latter finding suggests that the polypyrimidine tracts of these tissue-specific exons are longer or more pyrimidine-rich than their counterparts adjacent to constitutive exons.
Table 2. Nucleotide composition of introns.
| Letter | ϕ × 102 in experimental set | ϕ × 102 in control set | Contrast (× 102) |
|---|---|---|---|
| Upstream, 0–100 | |||
| U | 35.71 | 28.81 | 6.71 |
| C | 28.20 | 26.05 | 2.03 |
| A | 18.59 | 21.63 | –3.01 |
| G | 17.49 | 23.47 | –5.70 |
| Upstream, 0–1000 | |||
| U | 27.52 | 26.00 | 1.51 |
| C | 24.65 | 24.61 | 0.04 |
| G | 24.38 | 25.13 | –0.72 |
| A | 23.43 | 24.25 | –0.81 |
| Downstream, 0–100 | |||
| U | 30.55 | 26.63 | 3.67 |
| C | 25.74 | 23.58 | 2.14 |
| A | 20.69 | 22.90 | –1.96 |
| G | 23.03 | 26.88 | –3.84 |
| Downstream, 0–1000 | |||
| U | 27.76 | 26.68 | 1.07 |
| G | 24.65 | 24.64 | –0.01 |
| C | 23.60 | 24.01 | –0.40 |
| A | 23.96 | 24.64 | –0.68 |
Putative splicing regulatory elements were sought by counting all possible short oligonucleotide sequences and identifying those specific sequences that are statistically over-represented in the introns flanking regulated alternative exons, relative to their frequency adjacent to the control exon sample. In general, higher significance scores were noted in the regions closest to the exons, within 100–300 nt upstream and downstream, than in the regions more distal to the exon. These observations are consistent with the hypothesis that binding sites functionally important for alternative splicing regulation tend to be localized close to the exon.
We first used this algorithm to test the commonly accepted dogma that alternative exons often have ‘weak’ 5′ splice sites that diverge from the consensus sequence GURAGU found at the beginning of the intron. To aid in the comparison of control versus alternative 5′ splice sites we classified sequences into one of three groups: (i) consensus splice sites that match the sequence GURAGU; (ii) rare splice sites, defined as those occuring in <1% of the control sample; (iii) others, defined as non-consensus sites that occur with moderate frequency (>1%) in the control sample (Table 3). Interestingly, the alternative sample was highly enriched in rare variations of the consensus 5′ splice site sequence, as 56% of brain samples possessed splice sites that were very uncommon in the control sample, including 16% that were never found in the control sample of 1509 exons. Consensus splice sites, in contrast, were found somewhat less frequently in the alternative brain sample (28%) than in the control sample (33%). These results illustrate two important points. First, a significant number of alternative exons possess 5′ splice sites that match the intron consensus GURAGU. Presumably the failure of this class to be constitutively spliced must be attributed to another factor(s). The second conclusion is that sites that diverge from consensus appear to represent highly unfavorable sequences, based on their rarity in the control sample.
Table 3. Classification of the 5′ splice sites.
| Class | Number (%) in alternative set | Number (%) in control set | Difference in percentages |
|---|---|---|---|
| Consensusa | 7 (28.0%) | 507 (33.6%) | –6.6 |
| Rareb | 14 (56.0%) | 427 (28.3%) | +27.7 |
| Other | 4 (16.0%) | 575 (38.1%) | –22.1 |
Computational analysis of downstream sequence elements
Computational analysis of downstream introns in the brain-specific sample revealed significant differences from the control sample with regard to the relative abundance of various oligonucleotide sequences. Table 4 shows the most highly over-represented hexamer and pentamer sequences in the downstream intron regions adjacent to the brain-specific alternative exons, ranked in order of their statistical significance (see Materials and Methods). The hexanucleotide ‘winner’ sequence in the proximal downstream intron of the brain sample (0–100 nt from the exon–intron boundary) was UGCAUG. Out of all possible 4096 hexanucleotide elements this was the most frequent hexanucleotide sequence in terms of absolute counts. Moreover, UGCAUG also exhibited the most statistically significant contrast score relative to its lower frequency in the control sample. This latter point is illustrated by the high contrast score of 3.9 × 10–3, which is nearly 1.7 times greater than that of the second most over-expressed oligonucleotide. Interestingly, even the second (GCAUGC) and third (AUGCAU) most over-represented hexamers were closely related to the winner sequence, as were the pentamer winner sequences GCAUG and UGCAU (Table 4). By comparison, the frequency of UGCAUG in the downstream 0–100 nt region of the control sample was only slightly above the expected random occurrence (44 observed, 37 expected in 1509 control introns). As indicated in Table 4, the closely related pentamer and hexamer winners were also represented in the control sample at approximately the frequency expected for a random distribution.
