Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology (original) (raw)
Abstract
Interleukin-10 (IL-10) is a key inhibitory signal of inflammatory responses that regulates the production of potentially pathogenic cytokines like tumor necrosis factor (TNF). We show here that the development of chronic intestinal inflammation in IL-10-deficient mice requires the function of TNF, indicating that the IL-10/TNF axis regulates mucosal immunity. We further show that IL-10 targets the 3′ AU-rich elements (ARE) of TNF mRNA to inhibit its translation. Moreover, IL-10 does not alter TNF mRNA stability, and its action does not require the presence of the stability-regulating ARE binding factor tristetraprolin, indicating a differential assembly of stability and translation determinants on the TNF ARE. Inhibition of TNF translation by IL-10 is exerted mainly by inhibition of the activating p38/MAPK-activated protein kinase-2 pathway. These results demonstrate a physiologically significant cross-talk between the IL-10 receptor and the stress-activated protein kinase modules targeting TNF mRNA translation. This cross-talk is necessary for optimal TNF production and for the maintenance of immune homeostasis in the gut.
Keywords: animal model/anti-inflammatory/Crohn’s disease/gene expression/knockout
Introduction
Tumor necrosis factor (TNF) plays a central role in diverse immune and inflammatory processes (Kollias et al., 1999). Although, physiologically, TNF actions are beneficial, it is clear that aberrations in TNF production in vivo may be pathological. The potential of TNF to induce chronic inflammatory disease has been exemplified in animal models, where deregulation of its production leads to the development of various pathologies (Kollias et al., 1999). Conversely, interleukin-10 (IL-10) has emerged as a macrophage deactivator competent to suppress the expression of inflammatory mediators as well as the macrophages’ ability to support accessory functions to adaptive immunity (for review see Moore et al., 1993). The anti-inflammatory potential of IL-10 has been repeatedly demonstrated in vivo, in preventing endotoxemia (Gerard et al., 1993; Marchant et al., 1994) and suppressing the development of intestinal inflammation (Kuhn et al., 1993). In particular, an indication of a homeostatic TNF/IL-10 axis against inflammatory bowel disease (IBD) is provided by the fact that mice deficient in IL-10, or in macrophage signaling molecules that mediate IL-10 receptor signals, overproduce TNF and develop IBD (Kuhn et al., 1993; Takeda et al., 1999). Activation of macrophages by lipopolysaccharide (LPS) results in the rapid production of both TNF and IL-10, with similar kinetics (Marchant et al., 1994). However, the mechanism by which IL-10 suppresses TNF expression remains elusive. It is possible that IL-10 may affect the same pathways that are required for LPS-induced TNF production (Geng et al., 1994; Wang et al., 1995). However, there is a great degree of contradictory data as to what level(s) IL-10 exerts its inhibitory action (Bogdan et al., 1991, 1992; de Waal et al., 1991; Wang et al., 1995; Brown et al., 1996; Kishore et al., 1999).
Homeostatic control of TNF biosynthesis is exerted at multiple levels and proceeds through a multitude of signals. For example, binding of LPS to its Toll-receptor complex on macrophages transmits signals through tyrosine kinases, protein kinase C, NFκB, and the mitogen- and stress-activated protein kinases (MAPK/SAPK) ERK, JNK and p38 (reviewed by Beutler, 2000). These signals may affect TNF gene expression directly, at the level of transcription (Collart et al., 1990), splicing (Osman et al., 1999), mRNA stability and translation (Carballo et al., 1998; Kontoyiannis et al., 1999), and protein processing (Peschon et al., 1998). We have recently demonstrated that the AU-rich elements (ARE) residing in the 3′UTR of TNF mRNA are important in controlling message stability and translational activation (Kontoyiannis et al., 1999). The latter appears to rely on the activity of the MAPK/SAPK pathways (Kontoyiannis et al., 1999). Although data on TNF ARE binding factors remain scarce, recent evidence indicated that the prototype member of a zinc finger family of RNA binding proteins, tristetraprolin (TTP), has the capacity to modulate TNF expression in vivo by destabilizing TNF mRNA in an ARE-dependent fashion (Carballo et al., 1998). Whether TTP exclusively modulates TNF mRNA stability, and with what signaling requirements, remains to be demonstrated.
In this report we demonstrate that, in mouse macrophages, IL-10 exerts its anti-inflammatory and immunosuppressive action mainly by targeting ARE-mediated translation of the TNF message through modulation of activating p38/SAPK signals. Aberrations in TNF production due to the absence of IL-10 function in mutant mice are shown here to be causal for intestinal inflammation, indicating that correct regulation of the IL-10/TNF axis is of physiological significance to intestinal immune homeostasis.
Results
Absence of TNF attenuates development of IBD in IL-10-deficient mice
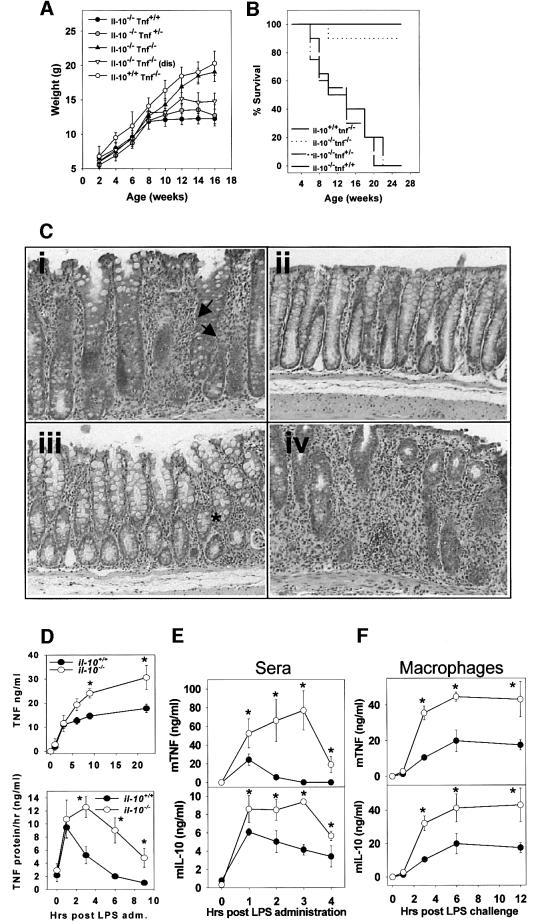
Examination of TNF production in LPS-induced macrophages from normal or IL-10-deficient mice demonstrated that TNF was overexpressed in the latter (Figure 1D). To substantiate whether a defective TNF/IL-10 axis is a general criterion for the development of intestinal inflammation, we assessed the role of TNF in the development of enterocolitis in IL-10-deficient mice by generating double-mutant (_Il-10_–/– _Tnf_–/–) mice. As shown in Figure 1B, the macroscopic disease developing in _Il-10_–/– and _Il-10_–/– Tnf+/– mice was similar, and resulted in lethality for ∼40–50% (n = 27 and 24 mice, respectively) of the animals by the age of 10 weeks. Clinical symptoms included gradual weight loss (Figure 1A) and an increased incidence of colorectal prolapses (in ∼70–80% of mice examined). In sharp contrast, 80% (18/21) of _Il-10_–/– _Tnf_–/– mice showed no symptoms of macroscopic disease and appeared normal throughout the period examined (20 weeks). However, three out of the 21 double-homozygous mice developed clinical symptoms similar to those presented by control groups. Histological evaluation of the large intestine of the control groups revealed moderate to severe inflammatory occurrences (Table I). In sharp contrast, colitis was completely prevented in 77% of the double-homozygous mice (Figure 1C; Table I). However, in the three diseased as well as in two asymptomatic _Il-10_–/– _Tnf_–/– mice, the histological hallmarks of colitis developed with differential onset. These results demonstrate that TNF is dominantly involved in the development of colitis in IL-10-deficient mice.
