Identification and Characterization of a Peptide That Specifically Binds the Human, Broadly Neutralizing Anti-Human Immunodeficiency Virus Type 1 Antibody b12 (original) (raw)
Abstract
Human monoclonal antibody (MAb) b12 recognizes a conformational epitope that overlaps the CD-4-binding site of the human immunodeficiency virus type 1 (HIV-1) envelope. MAb b12 neutralizes a broad range of HIV-1 primary isolates and protects against primary virus challenge in animal models. We report here the discovery and characterization of B2.1, a peptide that binds specifically to MAb b12. B2.1 was selected from a phage-displayed peptide library by using immunoglobulin G1 b12 as the selecting agent. The peptide is a homodimer whose activity depends on an intact disulfide bridge joining its polypeptide chains. Competition studies with gp120 indicate that B2.1 occupies the b12 antigen-binding site. The affinity of b12 for B2.1 depends on the form in which the peptide is presented; b12 binds best to the homodimer as a recombinant polypeptide fused to the phage coat. Originally, b12 was isolated from a phage-displayed Fab library constructed from the bone marrow of an HIV-1-infected donor. The B2.1 peptide is highly specific for b12 since it selected only phage bearing b12 Fab from this large and diverse antibody library.
Anti-human immunodeficiency virus type 1 (HIV-1) neutralizing antibodies (Abs) first appear months after the viremia that follows initial infection (1, 18, 27). This response, however, is highly type specific. Neutralizing Ab responses may broaden later in the infection (5, 24) but usually remain poor and occur sporadically in the majority of patients, including long-term-infected individuals (11, 23).
Only three broadly conserved neutralizing epitopes have been identified thus far on the viral envelope; they are defined by human monoclonal Abs (MAbs) b12, 2G12, and 2F5. MAb b12 binds to a discontinuous epitope that overlaps the CD4-binding site on gp120. MAb 2G12 recognizes a complex discontinuous epitope involving the C3-V4 region of gp120 and carbohydrate (34). MAb 2F5 binds to a linear epitope on the ectodomain of gp41 (8, 26, 32); however, the simplicity of this epitope is deceptive, since immunizations with recombinant influenza virus (25) or fusion proteins bearing this epitope (13, 17) have failed to produce significant 2F5-like neutralizing Ab responses, indicating that the native epitope on gp41 is more complex than the six-residue linear sequence. MAbs b12, 2G12, and 2F5 have shown in vitro neutralizing activity against a wide variety of primary isolates (7, 8, 12, 29, 33). Moreover, passive transfer of b12, 2F5, and 2G12 can provide sterile protection if adequate concentrations are achieved before HIV-1 exposure. Studies with 2F5, 2G12, and HIVIG showed that macaques were protected from intravenous (19) and vaginal (21) challenges with pathogenic SHIV 89.6PD (30). Passive immunization with IgG1 b12 protects hu-PBL-SCID mice from an HIV-1 primary-isolate challenge before and shortly after an intravenous viral challenge; (14) and macaques from a vaginal challenge with pathogenic R5 SHIV 162P (P. W. H. I. Parren, P. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton, submitted for publication).
The success of these passive-immunization studies indicates an obvious goal in the development of a prophylactic vaccine: to elicit Abs having neutralizing activities similar to those of the currently known, broadly neutralizing MAbs (b12, 2G12, and 2F5). Yet, all of the recombinant envelope-based vaccine candidates tested so far in clinical trials have been unable to elicit significant neutralizing responses against HIV-1 primary isolates (9, 20, 22), even in cases in which b12, 2F5, and 2G12 bound well to the immunizing subunit antigen, indicating that their respective epitopes are antigenic on these forms of the envelope proteins. Furthermore, these neutralizing epitopes are not recognized to any significant degree during natural infection; instead, as mentioned above, serum Abs having only weak cross-neutralizing titers are typically produced. Of the large number of MAbs cloned from infected donors, b12, 2G12, and 2F5 are the only ones reported so far that neutralize a broad spectrum of primary HIV-1 isolates. Thus, although the epitopes known to mediate broad neutralization are present on recombinant envelope proteins and on envelope proteins produced during natural infection, they do not elicit significant neutralizing Ab responses against primary isolates.
The low apparent immunogenicity of these neutralizing epitopes on the envelope proteins may be circumvented if suitable small molecules mimicking them can be generated (i.e., molecules that bind tightly to the combining sites of the neutralizing MAbs) and then presented in such a form that they elicit the cognate Abs. Our approach in developing a vaccine against HIV-1 has been to identify peptides that are specific for b12, 2F5, and 2G12 and to develop these into a vaccine that will actively target the production of broadly neutralizing Ab responses having specificities that are similar to these MAbs. This report describes the identification and characterization of a peptide that binds specifically to MAb b12.
We used biotinylated IgG1 b12 (6, 7) to screen a panel of 11 peptide libraries displayed on the major coat protein of filamentous bacteriophage (pVIII) as described in reference 4. Two clones, Ed1 and Ed2, were identified that bound b12; DNA sequencing revealed the amino-acid sequences of their displayed peptides, as shown in Table 1. The peptides displayed by these clones share the motif: SDLX3CI; however, the Ed1 sequence bears two Cys residues whereas Ed2 bears only a single Cys, whose position is the same in both clones. Thus, a set of two phage sublibraries displaying the shared residues and reflecting the Cys content of the two Ed clones was constructed as described in reference 4. The resulting sublibraries bear the random-peptide sequences XCX3SDLX3CI (B1 sublibrary, two fixed Cys residues) and X7SDLX3CI (B2 sublibrary, one fixed Cys residue), respectively. These sublibraries were screened with biotinylated IgG1 b12, yielding phage bearing the B1 and B2 peptide sequence families shown in Table 1. All but one of the selected phage clones bear two Cys residues, and all of the clones bound IgG1 b12, as shown by a direct phage enzyme-linked immunosorbent assay (ELISA) performed as described in reference 4.
