Specific cleavage of hyper-edited dsRNAs (original) (raw)
Abstract
Extended double-stranded DNA (dsRNA) duplexes can be hyper-edited by adenosine deaminases that act on RNA (ADARs). Long uninterrupted dsRNA is relatively uncommon in cells, and is frequently associated with infection by DNA or RNA viruses. Moreover, extensive adenosine to inosine editing has been reported for various viruses. A number of cellular antiviral defence strategies are stimulated by dsRNA. An additional mechanism to remove dsRNA from cells may involve hyper-editing of dsRNA by ADARs, followed by targeted cleavage. We describe here a cytoplasmic endonuclease activity that specifically cleaves hyper-edited dsRNA. Cleavage occurs at specific sites consisting of alternating IU and UI base pairs. In contrast, unmodified dsRNA and even deaminated dsRNAs that contain four consecutive IU base pairs are not cleaved. Moreover, dsRNAs in which alternating IU and UI base pairs are replaced by isomorphic GU and UG base pairs are not cleaved. Thus, the cleavage of deaminated dsRNA appears to require an RNA structure that is unique to hyper-edited RNA, providing a molecular target for the disposal of hyper-edited viral RNA.
Keywords: antisense/antiviral/inosine/ribonuclease/RNA editing
Introduction
Eukaryotic cells possess a number of sensitive mechanisms to detect and respond to the presence of extended double-stranded RNA (dsRNA) (Maquat and Carmichael, 2001). While short dsRNA duplexes (<10 bp) play important roles in a variety of cellular processes, there are relatively few examples of long uninterrupted dsRNA duplexes in cells. One source is the presence of antisense RNA transcripts, which have been implicated in the regulation of gene expression by forming duplexes with the sense strand mRNA (reviewed by Kumar and Carmichael, 1998). However, the presence of dsRNA is more commonly indicative of viral infection or of the presence of other invading nucleic acid elements.
The presence of dsRNA in mammalian cells is sufficient to elicit an antiviral response mediated by enzymes such as dsRNA-dependent protein kinase (PKR) (Samuel, 1979, 1998) or 2′-5′-oligoadenylate synthetase/RNase L (Zhou et al., 1993; Silverman et al., 1997), which mediate global down-regulation of translation and mRNA stability, respectively. In contrast, the more recently characterized phenomenon of RNA interference (RNAi) involves targeted post-transcriptional repression only of genes whose RNAs are homologous to a specific dsRNA in the cell (Bass, 2000). Another targeted response is the specific modification of dsRNA by adenosine deamination.
Extended dsRNA duplexes can be hyper-edited by members of the ADAR (adenosine deaminases that act on RNA) family of enzymes (Hough and Bass, 2000). ADARs modify dsRNA by hydrolytic deamination of adenosine (A) to inosine (I), and carry out either selective editing or hyper-editing (Emeson and Singh, 2000; Hough and Bass, 2000). Selective editing, in which a limited number of A to I conversions occur in an mRNA, is a well established mechanism of gene regulation. As inosine residues are recognized by both the translation and splicing machinery as guanosine (G), A to I editing changes the coding potential of mRNAs, and can affect pre-mRNA splicing by introducing new splice sites (Rueter et al., 1999). Hyper-editing occurs on extended RNA duplexes, resulting in up to 50% A to I conversion (Nishikura et al., 1991; Polson and Bass, 1994). Hyper-editing of dsRNAs arising from antisense transcription may be involved in gene regulation, as has been suggested for the Xenopus bFGF gene (Kimelman and Kirschner, 1989; Saccomanno and Bass, 1999) and for polyoma virus (Kumar and Carmichael, 1997). Hyper-editing has also been detected in Drosophila 4f-rnp cDNAs (Petschek et al., 1996), and in hairpin structures in poly(A)+ RNA isolated from Caenorhabditis elegans (Morse and Bass, 1999). Hyper-editing of a number of viral RNAs has also been reported (Emeson and Singh, 2000), including measles virus (Bass et al., 1989; Cattaneo et al., 1989), human parainfluenza virus (Murphy et al., 1991), vesicular stomatitis virus (O’Hara et al., 1984), avian leukosis virus (Hajjar and Linial, 1995), respiratory syncitial virus (Rueda et al., 1994), Rous-associated virus (Felder et al., 1994) and polyoma virus (Kumar and Carmichael, 1997). The cytoplasmic isoform of ADAR1 is inducible by interferon (Patterson and Samuel, 1995), a characteristic of other enzymes that are involved in antiviral defence [e.g. PKR and 2-5A system (RNase L)]. These observations lend weight to the idea that A to I hyper-modification is an antiviral mechanism. Although hyper-editing alone may be efficient in destroying the sense of viral dsRNA, it would still be engaged by the cellular translation machinery and would thus compete with normal cellular protein synthesis. A mechanism to dispose of hyper-edited dsRNAs might enable cells to combat viral infection more effectively.
We previously reported a ribonuclease activity in various protein extracts that specifically degraded inosine-containing RNA (I-RNA) (Scadden and Smith, 1997). To study this activity, referred to as I-RNase, we utilized I-RNA substrates where the inosine residues were incorporated by in vitro transcription, rather than by deamination (i.e. involving G to I rather than A to I substitutions). We subsequently found that a number of ribonucleases [e.g. RNase A, S1 nuclease, Rrp41p, Rrp4p (Allmang et al, 1999)] were able to degrade I-RNA much more rapidly than the equivalent guanosine-containing RNA. This suggested that the observed ‘I-RNase’ activity resulted from destabilization of intramolecular secondary structure within the single-stranded substrate RNAs, making them more generally accessible to a wide variety of ribonucleases.
We have now carried out a series of experiments to investigate the fate of deaminated-dsRNA (d-dsRNA) that contains multiple inosine substitutions as a result of in vitro hyper-editing by ADAR2. These RNA substrates were thus essentially equivalent to hyper-edited dsRNA detected in vivo. Upon incubation in various cytoplasmic extracts, d-dsRNA is susceptible to cleavage at specific sites consisting of alternating IU and UI base pairs. In contrast, unmodified dsRNA and even d-dsRNAs that contain four consecutive IU base pairs are not cleaved. Strikingly, dsRNAs in which alternating IU and UI base pairs are replaced by isomorphic GU and UG base pairs (Masquida and Westhof, 2000) are not cleaved. Hyper-editing therefore appears to generate specific RNA structures that might constitute a suitable unique tag for the disposal of viral dsRNA.