Fig. 1. Evidence for a defective TNF/IL-10 axis in IL-10 knockout and TNFΔARE mutant mice. (A) Weight distribution and (B) cumulative percent survival of _Il-10_–/– _Tnf_–/– and control mice. (C) Representative photomicrographs (×200) of the proximal colon of _Il-10_–/– _Tnf_–/– from: (i) 8-week-old _Il-10_–/– Tnf+/– control mice showing prominent transmural inflammation (arrows), epithelial cell hyperplasia and goblet cell loss; (ii) 14-week-old _Il-10_–/– _Tnf_–/– colon showing normal epithelial integrity; (iii) 12-week-old _Il-10_–/– _Tnf_–/– asymptomatic mouse showing mild signs of submucosal inflammation (asterisk); (iv) 12-week-old _Il-10_–/– _Tnf_–/– diseased mouse showing severe inflammation. (D) Kinetics of TNF protein accumulation (upper) and production per hour (lower) in Il-10+/+ and _Il-10_–/– TEPM following LPS stimulation in vitro. TNF and IL-10 protein levels in collected supernatants were determined by ELISA. Results shown as mean ± SD values from five cultures/group. (E) Kinetics of TNF and IL-10 protein production in Tnf+/+ (closed circles) and _Tnf_ΔARE/+ (open circles) mice following LPS challenge in vivo. Mice were challenged with 100 µg/ml LPS intraperitoneally and subsequently exsanguinated via cardiac puncture at the indicated time points. TNF and IL-10 protein levels in sera were determined by ELISA. Results shown as mean ± SD values from four mice/time point. (F) Kinetics of TNF and IL-10 in Tnf+/+ (closed circles) and _Tnf_ΔARE/+ (open circles) TEPM and following LPS stimulation, in vitro. TEPM were cultured as before and subsequently stimulated with LPS (1 µg/ml) for 12 h.
Table I. Effects of TNF absence on the incidence and severity of colitis in IL-10–/– mice.
| Genotype | No. of mice affected | Disease score distributiona | Mean disease score (0–20)a | ||||
|---|---|---|---|---|---|---|---|
| 0 | >0–5 | >5–10 | >10–15 | >15–20 | |||
| _Il-10_–/– | 14/14 (100%)† | 0 | 0 | 4 | 7 | 3 | 13.1 ± 4.1* |
| _Tnf_–/– | 0/5 (0%) | 5 | 0 | 0 | 0 | 0 | nil |
| _Il-10_–/– Tnf+/– | 13/15 (87%)†† | 2 | 2 | 3 | 6 | 3 | 10.6 ± 5.9** |
| _Il-10_–/– _Tnf_–/– | 5/21 (23%) | 16 | 2 | 0 | 1 | 2 | 3.47 ± 5.71 |
We next tested in the _Tnf_ΔARE mouse whether defective IL-10 production could be associated with the development of IBD in this system. Interestingly, TNF overexpression in _Tnf_ΔARE mice is accompanied by exacerbated production of IL-10 both in vivo and in vitro (Figure 1E and F). Thus, IL-10 production is enhanced in _Tnf_ΔARE mice, yet its action appears ineffective in suppressing the high TNF load.
IL-10 inhibits TNF production by macrophages at multiple levels
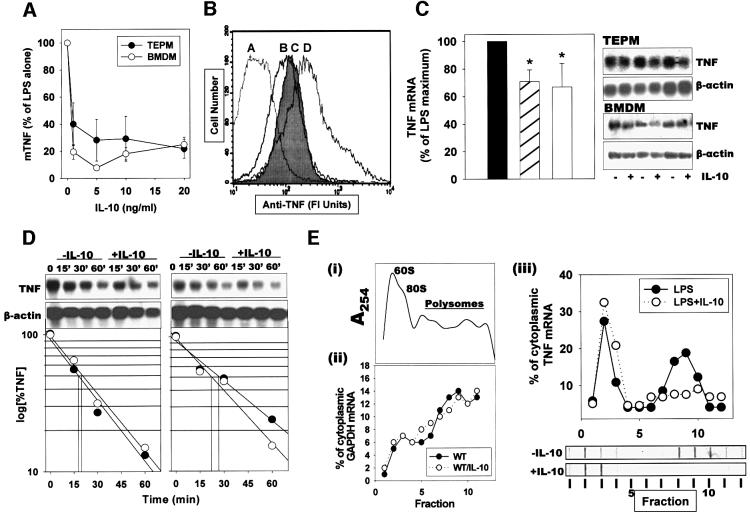
We examined in detail the effect of IL-10 on LPS-stimulated TNF production by murine macrophages. Since IL-10 production coincides with TNF production (Figure 1E and F), we hypothesized that, physiologically, IL-10 targets TNF expression concurrently with the LPS signals. IL-10 inhibited, in a dose-dependent manner, the secretion of TNF by thioglycolate-elicited peritoneal macrophages (TEPM) and bone marrow-derived (BMDM) macrophage cultures, reaching a maximal value of >80% inhibition at 5 ng (Figure 2A). At the same dose, IL-10 also reduced the levels of LPS-induced transmembrane TNF protein accumulation by 80–90%, as indicated by flow cytometry (Figure 2B). In contrast, IL-10 reduced the levels of steady-state TNF mRNA by <20–30% (Figure 2C). To verify whether this reduction in TNF mRNA levels occurred due to a decrease in transcription rates or to a decrease in message stability, we measured the effect of IL-10 on the decay of TNF mRNA in the presence of actinomycin D added 1 h following LPS stimulation. Remnant TNF mRNA levels were measured at 15, 30 and 60 min intervals using northern analysis (Figure 2D). From the estimated half-lives (TEPM: –IL-10, _t_1/2 = 17.8 min; +IL-10, _t_1/2 = 19 min; BMDM: –IL-10, _t_1/2 = 26 min; +IL-10, _t_1/2 = 22 min), it is evident that IL-10 does not significantly affect TNF mRNA decay in mouse macrophages. Similar conclusions were reached following transcriptional arrest at 3 and 6 h post-stimulation (data not shown). Therefore, following LPS stimulation, the IL-10-induced reduction in TNF mRNA levels probably reflects a change in TNF transcription. Consequently, the additional ∼60% reduction in the levels of soluble and transmembrane TNF protein may result from a direct suppressive effect of IL-10 on TNF mRNA translation. To verify this hypothesis, we fractionated cytoplasmic extracts from LPS- and LPS/IL-10-stimulated TEPM over sucrose gradients to compare the polysome profiles of TNF and GAPDH mRNAs (Figure 2E). A clear reduction in the number of polysomal fractions containing TNF mRNA in IL-10-treated macrophages (–IL-10: 3.75 ± 0.95; +IL-10: 1.5 ± 0.57; p = 0.006; n = 4) was noted. In contrast, the fraction of GAPDH transcripts associated with polysomes was not different in LPS- and LPS + IL-10-treated macrophages (Figure 2E). Overall, our data demonstrate that IL-10 inhibits macrophage-produced TNF primarily at the level of its mRNA translation.