TABLE 1.
Sequences and ELISA signals of peptide phage clones affinity selected by biotinylated IgG1 b12
| Clone | Peptide sequencea | OD405–490e | ||
|---|---|---|---|---|
| IgG1 b12 | Fab b12 | |||
| 37°C | 4°C | 37°C | ||
| Ed1 | T C LW SDL RAQ CI | NDb | ND | ND |
| B1 library | X C XX SDL XXX CI | |||
| B1.2 | G C LY SDL LAT CI | 1.321 | 0.013 | 0.019 |
| B1.11 | N C LY SDL TQS CI | 1.236 | 0.016 | 0.018 |
| B1.9 | N C LY SDL YAR CI | 1.223 | 0.013 | 0.015 |
| B1.20 | K C MY SDL LGI CI | 1.153 | 0.012 | 0.021 |
| B1.10 | D C LY SDL ESR CI | 0.818 | 0.015 | 0.021 |
| B1.4 | S C LY SDL LEL CI | 0.750 | ND | ND |
| B1.3 | E C MW SDL ELR CI | 0.571 | ND | ND |
| B1.12 | N C LW SDL EQF CI | 0.343 | ND | ND |
| Ed2 | REKRWIF SDL THT CI | ND | ND | ND |
| B2 library | XXXXXXX SDL XXX CI | |||
| B2.1 | HERSYMF SDL ENR CI | 1.236 | 1.071 | 0.219 |
| B2.11 | CSRNQLW SDL HGS CI | 1.207 | 0.012 | 0.016 |
| B2.12 | NNQGCLW SDL TAS CI | 1.189 | 0.013 | 0.017 |
| B2.18 | STTRCTW SDL YDS CI | 1.141 | 0.011 | 0.016 |
| B2.8 | QSSSCMW SDL FQQ CI | 0.992 | 0.016 | 0.018 |
| B2.6 | AQKQCTW SDL LSR CI | 0.903 | 0.019 | 0.018 |
| B2.7 | RPCRGVY SDL LDK CI | 0.886 | 0.016 | 0.020 |
| B2.10 | SSDHCLW SDL TMT CI | 0.644 | ND | ND |
| B2.3 | LPSSCSW SDL LNR CI | 0.276 | ND | ND |
| B2.15 | HTCAGTW SDL LST CI | 0.252 | ND | ND |
| gp120c | 1.121 | 0.863 | 1.166 | |
| f88-4d | 0.075 | 0.014 | 0.019 |
The deduced amino acid sequences of the peptides displayed by the phage clones isolated from the sublibraries revealed a more detailed consensus for both the B1 peptides alone and the B1 and B2 peptides. Almost all of the clones selected from the B1 library contain Leu followed by an aromatic amino acid (usually Tyr) N terminal to the fixed Ser-Asp-Leu sequence. Similarly, clones from the B2 library most often bear a hydrophobic residue (usually Leu), followed by an aromatic one (usually Trp), at this site. Most of the clones from the B2 sublibrary screening have a second Cys (selected for in the screening). The peptide displayed by only one clone, B2.1, contains a single Cys residue; this peptide sequence shares similarities with those of other clones from the B2 sublibrary, and even more similarity with the Ed2 sequence, in the region N terminal to the fixed Ser-Asp-Leu sequence. The B2.1 phage was significant in binding more tightly to b12 than the other clones. Signals for almost all of the clones were strong in ELISAs performed with IgG1 b12, whereas binding was much reduced in assays using Fab b12 when it was reacted with phage at 4°C and still lower when it was reacted at 37°C (Table 1). Fab b12 bound only the B2.1 and Ed2 peptides with signals above the background, and in a side-by-side titration experiment, it was further demonstrated that the binding of b12 to B2.1 was significantly stronger than that to Ed2 (data not shown); therefore, the Ed2 peptide was not further characterized.
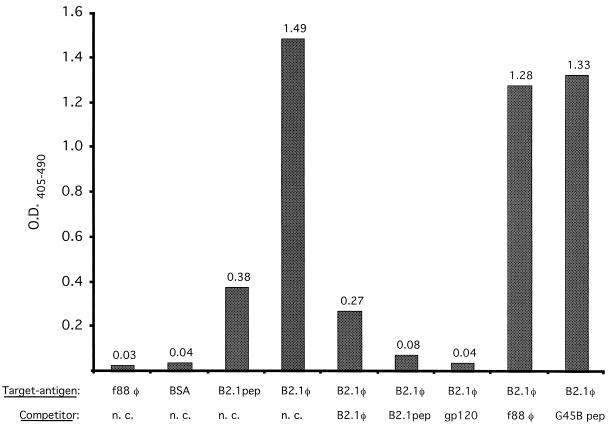
The ability of the B2.1 peptide to bind to the antigen-binding site of b12 was assessed in a competition ELISA. Biotinylated IgG1 b12 (1 nM) was preincubated with gp120Ba-L (100 nM) and reacted with plate-adsorbed B2.1 phage. Results in Fig. 1 show that gp120 blocked the binding of IgG1 b12 to immobilized B2.1 phage, indicating that the peptide binds to the antigen-binding site of IgG1 b12. The binding to B2.1 phage was also blocked by B2.1 synthetic peptide (300 μM), nonbiotinylated IgG1 (100 nM), and the recombinant B2.1 phage but not by f88-4 phage or the unrelated synthetic peptide G45B.