Results
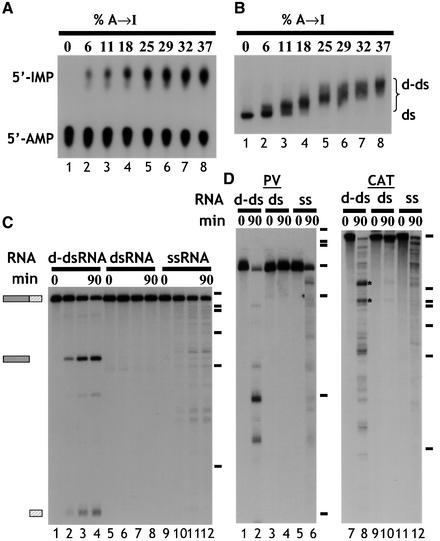
The dsRNA substrate that we initially used to analyse the fate of d-dsRNA was ΔKP, which comprises exons 2 and 3 of the rat α-tropomyosin gene with no intron between (Scadden and Smith, 1997). In each experiment, only one strand of each dsRNA substrate was labelled, and the single-strand RNA (ssRNA) control corresponded to the same strand. The ΔKP dsRNA was deaminated using ADAR2, typically resulting in up to ∼40% A to I conversion (Figure 1A). All substrate RNAs used in this study had a comparable level of deamination (∼40%) unless otherwise stated. Analysis of d-dsRNA by native gel electrophoresis showed that all of the input RNA was deaminated to a similar extent, as indicated by the relatively tight band with lower mobility than unmodified dsRNA (Figure 1B, compare lanes 1 and 8). When ΔKP d-dsRNA was incubated in Xenopus oocyte extract, it was cleaved at a unique position to give two products (Figure 1C, lanes 1–4). In contrast, unmodified ΔKP dsRNA was stable (lanes 5–8), while the equivalent ssRNA was degraded more slowly, yielding numerous degradation products (lanes 9–12). Moreover, the rate of cleavage of d-dsRNA was equal to or often greater than the rate of ssRNA degradation, depending on the individual extract (lanes 1–4, 9–12). The products of cleavage appeared to be stable, which suggested that they remained double stranded. When other d-dsRNA substrates were tested, similar results were obtained, although a more complex pattern of cleavage products was usually observed. Figure 1D shows experiments with polyoma virus (PV) and chloramphenicol acetyl transferase (CAT) RNAs, respectively. When PV d-dsRNA was incubated in the Xenopus extract, it was also less stable than the equivalent dsRNA (compare lanes 2 and 4 of Figure 1D). Similarly, it was degraded more rapidly than the ssRNA equivalent (compare lanes 2 and 6). Although cleavage of PV d-dsRNA yielded more products than ΔKP, they were again stable. Cleavage of the CAT d-dsRNA was more vigorous, giving a complex pattern of cleavage products (lane 8). Nevertheless, the equivalent dsRNA was relatively stable, and the ssRNA was degraded more slowly (compare lanes 8, 10 and 12). Note that in this particular assay a very small amount of specific cleavage of CAT dsRNA was detectable after 90 min (lane 10). This cleavage probably reflects the small amount of Xenopus ADAR activity present in the extract. When a time course of cleavage of CAT RNA was performed, two bands (indicated by asterisks in Figure 1D) were the first cleavage products detected. These then gave rise to other cleavage products (data not shown). Similar data showing specific cleavage were obtained with two other substrates (data not shown). In summary, for all substrates tested, cleavage of the d-dsRNA was enhanced compared with the equivalent dsRNA, yielding a limited number of discrete products. As cleavage of ΔKP was most easily interpreted due to its single cleavage site, this substrate was used as a model to study the specificity of d-dsRNA cleavage.
Fig. 1. (A) Quantitation of deamination. ΔKP dsRNA was deaminated to varying degrees using ADAR2. Digestion of the modified RNA using RNase P1, followed by TLC, enabled quantitation of the amount of A to I conversion. The RNAs used in the assays shown typically contained ∼40% A to I conversion (lane 8). (B) Deamination of ΔKP results in a homogenous population of RNA. As the level of deamination of ΔKP increased, there was a corresponding reduction in the mobility of the RNA on a native gel (lanes 1–8). For dsRNAs with ∼40% A to I conversion (lane 8), the RNA migrated as a relatively compact band (‘d-ds’), indicating that all RNAs in the population were modified. (C) Deaminated dsRNA is specifically cleaved. Incubation of ΔKP d-dsRNA in Xenopus oocyte extract gave rise to two discrete cleavage products (lanes 1–4). In contrast, ΔKP dsRNA was stable (lanes 5–8), and ΔKP ssRNA was degraded slowly yielding numerous products (lanes 9–12). The time course used in each assay was 0, 15, 60 and 90 min. Positions of DNA molecular weight markers are shown to the right of the figure (φX174 _Hae_III, 310, 281, 271, 234, 194, 118, and 72 nt). (D) Incubation of both PV and CAT d-dsRNAs for 90 min in Xenopus oocyte extracts also gave rise to discrete cleavage products (lanes 1–6 and 7–12, respectively). The equivalent dsRNAs were stable (compare lanes 2 and 4, and lanes 8 and 10), while degradation of the ssRNAs gave more numerous products (lanes 6 and 12).
Cleavage by a cytoplasmic ribonuclease
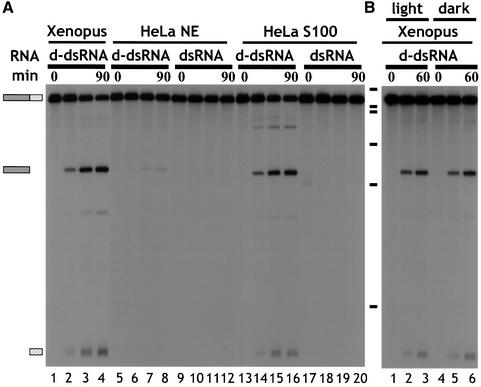
Cleavage of d-dsRNA was tested in HeLa cell nuclear (NE) and cytoplasmic (S100) extracts (Figure 2A). Cleavage of ΔKP d-dsRNA occurred in HeLa cell S100 extract, yielding the same major products as in the Xenopus oocyte extract (compare lanes 1–4 with 13–16). In addition, a minor cleavage product was detected (lanes 13–16). No cleavage of dsRNA was detected (lanes 17–20). In contrast, incubation of ΔKP d-dsRNA in HeLa cell NE resulted in negligible cleavage compared with HeLa cell S100 (compare lanes 8 and 16). Equivalent amounts of NE and S100 protein were used and the _A_260/_A_280 ratios were identical. Therefore, the difference in activity could not be accounted for by differences in either protein concentration or differential competition by nucleic acids. The minor amount of activity seen in NE (lanes 5–8) can probably be accounted for by cytoplasmic contamination during extract preparation. The ribonuclease activity responsible for specifically cleaving d-dsRNA therefore appears to be cytoplasmic. This is consistent with the previous observation that hyper-edited PV RNA could be detected in the nuclear, but not cytoplasmic, compartment (Kumar and Carmichael, 1997).
