Analysis of the Cat Eye Syndrome Critical Region in Humans and the Region of Conserved Synteny in Mice: A Search for Candidate Genes at or near the Human Chromosome 22 Pericentromere (original) (raw)
Abstract
We have sequenced a 1.1-Mb region of human chromosome 22q containing the dosage-sensitive gene(s) responsible for cat eye syndrome (CES) as well as the 450-kb homologous region on mouse chromosome 6. Fourteen putative genes were identified within or adjacent to the human CES critical region (CESCR), including three known genes (IL-17R,ATP6E, and BID) and nine novel genes, based on EST identity. Two putative genes (CECR3 and CECR9) were identified, in the absence of EST hits, by comparing segments of human and mouse genomic sequence around two solitary amplified exons, thus showing the utility of comparative genomic sequence analysis in identifying transcripts. Of the 14 genes, 10 were confirmed to be present in the mouse genomic sequence in the same order and orientation as in human. Absent from the mouse region of conserved synteny are_CECR1_, a promising CES candidate gene from the center of the contig, neighboring CECR4, and CECR7 and_CECR8_, which are located in the gene-poor proximal 400 kb of the contig. This latter proximal region, located ∼1 Mb from the centromere, shows abundant duplicated gene fragments typical of pericentromeric DNA. The margin of this region also delineates the boundary of conserved synteny between the CESCR and mouse chromosome 6. Because the proximal CESCR appears abundant in duplicated segments and, therefore, is likely to be gene poor, we consider the putative genes identified in the distal CESCR to represent the majority of candidate genes for involvement in CES.
Cat eye syndrome (CES, Online Mendelian Inheritance in Man (OMIM) no. 115470) is a rare developmental disorder in humans associated with the presence of three or four copies of a segment of chromosome 22q11.2, usually in the form of a bisatellited, isodicentric supernumerary chromosome (Schinzel et al. 1981; McDermid et al. 1986). CES is characterized by a variety of congenital defects including ocular coloboma, anal atresia, preauricular tags/pits, heart and kidney defects, dysmorphic facial features, and mental retardation (Schinzel et al. 1981). The penetrance and severity of these phenotypic features are highly variable. The smallest region of duplication required to produce the CES phenotype is the first 2 Mb of 22q (from the centromere to marker D22S57), as determined by analysis of a patient (25105) with all major features of the syndrome and an unusually small supernumerary dicentric ring chromosome (Mears et al. 1995). The 2-Mb CES critical region (CESCR) can be subdivided into ∼1-Mb proximal and distal halves (between markers D22S795 and D22S543) based on the proximal breakpoint of an interstitial duplication in patient “SK” (H. McDermid et al., in prep.). The CES features present in this patient (facial features, ear pits/tags, heart and kidney malformations, and mental retardation) (Knoll et al. 1995) should, therefore, map to the distal half of the CESCR. Absence of coloboma and anal atresia in this patient may be due to the phenotypic variability of the syndrome.
To identify candidate genes for CES, the distal half of the CESCR was cloned in a contiguous array of bacterial and P1-based artificial chromosomes (BACs/PACs) (Johnson et al. 1999). A minimal overlapping set of clones from the region now has been sequenced and partially annotated with various ESTs and the genes IL-17R,ATP6E, and BID (Dunham et al. 1999). Here, we report the analysis of 14 putative genes within and adjacent to the distal 1.1 Mb of the CESCR. In addition, the sequence of a contiguous array of orthologous mouse genomic BAC clones revealed that 10 of these genes are present as a single linkage group on mouse chromosome 6. Sequence analysis of the proximal 400 kb of the human CESCR region revealed a complex mosaic of duplicated segments originating from elsewhere in the genome, similar to the pericentromeric regions of human chromosomes 16 (Horvath et al. 2000) and 10 (Jackson et al. 1999). The discovery of a 22q pericentromeric region transcript (CECR7), in which individual exons appear to be derived via duplication of gene sequences from different chromosomes, indicates this region may be the birth site of novel genes produced by shuffling of existing sequences.
RESULTS
Physical and Transcript Map of the CESCR and Orthologous Mouse Region
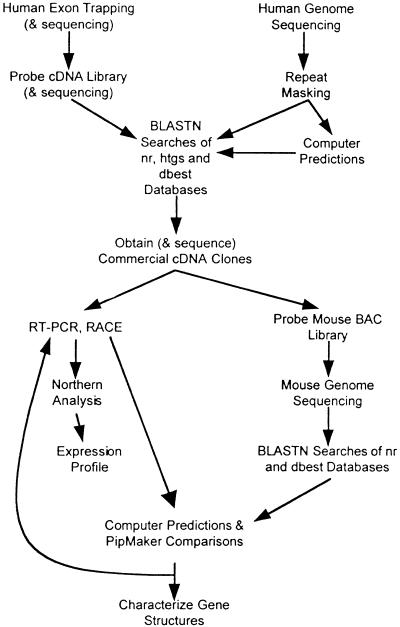
Figure 1 outlines our general strategy for combining standard positional cloning techniques with annotation of large-scale genomic sequence to establish comprehensive transcript maps of the distal 1.1 Mb of the CESCR and the 450-kb orthologous region of mouse chromosome 6. Previous mapping of Bid (Footz et al. 1998), Atp6e (Puech et al. 1997; Footz et al. 1998), and_Il-17r_ (Yao et al. 1997) to mouse chromosome 6 suggested that much of the region would show a conservation of synteny.
Figure 1.
Molecular and computer-based techniques used to identify genes in the CES critical region.
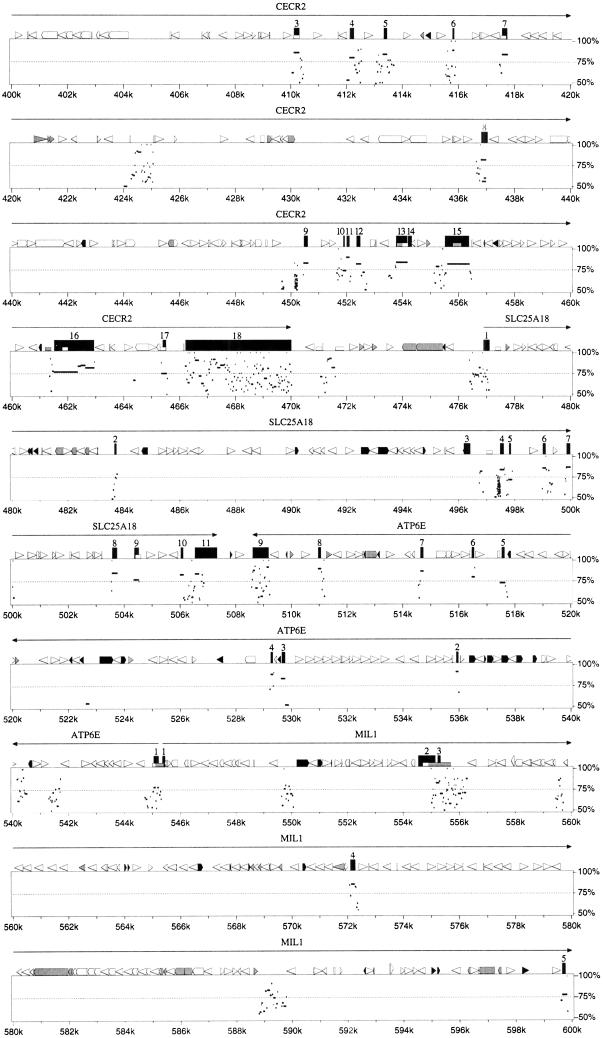
A minimal set of genomic BAC/PAC clones from human chromosome 22q11.2 and mouse chromosome 6 were subjected to shotgun sequencing and the relative position of putative genes identified are illustrated in Figure 2. The cloning, sequencing, and initial analysis of the human BAC/PAC contig for this region was described previously (Dunham et al. 1999; Johnson et al. 1999). The mouse contig, consisting of 13 clones (only seven are shown in Fig. 2), was constructed by probing a mouse BAC library with mouse cDNAs orthologous to human CESCR genes (identified throughBLASTN dbEST searches) and with a 5′ RACE–PCR product of_CECR2_. The sequence of the contigs of human and mouse clones was used to identify putative genes, described in detail below. A comparison of human and murine sequence using the PipMakerprogram (Schwartz et al. 2000) indicated that 10 genes from_IL-17R_ to BID on 22q11.2 have orthologs in a single linkage group on mouse chromosome 6, with preservation of both gene order and orientation, although two central genes are absent (Fig.3).