Table 4. Analysis of downstream intron sequences, 0–100.
| Word | ϕ × 103 (rank) in alternative set | ϕ × 103 (rank) in control set | Contrast (× 103) | P value |
|---|---|---|---|---|
| UGCAUG | 4.255 (1) | 0.310 (1117) | 3.945 | <0.01 |
| GCAUGC | 2.553 (2) | 0.225 (1827) | 2.328 | <0.01 |
| AUGCAU | 2.128 (6) | 0.155 (2585) | 1.973 | <0.01 |
| CUGCUA | 2.128 (6) | 0.190 (2194) | 1.937 | <0.01 |
| UCUCUG | 2.553 (2) | 0.662 (148) | 1.891 | 0.012 |
| CAUGCU | 2.128 (6) | 0.331 (1017) | 1.796 | 0.013 |
| UGCUUC | 2.128 (6) | 0.394 (678) | 1.733 | 0.014 |
| CCAUCC | 2.128 (6) | 0.444 (534) | 1.684 | 0.029 |
| CCUCCU | 2.553 (2) | 0.895 (41) | 1.658 | 0.029 |
| UGUCUG | 2.128 (6) | 0.508 (383) | 1.620 | 0.038 |
| GCAUG | 5.895 (1) | 0.788 (590) | 5.106 | <0.01 |
| UGCAU | 5.895 (1) | 0.886 (506) | 5.008 | <0.01 |
| CAUGC | 4.211 (4) | 0.907 (493) | 3.304 | <0.01 |
| CAUGG | 4.211 (4) | 1.144 (313) | 3.067 | <0.01 |
| UGCUU | 4.632 (3) | 1.660 (141) | 2.971 | <0.01 |
| UGCUA | 3.368 (18) | 0.698 (645) | 2.671 | 0.011 |
| CUGUC | 4.211 (4) | 1.542 (175) | 2.669 | 0.011 |
| UUUGC | 3.789 (12) | 1.151 (308) | 2.638 | 0.011 |
| AUGCU | 3.789 (12) | 1.200 (282) | 2.590 | 0.021 |
| AUGCA | 3.368 (18) | 0.788 (590) | 2.580 | 0.021 |
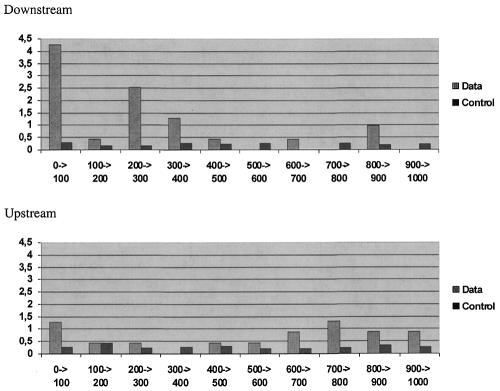
UGCAUG and the related pentamers have been reported previously to be important for the regulation of splicing in a few selected alternative exons (24–27). In those studies UGCAUG elements appeared to influence splicing from as far as 1.5 kb downstream (26). The distribution of UGCAUG among and within individual intron regions was therefore examined to characterize its frequency as a function of distance from the 5′ splice site at the exon–intron boundary. Among the 25 brain-specific exons examined here 18 (72%) possessed UGCAUG within 1000 nt downstream and one to three copies of UGCAUG were located within individual intron regions. These UGCAUG elements exhibited a frequency gradient, being highest in the proximal 100 nt region downstream of the exon (10/25 samples; Fig. 1). Interestingly, three of the seven introns lacking a downstream UGCAUG instead possessed UGCAUG in the upstream intron. An additional three possessed the pentamer winner GCAUG or UGCAU within 100 nt of the exon. Only one exon (agrin exon 28) lacked the hexamer winner within 1 kb or the pentamer winners within 100 nt. Taken together these data demonstrate that UGCAUG is widely distributed among the exons in this class, ruling out the possibility that the statistical significance was biased by the presence of a repeating element in one or a few introns. A >10-fold enrichment in UGCAUG was observed in the exon-proximal 100 nt of the downstream intron, suggesting that this element and/or closely related sequences are likely to be of general importance in regulation of alternative splicing in the brain.
Figure 1.
Distribution of UGCAUG elements in flanking introns downstream and upstream of constitutive exons (control) and brain-specific exons (data). The frequency of occurrence of UGCAUG in each 100 nt interval of the introns is shown.
In order to verify the statistical significance of these findings two additional analyses were performed. The first examined the likelihood that a random selection of downstream introns could by chance exhibit an over-representation of the specific UGCAUG sequence with a contrast score ≥3.9 × 10–3 observed for the brain versus control comparison. Repeated sampling (5000 times) of random subsets of 25 introns from the control group of 1509 introns revealed that UGCAUG was never more frequent than observed in the specialized brain sample. A second computational analysis was performed using a similar iterative sampling process to estimate the probability that any hexamer could be quantitatively over-expressed with a contrast value of 3.9 × 10–3. This latter statistical test indicated that the probability of achieving a higher contrast score by chance is <10–2 (P < 0.01). Intriguingly, the biological significance of this computational analysis is supported by previous studies showing that UGCAUG plays a role in the regulated splicing of a few selected alternative exons (24–27).