FIG. 1.
Analysis of binding of biotinylated IgG1 b12 to B2.1 phage (B2.1φ) and B2.1 synthetic peptide (B2.1pep) by ELISA. Competition for IgG1 b12 binding to plate-adsorbed B2.1 phage by the following in-solution competitors is shown: 2 × 1010 B2.1 phage, 300 μM B2.1 synthetic peptide, 100 nM gp120Ba-L (gp120), f88-4 phage (f88 φ), and unrelated peptide G45B, whose sequence is VERSKAFSNCYPYDVPDYASLRS. BSA is bovine serum albumin, and n. c. indicates no in-solution competitor. O.D.405–490, optical density at 405 minus 490 nm.
The specificity of the B2.1 peptide for b12 was also assessed. MAb b12 was originally isolated from a phage-displayed Fab library constructed from the bone marrow of HIV-1-infected donor M and subsequently screened with recombinant gp120 (6). To study the specificity of the B2.1 peptide for b12, we tested whether B2.1 would select phage bearing b12 out of the repertoire of expressed Fabs from donor M. Table 2 shows that yields of 10−1% were obtained after four rounds of panning of the M phage library on B2.1 phage. Moreover, the Fabs from all 12 independent phage clones that were sequenced from this phage pool were identical to b12. Thus, even though the M library contains a large number of other Fabs that recognize the CD4-binding site of gp120 (2), B2.1 selected only phage bearing Fab b12.
TABLE 2.
Percent yields of four successive rounds of affinity selection of phage-displayed Fab library M with B2.1 phagea
| Round and phage or protein immobilized on plate | Input (TU, 109) | Output (TU, 104) | % Yield |
|---|---|---|---|
| 1 | |||
| B2.1 | 62 | 4.8 | 9.2 × 10−5 |
| f88-4 | 62 | 4.0 | 7.8 × 10−5 |
| gp120 | 62 | 9.6 | 1.8 × 10−4 |
| 2 | |||
| B2.1 | 6.6 | 2.4 | 3.6 × 10−4 |
| f88-4 | 6.6 | 1.6 | 2.4 × 10−4 |
| gp120 | 6.6 | 140 | 2.1 × 10−2 |
| 3 | |||
| B2.1 | 2.1 | 14 | 6.8 × 10−3 |
| f88-4 | 2.1 | 14 | 6.8 × 10−3 |
| gp120 | 2.1 | 3,200 | 1.5 |
| 4 | |||
| B2.1 | 2.7 | 400 | 1.5 × 10−1 |
| f88-4 | 2.7 | 19 | 8.8 × 10−3 |
| gp120 | 2.7 | 110 | 4.1 × 10−1 |
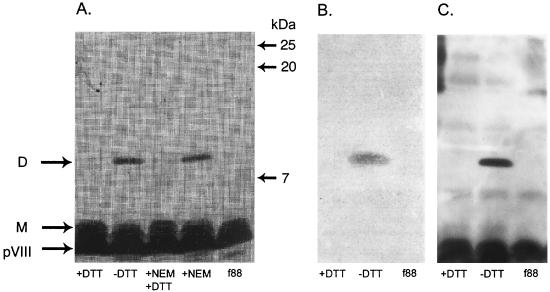
To produce synthetic peptides bearing the B2.1 sequence, we investigated the condition of the thiol group of the single Cys residue that is present in the B2.1 sequence. As multiple copies of the peptide-pVIII fusion protein are incorporated into the phage coat, the single Cys residue of B2.1-pVIII may potentially be in a reduced form (as a reduced thiol group) or disulfide bridged to a second copy of the B2.1-pVIII fusion protein. If the B2.1 peptide-pVIII fusion protein existed as a homodimer on the phage surface, it would have roughly twice the molecular weight of the pVIII monomer. Thus, B2.1 phage were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) using Tris-Tricine buffer as previously described (35). Phage samples were initially treated with the thiol-reactive reagent _N_-ethylmaleimide (NEM) (Fig. 2 A), which blocks free thiols that might be present on the phage coat and would prevent the formation of pVIII dimers after solubilization of the phage coat proteins with heat and SDS. Hence, if B2.1-pVIII fusions bear free thiols and are monomeric, reaction with NEM should prevent them from dimerizing after dissociation of the phage. Alternatively, if the B2.1-pVIII fusions exist on the phage coat as dimers (produced by disulfide bridging between displayed B2.1 peptides), treatment of the phage with NEM, followed by boiling in the presence of SDS, should not affect their migration as dimers. The results shown in Fig. 2A reveal that the recombinant pVIII from B2.1 phage migrates as a dimer that is not affected by NEM treatment, whereas it migrated as a monomer in samples treated with the reducing agent dithiothreitol (DTT). Samples sequentially treated with NEM and DTT also behaved as monomers. This proves that most or all of the B2.1 peptide displayed on the phage surface is homodimeric.
FIG. 2.
SDS-PAGE analysis of the f88-4 wild-type phage (f88) and recombinant B2.1 phage (all others). Phage were left untreated or treated with DTT, NEM, or NEM followed by DTT and then analyzed by SDS-PAGE. Monomeric (M) and dimeric (D) recombinant pVIII proteins are shown. Proteins in similar gels were either silver stained or transferred to a membrane and subjected to Western blotting with anti-phage Ab or IgG1 b12. Panels: A, silver-stained gel; B, Western blot using IgG1 b12 to show the reactive dimer; C, Western blot using rabbit anti-phage Ab to show the wild-type and recombinant pVIII proteins.