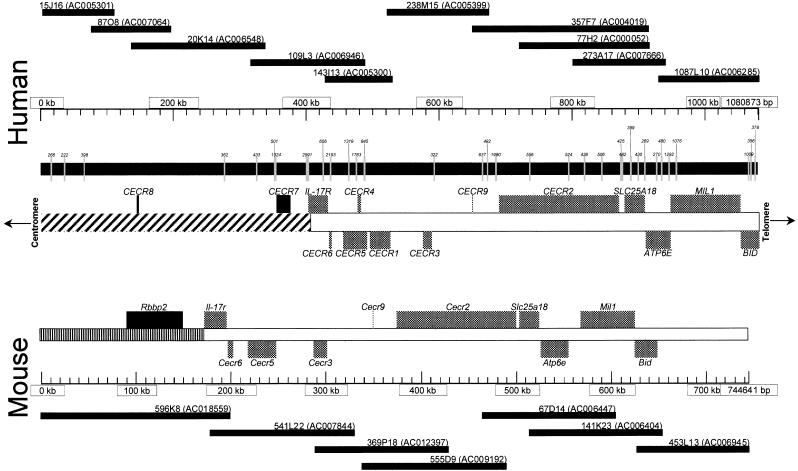
Figure 2.
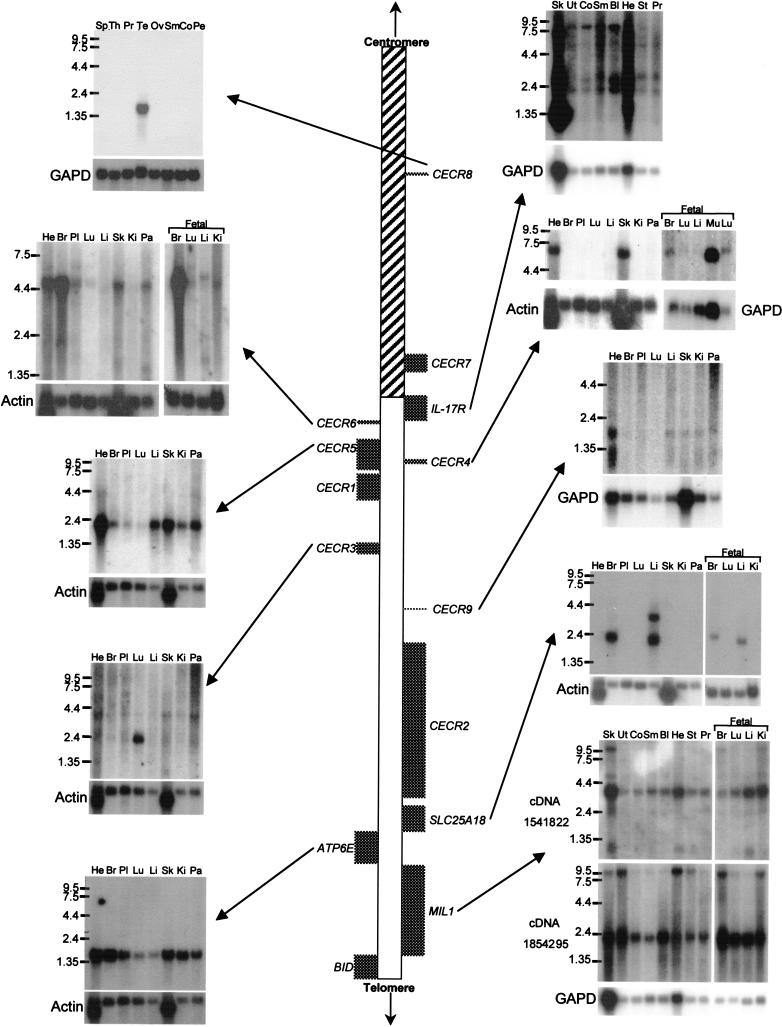
Putative genes identified in the CES critical region (CESCR) and region of conserved synteny in mouse. Sequenced BACs and PACs (with GenBank accession nos.) are shown above (human) or below (mouse) the size scales. CpG islands in the human sequence, with their size in base pairs, are shown directly below the human size scale. Below this are the identified genes, with genes transcribed centromere to telomere above the chromosome, and genes transcribed telomere to centromere below the chromosome. The hatched section of chromosome 22 represents the region rich in duplications from other regions of the genome. The mouse genes are shown above the mouse size scale and oriented as described above. The banded section represents the portion of mouse chromosome 6 orthologous to human chromosome 12p13.
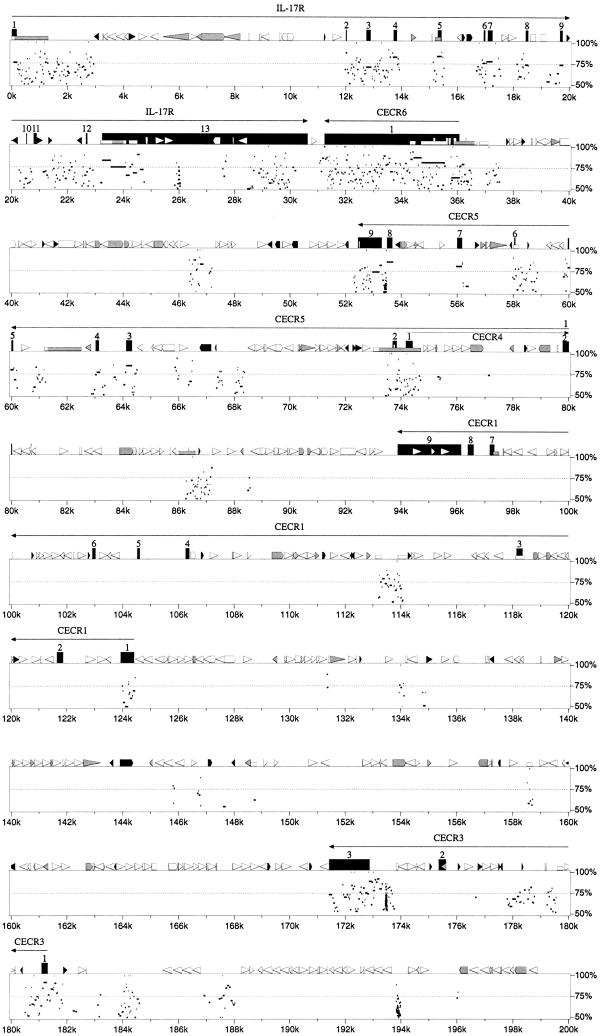
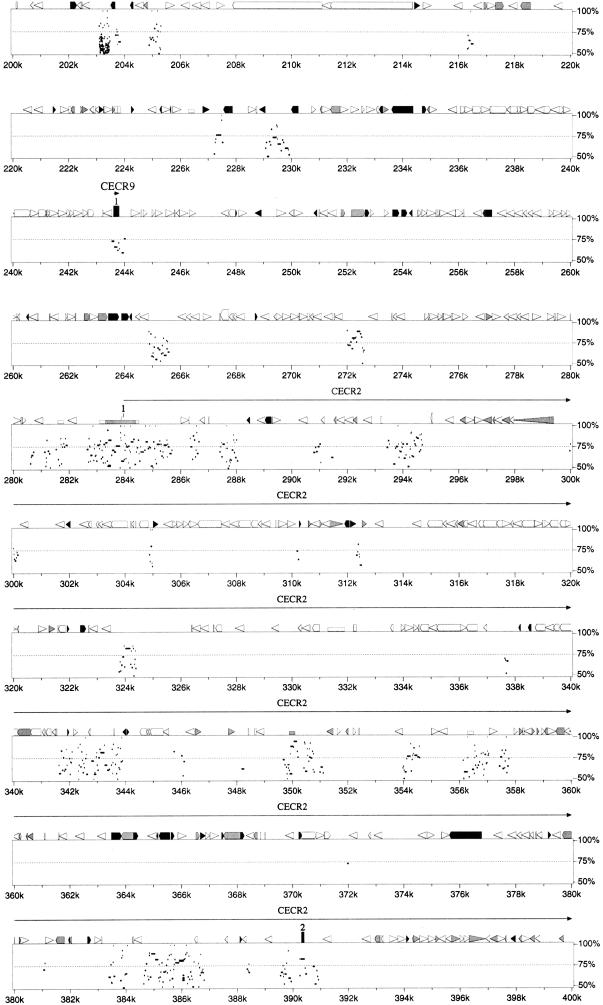
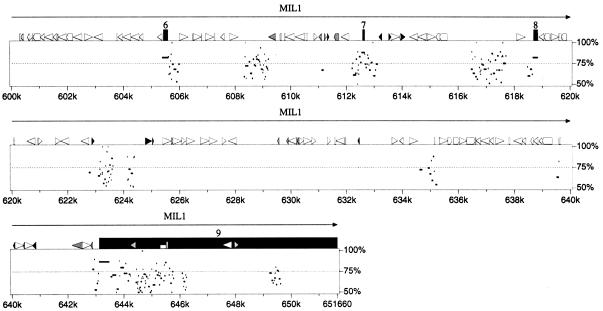
Figure 3.
Percent identity plot calculated by PipMaker (see Methods) for the human interval of IL-17R to MIL1 compared with the sequence of the region of conserved synteny from mouse chromosome 6. Gap-free segments demonstrating >50% nucleotide identity are indicated by horizontal black bars below the graphical depictions of interspersed repeats, CpG islands, and gene structures. Exons are numbered from the 5′-most cloned exon. A single gap-free alignment underneath a protein-coding exon indicates the mouse exon size is conserved, and thus the mouse locus maintains a homologous ORF.