Additional analyses of the brain exon sample were performed in order to test for possible correlations of the presence or absence of UGCAUG with other parameters such as exon size, strength of the 5′ splice site or strength of the 3′ splice site. These parameters are presented in Table 1 and organized with respect to the size of the exon. No obvious correlation is observed between exon size and presence of the UGCAUG element in the proximal downstream intron, since large exons and small exons appear equally likely to possess a closely linked UGCAUG. Similarly, no correlation between the presence of UGCAUG and 5′ splice site strength, 3′ splice site strength or a combination of 5′ and 3′ splice site strengths was detectable in the small sample available for analysis.
The results presented above suggest that UGCAUG functions in the regulated splicing of many alternative exons in the brain. It was of great interest to determine whether UGCAUG actually represents a key element responsible for brain specificity or whether UGCAUG functions more generally in conjunction with regulated alternative exons in other tissues. Therefore, the frequency of UGCAUG was assessed in a small sample of muscle-specific alternative exons. Among 12 muscle-specific internal cassette exons that exhibit clear inclusion in muscle and exclusion in other tissues, UGCAUG was the most over-represented hexamer in the proximal downstream intron region (preliminary contrast score in this small sample 5.4 × 10–3). Similar to the observation with the brain-specific exons, the distribution of elements was concentrated in the exon-proximal region. In contrast, examination of a number of exons that are alternatively spliced in many different cell types (i.e. they do not exhibit strong specificity for a single tissue type) revealed no over-representation of UGCAUG relative to the control group (data not shown). Together these data suggest that UGCAUG is important for the regulated splicing of many exons, but it is unlikely to function by itself as the determinant for splicing in a specific cell type.
A second significant difference between the introns flanking alternative brain exons versus control constitutive exons was found in the frequency of triple G sequences in the proximal downstream intron (nt 0–100). This was initially suggested by the data in Table 5, where five of the 10 most under-represented pentamers in the brain sample contained three or more consecutive G residues. In contrast, GGG-containing pentamers were abundant in the control sample (Table 5), consistent with previous studies (15,16). Similar results were obtained by examination of trinucleotides and tetranucleotides: GGG was the fifth most abundant trinucleotide in the control sample and four of the 10 most abundant tetramers contained a triple G; these GGG-containing trinucleotides and tetranucleotides were strongly under-represented in the brain sample (results not shown). Together these data are consistent with the proposed role of GGG in positive regulation of splicing of constitutive exons (15,16). However, these observations also indicate a significantly reduced role for GGG elements in splicing of regulated alternative exons.
Table 5. Most under-represented pentamers.
| Word | ϕ × 103 (rank) in alternative set | ϕ × 103 (rank) in control set | Contrast × 103 |
|---|---|---|---|
| Downstream | |||
| GUGAG | 1.684 (156) | 4.946 (1) | –3.261 |
| AGGGG | 0.421 (564) | 2.574 (22) | –2.152 |
| UGGGG | 1.684 (156) | 3.823 (2) | –2.138 |
| AGGAG | 0 (811) | 2.093 (58) | –2.093 |
| CAGAG | 0 (811) | 2.044 (62) | –2.044 |
| GGGUG | 0.842 (365) | 2.846 (12) | –2.004 |
| AAGGG | 0 (811) | 1.960 (75) | –1.960 |
| UGAGA | 0 (811) | 1.953 (76) | –1.953 |
| UGAGG | 0.421 (564) | 2.344 (34) | –1.923 |
| GGGGC | 0.842 (365) | 2.741 (16) | –1.899 |
| Upstream | |||
| GGCUG | 0 (737) | 2.309 (58) | –2.309 |
| CACAG | 0.421 (525) | 2.183 (65) | –1.762 |
| GGGGU | 0 (737) | 1.737 (123) | –1.737 |
| UGGGU | 0 (737) | 1.632 (143) | –1.632 |
| GCUGA | 0 (737) | 1.590 (154) | –1.590 |
| GCCCC | 0.842 (308) | 2.379 (52) | –1.537 |
| TGGGG | 1.263 (186) | 2.769 (33) | –1.506 |
| GGGUG | 0.421 (488) | 1.918 (93) | –1.497 |
| AGGGA | 0.421 (488) | 1.918 (93) | –1.497 |
| CAGGG | 0.421 (488) | 1.918 (93) | –1.497 |
Analysis of upstream intronic sequences
In contrast to the situation for the downstream intronic sequences, comparison of the proximal upstream introns revealed a high degree of similarity between the control and brain samples with regard to the identity of the abundant sequence elements. Essentially all of the frequently occurring hexamers and pentamers in both samples consisted of sequences highly enriched in pyrimidines. The apparent winner sequences were UUUUUU (contrast score 7.2 × 10–3, P < 0.01) and UUUUU (contrast 10.9 × 10–3, P < 0.01). Even after correcting for a biased distribution (eight of the 17 UUUUUU elements in the brain sample were located in one sample that possessed a long polyuridine tract), elements were statistically over-represented in the brain sample (UUUUUU contrast score 2.8 × 10–3, P = 0.041).