In contrast to the B2.1 dimers, the clones that display peptides containing two Cys residues produced monomers or, less often, a mixture of monomers and dimers, with monomers predominating (data not shown). This result suggests that, as opposed to B2.1, these clones bear mostly intrachain disulfide bridges, consistent with the results of Zwick et al. (35). Their survey of phage-displayed peptides bearing one and two Cys residues showed that almost all containing two Cys residues are cyclic whereas all of those bearing a single Cys residue form homodimers.
The requirement for an intact disulfide bridge for the antigenicity of B2.1, and of clones bearing cyclic peptides, was assessed by Western blot experiments (15) using IgG1 b12 or a rabbit polyclonal anti-phage Ab for detection. Figure 2B shows that IgG1 b12 binds only to the B2.1-pVIII fusion in its dimeric form. Staining with IgG1 b12 was present at the site of the dimer but not at the monomer, whereas both forms were detected by the anti-phage Ab (Fig. 2C). A clone selected from the B1 sublibrary, bearing a peptide-pVIII fusion containing two Cys residues, was also tested by a Western blot assay with IgG1 b12. It produced a band much weaker than that of the B2.1 phage, whereas blotting with the anti-phage Ab produced a recombinant band with an intensity similar to that of the B2.1 clone (data not shown). This supports the conclusions drawn from the ELISA data (Table 1) indicating that IgG1 b12 does not bind as tightly to peptides containing two Cys residues as it does to the B2.1 homodimer. Moreover, as with the B2.1 homodimer, reduction by DTT of the intrachain disulfide bridge of clones containing peptides bearing two Cys residues ablated b12 binding in the ELISA; thus, disulfide-bridging is also required for their antigenicity (data not shown).
The location of the Cys residue (and hence the disulfide bridge) in the B2.1 sequence is crucial to its reactivity with b12. Phage bearing mutations in the B2.1 peptide sequence were prepared and assayed for the abilities to bind IgG1 b12 and produce homodimer and/or monomer bands on analysis by SDS-PAGE. As shown in Table 3, replacement of Cys14 with Ser ablated dimer formation and Ab binding. Interestingly, replacement of Ser4 with Cys ablated binding, regardless of whether the residue at position 14 was Cys; even the dimeric form of this mutant peptide did not bind b12 significantly. Thus, the antigenicity of B2.1 is strongly affected by the presence and location of the disulfide bridge that produces homodimers.
TABLE 3.
Binding of b12 IgG to B2.1 phage mutants
| Phage clone | Peptide sequencee | IgG1 b12a | SDS-PAGEb | Western blotc b12 binding | ||
|---|---|---|---|---|---|---|
| 3 nM | 30 nM | Dimer | Monomer | |||
| B2.1 | HERSYMFSDLENRCI | 1.00 | 1.04 | + | + | + |
| B2.1-Δ Cys | HERSYMFSDLENRSI | 0.02 | 0.04 | − | + | − |
| B2.1-5′Cys | HERCYMFSDLENRSI | 0.02 | 0.05 | + | + | − |
| B2.1-CC | HERCYMFSDLENRCI | 0.03 | 0.13 | − | + | − |
| f88-4 | 0.02 | 0.03 | − | − | − | |
| None | 0.02 | 0.03 | NAd | NA | NA |
To study the affinity of the B2.1 homodimer out of the context of the phage coat, a synthetic version of the B2.1 peptide was prepared as a disulfide-bridged homodimer with the sequence NH3-HERSYMFSDLENRCIAAEGK-NH2 (Multiple Peptide Systems, San Diego, Calif.; monomer molecular weight, 2,354.6; >95% pure and >95% dimer). This synthetic B2.1 peptide was used as a target with which to isolate phage bearing b12 Fab from the M library; but no phage were selected (data not shown), indicating that the synthetic peptide does not bind b12 as tightly as the phage-borne one. To verify the relatively weak interaction of the synthetic peptide with b12 compared to phage-borne B2.1, a panning reconstruction experiment was performed in which phage bearing Fab b12 were mixed with various amounts of phage bearing unrelated Fab AD27/A47 (as a background control phage). The Fab phage were panned side by side in wells coated with gp120, B2.1 phage, or B2.1 peptide. The results in Table 4 show that gp120 and B2.1 phage enriched b12 phage 50- to 100-fold better than did the synthetic B2.1 peptide. Thus, the affinity of the phage-borne B2.1 for the b12 Fab appears to be stronger than that of the synthetic peptide.
TABLE 4.