A unique distribution of putative genes was discovered within the 1.1-Mb region of the CESCR. Only 2 genes (CECR7 and_CECR8_) localize to the ∼400 kb proximal to IL-17R, whereas the distal ∼700 kb contains 11 genes, including_IL-17R_ (Fig. 2). This gives an average gene density of 1 per 200 kb in the proximal region and 1 per 64 kb in the distal region. Other features suggest a basic difference between these two regions. The proximal 400 kb is 41.1% GC with only eight predicted CpG islands (∼1 per 50 kb), whereas the distal 700 kb is 46.4% GC and contains 28 predicted CpG islands (∼1 per 25 kb; Fig. 2). Of the eight CpG islands in the proximal 400 kb, only two are >1 kb in length, and these correspond to the 5′ end of CECR7 and the region just upstream of IL-17R. The remaining six CpG islands are ∼500 bp or less in size and none fall near the other pericentromeric gene,CECR8. In the distal 700 kb, CpG islands of >1 kb were found to correspond to the 5′ ends of CECR2, CECR6,ATP6E, and each of the alternate 5′ ends of MIL1 and_CECR5_. The remaining 20 CpG islands are all <900 bp in length and appear to be scattered throughout the distal region. The distribution of interspersed repeats also differs between the proximal and distal regions (Table 1). The 400 kb proximal to IL-17R is relatively rich in LINE and LTR elements but not in SINEs, whereas the distal 700 kb is SINE rich and LINE and LTR poor. Both regions, however, share similar fractions of total interspersed repeats. A preponderance of paralogous segments in the proximal 400 kb is reminiscent of the organization of other pericentromeres (Eichler et al. 1996, 1997; Régnier et al. 1997;Ritchie et al. 1998; Jackson et al. 1999; Horvath et al. 2000). A more detailed analysis of the sequences in the 22q pericentromere is described below. These differences suggest a boundary exists just proximal to IL-17R delineating the gene-poor and duplication-rich pericentromeric region from the rest of the q arm of chromosome 22 (Fig. 2). This boundary also corresponds to the divergence of conserved synteny on mouse chromosome 6, which is disrupted at mouse Il-17r, adjacent to the gene_Rbbp2_, whose human ortholog maps to the most telomeric known locus on 12p13.3 (http://www.ncbi.nlm.nih.gov/genemap99/map.cgi?BIN=406&MAP=G3).Bid and Atp6e have been genetically mapped proximal to a region of conserved synteny with human 12p13.3 (Puech et al. 1997;Footz et al. 1998). This suggests that the CESCR-homologous interval is oriented with Il-17r towards the telomeric end of mouse chromosome 6. The mouse is not expected to possess genes orthologous to_CECR7_ and CECR8 because of their high degree of similarity to other regions of the human genome, suggesting these duplication–divergence events occurred recently (see below).
Table 1.
Summary of RepeatMasker Output Analyzing Sequence Features of the 1.1 Mb Distal CESCR
| Proximal of IL-17R | IL-17R and Distal | |
|---|---|---|
| Length (in bp) | 402165 | 678708 |
| % GC | 41.11 | 46.44 |
| % SINES | 10.53 | 36.13 |
| % LINES | 29.30 | 9.20 |
| % LTR elements | 8.52 | 2.80 |
| % DNA elements | 1.44 | 2.48 |
| % Unclassified repeats | 0.22 | 0 |
| % Total repeats | 50.01 | 50.61 |
Further Delineation of the Distal Boundary of the CESCR
Previously, we demonstrated that BID maps telomeric to the CESCR (Footz et al. 1998) and that at least part of ATP6E is within the CESCR region (Mears et al. 1995). To refine the boundary of the CESCR, we performed dosage analysis with probes corresponding to candidate genes in the region. The intensity of fragments on autoradiographs was compared using DNA from patient 25105, whose dicentric r(22) chromosome defines the CESCR; a CES patient (CES01) with a typical supernumerary chromosome that should contain four copies of the region; and a normal control. A probe from MIL1 exon 9 (Fig. 4) showed the presence of two copies in patient 25105 and the control, as well as four copies in CES01, indicating that the 3′-most coding region maps outside the CESCR (data not shown). This does not eliminate the possibility of the shorter_MIL1_ transcript (exons 1–8) being present on the ring chromosome. However, as the predicted 5′ ends of MIL1 and_ATP6E_ are close (142 bp), this result also suggested that the CESCR distal boundary may be within ATP6E. Unexpectedly, both a cDNA probe to ATP6E exons 1–8 and a probe within the first intron showed an apparent two copies in all three individuals. Two copies of the cDNA probe were confirmed in a second CES patient known to contain four copies of the region distal to ATP6E (McDermid et al. 1996). These results suggested the presence of additional unprocessed copies of ATP6E in the genome, similar enough in sequence to produce identical bands on an autoradiograph. This would interfere with dosage analysis by increasing the overall copy number of the band in the normal genome, making an increase of one or two copies in a patient difficult to detect. BLASTN searches of the_ATP6E_ cDNA against the high-throughput genomic sequence database (htgs) revealed the presence of apparently processed pseudogenes of ATP6E, but no unprocessed copies. We hypothesized that there are unprocessed copies of the region around_ATP6E_ in areas of the genome not yet sequenced, possibly in pericentromeric regions of other chromosomes. Therefore, primers from the first intron of ATP6E were used to amplify products from a monochromosomal human/rodent hybrid panel. Products of 328 bp were seen in hybrid cell lines containing human chromosomes 12, 14, and 22 (results not shown). These products were directly sequenced, revealing a 1-bp difference between the copies: At position 65 the chromosome 22 copy has a C, whereas the chromosome 12 and 14 copies have a G. This supports our hypothesis that at least part of the region between the 3′ ends of ATP6E and MIL1 has undergone recent duplication and transposition to elsewhere in the genome. Because these additional copies cross-hybridize with the gene on chromosome 22, it is not possible to determine by dosage analysis the exact location of the 25105 breakpoint, which defines the CESCR.
Figure 4.
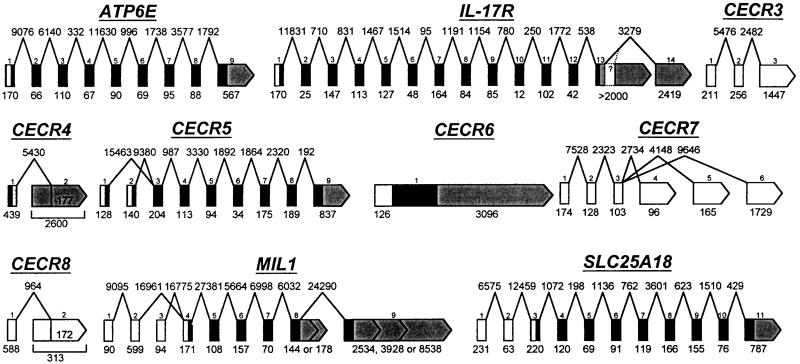
Genomic structure of the genes in the CES critical region (CESCR). Exons and introns are not shown to scale, but sizes in bp are given below the exons and above the introns. Exons are numbered from the 5′-most cloned exon; additional undiscovered exons may exist. ORFs are shown in black, 5′ UTRs in white, 3′ UTRs in grey. No significant ORFs have yet been predicted for CECR3, CECR7, or_CECR8_, hence all the exons are shown in white. Only one exon of CECR9 is currently known, therefore it was not included in this figure. CECR1 and BID were published previously (Footz et al. 1998; Riazi et al. 2000). CECR2 will be published elsewhere.
Characterization of Genes in the Distal 700 kb
For each gene, GenBank accession numbers and structural information are given in Table 2, and gene structure is diagrammed in Figure 4. Expression data is given in Table3 and illustrated in Figure5. Primers are given in Table 4 below.
Table 2.
Putative Genes Identified in the CESCR, Listed in Order of Location from the Centromere
Table 3.
Summary of Northern Blot Expression Profiles of Putative Genes in the CESCR, Listed in Order of Location from the Centromere
| Gene name | Probe used | Size of transcript(s) in kb | Tissues showing expressiona |
|---|---|---|---|
| CECR8 | cDNA 1840041 | 1.4 | Testis |
| IL-17R | 697 bp PCR fragment (primers IL17-F1 and R1), 3′ end of coding region | 1.05, 1.45, 1.9, 2.6, 5.0, 6.3, 8.8, and 10.5 | All |
| cDNA 310354, most proximal EST cluster | 1.05 and 6.3 | All | |
| cDNA 366663, most distal EST cluster | 1.05, 2.6 and 8.8 | All | |
| CECR6 | cDNA 46414 | 5 | All, especially adult heart, brain, prostate, testes, peripheral blood leukocytes, and fetal brain (not all shown in Fig. 5) |
| CECR5 | Single-stranded antisense of cDNA 1953625 | 1.9 | All |
| CECR4 | cDNA 321686 | 7 | Adult heart and skeletal muscle, all fetal tissues tested |
| CECR3 | cDNA | 2.4 and 3.7 | 2.4 kb—predominantly in lung; 3.7 kg—weakly in all tissues |
| CECR9 | Amplified exon | 2 | All, especially heart |
| SLC25A18 | cDNA 28949 | 2.3, 2.2, and 3.6 | 2.3 kb—adult and fetal brain; 2.2 kb—adult and fetal liver; 3.6 kb—adult liver |
| ATP6E | cDNA 61EW | 1.5 | All |
| MIL1 | cDNA 1541822 | 1.3, 2.4, 4.0, 5.0 and 10 | All |
Figure 5.