More interestingly, several of the over-represented pyrimidine-rich hexamers in the brain sample resemble binding sites for PTB, a known splicing regulatory protein. In the upstream proximal 100 nt, potential PTB binding elements CUCUCU and UUCUCU exhibited high absolute frequencies as well as high contrast scores (Table 6). The shorter core sequence UCUU, when situated in a high pyrimidine context, has also been identified in iterative selection experiments as a preferred PTB binding site (34). UCUU was already very abundant in the control sample (seventh most frequent tetramer) and five of the 15 most frequent hexamers in the control sample contained UCUU; nevertheless, all of these elements had positive contrast scores, indicating an even higher frequency in the corresponding intronic region flanking brain-specific exons. Examination of the individual upstream intronic regions revealed that 21 of 25 samples possessed at least one of these putative PTB binding sites. Thus, the data is consistent with a general role for PTB in regulation of many brain-specific exons. It is worth noting, however, that none of the other brain-specific samples exhibited the pattern described earlier for c-src, in which CUCUCU elements were present both upstream and downstream of the regulated exon (32).
Table 6. Analysis of upstream intron sequences, 0–100.
| Word | ϕ × 103 (rank) in alternative set | ϕ × 103 (rank) in control set | Contrast (× 103) | P value |
|---|---|---|---|---|
| UUUUUU | 7.234 (1) | 1.191 (24) | 6.042 | <0.01 |
| CUCUCU | 5.106 (2) | 1.586 (3) | 3.520 | <0.01 |
| UCUCUC | 4.681 (3) | 1.516 (6) | 3.165 | 0.016 |
| CUCCCU | 4.255 (4) | 1.227 (20) | 3.029 | 0.021 |
| UCUGUU | 3.404 (9) | 0.705 (176) | 2.699 | 0.035 |
| CCUCUC | 3.830 (5) | 1.163 (30) | 2.667 | 0.039 |
| CUGUGU | 3.404 (9) | 0.747 (149) | 2.657 | 0.040 |
| UUCUGU | 3.404 (9) | 0.790 (117) | 2.615 | 0.042 |
| CUGUCU | 3.404 (9) | 0.839 (103) | 2.565 | 0.042 |
| UUUUU | 10.947 (1) | 3.048 (23) | 7.899 | <0.01 |
| UCUCU | 10.526 (2) | 4.004 (2) | 6.522 | <0.01 |
| CUCUC | 9.263 (3) | 3.355 (8) | 5.908 | <0.01 |
| UCUGU | 8.421 (4) | 2.581 (41) | 5.840 | <0.01 |
| UCCCU | 7.579 (5) | 3.055 (22) | 4.524 | 0.013 |
| CAUUU | 5.895 (12) | 2.058 (79) | 3.837 | 0.038 |
| CCUCC | 6.737 (8) | 3.118 (17) | 3.619. | 0.058 |
| CUCUG | 6.737 (8) | 3.167 (16) | 3.570 | 0.059 |
| UGUCU | 5.895 (12) | 2.365 (54) | 3.530 | 0.059 |
| UUUUC | 7.158 (6) | 3.753 (3) | 3.405 | 0.071 |
The brain-specific RNA binding protein NOVA-1 has been reported to bind UCAY sequences and to positively regulate inclusion of flanking alternative exons in brain (22,31). It was therefore of interest to explore whether any of these elements might play a more general role in regulating brain-specific splicing in the larger exon sample. The results show that UCAY sequences are moderately common in the proximal upstream introns, of the control group (ranks 77 and 84), but are slightly under-represented in the brain upstream intron region, as indicated by the negative contrast scores (Table 7). In the downstream introns, UCAU and UCAC were represented with average abundance in the control sample (ranks 113 and 158) and exhibited positive contrast scores, indicating modest over-representation in the brain sample. Although analysis of the isolated tetramers in this way does not yield a statistically significant P value, it is possible that a more subtle feature of UCAY distribution or sequence context is important in regulation of a subclass of brain-specific alternative splicing events.