Fig. 2. (A) d-dsRNA is cleaved by a cytoplasmic ribonuclease. ΔKP d-dsRNA was specifically cleaved to give two major products when incubated in either Xenopus oocyte extract or HeLa cell S100 (lanes 1–4 and 13–16, respectively). In contrast, ΔKP d-dsRNA was predominantly stable in HeLa cell nuclear extract (NE; lanes 5–8). The minor amount of cleavage detected could probably be accounted for by cytoplasmic contamination of the nuclear extract. ΔKP dsRNA was stable in both HeLa cell S100 and NE extracts (lanes 17–20 and 9–12, respectively). A cytoplasmic activity thus appears to be responsible for cleavage of d-dsRNA. The time course used in each assay was 0, 15, 60 and 90 min. (B) ΔKP d-dsRNA does not undergo photocleavage. Cleavage of ΔKP d-dsRNA in Xenopus oocyte extract was identical in the light and dark (compare lanes 1–3 and 4–6, respectively). This indicated that photocleavage was not responsible for the observed cleavage. The time course used in each assay was 0, 30 and 60 min.
RNA duplexes containing a GU base pair can be photocleaved in the presence of small organic molecules such as flavin mononucleotide (FMN) or riboflavin (Burgstaller et al., 1997). Since ADAR treatment results in replacement of A–U by IU base pairs, which are isomorphic with GU pairs, it was possible that the d-dsRNAs were being attacked by a similar mechanism. To test this we carried out assays in the dark. Cleavage was identical when carried out in the light or dark (Figure 2B). Moreover, d-dsRNAs were stable in a Xenopus extract that had been heated at 60°C for 10 min prior to incubation or in an assay where the extract was omitted (data not shown). This demonstrates that photocleavage was not responsible for the observed cleavage of d-dsRNAs, but that a ribonuclease was involved.
Editing of a specific sequence is required for cleavage
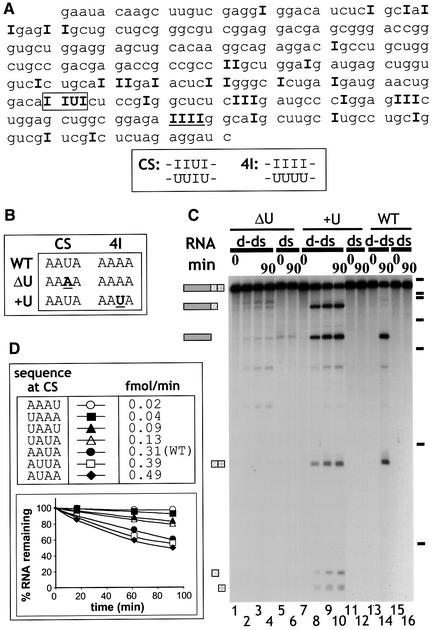
We used RT–PCR to amplify the sequences of deaminated ΔKP RNAs. Subsequent sequencing of the cloned RT–PCR products enabled the identification of edited positions in the RNA. The positions of deamination in the sense strand of ΔKP RNA deaminated to 40% are given in bold type in Figure 3A. As cleavage of ΔKP d-dsRNA was limited to one position (yielding two products), it was possible to map the site of cleavage by primer extension using a primer complementary to the 3′ end of the RNA (data not shown). This revealed that cleavage occurred within a sequence that potentially comprised both IU and UI base pairs (boxed; sequence CS below Figure 3A). Subsequent analyses of RT–PCR products corresponding to the antisense strand revealed that the base paired with U in the CS sequence was also inosine, even in d-dsRNAs with only 5% A to I conversion. Thus, cleavage of the d-dsRNA occurred within a sequence comprising four consecutive IU/UI wobble base pairs.
Fig. 3. (A) Identification of the sites of deamination in ΔKP d-dsRNA. Cloned RT–PCR products derived from a ΔKP d-dsRNA template deaminated to 40% were sequenced to identify positions of deamination. The sequence shown corresponds to the sense strand of ΔKP, and positions of A to I conversion are indicated by ‘I’. This sequence is typical of a number of sequenced clones. Cleavage of the d-dsRNA occurred at the sequence IIUI (boxed; referred to as CS). The sequence underlined (IIII) is referred to as 4I. The double-stranded sequence of the CS and 4I sites is indicated. (B) Two mutants of ΔKP were generated, where mutations were made in either the CS (ΔU mutant) or 4I (+U mutant) sequences, as shown (sense strand). ΔU potentially contained no cleavage sites while +U potentially contained two cleavage sites (compared with the single site in wild-type (WT) ΔKP). (C) Alternating IU and UI base pairs results in cleavage. ΔKP d-dsRNA (WT) was cleaved at the sequence CS in Xenopus oocyte extract to give two products (lane 14). In contrast, incubation of ΔU d-dsRNA yielded no cleavage products (lanes 1–4). Cleavage of +U d-dsRNA resulted in several products that corresponded to cleavage at both the CS and modified 4I sequences (lanes 7–10). The equivalent WT, ΔU and +U dsRNAs were stable (lanes 16, 6 and 12). The time course used to assay ΔU and +U d-dsRNAs was 0, 15, 60 and 90 min. (D) Mutants of ΔKP were generated where the WT CS sequence (AAUA) was replaced by the given sequences. The corresponding d-dsRNAs were incubated in Xenopus oocyte extract, and the cleavage was analysed by phosphoimaging. The initial rate of cleavage for each mutant (fmol full-length RNA cleaved/min) was calculated, as given in the table. The graph shows the cleavage of each d-dsRNA over a 90 min time course. These data indicate that sequences which potentially contain different arrangements of IU and UI base pairs are not cleaved with equal efficiently following hyper-editing.