Fig. 2. IL-10 targets TNF mRNA translation, but not TNF mRNA decay and transmembrane TNF processing. (A) Mouse TEPM and BMDM (5 × 105 adherent cells/ml) were stimulated with LPS (1 µg/ml) for 12 h in the presence of various concentrations of rmIL-10; TNF levels in cultured supernatants were determined by ELISA. Results shown as mean percentages of LPS values from at least three experiments (n = 5 mice/group/experiment). (B) Immunocytometric detection of transmembrane TNF on the surface of LPS-stimulated TEPM, in the presence or absence of IL-10. A, _Tnf_–/– control; B, non-stimulated; C, LPS (1 µg/ml) + rmIL-10 (5 ng/ml) for 2 h; D, LPS (1 µg/ml) for 2 h. (C) Northern analysis and quantitation of TNF mRNA isolated from TEPM (hatched bar) or BMDM (white bar) following stimulation with LPS (1 µg/ml) for 2 h, in the presence or absence of IL-10 (5 ng/ml). Results shown as percentages of the LPS values from three different experiments. (D) Decay analysis. TNF and β-actin mRNA from TEPM and BMDM stimulated with LPS for 1 h and then with actinomycin D, in the absence (open circles) or presence (closed circles) of IL-10. Northern analysis and semilogarithmic plots of data values obtained from densitometric analysis of the corresponding autoradiographs. (E) Sucrose gradient analysis. TEPM were activated with LPS in the presence or absence of IL-10 (5 ng/ml) for 2 h, and cell lysates were analyzed by sucrose gradient centrifugation. (i) Representative profile of the 254 nm UV absorption across the gradients, indicating the peaks corresponding to the 60S, 80S and the polysome containing fractions. Quantitation of GAPDH (ii) and TNF (iii) transcripts in individual fractions was determined by hybridization and autoradiography using a phosphoimager device. Closed circles, LPS-treated macrophages; open circles, LPS plus IL-10 treated macrophages. Representative dot-blots of fractions hybridized with a TNF probe are also presented.
Effective inhibition of TNF production by IL-10 requires the presence of TNF AU-rich elements
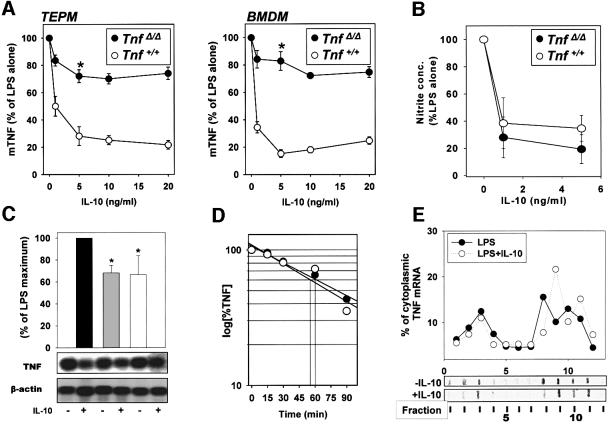
To elucidate whether IL-10-mediated inhibition of TNF gene expression required the 3′ARE, we examined the effect of IL-10 on TNF expression in _Tnf_ΔARE macrophages. TEPM and BMDM were isolated from a disease-free _TnfRI_–/– background (Kontoyiannis et al., 1999) to eliminate the possible effects of chronic inflammation occurring in the original _Tnf_ΔARE mice. IL-10 treatment of _TnfRI_–/– Tnf+/+ macrophages resulted in a reduction of LPS-induced TNF mRNA accumulation, polysome- associated TNF mRNA (data not shown) and TNF protein secretion (Figure 3A) similar to those observed in Tnf+/+ macrophages. _TnfRI_–/– _Tnf_ΔARE/ΔARE macrophages produce 3-fold higher levels of TNF mRNA than _TnfRI_–/– Tnf+/+ macrophages (Kontoyiannis et al., 1999). Nonetheless, similarly to the Tnf+/+ controls, _TnfRI_–/– _Tnf_ΔARE/ΔARE macrophages showed a decrease in mutant mRNA values of ∼30% (Figure 3C). Actinomycin D treatment of _TnfRI_–/– _Tnf_ΔARE/ΔARE macrophages (Figure 3D) indicated that TNFΔARE mRNA stability, albeit higher than that of the wild-type TNF mRNA, remained unaffected in the presence of IL-10 (TEPM: –IL-10, _t_1/2 = 54.6 min; +IL-10, _t_1/2 = 59.8 min; BMDM: –IL-10, _t_1/2 = 67.4 min; +IL-10, _t_1/2 = 71.2 min). In striking contrast, TNF protein secretion by the mutant _TnfRI_–/– _Tnf_ΔARE/ΔARE macrophages was only minimally inhibited by 20–30%, even at a maximal dose of 10 ng/ml (Figure 3A). The levels of protein reduction were comparable to the decrease in TNF mRNA accumulation (Figure 3C). The inability of IL-10 to suppress protein production efficiently in _TnfRI_–/– _Tnf_ΔARE/ΔARE macrophages was specific, since the level of nitric oxide (NO) production following LPS stimulation was inhibited equally in both wild-type and mutant macrophages (Figure 3B). Cytoplasmic extract fractionation and polysomal analysis revealed a wide distribution of TNFΔARE mRNA in the polysomal fractions, which was not significantly altered in the presence of IL-10 (–IL-10: 4.33 ± 0.57; +IL-10: 4.01 ± 1.0; n = 3) (Figure 3E). These results demonstrate that in the absence of an intact TNF 3′ARE, IL-10 cannot inhibit LPS-induced translation, and suggest that IL-10-mediated suppression of TNF translation requires an intact ARE.
Fig. 3. TNF 3′ARE absence desensitizes TNF translation to IL-10-mediated suppression. (A) TEPM and BMDM (5 × 105 adherent cells/ml) from Tnf+/+ and _Tnf_ΔARE/ΔARE mice were stimulated with LPS (1 µg/ml) for 12 h in the presence of various concentrations of IL-10; TNF levels in cultured supernatants were determined by ELISA. Results shown as mean percentages of LPS values from at least three experiments with cells derived from individual mice (n = 5 mice/group/experiment). (B) Detection of nitrite release using cultured Tnf+/+ and _Tnf_ΔARE/ΔARE TEPM stimulated for 24 h with LPS (1 µg/ml) plus IL-10 (5 ng/ml). Results shown as mean ± SD percentages from the LPS values from two experiments with TEPM derived from five individual mice/group. (C) Northern analysis (autoradiographs) and quantitation of TNF and β-actin mRNA isolated from Tnf+/+ (gray bar) and _Tnf_ΔARE/ΔARE (white bar) TEPM, activated with LPS in the presence or absence of IL-10 (5 ng/ml) for 2 h. Results shown as mean ± SD percentages of the LPS values (black bar) normalized to the GAPDH values from three different experiments. (D) mRNA decay analysis. Semi logarithmic plots of data values obtained from densitometric analysis of the corresponding autoradiographs following hybridization with TNF and β-actin probes before and after IL-10 treatment of Tnf+/+ (open circles) and _Tnf_ΔARE/ΔARE (closed circles) TEPM stimulated with LPS for 3 h, in the presence of actinomycin D. (E) Sucrose gradient analysis. _Tnf_ΔARE/ΔARE TEPM were activated with LPS in the presence or absence of IL-10 (5 ng/ml) for 2 h. Quantitation of labeled TNF transcripts in individual gradient fractions was determined using a phosphoimager device. Closed circles, LPS-treated macrophages; open circles, LPS plus IL-10 treated macrophages. Representative dot-blot of fractions hybridized with a TNF probe are presented.