Expression analysis of genes in the CES critical region (CESCR). Genes are positioned in order along the chromosome, with Northern blots adjacent to them. The hatched section of chromosome 22 represents the region rich in duplications from other regions of the genome. For each Northern blot, a control probing with β-actin or GAPD is shown. Numbers beside the Northern blots indicate sizes in kb. (He) Heart; (Br) brain; (Pl) placenta; (Lu) lung; (Li) liver; (Sk) skeletal muscle; (Ki) kidney; (Pa) pancreas; (Mu) muscle; (Ut) uterus; (Co) colon; (Sm) small intestine; (Bl) bladder; (St) stomach; (Sp) spleen; (Th) thyroid; (Pr) prostate; (Te) testis; (Ov) ovary; (Pe) peripheral blood leukocytes.
Table 4.
Primers Used in This Study
| Gene | Primer name | 5′–3′ Sequence |
|---|---|---|
| IL-17R | IL17-F1 | GCG CTG GGC GAA ATA GCG TC |
| IL-17R | IL17-R1 | TGG GAG CGG GCT GTG TGG AT |
| IL-17R | IL17-F2 | CAT GGC GTC TCC TGA CCT CCT T |
| IL-17R | IL17-R5 | CCG GGT GAC TGC CTG CTT TCA |
| ATP6E | ATP6EI1 | GGC TGT TTT CAA TCC TTG CAC AG |
| ATP6E | ATP6EI2 | GGG CTT GGT TGG GTC TTA CGA A |
| CECR3 | CES38-F1 | CCT GAG AGA GAG ACA GAA GCA GC |
| CECR3 | CES38-F4 | CTG CCG AGA GTG TCT TCA GC |
| CECR3 | CES38-R1 | CAT GAA TCA CTC TCT GGT GGT TTT GC |
| CECR3 | CES38-R3 | CTC TGG AGA AGG AAA CAG GCC AC |
| Cecr3 | M38F | CAT TGG GGC TGG GGA CCT AT |
| Cecr3 | M38R | CCC TTT GTC AGC GGA GCC AT |
| CECR5 | PSL-F6 | ACG GCG ACG GCC GGA TGG |
| CECR5 | PSL-R1 | CCA TCG ATG TCC AAC AGG AAC C |
| CECR5 | EX86-3 | GAC AGG CAG AAC ACA GAC TT |
| CECR6 | BTRT-F10 | ACC TGC TCG ACA GCT TCA CGC |
| CECR6 | BTRT-F9 | AGG TGC GAG TCC CCA CTG CT |
| CECR6 | BTRT-F11 | CTG TAC CTC ATC GCC GTC ACC |
| CECR6 | BTRT-R1 | GAG TGA CAT TCC ACA CCG ACT G |
| CECR6 | BTRT-R5 | GCG AGG GTG AGG AAG TAG ACG |
| CECR6 | BTRT-R6 | TGA CGG CGA TGA GGT ACA GG |
| Cecr6 | mBTRT-F5 | CTG GGC CAT CTT CTT CGC C |
| Cecr6 | mBTRT-R1 | CAA GTC CAC CTG GAC AGT TCC |
| SLC25A18 | R2 | CAC TGC CTA CTC AGT CTC TTC T |
| SLC25A18 | R1 | CTT TCG GTA AGA ACC TCT GC |
Known Genes
IL-17R
A novel cytokine receptor, IL-17R, was previously mapped to the 22q11.2 region using a radiation hybrid panel (Yao et al. 1997), but the published mRNA sequence did not define the 3′ end of the gene. Our Northern blot analysis using a probe from the 3′ end of the coding region showed eight different transcripts, suggesting alternative splicing or polyadenylation. Two clusters of ESTs distal to the coding sequence suggested alternative 3′ ends for IL-17R. This was confirmed by Northern blot analysis using cDNA probes from each of the EST clusters (Table 3). Also, RT-PCR using primers designed from the 3′ end of the coding region and the distal EST cluster (primers IL17F22 and R5) produced a product linking the two, as expected.
The comparison of human and mouse IL-17R proteins previously revealed 69% identity and 82% similarity at the amino acid level (Yao et al. 1997). The PipMaker prediction of the genomic organization of Il-17r demonstrated conservation of exon size and identified extensive sequence similarities within introns 1, 2, 3, and 11 and in the 3′ UTR (Fig. 3).
ATP6E
The ɛ subunit of the vacuolar H+-ATPase originally was cloned from selected hnRNA from a chromosome 22 somatic cell hybrid (Baud et al. 1994). PipMaker analysis revealed murine sequence conservation restricted to the exons and small regions of flanking introns, as well as two small regions in the middle of intron 1 (Fig. 3).
Genes Identified Through Exon Amplification and EST Analysis
SLC25A18
SLC25A18 (solute carrier family 25, mitochondrial carrier, member 18) was identified originally by an EST cluster. Northern blot analysis suggested tissue- and development-specific alternative splicing (Table 3; Fig. 5). The predicted ORF shows 43% identity and 53%–54% similarity to mitochondrial transport proteins citrin (SLC25A13, accession no. AF118838) and Aralar 1 and 2 (NP_003696; CAB62206.1). Although the sequence identifies this gene as a member of the mitochondrial carrier protein superfamily preserving the six membrane-spanning domains and the internal tripartite structure (Nelson et al. 1993), the carried solute is unknown.PipMaker analysis shows sequence conservation restricted to the coding exons (4–11) and around exons 1 and 2 in the 5′ UTR (Fig. 3).
CECR1
Details of the structure and expression of CECR1 were published earlier (Riazi et al. 2000). Comparison of human and mouse sequence in this region using PipMaker revealed an absence of sequence conservation in the corresponding position of the mouse contig (Fig. 3). Similarity was seen around the first exon of_CECR1_ (44% identity, 488 bp of human sequence to 737 bp of mouse sequence), but the ORF was not conserved. There was also 63% identity (904 bp of human sequence vs. 859 bp of mouse sequence) in the region of the CECR1 third intron. Fragments can be faintly visualized on an autoradiograph from a mouse genomic Southern blot hybridized with the human exon 1 as probe at reduced stringency (60°C hybridization and wash), but whether this represents hybridization to the exon 1 remnant or to Cecr1 in another region of the mouse genome is not clear. It is possible that this gene may not exist in mouse.
CECR2
CECR2, which encodes a bromodomain, was identified by a combination of GENSCAN predictions, EST-identified cDNA sequencing, and 5′/3′ RACE. An in-depth analysis of this gene is underway. PipMaker analysis demonstrates sequence conservation of the CECR2 exons of the human and mouse orthologs. It also reveals many regions of sequence conservation between the human and mouse genomes within intron 1, which extends ∼106 kb (between 284 and 390k in Fig. 3). It is unlikely that this represents another gene within the intron, as computer analysis of the intron alone shows no ESTs or predicted genes in either orientation. These conserved regions likely represent either alternate 5′ ends to the gene or regulatory sequences.
CECR4
CECR4 was recognized both through exon amplification and cDNAs identified by analysis of genomic sequence. Sequencing of two cDNAs (321686 and 462605) indicated possible alternative splicing (Fig.4). The cDNA 321686 encodes part of a possible incomplete ORF of 108 amino acids that shows no similarity to genes identified previously. However, cloning the remainder of this putative gene by 5′ RACE and RT-PCR has been unsuccessful, possibly because of the presence of a CpG island covering the most 5′-identified exon. At least 99 bp of overlap exists between CECR4 and the proximally adjacent gene,CECR5, which is transcribed in the opposite direction (Fig.2). The shared region contains an alternative start codon of_CECR5_ but is located within the 3′ UTR of CECR4. We have considered the possibility that CECR4 is an unprocessed pseudogene, however we have been unable to find evidence of a paralogous second location in the human genome. Southern blot analysis of genomic DNA digested with _Bgl_II, _Bsr_GI,_Eco_RV, _Hin_dIII, _Pvu_II, _Ssp_I, or_Sst_I showed only one band of the size expected from the sequence of the 22q11.2 region (data not shown), suggesting a single location in the genome or a very recent duplication. Digestion with_Pst_I and _Taq_I identified RFLPs.
Comparison of human and mouse sequence in this region usingBLAST2 sequences and the PipMaker program revealed sequence conservation of CECR5, but only exon 1 of_CECR4_ (Fig. 3).