Reconstruction panning of Fab b12 phage versus B2.1 peptide, B2.1 phage, and gp120 Ba-La
| Phage reconstruction | B2.1 peptide | B2.1 phage | gp120 Ba-L | |
|---|---|---|---|---|
| Fab b12 | DP47/AD27 | |||
| 1010 | 2.7 × 10−3 | 2.1 × 10−1 | 3.2 × 10−1 | |
| 109 | 9 × 109 | 3.0 × 10−3 | 1.4 × 10−1 | 2.0 × 10−1 |
| 108 | 1010 | 1.5 × 10−4 | 7.7 × 10−2 | 2.8 × 10−2 |
| 107 | 1010 | 1.6 × 10−4 | 3.1 × 10−4 | 1.0 × 10−3 |
| 106 | 1010 | 4.5 × 10−4 | 5.8 × 10−5 | 2.7 × 10−4 |
| 105 | 1010 | 1.2 × 10−4 | 1.3 × 10−4 | 1.2 × 10−4 |
| 104 | 1010 | 3.4 × 10−4 | 2.7 × 10−4 | 3.1 × 10−4 |
| 1010 | 1.6 × 10−4 | 2.0 × 10−4 | 4.9 × 10−4 |
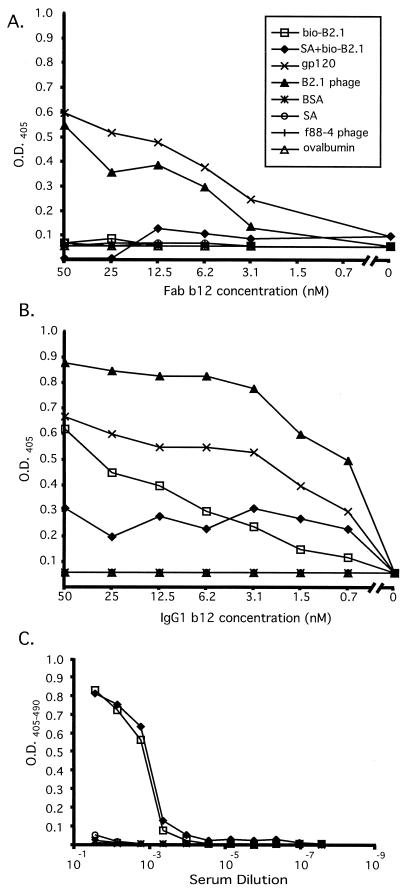
We also prepared a biotinylated, synthetic version of the B2.1 peptide having the sequence NH3-HERSYMFSDLENRCIAAE-Orn(biotin)-KK-NH2 (Multiple Peptide Systems; monomer molecular weight, 2,767.6; >95% pure and 80% dimer). This peptide (bio-B2.1) was biotinylated so that it could be bound to immobilized streptavidin in ELISA wells and directly detected during the production of conjugates for immunization, regardless of its IgG1 b12 antigenicity. The relative affinity of MAb b12 for the B2.1 sequence presented in different forms was assessed by direct titrations using Fab and IgG1 b12. The titrations were performed on streptavidin-captured and plate-immobilized bio-B2.1 peptide, as well as with recombinant B2.1 phage (and gp120 as a positive control). Figure 3A shows that the binding of Fab b12 to both plate-immobilized and streptavidin-captured synthetic B2.1 peptide was almost undetectable over the background. In contrast, Fab binding to recombinant B2.1 phage was strong and followed a titration curve similar to that of gp120 (Fig. 3A), suggesting that the affinities of b12 for gp120 and phage-displayed peptide are similar (_Kd_s of 3 nM [gp120MN] and 9.1 nM [gp120LAI] have been reported by Roben et al. [31] and Parren et al. [28], respectively). Although the results were somewhat different when IgG1 b12 was used instead of Fab for the titration ELISA (which was most likely due to the inherent avidity of the IgG), a similar trend was observed (Fig. 3B). IgG1 b12 reacted with both phage-displayed and synthetic B2.1 peptide; however, it bound more tightly to recombinant B2.1 phage than to either form of the synthetic peptide. Moreover, the Ab showed better binding to the plate-adsorbed peptide than to the streptavidin-captured one; thus, it was able to discriminate between these two means of presenting the peptide. In contrast to IgG1 b12, the IgG from a mouse that had been immunized with a B2.1 conjugate vaccine (see below) showed no discrimination between the streptavidin-bound and plate-adsorbed forms of bio-B2.1 (Fig. 3C) and binding to gp120 was undetectable. These results indicate that the ability of b12 to discriminate between plate-immobilized peptide and streptavidin-captured peptide is linked with its capacity to bind gp120, again suggesting that a specific B2.1 structure (or set of structures) is responsible for its antigenicity for b12. It is apparent from these titration experiments that the most antigenic structure of the B2.1 sequence is best represented by the recombinant peptide in the context of the phage coat.
FIG. 3.
Titration of Fab b12 (A), IgG1 b12 (B), and murine anti-B2.1 peptide serum (C) on different immobilized antigens. Twofold dilutions of Fab and IgG1 b12 and fourfold mouse serum dilutions were reacted with biotinylated B2.1 directly adsorbed to ELISA wells (bio-B2.1), biotinylated B2.1 bound to immobilized streptavidin (SA+bio-B2.1), gp120Ba-L, B2.1 recombinant phage, f88-4 phage, bovine serum albumin (BSA), ovalbumin, and streptavidin (SA). O.D., optical density.
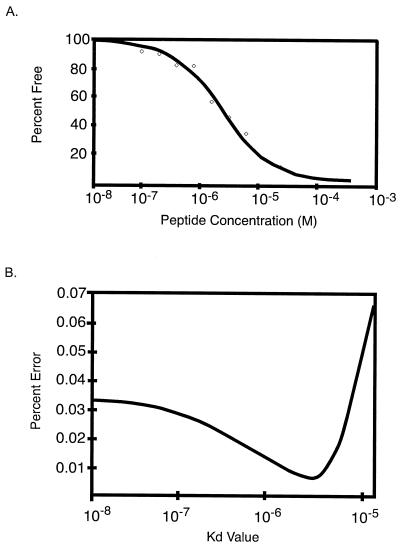
To assess the range of affinities of the different peptides for b12, the in-solution binding affinity of IgG1 b12 for the B2.1 peptide was determined by using a KinExA 3000 (Kinetic Exclusion Assay) instrument (Sapidyne Instruments, Inc., Boise, Idaho) (3) as described in reference 10. KinExA measurements involving in-solution monovalent antigen yields affinity constants that are independent of the Ab valency. The data in Fig. 4 show that the interaction between IgG1 b12 and the free peptide closely follow a 2.5-μM Kd best-fit theoretical curve derived from a simple second-order kinetic model (Fig. 4A). Comparison of the percent root mean square deviation errors (Fig. 4B) produced from the fit of these data to the best-fit curves calculated for a range of _Kd_s revealed the accuracy of the Kd found for b12 and B2.1 in solution. This Kd is ≈200-fold higher than the 9.1-nM Kd measured for the interaction between Fab b12 and recombinant gp120 from HIV-1LAI, as determined by surface plasmon resonance (31). However, the Kd is lower than the ≈100 μM found for a synthetic cyclic peptide made from one of the clones isolated from the B1 library by competition ELISA of that peptide with Fab b12 (A. Satterthwait and J. K. Scott, unpublished data).