Table 7. Analysis of UCAY and UCUU in proximal intron regions.
| Word | ϕ × 103 (rank) in alternative set | ϕ × 103 (rank) in control set | Contrast (× 103) (rank) |
|---|---|---|---|
| Upstream introns 0–100 | |||
| UCUU | 12.08 (11) | 8.91 (7) | 2.84 (18) |
| UCAC | 4.17 (73) | 4.59 (77) | –0.61 (144) |
| UCAU | 3.75 (84) | 4.41 (84) | –0.83 (161) |
| Downstream introns 0–100 | |||
| UCUU | 5.00 (63) | 6.22 (34) | –1.23 (205) |
| UCAU | 6.66 (34) | 3.88 (113) | 2.78 (15) |
| UCAC | 4.41 (89) | 3.19 (158) | 0.97 (64) |
DISCUSSION
Tissue-specific alternative pre-mRNA splicing is governed by the interactions between _cis_-acting regulatory sequences in the pre-mRNA and _trans_-acting RNA binding proteins/splicing factors. The decision to switch splicing of specific exons ‘on’ or ‘off’ at the appropriate developmental stages and in appropriate cell types likely involves combinatorial recognition of multiple RNA sequences by multiple splicing factors (4,21). The current study was based on the hypothesis that critical regulatory elements would be most evident in a population of exons that exhibit a shared, highly specific pattern of regulation and that computational analysis should be a powerful tool to identify such elements. The advantages of using brain-specific exons to test this strategy are the relatively large number of brain-specific exons identified in the literature to date, together with the availability of intronic sequences retrievable from the genetic databases. An obvious disadvantage is that even the ‘brain-specific exons’ category is functionally heterogeneous due to differential gene expression in various subsets of neuronal populations.
Intriguingly, the computational approach identified a single hexanucleotide (UGCAUG) and two related pentanucleotides (GCAUG and UGCAU) as candidate regulatory elements for brain-specific alternative splicing. These sequence elements were greatly over-represented in the proximal intron sequence downstream of brain alternative exons, relative to their frequency of occurrence in the control samples. Several statistical tests verified that this over-representation was highly significant. Moreover, examination of individual intronic regions revealed a widespread, though not universal, occurrence of these elements downstream of the brain-specific exons. Computational analysis therefore strongly suggests that these particular sequences play a physiologically important role in the splicing process. Independently, the biological relevance of UGCAUG was demonstrated in earlier biochemical studies employing functional splicing assays. For example, UGCAUG is a critical component of the intronic enhancer element downstream of the neural-specific exon in c-src pre-mRNA (23). Together, the computational and biochemical approaches support the hypothesis that UGCAUG plays an important role in the regulation of splicing.
Non-neural exons also possess functionally important UGCAUG elements: UGCAUG and/or GCAUG have been reported to influence the efficiency of alternative splicing in the pre-mRNAs for fibronectin (24), calcitonin/CGRP (25) and non-muscle myosin II heavy chain-B (26). UGCAUG was also over-represented in the modest collection of muscle-specific exons analyzed in the current study, with a frequency and distribution pattern similar to that observed in the brain sample. Finally, we have identified a few developmentally regulated alternative exons with selective expression in testis, erythroid or epithelial cells that also possess UGCAUG elements in the proximal downstream intron (not shown). These data suggest that UGCAUG, rather than representing a brain-specific splicing element, may function in a more general way as a determinant or switch for regulated splicing in multiple differentiated cell types. However, little is yet known concerning the identity of a putative UGCAUG-binding protein(s) or the mechanism(s) by which such a protein may activate splicing switches independently in various differentiated cell types. It is possible that tissue-specific splicing events could in part be coupled to tissue-specific transcriptional initiation events (37,38) that might influence the accessibility of splicing factors to the regulated exons.
A second goal of the current study was to examine functionally important regulatory elements that were defined in earlier biochemical studies of individual pre-mRNAs, to test computationally whether the same elements are important more generally in a larger population of alternative exons. One obvious and expected difference in the brain sample is the marked divergence from the consensus 5′ splice site sequence GUGAGU. This sequence is under-represented in the brain sample, consistent with many previous findings that alternative exons generally have weak 5′ splice sites. A second and more novel finding is the difference in frequency of the GGG motif between control (highly over-represented) and brain samples (approximately expected frequency). The G-rich motif is an enhancer-like element common in introns downstream of constitutive exons (27,39) and has been suggested to be important for splicing of small vertebrate introns (40). The abundance of G-rich elements in downstream introns has been utilized in some gene finding algorithms to aid in the identification of exons in genomic sequences (see refs in 16,41) and a G-rich enhancer element(s) is found in the intron downstream of several alternative exons (28,42,43). The current study demonstrates a relative paucity of G-rich motifs in the brain alternative exon sample, suggesting that this element is not a general enhancer for splicing of alternative exons. It is possible that the absence of this motif contributes to the ‘weak’ recognition of many alternative exons, necessitating utilization of distinct enhancer mechanisms.
Brain-specific alternative splicing may also be promoted through interaction of the RNA binding protein NOVA-1 with the short consensus sequence UCAY (22,31). Comparison of the control constitutive introns with the brain introns in the current study revealed no increased frequency of UCAY in the brain alternative intron sample. The data suggest therefore that UCAY is only important for a minority of brain-specific exons or, alternatively, that UCAY elements function only in a specific sequence context that was not evident in the current analysis.