The ARE binding protein TTP is not required for the IL-10-mediated inhibition of TNF production
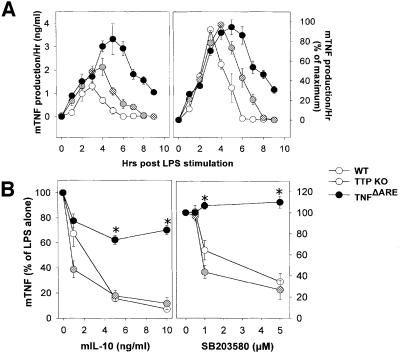
We have compared the kinetics of TNF protein production from LPS-stimulated wild-type, TTP-deficient and _Tnf_ΔARE BMDM. As has been described previously (Carballo et al., 1998; Kontoyiannis et al., 1999), both _TTP_–/– and _Tnf_ΔARE BMDM overproduce TNF following LPS stimulation relative to wild-type cultures. This correlated with an increase in the levels of TNF produced per hour by _TTP_–/– and, strikingly more so, by _Tnf_ΔARE macrophages (Figure 4A). Apart from the difference in the level of TNF protein produced, a difference in the duration of TNF production between the _TTP_–/– and the _Tnf_ΔARE macrophages could be revealed upon normalization of maximal values (Figure 4A). Wild-type TNF is produced for 5 h, whereas _TTP_–/– TNF is produced for 7 h and _Tnf_ΔARE TNF for >9 h. Comparison of the corresponding mRNA values showed a similar increase in TNF mRNA accumulation in LPS-stimulated _TTP_–/– and _Tnf_ΔARE BMDM relative to wild-type values, indicating a comparable effect of both mutations on TNF mRNA stability (data not shown). Thus, the difference in TNF protein production between TTP- and TNF ARE-deficient macrophages lies in the added modulation of TNF mRNA translation by the ARE. This was confirmed by the use of SB203580, which is known to inhibit the translation of TNF mRNA in a TNF ARE-dependent manner. SB203580 was as efficient in blocking TNF production by _TTP_–/– BMDM as it was in the case of TTP+/+ macrophages (Figure 4B). Similarly, IL-10-mediated suppression of TNF production, although minimal in the absence of TNF ARE, occurred normally in _TTP_–/– as well as control macrophages (Figure 4B). Taken together, these results show that TTP is not required for the ARE-dependent modulation of TNF mRNA translation in macrophages, indicating differential organization of TNF ARE-mediated stability and translational controls.
Fig. 4. Functional uncoupling of ARE-mediated functions: TTP does not interfere with ARE-dependent modulation of TNF mRNA translation. (A) Kinetics of TNF protein production/h. Supernatants from wild-type (wt), _TTP_–/– and _Tnf_ΔARE/+ BMDM (5 × 105 adherent cells/ml) were collected at hourly intervals after LPS stimulation. Following each sample collection, cells were washed and incubated with fresh medium. TNF protein levels were quantitated as before. Data shown as actual mean ± SD values (left) or as mean + SD percentages to each of the corresponding maximal values (right). (B) BMDM (5 × 105 adherent cells/ml) from wild-type (wt), _TTP_–/– and _Tnf_ΔARE/+ BMDM were stimulated with LPS (1 µg/ml) for 12 h in the presence of various concentrations of IL-10 or SB203580; TNF levels in cultured supernatants were as before. Results shown as mean percentages of LPS values; data from at least three experiments with cells derived from individual mice (n = 5 mice/group/experiment).
Reduced p38/SAPK activation in the presence of IL-10 in LPS-stimulated macrophages
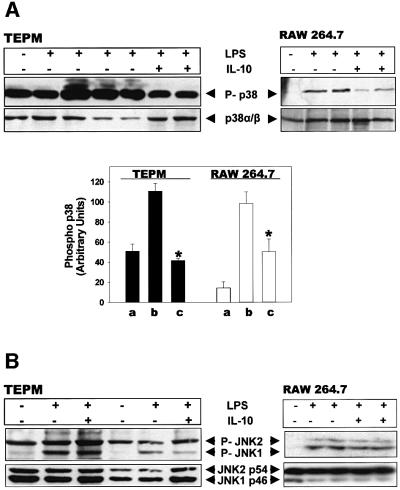
The p38, JNK and ERK MAPK/SAPK pathways have previously been demonstrated to modulate TNF biosynthesis (Lee et al., 1994; Swantek et al., 1997; Kontoyiannis et al., 1999; Srivastava et al., 1999; Dumitru et al., 2000). A paradigm of cross-talk between signals activated by the IL-10 receptor and MAPK/SAPK has also been demonstrated previously (Kallunki et al., 1994; Niiro et al., 1998; Turkson et al., 1999). However, other reports (reviewed by Donelly et al., 1999) have not been able to reproduce an effect of IL-10 on these signaling modules. We examined the effect of IL-10 on the LPS-induced activation of ERK, JNK and p38 MAPK in TEPM, and the mouse macrophage cell line RAW 264.7, by detecting the corresponding native and phosphorylated forms via western blotting. As can be readily seen in Figure 5A, IL-10 inhibited the LPS-induced phosphorylation of p38 MAPK by 90% in TEPM and by 50% in RAW 264.7 macrophages, while the levels of the native form of this protein remained unaltered. In contrast, no reduction was observed in the levels of the phosphorylated forms of JNK1/2 (Figure 5B) and ERK1/2 (not shown). These results suggest that IL-10 interferes with p38 signaling at the level of its phosphorylation by upstream kinases, but not at the level of p38 abundance.
Fig. 5. IL-10 inhibits p38 phosphorylation in LPS-induced macro phages. TEPM and RAW 264.7 macrophages were challenged with LPS (1 µg/ml) and IL-10 (5ng/ml) for 15 min. Total cell lysates were immunoblotted with antibody probes for the phosphorylated forms of p38/SAPK (A) and JNK/SAPK (B). Membranes were stripped and reprobed for the detection of the total protein content of each kinase. Representative blots are shown. Quantitation of phospho-p38 kinase levels, normalized to the total p38 content, is also shown (A). Results from one out of three experiments, shown as mean densitometric units (+ SD) from independent cultures.
IL-10 targets the p38/MAPKAP-2 pathway to modulate ARE-dependent TNF mRNA translation
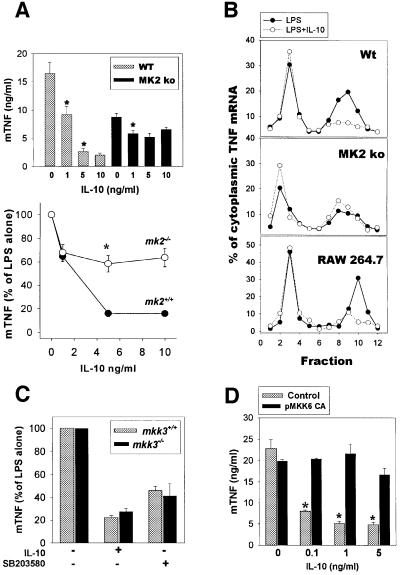
The p38/MAPK cascade (reviewed by Ono and Han, 2000) activates many downstream protein kinases, including the serine/threonine kinase MK2 [MAPK-activated protein (MAPKAP)-kinase 2]. MK2-deficient macrophages show reduced levels of TNF protein production, but not of the corresponding mRNA, following LPS challenge (Kotlyarov et al., 1999), indicating that the p38/MK2 pathway is required for activation of TNF translation by LPS. To examine whether MK2 modulates TNF translation in macrophages in an ARE-dependent manner, we have recently generated _TNF_ΔARE/+ _mk2_–/– mice (A.Kotlyarov, D.Kontoyiannis, A.Neininger, R.Winzen, R.Eckert, H.D.Volk, H.Holtmann, G.Kollias and M.Gaestel, in preparation). Macrophages from these mice show a pattern of TNF production similar to that observed in _TNF_ΔARE macrophages, indicating that MK2 targets TNF ARE. We further examined whether the absence of MK2 signals interferes with IL-10 targeting of TNF production. Exposure of LPS-stimulated _mk2_–/– TEPM to IL-10 reduced TNF protein production by 30–40% instead of 80% in mk2+/+ macrophages (Figure 6A). In addition, the LPS-induced, polysome-associated TNF mRNA profile in MK2-deficient TEPM, although it appears lower than in LPS-induced wild-type TEPM, is not significantly altered in the presence of IL-10 (–IL-10: 3.0 + 0.1; +IL-10: 2.5 + 0.70; n = 3) (Figure 6B). These observations demonstrate that MK2 is required for the ARE-dependent, IL-10-mediated inhibition of TNF production.