Inspection of the sequence in Figure 3A revealed several other positions with adjacent inosines that were not sites for cleavage. In particular, a run of four consecutive AU base pairs (underlined; sequence 4I below Figure 3A) was a hot spot for editing; even at 25% overall levels of deamination, this sequence contained four inosines in half of the sequenced clones and at least three inosines in 90% of clones. If the sole requirement for cleavage was extensive local deamination, one might expect the d-dsRNA to be cleaved at positions with three or four consecutive IU base pairs. This suggested that it was either the presence of alternating IU and UI base pairs that made the RNA susceptible for cleavage, or that the sequence context of the IU base pairs may be important for cleavage. To distinguish between these possibilities, two mutants were generated (Figure 3B). In the mutant ΔU, the U–A base pair in the cleavage site (CS) was mutated to A–U and the downstream site (4I) was unchanged. In the mutant +U, the downstream sequence was mutated to resemble the cleavage site (AAAA to AAUA), while the sequence at the cleavage site was unaltered. The two mutants potentially contained either no cleavage sites (ΔU) or two cleavage sites (+U). The three dsRNAs [wild type (WT), ΔU, +U] were deaminated to the same extent by ADAR2. Consistent with expectations, the ΔU d-dsRNA was not cleaved efficiently in Xenopus oocyte extract (Figure 3C, lanes 1–4), while +U d-dsRNA generated products corresponding to cleavage at both the original site and the mutated downstream site (lanes 7–10). The wild-type ΔKP was cleaved only at the previously identified site (lanes 13 and 14). Primer extension using a labelled primer complementary to the extreme 3′ end of the sense RNA strand was used to confirm the location of the cleavage sites in +U (data not shown). For both mutants no cleavage of dsRNA was observed (lanes 6 and 12). These data demonstrate that cleavage depends on the presence of alternating IU and UI base pairs rather than the position of IU pairs within the RNA. A further series of mutants were generated in which the order of A–U and U–A pairs in the unedited cleavage site were changed (Figure 3D). When each corresponding mutant d-dsRNA was incubated in Xenopus oocyte extract, they were cleaved with varying efficiencies (Figure 3D). Mutants AUAA and AUUA were cleaved even more efficiently than the wild-type AAUA. In contrast, AAAU and UAAA were cleaved to a very small extent. For all mutants, the equivalent dsRNAs were assayed in parallel and they were all stable as expected (data not shown). Although each of the RNAs were deaminated equally by ADAR2 (45% A to I), it is possible that the deamination efficiency of particular positions within the mutated cleavage sites were reduced, thereby reducing the likelihood of cleavage. On the other hand, none of the mutations introduced was likely to dramatically reduce the frequency of deamination within the cleavage site sequence according to either the 5′ or 3′ neighbour preferences of ADAR2 (Polson and Bass, 1994; Lehmann and Bass, 2000).
Following the determination of the requirements for cleavage in ΔKP, the sequences of the other test RNAs (Figure 1D) were reanalysed. For the PV RNA, where the cleavage pattern was relatively simple, it was possible to identify the precise sequences of alternating A–U and U–A base pairs that produced the cleavage site after editing. This analysis also revealed that both the A–U content of the RNAs and number of potential cleavage sites increased in the order ΔKP < PV1 < CAT. This order parallels the order of cleavage complexity (see Figure 1). It is possible that some sites are more likely to be deaminated than others according to the neighbour preferences of ADAR2 (Polson and Bass, 1994; Lehmann and Bass, 2000) and would therefore constitute better targets for cleavage.
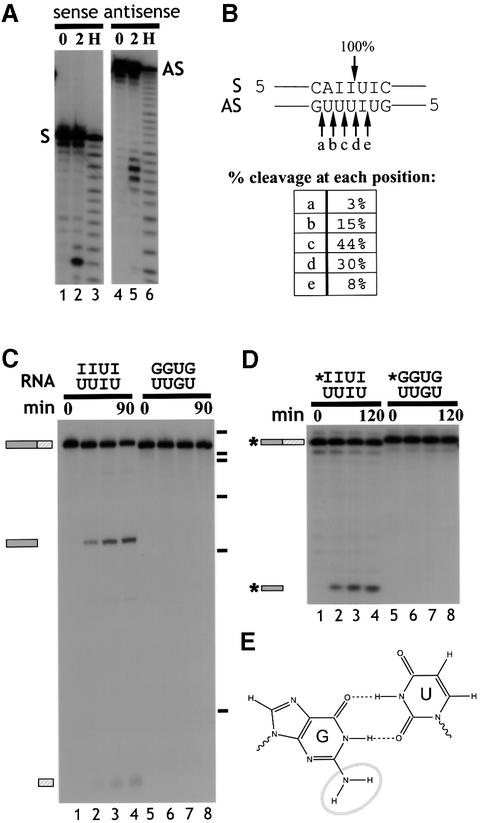
The precise cleavage site in d-dsRNA was mapped using 20 bp synthetic RNA duplexes that contained alternating IU base pairs corresponding to the cleavage site in ΔKP (Figure 4A and B; see Materials and methods). In each experiment, only one of the two strands was 5′ end-labelled. Complete hybridization of the labelled strand was confirmed by native gel electrophoresis (data not shown). Cleavage of the sense strand occurred 5′ of the U in the sequence 5′-IpU-3′, while the other strand was cleaved in five positions—predominantly 5′ of the two U residues opposite the two consecutive inosines (Figure 4B). The cleavage products appear to have 3′ phosphate ends since they co-migrated with the alkaline hydrolysis products (Figure 4A, lanes 3 and 6). This was confirmed by periodate oxidation and beta-elimination analysis. The full-length RNA was shortened by one nucleotide after beta-elimination, while the cleavage products were unaffected, showing that they do not possess a 3′ hydroxyl group (data not shown).
Fig. 4. (A) Cleavage of short synthetic dsRNA substrates. Short synthetic dsRNAs that contained the CS sequence were used to analyse cleavage. Incubation of short dsRNAs, 5′ end-labelled on either the sense (S) or antisense strand (AS), in Xenopus oocyte extract indicated that both RNA strands were cleaved (lanes 3 and 6). Electrophoresis alongside an alkaline hydrolysis ladder (H; lanes 1 and 4) enabled mapping of the cleavage sites on the sense and antisense strands. (B) Mapping the cleavage sites. The short synthetic RNAs were cleaved at the positions indicated. The sense strand was cleaved at one major position, 5′ of a U residue, and within the sequence of alternating IU and UI base pairs. In contrast, the antisense strand was cleaved at five positions, where ∼90% of the cleavage occurred 5′ of U residues. (C) Alternating GU and UG base pairs are not cleaved. ΔKP dsRNA was synthesized where the sequence of the cleavage site comprised GU and UG base pairs (as indicated above gel). In contrast with ΔKP d-dsRNA, this dsRNA was not cleaved when incubated in Xenopus oocyte extract (compare lanes 1–4 and 5–8). The time course used in each assay was 0, 15, 60 and 90 min. (D) Short dsRNAs that contained either alternating IU and UI base pairs or GU and UG base pairs (indicated above gel) were incubated in Xenopus oocyte extract. The time course used in each assay was 0, 30, 60 and 120 min. Each of the RNAs shown is labelled on the sense strand (indicated by *). While the dsRNAs containing alternating IU and UI base pairs were cleaved (lanes 1–4), the dsRNAs that contained GU and UG base pairs were stable (lanes 5–8). Thus, substitution of guanosine residues for inosine residues in the cleavage site inhibits cleavage of the RNA. (E) A non-Watson–Crick GU wobble base pair. An IU wobble base pair is isosteric with the GU base pair, but lacks the exocyclic amine group (circled) which projects into the minor groove.