CECR5
The 3′ end of CECR5 was identified by a cluster of ∼30 ESTs. The transcript was extended by sequencing cDNA 1953625, which contains exons 3–9 and part of exon 2. RT-PCR between the originally predicted first exon (exon 2; primer PSL-F6) and exon 3 (primer PSL-R1) confirmed the presence of a start codon with a partial Kozak consensus sequence (Kozak 1991, 1995), preceded by an in-frame stop codon. RT-PCR between exon 3 and an exon located 15 kb upstream (identified through exon amplification; primer EX86–3) revealed an alternate 5′ end with an alternate start codon. Each 5′ end is located within or near a CpG island. The two transcripts would not differ significantly in size. The predicted proteins of 393 and 423 amino acids show 30% identity and 51% similarity to Schizosaccharomyces pombe CDP–alcohol phosphatidyl transferase (accession no. Z99295), as well as 40% identity and 61% similarity to a phosphatidyl synthase-like gene in_Caenorhabditis elegans_ (accession no. AF125443). Comparison of human and mouse sequence in this region using BLAST 2sequences and the PipMaker program revealed sequence conservation around all coding exons except exon 1, as well as interspersed segments within introns 2–5 (Fig. 3).
CECR6
CECR6 was first identified by a cluster of ∼30 ESTs. Sequencing of selected cDNAs and RT-PCR products revealed two overlapping predicted ORFs in different frames (209 and 578 amino acids), neither of which shows similarity to any sequences available in public databases. The longer ORF contains a leucine zipper motif. Comparison of human CECR6 sequence with mouse sequence revealed 86% identity and 89% similarity for the predicted proteins. Only the larger, leucine zipper-containing predicted ORF was conserved in the mouse. PipMaker analysis revealed conservation of sequence around the entire CECR6/Cecr6 gene including the coding sequence and 5′ and 3′ UTRs (Fig. 3).
MIL1
MIL1 was identified by numerous EST clusters indicating the presence of at least four alternatively spliced transcripts. Five transcripts were identified by Northern blot analysis using a cDNA (1541822) from the most proximal cluster, which most likely represents the smallest, 1.3-kb transcript. The cDNA 1854295 from the most distal EST cluster hybridizes to the 2.4- and 10-kb transcripts. The 2.4-kb transcript identified by this double-stranded DNA probe represents an alternative transcript of BID, the distally adjacent gene (Footz et al. 1998) (Fig. 2). The transcripts from each gene overlap by 1030 bp, both within the 3′ UTRs on opposite strands (data not shown). No overlapping ESTs have been found for murine Mil1 and_Bid_. The human cDNA 663843 is derived from alternative splicing within exon 8 (Fig. 4), producing an extended ORF into exon 9, presumed to be common to all the larger transcripts of MIL1. The cDNA 663843 represents the 4.0-kb message and cDNA 34798 represents the 5.0-kb message. Additional EST analysis predicts an additional 2.4-kb transcript (Fig. 4) that uses an alternative 5′ end (exon 3) and an alternative-length terminal exon 8 (data not shown).
Comparison of human and mouse genomic sequence revealed extensive conservation of the MIL1 locus. PipMaker showed conservation of the coding exons (4–9), as well as regions of homology in the 3′ UTR and in every intron except intron 5 (Fig. 3). The amount and extent of conservation in noncoding regions of_MIL1_/Mil1 is unique compared with the adjacent CESCR genes, however sequence conservation at the amino acid level is somewhat lower for this gene. Analysis of the predicted MIL1_amino acid sequence did not reveal an obvious function for this gene. However, while this study was in progress, an mRNA sequence for this locus was deposited in GenBank (accession no. AF146568) naming the gene_MIL1 and indicating that it encodes a protein, localized to the mitochondria, that promotes cell survival. Thus, _MIL1_would have the opposite role of the overlapping gene, BID.
Genes Identified by Comparing Human and Mouse Sequence
CECR3
CECR3 originally was identified by exon amplification of a 123-bp clone from PAC 238M15. This putative exon identified a single cDNA from a fetal brain cDNA library. The sequence of this 526-bp cDNA and the surrounding genomic region showed no similarities to known genes or ESTs, nor could a gene be identified by gene or exon prediction programs. We therefore compared the human and mouse sequence of this region (Fig. 3) to predict possible conserved segments. This analysis revealed 12 segments, 32 to 259 bp in size, with sequence identities between 77% and 93%. These conserved regions extended from the putative polyadenylation signal in the 3′ UTR of the cDNA to 29,066 bp upstream. RT-PCR between these conserved regions using cDNA reverse transcribed from human lung RNA has confirmed three exons, representing a transcript of ∼1.9 kb. No ORF has been identified, suggesting that the reason this gene was not discovered by gene prediction programs is that it codes for a functional RNA product.
CECR9
An exon of 199 bp amplified from PAC 238M15 is located between_CECR3_ and CECR2 (Fig. 2). BLASTNsearches of the exon sequence and the genomic sequence surrounding it show no similarity to known genes or ESTs. The exon is 75% identical to the mouse genomic sequence (Fig. 3). The BLAST2sequences and PipMaker programs identify other regions of mouse/human similarity in this region, suggestive of exons for_CECR9_ or distant regulatory elements of the adjacent_CECR2_. GENSCAN predicts the structure of a gene, with moderate scores, which includes this exon (data not shown). The structure of this putative gene is being investigated.
Annotation of the Proximal 400 kb
Pericentromeric Repeat Pattern
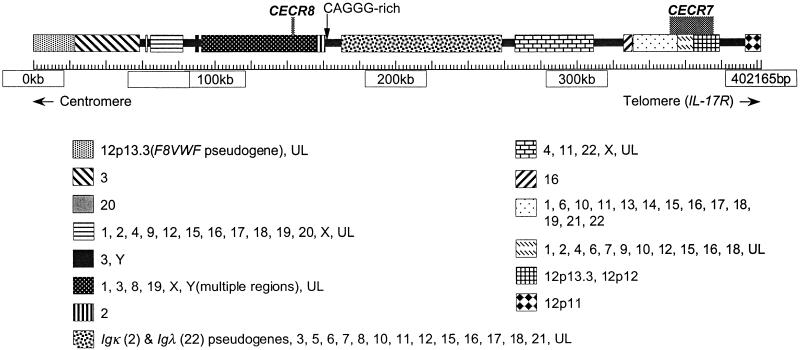
Figure 6 depicts the duplicated segments found in the proximal 400 kb of the 1.1-Mb contig of 22q11.2. Similarities between chromosome 22 sequences and other chromosomes were discovered through BLASTN queries of the nonredundant (nr) and htgs databases with repeat-masked sequence from the human contig. Throughout the region, similarity to segments from all other chromosomes was found. In many cases similarity is shared between more than two chromosomal locations. For example, a large portion of the IgΚ pseudogene cluster in the CESCR is shared with 2p11, whereas smaller portions are present on various other chromosomes (3, 5, 6, 7, 8, 10, 11, 12, 15, 16, 17, 18, and 21). Analysis of another region shared with chromosomes 1, 3, 8, 19, Xp22, and Y indicates that it is present in at least three copies on the Y chromosome.
Figure 6.
Analysis of the proximal 400 kb of the human contig, showing duplications indicative of pericentromeric regions. Repeat-masked genomic sequence was compared with the Homo sapiens subset of the nonredundant (nr) and high-throughput genomic sequence database (htgs) databases. Identity to fully or partially sequenced paralogous clones is indicated as blocks between regions apparently unique to chromosome 22, with the known chromosomal locations identified. An individual chromosome may not show paralogy over the entire block. (UL) Unlocalized clones. The analysis was performed on May 10, 2000, when 80.8% of the genome was represented by draft (61.9%) and/or finished (18.9%) sequence (http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F=HsHome.html&ORG=Hs). Additional paralogous segments may be found as the genome sequence is finished.
Two well-studied unprocessed pseudogenes were found in the 400-kb pericentromeric region: the von Willebrand factor pseudogene (accession no. M60676) (Mancuso et al. 1991) and the IgΚ pseudogene cluster (Lotscher et al. 1986, 1988). When the sequence of this region was used to search dbEST, numerous cDNAs were identified that were similar, but not identical, to chromosome 22. These presumably represent genes on other chromosomes that have homology to pericentromeric 22q11.2. However, two EST clusters showed identity to chromosome 22 rather than the homologous chromosomal regions and, therefore, represent putative chromosome 22-specific genes. A preliminary description is given below.
Putative Genes
CECR7
CECR7 was identified originally by two exons (exons 2 and 3, Fig. 4) trapped from a cosmid pool (containing cosmids c1A3, c1H9, and c88G3) mapping to PAC 109L3. cDNA clones 23249 and 1367959 and two additional cDNAs isolated from a CaCo cDNA library were used to elucidate gene structure, although the 5′ end of the gene remains to be cloned. Exons 2 and 3 show 86% and 84% identity to exons of the human homolog of a rat kidney-specific gene (accession no. AAC23497). The genomic sequence between exons 2 and 3 shows 86% identity to genomic sequence from chromosome 16 and is similar to sequence on various other chromosomes as well (Fig. 6). Exon 1 is 99% identical to sequence on chromosome 13 and 21. The paralogous region of chromosome 21 is part of the 3′ UTR of a GENSCAN-predicted gene. Three alternative 3′ ends of CECR7 are associated with three different exons. Exon 6, the 3′ end associated with the cDNA 1367959 and one of the CaCo cDNAs, is 84% identical to a region of chromosome 12. The cDNA 1461704 maps to this region of chromosome 12 with 100% identity and also shows 87% identity to the chromosome 22 exon 6. Exons 4 and 5 are rich in repeat sequences. This data indicates that the _CECR7_transcripts contain exons originating from several other chromosomes.