Finally, previous studies of neural-specific splicing events have demonstrated that at least part of the tissue specificity arises from negative regulation of splicing in non-neural cells (32,33,44). Neural-specific alternative exons in the c-src, _GABA_A receptor γ2 subunit, clathrin light chain B and NMDA receptor NR1 pre-mRNAs appear to be repressed in non-neural cells due to binding of PTB (33). Although PTB is also expressed in brain, recent data have demonstrated the existence of a neuronal homolog nPTB (21,45) that may allow splicing of brain exons to occur by virtue of its weaker splicing inhibitory activity. Computational analysis of PTB consensus binding sites revealed that these elements are frequently represented in the proximal upstream introns of the control sample, but they are nevertheless statistically 2- to 3-fold more over-represented in the brain sample. Thus, the computational data provides support for models, proposed on the basis of studies performed with a few selected brain-specific splicing events, invoking a general role for PTB as a negative factor in suppressing neural exons in non-neural cells.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to A.A.Mironov and V.Yu.Makeev for helpful discussions. This research was supported by NIH grant HL45821 to J.G.C., as well as grants to M.G. from the Russian Fund of Basic Research (99-04-48247 and 00-15-99362), the Russian State Scientific Program ‘Human Genome’, INTAS (99-1476) and the Howard Hughes Medical Institute (55000309). The work was also supported by a grant to S.S. and M.Z. from the Office of Biological and Environmental Research, Office of Science, Department of Energy.
References
- 1.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanke J., Brett,D., Zastrow,I., Aydin,A., Delbruck,S., Lehmann,G., Luft,F., Reich,J. and Bork,P. (1999) Alternative splicing of human genes: more the rule than the exception? Trends Genet., 15, 389–390. [DOI] [PubMed] [Google Scholar]
- 3.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 4.Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 5.Missler M. and Sudhof,T.C. (1998) Neurexins: three genes and 1001 products. Trends Genet., 14, 20–26. [DOI] [PubMed] [Google Scholar]
- 6.Breitbart R.E., Andreadis,A. and Nadal-Ginard,B. (1987) Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem., 56, 467–495. [DOI] [PubMed] [Google Scholar]
- 7.Gascard P., Lee,G., Coulombel,L., Auffray,I., Lum,M., Parra,M., Conboy,J.G., Mohandas,N. and Chasis,J.A. (1998) Characterization of multiple isoforms of protein 4.1R expressed during erythroid terminal differentiation. Blood, 92, 4404–4414. [PubMed] [Google Scholar]
- 8.MacDougall C., Harbison,D. and Bownes,M. (1995) The developmental consequences of alternate splicing in sex determination and differentiation in Drosophila. Dev. Biol., 172, 353–376. [DOI] [PubMed] [Google Scholar]
- 9.Munroe S.H. (1984) Secondary structure of splice sites in adenovirus mRNA precursors. Nucleic Acids Res., 12, 8437–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mount S.M. (1982) A catalogue of splice junction sequences. Nucleic Acids Res., 10, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds) The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 12.Gelfand M.S. (1995) Prediction of function in DNA sequence analysis. J. Comput. Biol., 2, 87–115. [DOI] [PubMed] [Google Scholar]
- 13.Burset M. and Guigo,R. (1996) Evaluation of gene structure prediction programs. Genomics, 34, 353–367. [DOI] [PubMed] [Google Scholar]
- 14.Burge C.B. and Karlin,S. (1998) Finding the genes in genomic DNA. Curr. Opin. Struct. Biol., 8, 346–354. [DOI] [PubMed] [Google Scholar]
- 15.Nussinov R. (1988) Conserved quartets near 5′ intron junctions in primate nuclear pre-mRNA. J. Theor. Biol., 133, 73–84. [DOI] [PubMed] [Google Scholar]
- 16.Engelbrecht J., Knudsen,S. and Brunak,S. (1992) G+C-rich tract in 5′ end of human introns. J. Mol. Biol., 227, 108–113. [DOI] [PubMed] [Google Scholar]
- 17.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 18.Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 19.Fu X.D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- 20.Manley J.L. and Tacke,R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 21.Markovtsov V., Nikolic,J.M., Goldman,J.A., Turck,C.W., Chou,M.Y. and Black,D.L. (2000) Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol., 20, 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen K.B., Dredge,B.K., Stefani,G., Zhong,R., Buckanovich,R.J., Okano,H.J., Yang,Y.Y. and Darnell,R.B. (2000) Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron, 25, 359–371. [DOI] [PubMed] [Google Scholar]