Sequences containing GU and UG base pairs are not cleaved
Inosine differs from guanosine only by the absence of the _N_2 exocyclic amine group (circled in Figure 4E). IU and UI wobble base pairs are therefore isomorphic with GU and UG wobble base pairs, respectively, although they are slightly less stable (Strobel et al., 1994). Due to the asymmetry of the glycosidic bond angle between the base and C1′ sugar atom in a GU pair compared with a Watson–Crick base pair, GU and UG pairs are not isomorphic with each other. In addition, the presence of tandem GU or UG pairs in a duplex causes overwinding or underwinding of the RNA helix depending on the order of the wobble base pairs. This causes large and distinctive conformational changes in the RNA duplex (Masquida and Westhof, 2000; Varani and McClain, 2000). It is likely that consecutive IU and UI base pairs would cause very similar distortions in the dsRNA structure. Such structural distortions could be responsible for the specific cleavage of d-dsRNA at alternating IU/UI base pairs, but not at runs of IU base pairs (Figure 3).
To test whether the cleavage of d-dsRNA is related only to the distortion of the dsRNA geometry, we made ΔKP dsRNA in which IU and UI were replaced by GU and UG base pairs (GU-dsRNA). The GU-dsRNAs were prepared by in vitro transcription using templates derived by RT–PCR from d-dsRNA (see Materials and methods). The d-dsRNA and GU-dsRNA were then assayed for cleavage in Xenopus oocyte extract (Figure 4C). While d-dsRNA was cleaved (lanes 1–4), the GU-dsRNA was stable (lanes 5–8). This observation is made even more significant by the fact that only a proportion of the d-dsRNA has the correct sequence necessary for cleavage (by deamination), while 100% of the GU-dsRNAs contain GU and UG base pairs at the cleavage site. Therefore, cleavage of the d-dsRNA cannot be replicated in dsRNAs containing equivalent GU distortions.
To analyse this further we compared cleavage of short synthetic RNA duplexes that contained either IU and UI base pairs or GU and UG base pairs (Figure 4D). The sequence of the short dsRNAs (20 bp duplex) comprised the cleavage site in ΔKP plus flanking nucleotides (see Materials and methods). When these RNAs were incubated in Xenopus oocyte extract, the IU duplex was cleaved while the GU-dsRNA was stable (Figure 4D; compare lanes 1–4 and 5–8). Moreover, both strands of the inosine-containing dsRNA were cleaved while neither strand of the guanosine-containing dsRNA was cut (data not shown). The cleavage of the short synthetic RNAs not only confirmed the results obtained with long RNA duplexes, but also demonstrated that inosine is required only at the cleavage site.
There are two obvious explanations for the resistance of the GU substrate to cleavage. First, the GU and IU duplexes may have an identical overall structure, but the additional minor groove amine groups on guanosine might interfere with cleavage. Alternatively, the IU duplex may adopt a different structure from the GU duplex, which at the extreme could be locally melted around the IU base pairs, giving rise to the differential cleavage. To address these possibilities, the stability of the 20 bp synthetic duplexes was analysed by RNA melting experiments (Loakes and Brown, 1994). Both duplexes showed a single _A_260 transition with melting temperatures of 63.3 and 60.0°C for the GU and IU duplexes, respectively (assays in 10 mM HEPES pH 7.5, 100 mM NaCl). This is consistent with previous reports that IU base pairs are less stable than GU (Strobel et al., 1994). The modest difference in melting temperature does not appear sufficient to explain the complete lack of cleavage of GU RNA at 25°C while IU RNA is efficiently cleaved. Thus, cleavage of hyper-edited RNA in cytoplasmic extracts appears to involve recognition of specific distorted dsRNA structures.
Discussion
The occurrence of extended dsRNA in cells is relatively uncommon, and is frequently associated with infection by DNA or RNA viruses. Antiviral defence mechanisms such as the PKR and 2′-5′-adenylate synthase/RNase L pathways are stimulated by the presence of dsRNA. An additional mechanism to remove dsRNA from cells may involve hyper-editing of dsRNA by ADARs, followed by targeted cleavage. The data presented here clearly demonstrate that various cell extracts contain an endoribonuclease activity that fulfils the requirements for the second step in this proposed pathway. Confinement of the novel ribonuclease to the cytoplasmic fraction (Figure 2) is consistent with previous data where PV d-dsRNA was detected only in the nucleus (Kumar and Carmichael, 1997). While the favoured interpretation of these data was that hyper-editing prevented export to the cytoplasm, an alternative explanation consistent with our data is that d-dsRNA deaminated by constitutive nuclear ADARs and exported from the nucleus may be cleaved rapidly and thus not detected by RT–PCR. Alternatively, viral dsRNA in the cytoplasm may be edited by the interferon-inducible isoform of ADAR1 (Patterson and Samuel, 1995) and then cleaved.
Early investigations upon the effects of ADARs indicated that the modified RNA became more generally vulnerable to attack by single-stranded RNases, indicative of general loosening of the dsRNA secondary structure (Bass and Weintraub, 1988). Our data now indicate that hyper-edited dsRNA becomes hypersensitive to endonuclease attack at very specific sites consisting of alternating IU and UI base pairs. The specificity for alternating wobble base pairs was emphatically demonstrated by changing a single UI base pair to IU, producing a sequence with four consecutive IU pairs that was resistant to cleavage (Figure 3). There is more than one possible explanation for the targeting of cleavage at this specific site. Formation of a specific base-paired structure that deviates significantly from the normal geometry of A-form dsRNA could facilitate recognition by a specific endonuclease. Alternatively, the substitution of A–U by IU base pairs could decrease the stability of the RNA duplex sufficiently to cause local melting of RNA structure, thus allowing greater access by single-strand-specific RNases. These two scenarios are not mutually exclusive; a non-canonical base-paired structure might allow specific recognition as well as enhanced accessibility due to breathing of the structure. Treatment of hyper-edited dsRNA with the single-strand-specific nucleases S1 and RNase A suggests that there is some general increased accessibility, but that specific recognition may play a more important role in the extracts. Degradation of d-dsRNA by S1 nuclease was accelerated relative to dsRNA, but there was no evidence of discrete cleavage products corresponding to those seen in the cytoplasmic extracts (see Supplementary data, available at The EMBO Journal Online), arguing against localized melting at the cleavage site. In contrast, treatment of IU and GU ΔKP RNAs with RNase A, which cleaves 3′ of accessible U residues, indicated that the alternating wobble base pairs do provide a target for this single-stranded nuclease (Supplementary data). However, RNase A cleaved the IU-dsRNA ∼300-fold slower than the corresponding ssRNA. This is in strong contrast to the activity in the cytoplasmic extracts, where the d-dsRNA was usually degraded at a comparable or greater rate than the control ssRNA, even though the ssRNA contains many more potential cleavage sites for single-strand RNases (Figure 1C and D). Another important insight from experiments with RNase A is that the GU RNA was also cleaved at the same site as the d-dsRNA at a 2.5-fold lower rate (Supplementary data). The cleavage rates of the IU RNA and the equivalent GU RNA by RNase A give an indication of the accessibility of the IU and GU substrates due to structural breathing. The 2.5-fold greater cleavage rate of the IU compared with the GU substrate is consistent with the small difference in melting temperature of the 20 bp GU and IU duplexes (63.3 and 60.0°C, respectively). The complete lack of cleavage in extracts when IU base pairs were replaced by GU (Figure 4C and D) therefore argues that the enzyme in the extracts does not recognize the d-dsRNA simply due to localized melting. Instead, a specific RNA secondary structure may be recognized, and the presence of the 2-amino groups of G in the minor groove is sufficient to interfere with this recognition.