Fig. 6. IL-10 inhibits p38-mediated signals that activate TNF mRNA translation. (A) Impaired efficacy of IL-10 to suppress TNF production in mk2-deficient macrophages. LPS-stimulated TEPM from mk2+/+ and _mk2_–/– mice were cultured with various concentrations of rmIL-10 for 12 h, and TNF protein was detected in culture supernatants by ELISA. Results are from a representative of two experiments with cells derived from five mice/group. Data are as absolute values (upper) as well as percentages (+ SD) of the corresponding LPS values (lower). (B) Defective inhibition of polysome TNF mRNA in the absence of MK2. Sucrose gradient analysis of monosomal/polysomal-associated TNF mRNA in LPS plus IL-10-stimulated mk2+/+, _mk2_–/– TEPM as well as RAW 264.7 macrophages. Results from representative experiments shown as percentages of total cytoplasmic fractions. (C) Normal IL-10 targeting of TNF production in MKK3-deficient macrophages. TEPM from mkk3+/+ and _mkk3_–/– macrophages were stimulated with LPS (1 µg/ml) in the presence of 5 ng/ml rmIL-10. Data shown as mean (± SD) of the corresponding LPS values. Results from two experiments with macrophages from three mice/group. (D) Transfection of a constitutively active form of MKK6 inhibits IL-10-mediated targeting of TNF production. RAW 264.7 cells were transfected via electroporation with control plasmid or the expression plasmid for the mutated MKK6(Glu) (MKK6 CA). Following 18 h starvation, cells were challenged with LPS supplemented with the indicated quantities of IL-10. TNF protein quantitation was performed as before. Data shown as mean (± SEM) percentages of LPS values. Results from three independent transfected cultures for each group. *p <0.01.
p38/SAPK is activated predominantly by the MAP kinase kinases MKK3 and MKK6 (Raingeaud et al., 1996). Although LPS-induced mkk3_–/– macrophages secrete normal quantities of TNF protein, a reduction in the general p38 activity was indicated in these cells,_ which could affect the IL-10-instigated inhibition of TNF production. Nevertheless, TNF production by LPS-stimulated _mkk3_–/– macrophages in the presence of IL-10 was similar to that of control cultures (Figure 6C), indicating that MKK3 is not involved in the inhibition of TNF translation by IL-10.
To verify whether inhibition of the p38 pathway is required for efficient IL-10 targeting of TNF production by macrophages, we transiently transfected the mouse macrophage cell line RAW 264.7 with a constitutively active form of MKK6, MKK6(Glu) (Raingeaud et al., 1996). TNF translation by LPS-induced RAW 264.7 macrophages was affected by IL-10 in a manner similar to wild-type TEPM, as revealed by the corresponding cell fractionation studies (Figure 6B). Introduction of MKK6(Glu) into RAW 264.7 readily resulted in secretion of TNF (<2 ng), indicating functional interference of the exogenous MKK6(Glu) with TNF production (data not shown). However, LPS stimulation resulted in comparative values between MKK6(Glu) transfectants and controls. Most importantly, the ability of IL-10 to inhibit TNF production following LPS stimulation of RAW 264.7 was completely abrogated in the presence of MKK6(Glu) (Figure 6D). Overall, our results demonstrate that IL-10 interferes with the activation of the p38/MAPKAP-2 pathway to inhibit the ARE-dependent TNF translation in mouse macrophages.
Discussion
In this study, we show that IL-10-mediated signals primarily target TNF mRNA translation. This process is clearly dependent on TNF 3′ARE, since in its absence the efficacy of IL-10 in inhibiting TNF expression is severely compromised. However, we failed to detect any effect of IL-10 on TNF mRNA decay, either in the presence or absence of TNF ARE, suggesting that TNF mRNA stability may not be targeted by IL-10 in macrophages. Our data seem different from previous reports suggesting that IL-10 leads to ARE-dependent destabilization of granulocyte–macrophage colony-stimulating factor, granulocyte colony-stimulating factor and KC cytokine mRNAs (Brown et al., 1996; Kishore et al., 1999). This variance indicates differential properties imposed by the AREs on different mRNAs. Several additional observations support a functional separation between TNF 3′ARE-mediated modulation of translation and stability. Pharmacological inhibition of p38/MAPK affects TNF mRNA translation by inhibiting LPS-induced polysome coupling of TNF mRNA in macrophages, without affecting TNF mRNA accumulation (Prichett et al., 1995). Similarly, the absence of MK2 leads to a profound reduction of LPS-induced TNF production, yet TNF mRNA accumulation and stability are not affected (Kotlyarov et al., 1999). Thirdly, and most importantly, TTP (a TNF ARE binding protein regulating TNF biosynthesis) exerts a potent destabilizing activity on TNF mRNA (Carballo et al., 1998; Lai et al., 1999) without imposing translational control (present data). These observations are important in conceptualizing independent pathways of ARE-mediated translational and stability controls being exerted, presumably, by different ARE binding complexes and associated signals. Two RNA binding proteins that have recently been demonstrated to have TNF ARE binding capacities are TIAR and TIA-1 (Gueydan et al., 1999; Piecyk et al., 2000). Genetic ablation of TIA-1 leads to increased LPS-induced translation of TNF mRNA without affecting its abundance, making TIA-1 a putatively specific translational modulator.
Our results indicate that IL-10 interferes with the activation potential of the p38/MAPK pathway, which is required to activate TNF translation, as suggested by the MK2-deficient macrophages and by the MKK6 (Glu) transfection assays. The reduction in the phosphorylated form of p38 following IL-10 treatment of macrophages indicates that IL-10 inhibits upstream of p38 activation. However, IL-10 still affects TNF production in MKK3-deficient macrophages, in which a reduction in p38 activation was also noted (Lu et al., 1999), indicating that MKK3 is not required. The most likely alternative could be that IL-10 interferes with the MKK6 signals activating p38 MAPK; however, in the absence of the corresponding deficient mouse, we can only hypothesize on such a mechanism. Previous reports have provided controversial results on the effect of IL-10 on p38/SAPK (reviewed by Donelly et al., 1999). Furthermore, earlier studies point towards a role for p38 in modulating the stability of TNF mRNA (Wang et al., 1999; Brook et al., 2000). Our prediction is that the differences described in the literature result from different experimental conditions and systems utilized in each case. For example, in most previous reports, macrophages were pre-treated with IL-10 for at least 2 h prior to LPS stimulation. Under conditions of cytokine pre-treatment, however, altered responses have been reported to occur (Erwig et al., 1998). We selected to co-administer LPS and IL-10 since IL-10 production is concomitant with or consecutive to LPS-induced TNF production in mouse macrophages (Figure 1), and thus IL-10 pre-treatment of these cells may not be physiologically relevant. Finally, from the current data, we cannot exclude the possibility that other MAPK/SAPK-mediated signals may also be utilized for the maximal effect of IL-10 towards TNF. However, since the activation of the JNK and ERK pathways is not being affected by IL-10, it is currently unclear how these pathways may interfere.