- 23.Modafferi E.F. and Black,D.L. (1997) A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol., 17, 6537–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh G.S. and Hynes,R.O. (1994) Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev., 8, 1561–1574. [DOI] [PubMed] [Google Scholar]
- 25.Hedjran F., Yeakley,J.M., Huh,G.S., Hynes,R.O. and Rosenfeld,M.G. (1997) Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl Acad. Sci. USA, 94, 12343–12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamoto S. (1996) Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J. Biol. Chem., 271, 17613–17616. [PubMed] [Google Scholar]
- 27.Carlo T., Sterner,D.A. and Berget,S.M. (1996) An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA, 2, 342–353. [PMC free article] [PubMed] [Google Scholar]
- 28.Sirand-Pugnet P., Durosay,P., Brody,E. and Marie,J. (1995) An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken β-tropomyosin pre-mRNA. Nucleic Acids Res., 23, 3501–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan K.J. and Cooper,T.A. (1996) Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol. Cell. Biol., 16, 4014–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philips A.V., Timchenko,L.T. and Cooper,T.A. (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 31.Jensen K.B., Musunuru,K., Lewis,H.A., Burley,S.K. and Darnell,R.B. (2000) The tetranucleotide UCAY directs the specific recognition of RNA by the nova K-homology 3 domain. Proc. Natl Acad. Sci. USA, 97, 5740–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan R.C. and Black,D.L. (1995) Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol. Cell. Biol., 15, 6377–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Liu,W. and Grabowski,P.J. (1999) Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA, 5, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez I., Lin,C.H., McAfee,J.G. and Patton,J.G. (1997) Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA, 3, 764–778. [PMC free article] [PubMed] [Google Scholar]
- 35.Shapiro M.B. and Senapathy,P. (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res., 15, 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamm S., Zhang,M.Q., Marr,T.G. and Helfman,D.M. (1994) A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res., 22, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cramer P., Caceres,J.F., Cazalla,D., Kadener,S., Muro,A.F., Baralle,F.E. and Kornblihtt,A.R. (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell, 4, 251–258. [DOI] [PubMed] [Google Scholar]
- 38.Hirose Y. and Manley,J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- 39.Nussinov R. (1989) Conserved signals around the 5′ splice sites in eukaryotic nuclear precursor mRNAs: G-runs are frequent in the introns and C in the exons near both 5′ and 3′ splice sites. J. Biomol. Struct. Dyn., 6, 985–1000. [DOI] [PubMed] [Google Scholar]
- 40.McCullough A.J. and Berget,S.M. (1997) G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol., 17, 4562–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solovyev V.V., Salamov,A.A. and Lawrence,C.B. (1994) Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res., 22, 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min H., Chan,R.C. and Black,D.L. (1995) The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev., 9, 2659–2671. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy E.M. and Phillips,J.A. (1998) Characterization of an intron splice enhancer that regulates alternative splicing of human GH pre-mRNA. Hum. Mol. Genet., 7, 1491–1496. [DOI] [PubMed] [Google Scholar]
- 44.Ashiya M. and Grabowski,P.J. (1997) A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA, 3, 996–1015. [PMC free article] [PubMed] [Google Scholar]
- 45.Polydorides A.D., Okano,H.J., Yang,Y.Y., Stefani,G. and Darnell,R.B. (2000) A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of nova to regulate neuron-specific alternative splicing. Proc. Natl Acad. Sci. USA, 97, 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunimoto M. (1995) A neuron-specific isoform of brain ankyrin, 440-kD ankyrinB, is targeted to the axons of rat cerebellar neurons. J. Cell Biol., 131, 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S.M., Li,H.Y., Ng,E.K., Or,S.M., Chan,K.K., Kotaka,M., Chim,S.S., Tsui,S.K., Waye,M.M., Fung,K.P. and Lee,C.Y. (1999) Characterization of a brain-specific nuclear LIM domain protein (FHL1B) which is an alternatively spliced variant of FHL1. Gene, 237, 253–263. [DOI] [PubMed] [Google Scholar]
- 48.Keeton T.P. and Shull,G.E. (1995) Primary structure of rat plasma membrane Ca(2+)-ATPase isoform 4 and analysis of alternative splicing patterns at splice site A. Biochem. J., 306, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plummer N.W., McBurney,M.W. and Meisler,M.H. (1997) Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J. Biol. Chem., 272, 24008–24015. [DOI] [PubMed] [Google Scholar]