At present, we can only speculate upon the nature of the RNase responsible for cleaving d-dsRNA. It is possible that a previously identified dsRNA-specific or ssRNA-specific ribonuclease (or both) may be involved. The observed specificity of cleavage (5′ of U leaving a 3′ phosphate) is not characteristic of known dsRNases such as ribonuclease III (Nicholson, 1997; Bass, 2000), suggesting that a novel enzyme may be responsible. In addition to the major cleavage site, d-dsRNA is also cleaved at other sites, albeit much less efficiently. For example, a minor cleavage product of ΔKP was detected in HeLa cell S100 (Figure 2A, lanes 13–16) that appeared to map to the 4I site. This could represent less efficient cleavage by the same enzyme that acts on the major cleavage site, the activity of another enzyme specific for consecutive IU pairs, or simply partial access of single-strand specific RNases due to enhanced breathing at this site.
It is worth noting that the specificity of cleavage that we observe may arise not only from the intrinsic specificity of the endonuclease, but also from differential binding of cellular proteins to dsRNA and d-dsRNA, or even between different regions of d-dsRNA with different editing patterns. Such proteins could either protect RNA from cleavage or help to direct RNases to specific sites. In this regard, it is interesting that there are reports of cellular proteins that bind specifically to inosine-containing RNA (Maquat and Carmichael, 2001). The question of whether cleavage specificity derives from the RNase alone can only be addressed effectively by purifying and characterizing the cleavage enzyme. This endeavour is currently under way.
Irrespective of the identity of the nuclease, a major insight from our work is that hyper-edited RNAs become highly sensitive to cleavage at runs of alternating IU and UI base pairs. This specificity means that RNAs containing GU wobble base pairs or selectively edited mRNAs, which contain a limited number of unpaired inosines, will not be recognized and cleaved. None of the known mRNA editing sites generates an optimal cleavage site (Emeson and Singh, 2000). The cytoplasmic location of the cleavage enzyme also protects bone fide edited cellular mRNAs, since the intron–exon duplex that guides specific editing is not present when the mRNA is exported to the cytoplasm.
Cleavage of d-dsRNA will destroy its function. Although the cleaved fragments were relatively stable in our assays, it is likely that they would undergo further processing steps. These could involve the action of both RNA helicases, which may be able to unwind d-dsRNA more efficiently than unmodified dsRNA, and by other ribonucleases. We have shown previously (Scadden and Smith, 1997) that denatured d-dsRNA was degraded more rapidly than the equivalent unmodified ssRNA, so it is likely that unwound d-dsRNA is also preferentially degraded by various nucleases.
Finally, it is possible that our observations could be exploited for use in antisense technology. If the targeted RNA included sites that would be deaminated and then cleaved efficiently, down-regulation of gene expression may be enhanced.
Materials and methods
Preparation of ‘long’ dsRNA substrates
All of the long dsRNAs were prepared by in vitro transcription of both the sense and antisense RNAs, using a method based on that described by Melton et al. (1984). Only one strand of each RNA duplex was labelled internally with [α-32P]ATP (3000 Ci/mmol; Amersham), where 25 µCi of [α-32P]ATP was added per 50 µl transcription reaction. The ‘unlabelled’ strand of the RNA duplex was trace-labelled with [α-32P]UTP (3000 Ci/mmol; Amersham) at a specific activity at least 250-fold lower than that of the labelled RNA strand. All of the RNAs were initiated using an m7G(5′)ppp(5′)G dinucleotide primer. To make dsRNAs, the complementary RNAs were mixed at a molar ratio of 1:2 (sense:antisense) in buffer Q (50 mM Tris–HCl pH 7.9, 100 mM KCl, 5 mM EDTA, 10% glycerol), and then heated at 75°C for 10 min. The RNAs were allowed to cool slowly to 30°C. This method was used to prepare all dsRNA substrates.
ΔKP. The construct ΔKP was as described previously (Scadden and Smith, 1997). The sense RNA strand was synthesized using SP6 RNA polymerase (Stratagene) following linearization of ΔKP with _Bam_HI to give a 296-nt transcript. The antisense RNA was synthesized using T7 RNA polymerase from a template generated from ΔKP to give a 329-nt transcript. The template (348 bp) was made by PCR using primers to the SP6 and T7 promoter sequences. As the antisense RNA was 33 nt longer than the sense strand, there was an unpaired sequence flanking both the 5′ and 3′ ends of the duplex (16 and 17 nt, respectively).
PV1. The construct PV1 contains a sequence corresponding to part of the polyoma virus large T antigen (nucleotides 1653–1855 in the polyoma virus sequence). The plasmid containing the polyoma virus sequence was a gift from B.Griffin. The sense RNA strand was synthesized using T7 RNA polymerase following linearization of PV1 with _Bam_HI to give a 230-nt transcript. The antisense RNA strand was synthesized using SP6 RNA polymerase following linearization with _Eco_RI to give a 280-nt transcript.
CAT. The CAT plasmid used was as described previously (O’Connell and Keller, 1994). The sense RNA was synthesized using T7 RNA polymerase after linearizing the plasmid with _Hin_dIII to give a 605-nt transcript. The antisense RNA was synthesized using T3 RNA polymerase (Promega) after linearizing the plasmid with _Bam_HI to give a 594-nt transcript.
ΔKP mutants. Mutations were introduced into ΔKP using a PCR-based site-directed mutagenesis kit (Stratagene). The sense and antisense strands were generated using the same methods as used for ΔKP (above).
GU-dsRNA. The two strands that make GU-dsRNA are generated from two plasmids, pΔKP-dS and pΔKP-dAS (see below for details of plasmids). The sense strand was transcribed using SP6 RNA polymerase following linearization of pΔKP-dS with _Bam_HI. The antisense strand was transcribed using T7 RNA polymerase from a PCR template generated from pΔKP-dAS by the same method used for pΔKP (above).
Preparation of ‘short’ dsRNA substrates
2′-protected RNA oligonucleotides were purchased from Dharmacon Research, Inc., and were deprotected before use according to the manufacturer’s instructions. The sequences of the short RNAs used are as follows: 5′-ACUGGACAIIUICUCCGAGG (S–I), 5′-ACUGGACAGGUGCUCCGAGG (S–G), 5′-GAGAGCCUCGGAGUIUUUGUCCAGUUCAUC (AS–I), 5′-CCUCGGAGUGUUUGUCCAGU (AS–G). Oligonucleotides were 5′ end-labelled with [γ-32P]ATP (3000 Ci/mmol; Amersham) using T4 polynucleotide kinase by standard methods (Sambrook et al., 1989). In each short dsRNA, only one strand of the duplex was labelled. RNA oligonucleotides were hybridized to form dsRNAs using the same method as described for long dsRNAs.