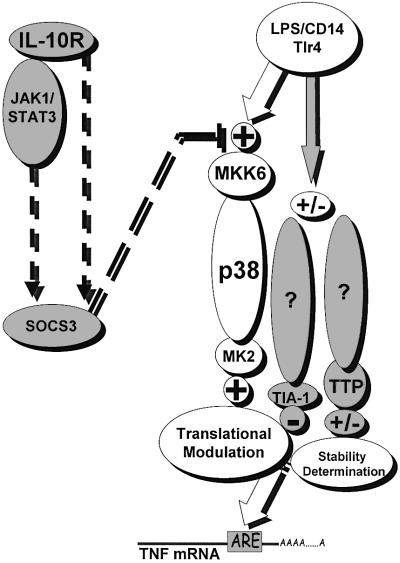
The detailed mechanism by which IL-10 targets MAPK/SAPK signals is currently unknown. It is possible that IL-10 signaling interferes directly with the activation of MAPKs. Engagement of the IL-10 receptor (IL-10R) has been shown to activate the JAK–STAT pathway. Specifically, IL-10R signaling requires the activation of Jak1 and Tyk2, and induces the activation of Stat1, Stat3 and Stat5 (Riley et al., 1999). There is evidence that points towards the requirement for Jak-1/STAT3 in TNF suppression (Riley et al., 1999; Takeda et al., 1999). Thus, one possibility would be that STAT3 activates factors capable of interfering with MAPK/SAPK signaling. One such example has been provided by the STAT3-dependent inhibition of inflammatory signals through the activation of SOCS-3 (Bode et al., 1999; Cassatella et al., 1999; Levings and Schrader, 1999; Turkson et al., 1999; Schmitz et al., 2000). However, since inhibition of p38/MAPK by IL-10 occurs very early (i.e. at 15 min), it is difficult to conceptualize a STAT3-dependent mechanism that would require de novo protein synthesis, e.g. of SOCS proteins. Interestingly, both STAT3 and SOCS3 can be activated by inflammatory stimuli like LPS and TNF and, most importantly, SOCS3 activation can be induced by STAT3-independent mechanisms (Stoiber et al., 1999; Ahmed and Ivashkiv, 2000). In addition, LPS can synergize with IL-10 to prolong SOCS3 mRNA stability in myeloid cells (Cassatella et al., 1999). Therefore, early post-transcriptional mechanisms may provide a pool of SOCS-3, which can be rapidly activated in the absence of de novo gene transcription. Consequently, SOCS-3 transcription may allow for the replenishment of a SOCS-3 pool to enforce its negative action in a temporal manner. This may also provide an answer on the seeming paradox of p38/MAPK dependency of both STAT3 and SOCS3 for their activation as well as for the apparent capacity of SOCS3 to suppress STAT3 activation itself (Ahmed and Ivashkiv, 2000; Suzuki et al., 2001). Taken together, this evidence points towards a complex temporal regulation of these factors, which will eventually tune, in a timely manner, the pro- and anti-inflammatory signals. Figure 7 summarizes the possible interactions of IL-10R signaling to modules affecting activation of TNF gene expression.
Fig. 7. Proposed scheme of interactions between IL-10R and TNF modulating signals. LPS binding/receptor complex leads to the transmission of p38/MAPK signals activating TNF mRNA translation (open circles and arrows). At similar or consecutive time points, it leads to the activation of stability and translation determining factors that act in a negative fashion, apparently via p38-independent mechanisms (gray shaded circles and arrows). On the other hand, IL-10 receptor engagement may transmit STAT-3-dependent and -independent signals towards SOCS-3 activation, building up a negative regime targeting p38/MK2 activating signals towards ARE-dependent TNF mRNA translation.
The functional significance of the current work is perhaps most convincingly exemplified by the pathologies developing in the models of compromised TNF modulation by IL-10R. _Tnf_ΔARE mice, as well as IL-10- and myeloid-specific STAT3-deficient mice, develop overt inflammatory pathologies in the intestine, indicating a bias towards pro-inflammatory/immune-activating action. In addition, it has recently been demonstrated that inhibition of SOCS-3 activation in transgenic mice renders them more susceptible to a model of induced colitis (Suzuki et al., 2001), verifying its potential to control inflammation. As demonstrated herein, TNF is required to a major extent for the development of IBD in IL-10-deficient mice. Our results may seem in contrast with earlier observations that anti-TNF treatment of IL-10-deficient mice was inefficient in controlling IBD in this model (Rennick et al., 1997). This discrepancy may reflect a developmental requirement for TNF in the pathology in the IL-10-deficient mice. Our current data provide mechanistic clues as to how the IL-10/TNF axis may be deviated in the course of mutations affecting the function of TNF ARE-mediated controls, and demonstrate that a homeostatic cross-talk between IL-10 and TNF is essential in preventing immune dysregulation and disease.
Materials and methods
Mice
_Il-10_–/– (Kuhn et al., 1993), _Tnf_–/– (Pasparakis et al., 1996), _Tnf_ΔARE/+, _Tnf_ΔARE/ΔÁRE _TnfRI_–/– (Kontoyiannis et al., 1999), _TTP_–/– (Taylor et al., 1996), _mk2_–/– (Kotlyarov et al., 1999) and _mkk3_–/– (Lu et al., 1999) mutant mice were bred and maintained on a C57BL/6J or a mixed C57BL/6J×129SvEm genetic background. All mice were housed under specific pathogen-free (SPF) conditions.
Histopathological evaluation
Paraffin-embedded colon sample tissues were sectioned and stained with hematoxylin and eosin. Inflammation in each region of the colon was graded semiquantitatively from 0 to 5 in a blinded fashion, using the following criteria: (0) no signs of inflammation to (5) most severe change including maximal degree of transmural inflammation, goblet cell loss, crypt abscesses and ulceration, and lamina propria fibrosis. The total disease score per mouse is calculated by the summation of the score for each segment of the colon, from 0 to 20, where 0 is no change, >0–5 is mild disease, >5–10 is moderate disease and >10–20 is severe disease. Data are represented as mean ± SD.
Cell isolation and culture
All cultures were grown and maintained at 37°C in 5% CO2. TEPM were isolated by peritoneal lavage from ≤10-week-old mice, 3 days after a single peritoneal injection of aged 4% thioglycolate broth (1 ml; Difco Laboratories). Cells were left to adhere for 1 h in Petri dishes, and washed and covered with fresh medium. TEPM cultures were left to rest for 4 h prior to experimentation. The ex vivo derivation of BMDM was performed as previously described (Kontoyiannis et al., 1999). For all macrophage experiments, cells were seeded at a density of 5 × 105 cells/well in 24-well tissue culture plates. Following resting, cultures were incubated in the presence or absence of the indicated quantities of LPS (Salmonella enteriditis; Sigma; L-6011), recombinant mouse IL-10 (R&D) or the p38 inhibitor SB203580 (Calbiochem) in RPMI + 5% fetal calf serum (Gibco).
TNF ELISA and NO detection assay
Mouse TNF and IL-10 levels in sera and supernatants were determined using a sandwich TNF (Kontoyiannis et al., 1999) and IL-10 (R&D) ELISAs. To evaluate NO, NO3– and NO2– were measured from cell culture supernatants using Griess’s assay (Green et al., 1982).
FACS analysis: detection of tmTNF
TEPM were washed extensively in phosphate-buffered saline (PBS) + 0.1% bovine serum albumin + 0.01% sodium azide (PBA) and saturated with blocking buffer (PBA + 5% normal rabbit serum). Cells were incubated with biotinylated rabbit anti-mouse and -rat TNF polyclonal antibodies (PharMingen), followed by incubation with streptavidin–phycoerythrin (PharMingen). Subsequently, cells were fixed with 4% paraformaldehyde for 10 min at 4°C and analyzed with a FACSCalibur™ cytometer.