FIG. 4.
Kinetics of binding of IgG1 b12 to B2.1 peptide in-solution. (A) Percent free Ab versus molar concentration of peptide; data (diamonds) and the best-fit theoretical curve are shown. (B) Percent error from fit of the data in Fig. 3A to the best-fit curves calculated for a range of K _d_s. The 95% confidence interval calculated for this experiment is 1.3 to 3.7 μM.
The Fab and IgG1 titration data and the in-solution affinities of b12 for B2.1 and gp120 may be used to provide very rough reference values from which the affinity of the plate-bound and phage-displayed peptides could be interpolated. Given the range of 9 nM for gp120LAI and ≈3 μM for the free peptide (and assuming that the affinity of free B2.1 is similar to that of bio-B2.1 captured on streptavidin), we speculate that the plate-adsorbed peptide binds with a Kd ranging between 20 and 500 nM. The phage-displayed recombinant peptide shows the highest affinity for binding to Fab b12, with the data suggesting a Kd value close to that of b12 for gp120.
Taken together, our results support the idea that the affinity of the B2.1-b12 interaction is dependent on the environment in which the peptide is presented to b12. The data suggest the existence of different structures of the B2.1 homodimer and indicate that the predominant structure of B2.1 in solution (and tethered, via biotin, to streptavidin) is either unfolded or unstable and different from the one(s) that it assumes in the context of the phage coat. Our results obtained with synthetic and recombinant B2.1 peptides indicate that the structure of the homodimer could be further optimized to maximize its antigenicity. B2.1 binds b12 preferentially when fused to the pVIII coat protein and displayed on the phage surface, perhaps because the highly structured phage coat provides a more rigid and/or stable environment for the peptide. This would be in keeping with the work of Jelinek et al. (16), which shows that antigenic peptides fused to pVIII produce nuclear magnetic resonance (NMR)-definable structures, even though the free peptide in solution does not.
The sequences of the peptides we have discovered (Table 1), especially B2.1, show significant homology to the D-loop region of gp120 (residues 273 to 285). The residues of the B2.1 sequence that are shared with D-loop sequences from a number of gp120s are bolded in HERSYMFSDLENRCI. The D loop region of gp120 contains a number of residues that are highly conserved among HIV-1 isolates from different clades. Frequencies in gp120s from all clades for the bolded residues in the B2.1 sequence are 96% for Arg273, 96% for Ser274, 52% for Phe277, 99% for Ser or Thr at position 278, 99% for Asp or Asn at position 279, and 98% for Ile285 (frequency data were taken from the Los Alamos Env sequence database at: http://hiv-web.lanl.gov/). As well, residue Asp279 of the D loop also makes contact with CD4 and thus forms part of the CD4 binding site on gp120 (16a).
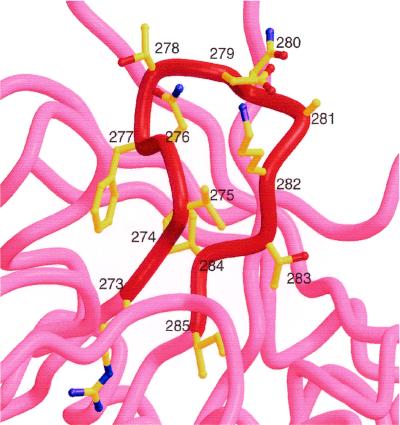
Figure 5 shows the sequence and structure of the D loop of HXB2 gp120; it appears to be partially stabilized by the interaction between the side chains of Val275 and Ile284. The b12-selected clones that contain two Cys residues most often have loop lengths of eight residues (Table 1). If the sequences of the cyclic peptides having 8-mer loops are overlaid on the D loop, with their N- and C-terminal Cys residues being placed where Val275 and Ile284 of the D loop are located, respectively, homologous residues shared between the two are perfectly aligned. The location of the FSD sequence aligns with that of the D loop's FTD sequence, and the N-terminal Ile of the peptides aligns with the gp120 Ile285.
FIG. 5.
Structure of the D loop of gp120, residues 273 to 285, taken from the HXB2 HIV-1 isolate. The sequence of this region, whose alpha-carbon backbone is shown in red, is RSVNFTDNAKTII. Residues shared with the B2.1 peptide, HERSYMFSDLENRCI, are in bold type.
The homologies between residues in B2.1 and conserved residues in the D loop, as well as the structural homologies with the cyclic peptides, lead us to predict that the D loop of gp120 is involved in binding the b12 Ab, with the residues RS, FSD, and I being of importance in maintaining a D-loop-like structure and/or in making direct contacts with the b12 Ab. The crystallographic structure of IgG1 b12 has been elucidated, both in the free form and bound to the B2.1 peptide (E. O. Saphire, personal communication). These structures should prove useful in directing further optimization of the B2.1 peptide as a b12 ligand and as a gp120 mimic and in characterizing the gp120 epitope for b12 at the atomic level.