Preparation of extracts
HeLa cell nuclear and cytoplasmic (S100) extracts were prepared according to the method described by Abmayr et al. (1988). The Xenopus oocyte extract was prepared by spin crushing following removal of the cells supporting the oocytes by collagenase treatment. To minimize the activity of non-specific proteases during this procedure, the ovary was washed thoroughly in OM2 buffer (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES–KOH pH 7.5) lacking Ca2+. The buffer was then exchanged for an ovary-equivalent volume of OM2 buffer containing 1 mg/ml collagenase B (Boehringer Mannheim) and the ovary left rotating at room temperature for ∼2 h or until most of the oocytes had separated. The treated ovary was washed 10 times in ∼30 ml OM2 followed by several washes in MBS [88 mM NaCl, 1 mM KCl, 0.41 mM CaCl2, 0.33 mM Ca(NO3)2, 0.82 mM MgSO4, 2.4 mM NaHCO3, 10 mM HEPES–KOH pH 7.4]. Oocytes were subsequently crushed by spinning for 20 min at 10 000 r.p.m. in a Sorvall HB-4 swing-out rotor. The entire aqueous phase was removing using a wide bore needle (18 gauge), avoiding the fat-rich layer and the bottom pellet of yolk and membrane debris. To minimize protein degradation, the extract was supplemented with Complete Inhibitors Solution lacking EDTA (Boehringer Mannheim). To remove all remaining debris, the extract was cleared by several short spins (1 min) at maximum speed in a microcentrifuge (at 4°C). The protein concentration was determined using a Bradford assay (Sambrook et al., 1989) with bovine serum albumin (BSA) as the reference protein.
Deamination reaction
dsRNA (300 fmol) was deaminated in a 25 µl reaction that contained the following: 50 mM Tris–HCl pH 7.9, 100 mM KCl, 5 mM EDTA, 10% glycerol, 3.75 µg of tRNA, 0.2 µg of BSA and 1 µl of hADAR2b. hADAR2b (His6-tagged) was expressed in Pichia pastoris (O’Connell et al., 1998) using a recombinant strain generously provided by André Gerber (University of Basel). The deamination reaction was carried out at 30°C for 1 h, then 25 µl of 2× proteinase K buffer [200 mM Tris–HCl pH 7.5, 25 mM EDTA, 300 mM NaCl, 2% (w/v) SDS] and 2 µl of proteinase K (20 mg/ml) were added. Following incubation at 37°C for 30 min, the deaminated RNA was extracted with phenol/chloroform, recovered by ethanol precipitation and resuspended in 6 µl of double-distilled water (50 fmol/µl). To analyse the efficiency of the deamination reactions, 50 fmol of RNA were digested in P1 buffer (30 mM KOAc pH 5.3, 10 mM ZnSO4) with 1.5 µg RNase P1 (Boehringer Mannheim) for 1 h at 50°C. The digestion products were fractionated on thin layer chromatography (TLC) plates (Polygram CEL 300 PEI/UV254; Machery and Nagel) using a chromatographic solvent that comprises saturated (NH4)2SO4, 0.1 M Na acetate (NaOAc) pH 6.0 and isopropanol (79:19:2). The identity of the spots corresponding to 5′-AMP and 5′-IMP were verified by the migration of unlabelled standards. Radioactivity in individual spots was quantified by phosphoimaging and volumetric analyses using Imagequant software (Molecular Dynamics).
Cleavage reaction
RNA substrates were assayed in a 10 µl reaction that contained 10 mM HEPES–KOH pH 7.9, 0.8 mM ATP, 50 fmol RNA and protein extract [either 1 µl of Xenopus extract (4 mg/ml), 1.8 µl of HeLa S100 (14 mg/ml) or 2.8 µl of HeLa nuclear extract (9 mg/ml)]. Reactions were incubated at 25°C for 0–4 h and then treated with 6 µl of proteinase K (20 mg/ml) in a total volume of 100 µl of Proteinase K buffer for 30 min at 37°C. The RNA was extracted once with phenol/chloroform and recovered by precipitation with ethanol. The RNA pellets were dried, resuspended in 50 µl of formamide dyes, and 10 µl samples were analysed by denaturing polyacrylamide gel electrophoresis. The gels were dried under vacuum and visualized by either autoradiography or phosphoimaging. Beta-elimination was used to investigate the nature of the 3′ ends of the cleaved RNA. RNA was incubated in 100 mM NaIO4 for 30 min at 25°C (in the dark). Excess periodate was removed by adding rhamnose (final concentration 100 mM) and then incubating for 30 min at 25°C. Beta-elimination was performed for 30 min at 45°C in the presence of 250 mM aniline and 50 mM NaOAc pH 5.
RT–PCR and cloning of edited RNAs
All oligonucleotides used were synthesized in the PNAC Facility, Department of Biochemistry, University of Cambridge. The oligonucleotides used for both RT and PCR of the sense strand contained some degenerate positions (A/G or C/T) to accommodate sequence changes resulting from deamination. The oligonucleotides were: ΔKP5′, 5′-GAATACAAGCTTGTCGA/GG, and ΔKP3′, 5′-GATCCTCT/CAGAGT/CCGAT/CCG. The oligonucleotides used for amplification of the antisense strand were complementary to the single-stranded overhangs at either end of the duplex. They were: ΔKPc5′, 5′-GAGACCGGAATTCGGATC, and ΔKPc3′, 5′-ATTTAGGTGACACTATAGAA. RT of both RNA strands was carried out using primers ΔKP 3′ and ΔKP c3′. PCR was then performed using the primer pairs ΔKP5′/ΔKP3′ and ΔKPc5′/ΔKPc3′. RT and PCR were performed using standard conditions (Ausubel et al., 1994). Following RT–PCR, the products were gel purified using Qiaex (Qiagen) and cloned into a pGEM-T vector (Promega). Clones were sequenced by the DNA Sequencing Facility, Department of Biochemistry, University of Cambridge.
One of the clones that contained an insert corresponding to RNA edited on the sense strand (with the sequence 5′-IIUI at the cleavage site) was named pΔKP-dS. Similarly, a clone that contained an insert corresponding to RNA edited on the antisense strand (with the sequence 5′-UIUU at the cleavage site) was named pΔKP-dAS.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank André Gerber, Walter Keller, Mary O’Connell, Anna Git and Beverly Griffin for gifts of reagents, and David Loakes for performing RNA melting experiments. We would also like to thank Mary O’Connell and Eric Westhof for helpful discussions. Finally, we would like to thank Justine Southby for critically reading the manuscript. This work was supported by a grant from the Wellcome Trust (grant number 052241). A.D.J.S. was also supported by a Research Fellowship from Newnham College, University of Cambridge.