CECR8
CECR8 was identified by 3′ ESTs of two cDNAs (1840041 and 1840206), which showed 98%–100% identity to chromosome 22 when sequenced. Both cDNAs originate from testis cDNA libraries. Two exons have been identified (Fig. 4) that are 83% and 89% identical to sequence on chromosome Y (accession no. AC006040). The cDNA 1840206 may represent an intronless alternative 3′ end of the gene. Northern analysis showed a band only in the testis (Fig. 5), which likely represents expression from both the chromosome 22 locus and at least one other locus on the Y chromosome. Sequencing of a second group of dbEST cDNAs (accession nos. AA435549, AI469188, AA394010, and AI798750) showed 98%–100% identity to the chromosome Y locus and only 86%–89% identity to chromosome 22. Additional ESTs similar to_CECR8_ match unmapped genomic clones. The_CECR8_-related sequence on the Y chromosome is part of aGENSCAN-predicted gene showing 47% amino acid identity to the LW-1 gene (accession no. AAD45879, L. Wang and S.N. Thibodeau, unpubl.) on the X chromosome.
DISCUSSION
The overexpression of genes, associated with trisomy or partial chromosome duplication, is a common cause of congenital malformations and mental retardation. Gene overexpression is, however, difficult to study at the molecular level because of the large region of the genome usually involved. CES provides an ideal model system for studying the effects of the overexpression of genes during human development, because of the small region of the genome duplicated and the multiple organ systems affected. Fourteen putative genes in and adjacent to the CESCR have now been identified through a combination of exon amplification, EST analysis, gene prediction, and the comparison of human and mouse genomic sequence. Of these 14 genes, only 3 previously were known to be present in this region (IL17R,ATP6E, and BID).
Although a number of these genes were identified initially by exon amplification, gene prediction and EST analysis were the main methods used for identification. However, it is striking that the primary means to establish the presence of CECR3 and CECR9, which were not represented by any ESTs in the database, was through homology to mouse sequence. Northern analysis of CECR3 shows it is strongly expressed in lung, but it is unclear why there are no database ESTs for the transcript. It is particularly interesting that_CECR3_ and CECR9 would have been overlooked by standard computer-based techniques if mouse sequence had not been available, underscoring the utility of such comparative sequence analyses. Comparison of human and mouse genomic DNA has proved to be a very sensitive method for detecting genes in other regions (Hood et al. 1993; Ansari-Lari et al. 1998; Jang et al. 1999; Lund et al. 2000). Conservation of noncoding regions can also indicate the presence of sequences involved in gene regulation (Hardison et al. 1997a,b; Oeltjen et al. 1997). Numerous stretches of homology in noncoding regions throughout the CESCR indicate the possible locations of regulatory regions that may prove important in the future study of these genes.
One goal of this study was to identify genes that would be reasonable candidates for involvement, either singly or in combination, in the production of the CES phenotypic features. Although at this point it is difficult to completely rule out any as candidate genes, some genes are unlikely to be involved. SLC25A18 is similar to_citrin_ (OMIM 603859 ), which causes the autosomal recessive disorder citrullinemia when mutated. Thus, based on similarity to_citrin, SLC25A18_ is unlikely to be sensitive to increased gene dose. The similarity of CECR5 to yeast phosphatidyl synthases suggests that it is an enzyme involved in fatty acid metabolism, possibly in the production/processing of membrane phospholipids (Antonsson 1997; Yamashita and Nikawa 1997) and, therefore, is unlikely to be sensitive to dosage changes. The testis-restricted expression of CECR8 makes it an unlikely candidate for CES. Finally, IL-17R, although expressed ubiquitously (Yao et al. 1997), functions in the immune response, which is not reported to be disrupted in CES patients.
Our understanding of several of the CESCR genes is limited by technical difficulties, which currently preclude prediction of whether they are reasonable CES candidate genes. It is not yet clear whether_CECR4_ is a pseudogene with a near-identical copy elsewhere, a spurious transcript from a bidirectional promoter of CECR5, or a gene that overlaps CECR5 in the opposite direction and its exons are resistant to prediction and cloning. The possible candidacy of ATP6E and the shortest transcript of MIL1 is confounded by uncertainty over whether each complete gene is present in the CESCR, as defined by dosage analysis with the r(22) 25105 CES patient. Resolving this uncertainty will require an alternate approach, such as fluorescence in situ hybridization of patient 25105 cells.
We consider CECR1 to be the most suitable CES candidate gene identified to date (Riazi et al. 2000). This gene is homologous to a novel family of growth factors first characterized in invertebrates (Sossin et al. 1989; Homma et al. 1996). Sites of expression in a 35-day human embryo include the outflow tract and atrium of the forming heart, as well as the VII/VIII cranial nerve ganglion, suggesting potential involvement in the heart and facial defects of CES (Riazi et al. 2000). The C terminus of the CECR1 protein shows homology to adenosine deaminase, suggesting that CECR1 may function by regulating the concentration of extracellular adenosine.
There are several other promising CES candidate genes. CECR2, with a leucine zipper and a bromodomain (G. Banting and H.E. McDermid, unpubl.), may be involved in chromatin remodeling (Jeanmougin et al. 1997; Collingwood et al. 1999; Winston and Allis 1999). _CECR6_is a single-exon gene encoding a leucine zipper motif, suggesting potential protein–protein interactions exhibited by many transcription factors and gene regulatory proteins (Busch and Sassone-Corsi 1990;Hagerman 1996). Genes involved in such interactions have the potential to be sensitive to dosage effects.
BID and the most telomeric coding exon of MIL1 are located outside the CESCR as defined by the r(22) 25105 CES patient, however they are present in four copies in cases of CES with a typical CES chromosome. Because the patient that defines the CESCR (25105) died at 17 days (Mears et al. 1995), further physical and mental development of this patient could not be determined. It is therefore possible that overexpression of MIL1 and BID may be involved in physical and mental development of CES patients. Interestingly, the overlap of the 3′ UTR of these two genes suggests the possibility of antisense regulation (Spencer et al. 1986).
We have established that the mouse genome carries a single linkage group containing genes orthologous to the CESCR. However, the ortholog of CECR1, the most promising candidate gene for involvement in CES features, is absent from our sequenced contig, although it may map elsewhere in the mouse genome. This precludes straightforward modeling of CES in mice by manipulation of murine ES cells to engineer a duplication of the homologous linkage group using the Cre/loxP system (Ramirez-Solis et al. 1995). Additionally, if CECR7 and_CECR8_ are involved in producing the CES phenotype, it would likely be impossible to model their effect in a mouse, as their evolution from apparently recent duplications argues against the existence of rodent orthologs (see below). CECR8 is expressed only in testis by Northern blot analysis and, therefore, is unlikely to be involved in CES. The expression pattern of CECR7 is currently unknown. Genes distal to CECR7, however, should be amenable to testing the effects of overexpression in a mouse model. We are therefore producing transgenic mice using the BAC and PAC clones of the human contig to determine whether overexpression leads to abnormalities similar to those seen in CES.
A unique junction was discovered within the distal half of the CESCR, between the gene-poor pericentromeric portion and the remaining gene-rich q arm of chromosome 22. Although a small amount of chromosome 22-specific sequence may be present in the proximal 400 kb of the distal CESCR, the majority of this region appears to represent duplications of syntenic and nonsyntenic loci in the genome. Preliminary analysis indicates this pattern of duplications extends over an additional 1 Mb to the centromere. Analysis of the pericentromeric regions of chromosome 10, 16, and others (Eichler et al. 1996, 1997; Régnier et al. 1997; Ritchie et al. 1998; Jackson et al. 1999; Horvath et al. 2000) has led to the hypothesis that genomic duplications are recruited to pericentromeres, possibly serving as a buffer between heterochromatin and euchromatin (Eichler et al. 1999;Horvath et al. 2000). Because many of these fragments contain portions of genes, the pericentromere appears to be a “junkyard” of gene fragments. However, our discovery of transcribed elements within the duplication segments also suggests that this region may serve as a nursery for the evolution of new genes (Eichler et al. 1997; Jackson et al. 1999; Horvath et al. 2000). Because these duplications appear to be evolutionarily recent, the creation of new genes in this region would likely be specific to human and higher primates (Horvath et al. 2000).
The mechanism by which gene duplication recruitment occurs has recently been speculated to be driven by recombinogenic structures composed of an array of CAGGG elements and various subtelomeric-like repeats (Eichler et al. 1999). This is not apparent in the duplication region examined in this study. Although a cluster of CAGGG repeats was discovered in the proximal 400 kb of the distal CESCR, its organization is not reminiscent of the previously described arrays (Eichler et al. 1999) as it does not contain the repeat exclusively on one strand. Instead, the 2100-bp interval at positions 161205–163304 bp (Fig. 6) contains 13 CAGGG elements intermixed with 16 CCCTG repeats (i.e., the reverse complement), and the 2100-bp CAGGG-rich cluster differs from the “consensus” clusters (Eichler et al. 1999) in that no known subtelomeric repeats were detected in the vicinity. The 22q duplication flanked by CAGGG repeats studied by Eichler et al. (1999) is situated ∼6–7 Mb from the centromere, within an Igλ cluster (accession no.D87003/018). Also within the Igλ cluster, ∼200 kb centromeric to this duplication, is a stretch of α-satellite repeats (Kawasaki et al. 1997). This region may therefore represent an isolated group of centromere-like repeats, distinct from the duplications directly adjacent to the centromere. The mechanism for recruiting gene duplications to these separate regions of 22q may turn out to be distinct.