Our goal in developing a peptide mimic of the b12 epitope is to use it in a vaccine against HIV-1 infection to elicit b12-like neutralizing Abs. We conjugated the biotinylated synthetic peptide to wild-type phage and ovalbumin using BS3 as a cross-linker. The conjugates bound IgG1 b12, as shown by Western blot assay, ELISA, and immunoprecipitation, indicating that the antigenic structure of B2.1 was conserved after the conjugation. We found that both conjugates were immunogenic in mice and rabbits but did not elicit significant gp120-cross-reactive Ab titers, indicating that b12-like Abs were not produced at detectable levels (data not shown). At least two reasons may account for this: (i) the relatively low affinity of b12 for the synthetic version of the peptide and/or (ii) the species barrier (b12 has an 18-residue-long H3, and Abs with such features are not produced in mice). We also carried out immunizations with B2.1 recombinant phage, which produced only moderate anti-peptide Ab titers, accompanied by high anti-phage Ab responses. In addition to the species problem mentioned above, we believe that the relatively low copy number of the B2.1 homodimer displayed on the phage surface (≈200 copies/phage) explains this result.
Thus, the isolation and characterization of the B2.1 peptide constitute the first stage of a new strategy for targeting the production of Abs against a single prespecified neutralizing epitope on HIV-1. However, the generation of a successful B2.1 immunogen requires further optimization at several levels. Our results indicate that the structure of the homodimer displayed on the phage coat is best for the binding of b12; thus, the recombinant phage is the primary target of our efforts at structure-based optimization of B2.1 as an antigen and immunogen. We are currently assessing the peptide residues that are critical for b12 binding in the context of the phage. These functional data, coupled with the crystallographic data mentioned above, should provide insight into further optimization of the B2.1 peptide. Such optimization also requires knowledge of the phage-borne structure of B2.1 Thus, we are also making efforts to raise the copy number of the B2.1 homodimer on the phage coat, to allow NMR-based analyses, and to generate soluble B2.1 fusion proteins so that the dimer can be studied in a “monovalent” protein format (compared to phage particles, which bear multiple copies of the dimer). Other immunization strategies (a prime-boost approach) and species (monkeys and XenoMouse) will also be explored.
Acknowledgments
This work was supported by grants from the NHRDP (J.K.S.), the MRC (MT-14562 to J.K.S.), and the NIH (R21-AI44395 to J.K.S., AI42653 to P.W.H.I.P., and AI33292 to D.R.B.). M.B.Z. was supported by a predoctoral scholarship from the NSERC, and J.K.S. was supported in part by a fellowship from the BCHRF.
We thank Edward Leong, Kelly Brown, Nienke van Houten, Firmin Hung, and Ann Hessell for excellent technical contributions to this work and Brett Vanderkist for help with the figures. We gratefully acknowledge Arnold Satterthwait for cyclic peptide studies and Tim Fouts for providing gp120. Figure 5 was kindly provided courtesy of Robyn Stanfield.
REFERENCES
- 1.Ariyoshi K, Harwood E, Chiengsong-Popov R, Weber J. Is clearance of HIV-1 viremia at seroconversion mediated by neutralising antibodies? Lancet. 1992;340:1257–1258. doi: 10.1016/0140-6736(92)92953-d. [DOI] [PubMed] [Google Scholar]
- 2.Barbas C F, III, Collet T A, Amberg W, Roben P, Binley J M, Hoekstra D, Cababa D, Jones T M, Williamson R A, Pilkington G R, Haigwood N L, Cabezas E, Satterthwait A C, Sanz I, Burton D R. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 3.Blake R C, II, Pavlov A R, Blake D A. Automated kinetic exclusion assays to quantify protein binding interactions in homogeneous solution. Anal Biochem. 1999;272:123–134. doi: 10.1006/abio.1999.4176. [DOI] [PubMed] [Google Scholar]
- 4.Bonnycastle L L C, Mehroke J S, Rashed M, Gong X, Scott J K. Probing the basis of antibody reactivity with a panel of constrained peptide libraries displayed by filamentous phage. J Mol Biol. 1996;258:747–762. doi: 10.1006/jmbi.1996.0284. [DOI] [PubMed] [Google Scholar]
- 5.Bradney A P, Scheer S, Crawford J M, Buchbinder S P, Montefiori D. Neutralization escape in human immunodeficiency virus type 1 infected long-term nonprogressors. J Infect Dis. 1999;179:1264–1267. doi: 10.1086/314711. [DOI] [PubMed] [Google Scholar]
- 6.Burton D R, Barbas III C F, Persson M A A, Koening S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton D R, Jayashree P, Kodury R, Sharp S J, Thornton G B, Parren P W H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P, Lamacchia M, Garrati E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F., III Efficient neutralization of primary isolates of HIV 1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 8.Conley A J, Kessler J A, I. I, Boots L J, Tung J S, Arnold B A, Keller P M, Shaw A R, Emini E A. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc Natl Acad Sci USA. 1994;91:3348–3352. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor R I, Korber B T, Graham B S, Hahn B H, Ho D D, Walker B D, Neumann A U, Vermund S H, Mestecky J, Jackson S, Fenamore E, Cao Y, Gao F, Kalams S, Kunstman K J, McDonald D, McWilliams N, Trkola A, Moore J P, Wolinsky S M. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J Virol. 1998;72:1552–1576. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig L, Sanschagrin P C, Rozek A, Lackie S, Kuhn L A, Scott J K. The role of structure in antibody cross-reactivity between peptides and folded proteins. J Mol Biol. 1998;281:183–201. doi: 10.1006/jmbi.1998.1907. [DOI] [PubMed] [Google Scholar]