References
- Abmayr S.M., Reed,R. and Maniatis,T. (1988) Identification of a functional mammalian spliceosome containing unspliced pre-mRNA. Proc. Natl Acad. Sci. USA, 85, 7216–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′–5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Bass B.L. and Weintraub,H. (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell, 55, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Bass B.L., Weintraub,H., Cattaneo,R. and Billeter,M.A. (1989) Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell, 56, 331. [DOI] [PubMed] [Google Scholar]
- Burgstaller P., Hermann,T., Huber,C., Westhof,E. and Famulok,M. (1997) Isoalloxazine derivatives promote photocleavage of natural RNAs at GU base pairs embedded within helices. Nucleic Acids Res., 25, 4018–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., Kaelin,K., Baczko,K. and Billeter,M.A. (1989) Measles virus editing provides an additional cysteine-rich protein. Cell, 56, 759–764. [DOI] [PubMed] [Google Scholar]
- Emeson R.B. and Singh,M. (2000) Adenosine-to-inosine RNA editing: substrates and consequences. In Bass,B. (ed.), RNA Editing. Oxford University Press, Oxford, UK, pp. 109–138.
- Felder M.P., Laugier,D., Yatsula,B., Dezelee,P., Calothy,G. and Marx,M. (1994) Functional and biological properties of an avian variant long terminal repeat containing multiple A-conversion to G-conversion in the U3 sequence. J. Virol., 68, 4759–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar A.M. and Linial,M.L. (1995) Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J. Virol., 69, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough R.F. and Bass,B.L. (2000) Adenosine deaminases that act on RNA. In Bass,B. (ed.), RNA Editing. Oxford University Press, Oxford, UK, pp. 77–108.
- Kimelman D. and Kirschner,M.W. (1989) An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell, 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Konarska M.M. and Sharp,P.A, (1986) Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell, 46, 845–855. [DOI] [PubMed] [Google Scholar]
- Kumar M. and Carmichael,G.G., (1997) Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl Acad. Sci. USA, 94, 3542–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. and Carmichael,G.G., (1998) Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev., 62, 1415–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.A. and Bass,B.L. (2000) Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry, 39, 12875–12884. [DOI] [PubMed] [Google Scholar]
- Loakes D. and Brown,D.M. (1994) 5-nitroindole as a universal base analogue. Nucleic Acids Res. 22, 4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. and Carmichael,G.G. (2001) Quality control of mRNA function. Cell, 104, 173–176. [DOI] [PubMed] [Google Scholar]
- Masquida B. and Westhof,E. (2000) On the wobble GU and related pairs. RNA, 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D.A., Krieg,P.A., Rebagliati,M.R., Maniatis,T., Zinn,K. and Green,M.R. (1984) Efficient in vitro synthesis of biologically active RNA and RNA hybridisation probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res., 12, 7035–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D.P. and Bass,B.L. (1999) Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+RNA. Proc. Natl Acad. Sci. USA, 96, 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.G., Dimock,K. and Kang,C.Y. (1991) Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology, 181, 760–763. [DOI] [PubMed] [Google Scholar]
- Nicholson A.W. (1997) Escherichia coli ribonucleases: paradigms for understanding cellular RNA metabolism and regulation. In D’Alessio,G. and Riordan,J.F. (eds), Ribonucleases: Structures and Functions. Academic Press, New York, NY, pp. 1–49.
- Nishikura K., Yoo,C., Kim,U., Murray,J.M., Estes,P.A., Cash,F.E. and Liebhaber,S.A. (1991) Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J., 10, 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.A. and Keller,W. (1994) Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc. Natl Acad. Sci. USA, 91, 10596–10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M.A., Gerber,A. and Keegan,L.P. (1998) Purification of native and recombinant double-stranded RNA-specific adenosine deaminases. Methods, 15, 51–62. [DOI] [PubMed] [Google Scholar]
- O’Hara P.J., Nichol,S.T., Horodyski,F.M. and Holland,J.J. (1984) Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell, 36, 915–924. [DOI] [PubMed] [Google Scholar]
- Patterson J.B. and Samuel,C.E. (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol., 15, 5376–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschek J.P., Mermer,M.J., Scheckelhoff,M.R., Simone,A.A. and Vaughn,J.C. (1996) RNA editing in Drosophila 4f-rnp gene nuclear transcripts by multiple A-to-G conversions. J. Mol. Biol., 259, 885–890. [DOI] [PubMed] [Google Scholar]
- Polson A.G. and Bass,B.L. (1994) Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J., 13, 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda P., Garciabarreno,B. and Melero,J.A. (1994) Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A–G substitutions (hypermutations). Virology, 198, 653–662. [DOI] [PubMed] [Google Scholar]
- Rueter S.M., Dawson,T.R. and Emeson,R.B. (1999) Regulation of alternative splicing by RNA editing. Nature, 399, 75–80. [DOI] [PubMed] [Google Scholar]
- Saccomanno L. and Bass,B.L. (1999) A minor fraction of basic fibroblast growth factor mRNA is deaminated in Xenopus stage VI and matured oocytes. RNA, 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. et al. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbour Laboratory Press, Cold Spring Harbor, NY.
- Samuel C.E. (1979) Mechanism of interferon action. Phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase possessing the specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc. Natl Acad. Sci. USA, 76, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. (1998) Protein–nucleic acid interactions and cellular responses to interferon. Methods, 15, 161–165. [DOI] [PubMed] [Google Scholar]
- Scadden A.D.J. and Smith,C.W.J. (1997) A ribonuclease specific for inosine-containing RNA: a potential role in antiviral defence. EMBO J., 16, 2140–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R.H. (1997) 2-5A dependent RNase L: a regulated endoribonuclease in the interferon system. In D’Alessio,G. and Riordan,J.F. (eds), Ribonucleases: Structure and Functions. Academic Press, New York, NY, pp. 515–551.
- Smith C.W.J., Chu,T.T. and Nadal Ginard,B. (1993) Scanning and competition between AGs are involved in 3′ splice-site selection in mammalian introns. Mol. Cell. Biol., 13, 4939–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S.A., Cech,T.R., Usman,N. and Beigelman,L. (1994). The 2,6-diaminopurine riboside5-methylisocytidine wobble base pair: an isoenergetic substitution for the study of GU base pairs in RNA. Biochemistry, 33, 13824–13835. [DOI] [PubMed] [Google Scholar]
- Varani G. and McClain,W.H. (2000) The GU wobble base pair—a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep., 1, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Hassel,B.A. and Silverman,R.H. (1993) Expression cloning of 2-5A-dependent RNase: a uniquely regulated mediator of interferon action. Cell, 72, 753–765. [DOI] [PubMed] [Google Scholar]