IL-17R is situated at the boundary between the duplicated segments and unique genes in the CESCR. Close inspection reveals that exon 1 and part of the first intron are actually shared with the pericentromere of 12p11 (accession no. AC010198). As this exon is orthologous with the BAC contig on mouse chromosome 6, it is likely that this portion of 22q11.2 was duplicated and transposed to 12p11. The juxtaposition of Il-17r and Rbbp2 on mouse chromosome 6 suggests that either these genes were separated in the human lineage following the divergence of mouse and man (with_IL-17R_ and RBBP2 being incorporated as the outermost unique genes on their respective human chromosomes, 22qcen and 12pter) or that in the mouse lineage there was a fusion of the linkage groups carrying Il-17r and Rbbp2.
Because the region of pericentromeric repeats on chromosome 22 is unlikely to produce many transcripts, most if not all of the genes associated with CES will be located between IL17R and_ATP6E_. Further characterization of these genes should shed light on the developmental processes that lead to the CES phenotype upon their overexpression.
METHODS
Hybrid Cell Lines
Cell lines from the NIGMS human/rodent somatic cell hybrid monochromosomal panel no. 2 (version 2) were grown under recommended conditions and harvested for DNA.
Exon Amplification
Exon amplification was performed using the Gibco BRL Exon Trapping System according to the manufacturer's instructions. Exons were trapped from 2 BACs (95A8, 233A2), 3 PACs (109L3, 238M15, and 120N18), and 27 cosmid clones that cover a portion of the distal CESCR.
Sequencing
Human and mouse genomic clones were sequenced by a random shotgun strategy (protocol available from http://www.genome.ou.edu). PCR products, exons, and cDNAs were sequenced either manually using a Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science) according to the manufacturer's instructions, or using ABI and LI-COR automated sequencers.
Hybridization
Hybridization of exon and cDNA probes to total human or bacterial clone DNA was carried out as described previously (Mears et al. 1994) with the following exceptions. DNA probes were purified in 0.7% or 1.0% low-melt agarose (SeaPlaque, FMC); transferred DNA was separated on 0.7%–2.0% agarose; the first post-hybridization wash consisted of 1.5× SSC/0.2% SDS; and all Southern blot washes ranged from 5–20 min. Human/human or mouse/mouse hybridizations were performed at 65°C, whereas interspecific hybridizations were done at 50°C–55°C with washes no higher than 60°C.
Mouse BAC clones were obtained by screening a library for strain 129SV (Research Genetics). Hybridizations to eight high-density membranes (containing >200,000 double-spotted unique clones in total) were performed in 5× SSC/5× Denhardt's/0.5% SDS with washes as described for Southern analysis above. Commercial cDNA probes were used for the following genes: Il-17r (accession no. W12281), Cecr6(accession no. AA030766), Mil1 (accession no. AA683716), and_Bid_ (accession no. AA104077). PCR products were used for_Cecr3_ (1.6-kb mouse genomic fragment with primers M38F + M38R) and Cecr2 (human exons 1 and 2, see Fig. 3; G. Banting and H.E. McDermid, unpubl.).
Northern Hybridizations
Human multiple tissue Northern blots were purchased from Clontech Laboratories and Invitrogen. Each Northern blot contains ∼2 μg of poly(A)+ RNA per lane from various different human adult or fetal tissues. Probes were labeled using the Strip-EZ DNA kit (Ambion).
Screening cDNA Libraries
A fetal brain cDNA library (Stratagene) and a cDNA library made from CaCo cell RNA (library kindly provided by Dr. Johanna Rommens, Hospital for Sick Children, Toronto) were screened to identify clones containing exons identified previously through the exon amplification procedure. Plaque lifts were performed using Hybond-N (Amersham Life Science) nylon membranes as per the manufacturer's instructions.
RT-PCR
RT-PCR was performed using the ThermoScript RT-PCR System (Life Technologies) on 2–5 μg of total RNA isolated from various human or mouse tissues. In cases where subsequent PCR was to be performed using primers not flanking intronic sequence, the RNA was pretreated with DNase following the manufacturer's instructions (either DNase I [10 U/μL, Boehringer Mannheim]; DNase I, amplification grade [1 U/μL, Life Technologies]; or DNA-free system [Ambion]). In these situations, a negative control reverse transcription was also performed as above but without the addition of the ThermoScript reverse transcriptase enzyme. Primers used for RT-PCR and PCR are given in Table 4.
PCR
PCR was performed using a PTC-100 programmable thermal controller (MJ Research). The PCR conditions typically consisted of a touchdown procedure: 95°C for 1–2 h (Tminitial, 0.5°C–1.0°C/cycle for 30 min; 72°C for 1 h per kb; 94°C for 30 min) 10 times; (Tmfinal, 30 min; 72°C for 1 h per kb; 95°C for 30 min) 20–30 times; Tmfinal, 30 min; 72°C for 10 h. When PCR was performed on CG-rich templates, the PCRx Enhancer System (Life Technologies) was used according to manufacturer's instructions or with 5% DMSO.
Rapid amplification of cDNA ends (Frohman et al. 1988) was performed using Marathon cDNA kits (Clontech) and RNA from human testis, brain, heart, and fetal heart. The kits used were prepared from poly(A)+ RNA from either human adult heart, testis, or brain, or fetal brain. A primary RACE was performed using an internal gene-specific primer and the Marathon Adapter Primer (AP1), followed by a nested PCR using a nested gene-specific primer and the nested Marathon Primer (AP2).
Computer Manipulations
GeneTool v1.0 for Macintosh (BioTools) was employed for sequence chromatograph interpretation, in silico gene modeling, CpG island prediction (parameters used a window size of 500 bp and a_Y_-value > 0.6, %GC > 0.5) and the annotation of genomic sequence used in Figures 2, 5, and 6.
Human and rodent interspersed repeats were detected and masked usingRepeatMasker (A.F.A. Smit and P. Green, unpubl. software) at the Web interfacehttp://ftp.genome.washington.edu/cgi-bin/RepeatMasker. Simple sequence repeats and low complexity regions were not masked.
The BLAST algorithm (Altschul et al. 1990) (http://www.ncbi.nlm.nih.gov/blast/) was used for DNA and protein similarity searches against the nr, expressed sequence tag (dbEST), and htgs databases using the default parameters (or without filtering). TheBLAST 2 sequences algorithm (Tatusova and Madden 1999) for pairwise alignments used the default parameters (or without filtering). All sequences were repeat-masked before querying a database. At least one sequence was repeat-masked when compared against another byBLAST 2 sequences.
Exon and gene structure predictions in repeat-masked sequence were made using GENSCAN (Burge and Karlin 1997 ) athttp://bioweb.pasteur.fr/seqanal/interfaces/GENSCAN.htm,GRAIL2 athttp://compbio.ornl.gov/Grail-1.3-bin/OrgForm.DoPost, MZEFat http://sciclio.cshl.org/genefinder/, Genie athttp://www.fruitfly.org/seq_tools/genie.html, andFGENES athttp://dot.imgen.bcm.tmc.edu:9331/seq-search/gene-search.html.
Mouse genomic DNA, corresponding to 453,257 bp of noncontinuous sequence in four contigs covering the region of Il-17r to_Mil_1 (Fig. 2), was compared with repeat-masked human sequence (from IL-17R to MIL1, 651,660 bp of continuous sequence) using the PipMaker program accessed at the web site http://bio.cse.psu.edu/pipmaker/. Individual gap-free alignments, ranging in size from 3 to 850 bp, exhibiting >50% identity are depicted in the percent identity plot (PIP; Schwartz et al. 2000) in Figure 3.
Genomic clones, paralogous to the pericentromeric portion of the CESCR, being sequenced by the Washington University Genome Sequencing Center, were searched at the web sitehttp://genome.wustl.edu/cgi-bin/ace/ctc_choices/ctc.ace to identify their chromosomal location.