- 11.Dreyer K, Kallas E G, Planelles V, Montefiori D, McDermott M P, Hasan M S, Evans T G. Primary isolate neutralization by HIV type 1-infected patient sera in the era of highly active antiretroviral therapy. AIDS Res Hum Retrovirol. 1999;15:1563–1571. doi: 10.1089/088922299309856. [DOI] [PubMed] [Google Scholar]
- 12.D'Souza, M. P., D. Livnat, J. A. S. H. Bradac. Bridges, The AIDS Clinical Trials Group Antibody Selection Working Group, and collaborating investigators. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. **197:**1056–1062. [DOI] [PubMed]
- 13.Eckhart L, Raffelsberger W, Ferko B, Klima A, Purtscher M, Katinger H, Ruker F. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J Gen Virol. 1996;77:2001–2008. doi: 10.1099/0022-1317-77-9-2001. [DOI] [PubMed] [Google Scholar]
- 14.Gauduin M C, Parren P W H I, Weir R, Barbas III C F, Burton D R, Koup R A. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 16a.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jelinek R, Terry T D, Gesell J J M P, Perham R N, Opella S J. NMR structure of the principal neutralizing determinant of HIV-1 displayed in filamentous bacteriophage coat protein. J Mol Biol. 1997;266:649–655. doi: 10.1006/jmbi.1996.0821. [DOI] [PubMed] [Google Scholar]
- 17.Liang X, Munshi S, Shendure J, Mark G, III, Davies M E, Freed D C, Montefiori D C, Shiver J W. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine. 1999;17:2862–2878. doi: 10.1016/s0264-410x(99)00125-5. [DOI] [PubMed] [Google Scholar]
- 18.Locher C P, Grant R M, Collisson E A, Reyes-Teran G, Elbeik T, Khan J O, Levy J A. Antibody and cellular immune responses in breakthrough infection subjects after HIV type 1 glycoprotein 120 vaccination. AIDS Res Hum Retrovir. 1999;15:1685–1689. doi: 10.1089/088922299309720. [DOI] [PubMed] [Google Scholar]
- 19.Mascola J R, Lewis M G, Tiegler G, Harris D, VanCott T C, Haynes D, Louder M K, Brown C R, Sapan C V, Frankel S S, Lu Y, Robb M L, Katinger H, Birx D. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascola J R, Snyder S W, Weislow O S, Belay S M, Belshe R B, Schwartz D H, Clements M L, Dolin R, Graham B S, Gorse G J, Keefer M C, McElrath M J, Walker M C, Wagner K F, McNeil J G, McCutchan F E, Burke D S. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 21.Mascola J R, Stiegler G, VanCott T C, Katinger H, Carpenter C B, Hanson C E, Beary H, Hayes D, Frankel S S, Birx D L, Lewis M G. Protection of macaques against vaginal transmission of a pathogenic HIV 1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 22.McCormack S, Tilzey A C, Jones A, G, F, K, J, N, A, G, Lister S, Beddows S, Cheingsong R, Rees A, Babiker A, Banatvala J, Bruck C, Darbyshire J, Tyrrell D, VanHoecke C, Weber J. A phase I trial in HIV negative healthy volunteers evaluating the effect of potent adjuvants on immunogenicity of a recombinant gp120W61D derived from dual tropic R5X4 HIV-1ACH320. Vaccine. 2000;18:1166–1177. doi: 10.1016/s0264-410x(99)00388-6. [DOI] [PubMed] [Google Scholar]
- 23.Montefiori D C, Pantaleo G, Fink L M, Zhou J T, Zhou J Y, Bilska M, Miralles G D, Fauci A S. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J Infect Dis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- 24.Moog C H, Fleury J A, Pellegrin I, Kirn A, Aubertin A M. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:3734–3741. doi: 10.1128/jvi.71.5.3734-3741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muster T, Ferko B, Klima A, Purtscher M, Trkola A, Schulz P, Grassauer A, Engelhardt O G, Garcia-Sastre A, Palese P, et al. Mucosal model of immunization against human immunodeficiency virus type 1 with a chimeric influenza virus. J Virol. 1995;69:6678–6686. doi: 10.1128/jvi.69.11.6678-6686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyambi P N, Lewi P, Peeters M, Janssens W, Heyndrickx L, Fransen K, Andries K, Vanden Haesevelde M, Heeney J, Piot P, van der Groen G. Study of the dynamics of neutralization escape mutants in a chimpanzee naturally infected with the simian immunodeficiency virus SIVcpz-ant. J Virol. 1997;71:2320–2330. doi: 10.1128/jvi.71.3.2320-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parren P W H I, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parren P W H I, Moore J P, Burton D R, Sattentau Q J. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13:S137–S162. [PubMed] [Google Scholar]
- 30.Reimann K A, Li J T, Veazey R, Halloran M, Park I-W, Karlsson G B, Sodroski J, Letvin N. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roben P, Moore J P, Thali M, Sodroski J, Barbas III C F, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trkola A, Grassauer A, Schulz P M, Klima A, Dopper S, Gruber G, Buchacher A, Muster T, Katinger H. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS. 1996;10:587–593. doi: 10.1097/00002030-199606000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P G, Allaway G P, Katinger H, Barbas III C F, Burton D R, Ho D D, Moore J P. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zwick M B, Shen J, Scott J K. Homodimeric peptides displayed by the major coat protein of filamentous phage. J Mol Biol. 2000;300:307–320. doi: 10.1006/jmbi.2000.3850. [DOI] [PubMed] [Google Scholar]