- 50.Butler M.H., David,C., Ochoa,G.C., Freyberg,Z., Daniell,L., Grabs,D., Cremona,O. and De Camilli,P. (1997) Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J. Cell Biol., 137, 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan J.Q. and Lipscombe,D. (2000) Alternative splicing in the cytoplasmic II-III loop of the N-type Ca channel alpha 1B subunit: functional differences are β subunit-specific. J. Neurosci., 20, 4769–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zukin R.S. and Bennett,M.V. (1995) Alternatively spliced isoforms of the NMDARI receptor subunit. Trends Neurosci., 18, 306–313. [Published erratum in Trends Neurosci., 1995, 18, 441] [DOI] [PubMed] [Google Scholar]
- 53.Stamm S., Casper,D., Dinsmore,J., Kaufmann,C.A., Brosius,J. and Helfman,D.M. (1992) Clathrin light chain B: gene structure and neuron-specific splicing. Nucleic Acids Res., 20, 5097–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miescher G.C., Lutzelschwab,R., Erne,B., Ferracin,F., Huber,S. and Steck,A.J. (1997) Reciprocal expression of myelin-associated glycoprotein splice variants in the adult human peripheral and central nervous systems. Brain Res. Mol. Brain Res., 52, 299–306. [DOI] [PubMed] [Google Scholar]
- 55.Huang J.P., Tang,C.J., Kou,G.H., Marchesi,V.T., Benz,E.J.,Jr and Tang,T.K. (1993) Genomic structure of the locus encoding protein 4.1. Structural basis for complex combinational patterns of tissue-specific alternative RNA splicing. J. Biol. Chem., 268, 3758–3766. [PubMed] [Google Scholar]
- 56.Muller J., Cacace,A.M., Lyons,W.E., McGill,C.B. and Morrison,D.K. (2000) Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol. Cell. Biol., 20, 5529–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walensky L.D., Blackshaw,S.S., Conboy,J.G., Mohandas,N. and Snyder,S.H. (1997) Molecular cloning of a novel neuron-specific homologue of the erythrocyte membrane skeletal protein 4.1. Soc. Neurosci. Abstr., 23, 1674. [Google Scholar]
- 58.Parra M., Gascard,P., Walensky,L.D., Gimm,J.A., Blackshaw,S., Chan,N., Takakuwa,Y., Berger,T., Lee,G., Chasis,J.A., Snyder,S.H., Mohandas,N. and Conboy,J.G. (2000) Molecular and functional characterization of protein 4.1B, a novel member of the protein 4.1 family with high level, focal expression in brain. J. Biol. Chem., 275, 3247–3255. [DOI] [PubMed] [Google Scholar]
- 59.Mori K., Iwao,K., Miyoshi,Y., Nakagawara,A., Kofu,K., Akiyama,T., Arita,N., Hayakawa,T. and Nakamura,Y. (1998) Identification of brain-specific splicing variants of the hDLG1 gene and altered splicing in neuroblastoma cell lines. J. Hum. Genet., 43, 123–127. [DOI] [PubMed] [Google Scholar]
- 60.Pan Y.X., Xu,J., Wan,B.L., Zuckerman,A. and Pasternak,G.W. (1998) Identification and differential regional expression of KOR-3/ORL-1 gene splice variants in mouse brain. FEBS Lett., 435, 65–68. [DOI] [PubMed] [Google Scholar]
- 61.Hoch W., Ferns,M., Campanelli,J.T., Hall,Z.W. and Scheller,R.H. (1993) Developmental regulation of highly active alternatively spliced forms of agrin. Neuron, 11, 479–490. [DOI] [PubMed] [Google Scholar]
- 62.Geist R.T. and Gutmann,D.H. (1996) Expression of a developmentally-regulated neuron-specific isoform of the neurofibromatosis 1 (NF1) gene. Neurosci. Lett., 211, 85–88. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J.S., Honkaniemi,J., Yang,T., Yeo,T.T. and Longo,F.M. (1998) LAR tyrosine phosphatase receptor: a developmental isoform is present in neurites and growth cones and its expression is regional- and cell-specific. Mol. Cell. Neurosci., 10, 271–286. [DOI] [PubMed] [Google Scholar]
- 64.Shoji H., Nakamura,T., van den Eijnden-van Raaij,A.J. and Sugino,H. (1998) Identification of a novel type II activin receptor, type IIA-N, induced during the neural differentiation of murine P19 embryonal carcinoma cells. Biochem. Biophys. Res. Commun., 246, 320–324. [DOI] [PubMed] [Google Scholar]
- 65.Whiting P., McKernan,R.M. and Iversen,L.L. (1990) Another mechanism for creating diversity in γ-aminobutyrate type A receptors: RNA splicing directs expression of two forms of γ 2 phosphorylation site. Proc. Natl Acad. Sci. USA, 87, 9966–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan R.C. and Black,D.L. (1997) Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol. Cell. Biol., 17, 2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei N., Lin,C.Q., Modafferi,E.F., Gomes,W.A. and Black,D.L. (1997) A unique intronic splicing enhancer controls the inclusion of the agrin Y exon. RNA, 3, 1275–1288. [PMC free article] [PubMed] [Google Scholar]
- 68.Hu Q., Hearn,M.G., Jin,L.W., Bressler,S.L. and Martin,G.M. (1999) Alternatively spliced isoforms of FE65 serve as neuron-specific and non-neuronal markers. J. Neurosci. Res., 58, 632–640. [DOI] [PubMed] [Google Scholar]