Acknowledgments
We thank Angela Johnson, Dana Shkolny, Lisa Lane, and Heather Wright for technical support. Dr. Roseline Godbout and Paul Grundy provided DNA from some somatic cell hybrid lines. This research was supported by grants from the Medical Research Council of Canada (H.E.M.) and NIH- NHGRI grant no. HG00313 (B.A.R.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hmcdermi@ualberta.ca; FAX (780) 492-9234.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.154901.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, Gibbs RA. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Antonsson B. Phosphatidylinositol synthase from mammalian tissues. Biochim Biophys Acta. 1997;1348:179–186. doi: 10.1016/s0005-2760(97)00105-7. [DOI] [PubMed] [Google Scholar]
- Baud V, Mears AJ, Lamour V, Scamps C, Duncan AM, McDermid HE, Lipinski M. The E subunit of vacuolar H(+)-ATPase localizes close to the centromere on human chromosome 22. Hum Mol Genet. 1994;3:335–339. doi: 10.1093/hmg/3.2.335. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Busch SJ, Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- Collingwood TN, Urnov FD, Wolffe AP. Nuclear receptors: Coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;23:255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet. 1996;5:899–912. doi: 10.1093/hmg/5.7.899. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum Mol Genet. 1997;6:991–1002. doi: 10.1093/hmg/6.7.991. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 1999;9:1048–1058. doi: 10.1101/gr.9.11.1048. [DOI] [PubMed] [Google Scholar]
- Footz TK, Birren B, Minoshima S, Asakawa S, Shimizu N, Riazi MA, McDermid HE. The gene for death agonist BID maps to the region of human 22q11.2 duplicated in cat eye syndrome chromosomes and to mouse chromosome 6. Genomics. 1998;51:472–475. doi: 10.1006/geno.1998.5392. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ. Do basic region-leucine zipper proteins bend their DNA targets … does it matter? Proc Natl Acad Sci. 1996;93:9993–9996. doi: 10.1073/pnas.93.19.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian beta-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997a;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997b;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Homma K, Matsushita T, Natori S. Purification, characterization, and cDNA cloning of a novel growth factor from the conditioned medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina(flesh fly) J Biol Chem. 1996;271:13770–13775. doi: 10.1074/jbc.271.23.13770. [DOI] [PubMed] [Google Scholar]
- Hood L, Koop BF, Rowen L, Wang K. Human and mouse T-cell-receptor loci: The importance of comparative large-scale DNA sequence analyses. Cold Spring Harb Symp Quant Biol. 1993;58:339–348. doi: 10.1101/sqb.1993.058.01.040. [DOI] [PubMed] [Google Scholar]
- Horvath JE, Viggiano L, Loftus BJ, Adams MD, Archidiacono N, Rocchi M, Eichler EE. Molecular structure and evolution of an alpha satellite/non-alpha satellite junction at 16p11. Hum Mol Genet. 2000;9:113–123. doi: 10.1093/hmg/9.1.113. [DOI] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, Marzella R, Viggiano L, Archidiacono N. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- Jang W, Hua A, Spilson SV, Miller W, Roe BA, Meisler MH. Comparative sequence of human and mouse BAC clones from the mnd2 region of chromosome 2p13. Genome Res. 1999;9:53–61. [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- Johnson A, Minoshima S, Asakawa S, Shimizu N, Shizuya H, Roe BA, McDermid HE. A 1.5-Mb contig within the cat eye syndrome critical region at human chromosome 22q11.2. Genomics. 1999;57:306–309. doi: 10.1006/geno.1999.5757. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, Wang J, Shimizu N. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Asamoah A, Pletcher BA, Wagstaff J. Interstitial duplication of proximal 22q: Phenotypic overlap with cat eye syndrome. Am J Med Genet. 1995;55:221–224. doi: 10.1002/ajmg.1320550214. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- ————— Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc Natl Acad Sci. 1995;92:2662–2666. doi: 10.1073/pnas.92.7.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotscher E, Grzeschik KH, Bauer HG, Pohlenz HD, Straubinger B, Zachau HG. Dispersed human immunoglobulin kappa light-chain genes. Nature. 1986;320:456–458. doi: 10.1038/320456a0. [DOI] [PubMed] [Google Scholar]
- Lotscher E, Zimmer FJ, Klopstock T, Grzeschik KH, Jaenichen R, Straubinger B, Zachau HG. Localization, analysis and evolution of transposed human immunoglobulin V kappa genes. Gene. 1988;69:215–223. doi: 10.1016/0378-1119(88)90432-5. [DOI] [PubMed] [Google Scholar]
- Lund J, Chen F, Hua A, Roe B, Budarf M, Emanuel BS, Reeves RH. Comparative sequence analysis of 634 kb of the mouse chromosome 16 region of conserved synteny with the human velocardiofacial syndrome region on chromosome 22q11.2. Genomics. 2000;63:374–383. doi: 10.1006/geno.1999.6044. [DOI] [PubMed] [Google Scholar]
- Mancuso DJ, Tuley EA, Westfield LA, Lester-Mancuso TL, Le Beau MM, Sorace JM, Sadler JE. Human von Willebrand factor gene and pseudogene: Structural analysis and differentiation by polymerase chain reaction. Biochemistry. 1991;30:253–269. doi: 10.1021/bi00215a036. [DOI] [PubMed] [Google Scholar]
- McDermid HE, Duncan AM, Brasch KR, Holden JJ, Magenis E, Sheehy R, Burn J, Kardon N, Noel B, Schinzel A, Teshima I, White BN. Characterization of the supernumerary chromosome in cat eye syndrome. Science. 1986;232:646–648. doi: 10.1126/science.3961499. [DOI] [PubMed] [Google Scholar]
- McDermid HE, McTaggart KE, Riazi MA, Hudson TJ, Budarf ML, Emanuel BS, Bell CJ. Long-range mapping and construction of a YAC contig within the cat eye syndrome critical region. Genome Res. 1996;6:1149–1159. doi: 10.1101/gr.6.12.1149. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Duncan AM, Budarf ML, Emanuel BS, Sellinger B, Siegel-Bartelt J, Greenberg CR, McDermid HE. Molecular characterization of the marker chromosome associated with cat eye syndrome. Am J Hum Genet. 1994;55:134–142. [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, el-Shanti H, Murray JC, McDermid HE, Patil SR. Minute supernumerary ring chromosome 22 associated with cat eye syndrome: Further delineation of the critical region. Am J Hum Genet. 1995;57:667–673. [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Lawson JE, Klingenberg M, Douglas MG. Site-directed mutagenesis of the yeast mitochondrial ADP/ATP translocator. Six arginines and one lysine are essential. J Mol Biol. 1993;230:1159–1170. doi: 10.1006/jmbi.1993.1233. [DOI] [PubMed] [Google Scholar]
- Oeltjen JC, Malley TM, Muzny DM, Miller W, Gibbs RA, Belmont JW. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 1997;7:315–329. doi: 10.1101/gr.7.4.315. [DOI] [PubMed] [Google Scholar]
- Puech A, Saint-Jore B, Funke B, Gilbert DJ, Sirotkin H, Copeland NG, Jenkins NA, Kucherlapati R, Morrow B, Skoultchi AI. Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization. Proc Natl Acad Sci. 1997;94:14608–14613. doi: 10.1073/pnas.94.26.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- Régnier V, Meddeb M, Lecointre G, Richard F, Duverger A, Nguyen VC, Dutrillaux B, Bernheim A, Danglot G. Emergence and scattering of mutliple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Hum Mol Genet. 1997;6:9–16. doi: 10.1093/hmg/6.1.9. [DOI] [PubMed] [Google Scholar]
- Riazi MA, Brinkman-Mills P, Nguyen T, Pan H, Phan S, Ying F, Roe BA, Tochigi J, Shimizu Y, Minoshima S, et al. The human homolog of insect-derived growth factor, CECR1, is a candidate gene for features of cat eye syndrome. Genomics. 2000;64:277–285. doi: 10.1006/geno.1999.6099. [DOI] [PubMed] [Google Scholar]
- Ritchie RJ, Mattei MG, Lalande M. A large polymorphic repeat in the pericentromeric region of human chromosome 15q contains three partial gene duplications. Hum Mol Genet. 1998;7:1253–1260. doi: 10.1093/hmg/7.8.1253. [DOI] [PubMed] [Google Scholar]
- Schinzel A, Schmid W, Fraccaro M, Tiepolo L, Zuffardi O, Opitz JM, Lindsten J, Zetterqvist P, Enell H, Baccichetti C, Tenconi R, Pagon RA. The ‘cat eye syndrome‘: Dicentric small marker chromosome probably derived from a no.22 (tetrasomy 22pter to q11) associated with a characteristic phenotype. Report of 11 patients and delineation of the clinical picture. Hum Genet. 1981;57:148–158. doi: 10.1007/BF00282012. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, Kreiner T, Barinaga M, Schilling J, Scheller RH. A dense core vesicle protein is restricted to the cortex of granules in the exocrine atrial gland of Aplysia california. J Biol Chem. 1989;264:16933–16940. [PubMed] [Google Scholar]
- Spencer CA, Gietz RD, Hodgetts RB. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature. 1986;322:279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Winston F, Allis CD. The bromodomain: A chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Nikawa J. Phosphatidylserine synthase from yeast. Biochem Biophys Acta. 1997;1348:228–235. doi: 10.1016/s0005-2760(97)00102-1. [DOI] [PubMed] [Google Scholar]
- Yao Z, Spriggs MK, Derry JM, Strockbine L, Park LS, VandenBos T, Zappone JD, Painter SL, Armitage RJ. Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine. 1997;9:794–800. doi: 10.1006/cyto.1997.0240. [DOI] [PubMed] [Google Scholar]