Cell fractionation and polysome analysis
Macrophages were collected and maintained as described above in 10 cm plates. LPS (1 µg/ml) in the absence or presence of IL-10 (5 ng/ml) was added for 2 h. Cells were washed in ice-cold PBS containing 150 µg/ml cyclohexamide and collected into 1.0 ml of cold hypotonic lysis buffer (10 mM KCl, 10 mM Tris pH 7.2, 10 mM MgCl2, 20 mM dithiothreitol, 150 µg/ml cycloheximide, 0.5 µg/ml heparin, 0.5% NP-40 and 100 U/ml RNasin) and mechanically disrupted by douncing. Nuclei and cell membranes were removed by microcentrifugation for 10 min. The supernatant was layered onto a 10–50% continuous sucrose gradient. Centrifugation was performed at 27 000 r.p.m. for 3 h using a SW27Ti rotor. Fractions (2 ml) were collected, starting from the top of the gradient, and UV absorbance was monitored at 254 nm to identify fractions containing monosomes and polysomes. Samples of total RNA were extracted with phenol/chloroform from each fraction, ethanol precipitated and resuspended in 30 µl of diethyl pyrocarbonate-treated ddH2O. Ten microliters of each sample were mixed with 20 µl of formamide, 7 µl of formaldehyde (37%) and 2 µl of 20× SSC, and applied to a nitrocellulose membrane using a dot-blot apparatus. Following hybridization, the relative amounts of 32P-labeled TNF and GAPDH mRNAs in each fraction were determined using a Storm phosphoimager.
Cell transfections
RAW 264.7 macrophages were grown and maintained in RPMI medium containing 5% fetal bovine serum at 37°C in 5% CO2. Cells were transfected by electroporation using a Bio-Rad Gene Pulser™ as previously described (Thompson et al., 1999). Briefly, cells were washed extensively in PBS. Cells (5 × 106) were then mixed with 5 µg of the expression plasmid for constitutively active MKK6(Glu) (Raingaud et al., 1996), or the control plasmid pGFP (Clontech, USA), into a final volume of 400 µl. Cells were incubated on ice for 10 min and then pulsed into 0.4 cm cuvettes at 260 V and 960 µF. Transfected cells were split into the desired number of plates to recover and adhere for 2 h, washed, and left to rest for 18 h. Transfection efficiency was verified using FACS analysis using a FACScan cytometer (Becton & Dickinson) for the detection of green fluorescent protein (GFP). Maximal GFP expression was detected at 24 h (∼70%), reducing to 30–40% at 48 h. The plates were then stimulated at 20 h with LPS (1 µg/ml), with or without the indicated quantities of IL-10, and incubated for 12 h. Cell survival was assessed via crystal violet staining (0.5%) for the cytokine production assays. Supernatants were harvested as previously described and used immediately for TNF ELISA.
Immunoblotting and RNA analysis
For western analysis, the probes used were monoclonal antibodies for p38α/β (A-12) and JNK1/2 (D-2), as well as polyclonal antibodies for the phosphorylated forms of JNK [specific for Thr-(P)-183 and Tyr-(P)-185 of human JNK1 & 2] and p38 [specific for Tyr-(P)-182 of human p38]. All antibodies were purchased from Santa Cruz Biotechnologies. For northern analysis of RNA samples, a 0.9 kb _Nar_I–_Bgl_II genomic probe containing part of the first exon of mTNF gene was used. A 0.9 kb mouse _Pst_I β-actin fragment was used as a quantitative control.
Statistics
Fisher’s exact test for non-parametric data for statistical significance was used to compare results between different populations of mice. The unpaired Student’s _t_-test for statistical significance was used to compare protein and mRNA values.
Acknowledgments
Acknowledgements
We would like to thank Wim Buurman for the TNF-specific ELISAs, Vassilios Aidinis for his assistance on the fractionation assays and Spiros Lalos for technical assistance in histopathology. This work was supported in part by the Hellenic Secretariat for Research and Technology and European Commission grants QLG1-CT1999-00202 and QLK6-1999-02203.
References
- Ahmed S.T. and Ivashkiv,L.B. (2000) Inhibition of IL-6 and IL-10 signaling and Stat activation by inflammatory and stress pathways. J. Immunol., 165, 5227–5237. [DOI] [PubMed] [Google Scholar]
- Beutler B. (2000) Tlr4: central component of the sole mammalian LPS sensor. Curr. Opin. Immunol., 12, 20–26. [DOI] [PubMed] [Google Scholar]
- Bode J.G., Nimmesgern,A., Schmitz,J., Schaper,F., Schmitt,M., Frisch,W., Haussinger,D., Heinrich,P.C. and Graeve,L. (1999) LPS and TNFα induce SOCS3 mRNA and inhibit IL-6-induced activation of STAT3 in macrophages. FEBS Lett., 463, 365–370. [DOI] [PubMed] [Google Scholar]
- Bogdan C., Vodovotz,Y. and Nathan,C. (1991) Macrophage deactivation by interleukin 10. J. Exp. Med., 174, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C., Paik,J., Vodovotz,Y. and Nathan,C. (1992) Contrasting mechanisms for suppression of macrophage cytokine release by transforming growth factor-β and interleukin-10. J. Biol. Chem., 267, 23301–23308. [PubMed] [Google Scholar]
- Brook M., Sully,G., Clark,A.R. and Saklatvala,J. (2000) Regulation of tumour necrosis factor α mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. FEBS Lett., 483, 57–61. [DOI] [PubMed] [Google Scholar]
- Brown C.Y., Lagnado,C.A., Vadas,M.A. and Goodall,G.J. (1996) Differential regulation of the stability of cytokine mRNAs in lipopolysaccharide-activated blood monocytes in response to interleukin-10. J. Biol. Chem., 271, 20108–20112. [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai,W.S. and Blackshear,P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science, 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Cassatella M.A. et al. (1999) Interleukin-10 (IL-10) selectively enhances CIS3/SOCS3 mRNA expression in human neutrophils: evidence for an IL-10-induced pathway that is independent of STAT protein activation. Blood, 94, 2880–2889. [PubMed] [Google Scholar]
- Collart M.A., Baeuerle,P. and Vassalli,P. (1990) Regulation of tumor necrosis factor α transcription in macrophages: involvement of four κB-like motifs and of constitutive and inducible forms of NF-κB. Mol. Cell. Biol., 10, 1498–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal M., Abrams,J., Bennett,B., Figdor,C.G. and de Vries,J.E. (1991) Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J. Exp. Med., 174, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelly R.P., Dickensheets,H. and Finbloom,D.S. (1999) The IL-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J. Interferon Cytokine Res., 19, 563–573. [DOI] [PubMed] [Google Scholar]
- Dumitru C.D. et al. (2000) TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell, 103, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Erwig L.P., Kluth,D.C., Walsh,G.M. and Rees,A.J. (1998) Initial cytokine exposure determines function of macrophages and renders them unresponsive to other cytokines. J. Immunol., 161, 1983–1988. [PubMed] [Google Scholar]
- Geng Y., Gulbins,E., Altman,A. and Lotz,M. (1994) Monocyte deactivation by interleukin 10 via inhibition of tyrosine kinase activity and the Ras signaling pathway. Proc. Natl Acad. Sci. USA, 91, 8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard C., Bruyns,C., Marchant,A., Abramowicz,D., Vandenabeele,P., Delvaux,A., Fiers,W., Goldman,M. and Velu,T. (1993) Interleukin 10 reduces the release of tumor necrosis factor and prevents lethality in experimental endotoxemia. J. Exp. Med., 177, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L.C., Wagner,D.A., Glogowski,J., Skipper,P.L., Wishnok,J.S. and Tannenbaum,S.R. (1982) Analysis of nitrate, nitrite and [15N]nitrate in biological fluids. Anal. Biochem., 126, 131–138. [DOI] [PubMed] [Google Scholar]
- Gueydan C., Droogmans,L., Chalon,P., Huez,G. and Kruys,V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem., 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Kallunki T., Su,B., Tsigelny,I., Sluss,H.K., Derijard,B., Moore,G., Davis,R. and Karin,M. (1994) JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev., 8, 2996–3007. [DOI] [PubMed] [Google Scholar]
- Kishore R., Tebo,J.M., Kolosov,M. and Hamilton,T.A. (1999) Cutting edge: clustered AU-rich elements are the target of IL-10-mediated mRNA destabilization in mouse macrophages. J. Immunol., 162, 2457–2461. [PubMed] [Google Scholar]
- Kollias G., Douni,E., Kassiotis,G. and Kontoyiannis,D. (1999) On the role of tumor necrosis factor and receptors in models of multiorgan failure, rheumatoid arthritis, multiple sclerosis and inflammatory bowel disease. Immunol. Rev., 169, 175–194. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis,M., Pizarro,T.T., Cominelli,F. and Kollias,G. (1999) Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity, 10, 387–398. [DOI] [PubMed] [Google Scholar]
- Kotlyarov A., Neininger,A., Schubert,C., Eckert,R., Birchmeier,C., Volk,H.D. and Gaestel,M. (1999) MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nature Cell Biol., 1, 94–97. [DOI] [PubMed] [Google Scholar]
- Kuhn R., Lohler,J., Rennick,D., Rajewsky,K. and Muller,W. (1993) Interleukin-10-deficient mice develop chronic enterocolitis. Cell, 75, 263–274. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.C. et al. (1994) A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature, 372, 739–746. [DOI] [PubMed] [Google Scholar]
- Levings M.K. and Schrader,J.W. (1999) IL-4 inhibits the production of TNF-α and IL-12 by STAT6-dependent and -independent mechanisms. J. Immunol., 162, 5224–5229. [PubMed] [Google Scholar]
- Lu H.T., Yang,D.D., Wysk,M., Gatti,E., Mellman,I., Davis,R.J. and Flavell,R.A. (1999) Defective IL-12 production in mitogen-activated protein (MAP) kinase kinase 3 (Mkk3)-deficient mice. EMBO J., 18, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A. et al. (1994) Interleukin-10 controls interferon-γ and tumor necrosis factor production during experimental endotoxemia. Eur. J. Immunol., 24, 1167–1171. [DOI] [PubMed] [Google Scholar]
- Moore K.W., O’Garra,A., de Waal,M., Vieira,P. and Mosmann,T.R. (1993) Interleukin-10. Annu. Rev. Immunol., 11, 165–190. [DOI] [PubMed] [Google Scholar]
- Niiro H. et al. (1998) MAP kinase pathways as a route for regulatory mechanisms of IL-10 and IL-4 which inhibit COX-2 expression in human monocytes. Biochem. Biophys. Res. Commun., 250, 200–205. [DOI] [PubMed] [Google Scholar]
- Ono K. and Han,J. (2000) The p38 signal transduction pathway—activation and function. Cell. Signalling, 12, 1–13. [DOI] [PubMed] [Google Scholar]
- Osman F., Jarrous,N., Ben-Asouli,Y. and Kaempfer,R. (1999) A _cis_-acting element in the 3′-untranslated region of human TNF-α mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev., 13, 3280–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., Alexopoulou,L., Episkopou,V. and Kollias,G. (1996) Immune and inflammatory responses in TNF α-deficient mice: a critical requirement for TNF α in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers and in the maturation of the humoral immune response. J. Exp. Med., 184, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon J.J. et al. (1998) An essential role for ectodomain shedding in mammalian development. Science, 282, 1281–1284. [DOI] [PubMed] [Google Scholar]
- Piecyk M. et al. (2000) TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J., 19, 4154–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichett W., Hand,A., Sheilds,J. and Dunnington,D. (1995) Mechanism of action of bicyclic imidazoles defines a translational regulatory pathway for tumor necrosis factor α. J. Inflamm., 45, 97–105. [PubMed] [Google Scholar]
- Raingeaud J., Whitmarsh,A.J., Barrett,T., Derijard,B. and Davis,R.J. (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol., 16, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennick D.M., Fort,M.M. and Davidson,N.J. (1997) Studies with IL-10–/– mice: an overview. J. Leukoc. Biol., 61, 389–396. [DOI] [PubMed] [Google Scholar]
- Riley J.K., Takeda,K., Akira,S. and Schreiber,R.D. (1999) Interleukin-10 receptor signaling through the JAK–STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J. Biol. Chem., 274, 16513–16521. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Weissenbach,M., Haan,S., Heinrich,P.C. and Schaper,F. (2000) SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. J. Biol. Chem., 275, 12848–12856. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Weitzmann,M.N., Cenci,S., Ross,P.F., Adler,S. and Pacifici,R. (1999) Estrogen decreases TNF gene expression by blocking JNK activity and the resulting production of c-Jun and JunD. J. Clin. Invest., 104, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber D., Kovarik,P., Cohney,S., Johnston,J.A., Steinlein,P. and Decker,T. (1999) Lipopolysaccharide induces in macrophages the synthesis of the suppressor of cytokine signalling 3 and suppresses signal transduction in response to the activating factor IFN-α. J. Immunol., 163, 2640–2647. [PubMed] [Google Scholar]
- Suzuki A. et al. (2001) CIS/SOCS3/SSI3 plays a negative role in STAT3 activation and intestinal inflammation. J. Exp. Med., 193, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swantek J.L., Cobb,M.H. and Geppert,T.D. (1997) Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) is required for lipopolysaccharide stimulation of tumor necrosis factor α (TNF-α) translation: glucocorticoids inhibit TNF-α translation by blocking JNK/SAPK. Mol. Cell. Biol., 17, 6274–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Clausen,B.E., Kaisho,T., Tsujimura,T., Terada,N., Forster,I. and Akira,S. (1999) Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity, 10, 39–49. [DOI] [PubMed] [Google Scholar]
- Taylor G.A. et al. (1996) A pathogenetic role for TNF α in the syndrome of cachexia, arthritis and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity, 4, 445–454. [DOI] [PubMed] [Google Scholar]
- Thompson C.D., Frazier-Jessen,M.R., Rawat,R., Nordan,R.P. and Brown,R.T. (1999) Evaluation of methods for transient transfection of a murine macrophage cell line, RAW 264.7. Biotechniques, 27, 824–830, 832. [DOI] [PubMed] [Google Scholar]
- Turkson J. et al. (1999) Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol. Cell. Biol., 19, 7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Wu,P., Siegel,M.I., Egan,R.W. and Billah,M.M. (1995) Interleukin (IL)-10 inhibits nuclear factor κ B (NFκB) activation in human monocytes. IL-10 and IL-4 suppress cytokine synthesis by different mechanisms. J. Biol. Chem., 270, 9558–9563. [DOI] [PubMed] [Google Scholar]
- Wang S.W., Pawlowski,J., Wathen,S.T., Kinney,S.D., Lichenstein,H.S. and Manthey,C.L. (1999) Cytokine mRNA decay is accelerated by an inhibitor of p38-mitogen-activated protein kinase. Inflamm. Res., 48, 533–538. [DOI] [PubMed] [Google Scholar]