T-DNA-Associated Duplication/Translocations in Arabidopsis. Implications for Mutant Analysis and Functional Genomics (original) (raw)
Abstract
T-DNA insertion mutants have become a valuable resource for studies of gene function in Arabidopsis. In the course of both forward and reverse genetic projects, we have identified novel interchromosomal rearrangements in two Arabidopsis T-DNA insertion lines. Both rearrangements were unilateral translocations associated with the left borders of T-DNA inserts that exhibited normal Mendelian segregation. In one study, we characterized the _embryo-defective_88 mutation. Although _emb_88 had been mapped to chromosome I, molecular analysis of DNA adjacent to the T-DNA left border revealed sequence from chromosome V. Simple sequence length polymorphism mapping of the T-DNA insertion demonstrated that a >40-kbp region of chromosome V had inserted with the T-DNA into the _emb_88 locus on chromosome I. A similar scenario was observed with a prospective T-DNA knockout allele of the LIGHT-REGULATED RECEPTOR PROTEIN KINASE (LRRPK) gene. Whereas wild-type LRRPK is on lower chromosome IV, mapping of the T-DNA localized the disrupted LRRPK allele to chromosome V. In both these cases, the sequence of a single T-DNA-flanking region did not provide an accurate picture of DNA disruption because flanking sequences had duplicated and inserted, with the T-DNA, into other chromosomal locations. Our results indicate that T-DNA insertion lines—even those that exhibit straightforward genetic behavior—may contain an unexpectedly high frequency of rearrangements. Such duplication/translocations can interfere with reverse genetic analyses and provide misleading information about the molecular basis of mutant phenotypes. Simple mapping and polymerase chain reaction methods for detecting such rearrangements should be included as a standard step in T-DNA mutant analysis.
The ability of Agrobacterium tumefaciens to genetically transform host cells has revolutionized plant biology and helped to usher in the era of plant functional genomics and biotechnology. In nature, A. tumefaciens induces crown galls by transferring a portion of the tumor-inducing (Ti) plasmid into the nucleus of plant cells (Van Larebeke et al., 1975). A region of the Ti plasmid, the T-DNA, encodes functions that induce gall formation. Upon infection, the T-DNA is transferred into host cells and inserts into the nuclear genome. Two short regions on the ends of the T-DNA, known as the left border (LB) and right border (RB), are necessary for the transfer of the T-DNA into the nucleus and for subsequent integration (for review, see Gordon, 1998). Proteins essential for T-DNA transfer and host integration are encoded by the vir operon elsewhere on the Ti plasmid; their identities and functions have been reviewed by Zambryski et al. (1989). A definitive model for T-DNA integration has not yet been established, but likely involves regions of microhomology between the T-DNA borders and the plant genome and possibly occurs by illegitimate recombination (Gheysen et al., 1991). Host DNA repair mechanisms may be recruited to assist in integration (Tinland, 1996, Nacry et al., 1998). T-DNA appears to insert randomly within the genome (Thomas et al., 1994; for review, see Azpiroz-Leehan and Feldmann, 1997) although some bias for transcribed regions has been suggested (Tinland, 1996).
T-DNA has proven to be a valuable vector for making transgenic plants, with two different kinds of applications. The placement of intact genes and selectable markers such as the neomycin phosphotransferase gene, providing kanamycin resistance, between the LB and RB of the Ti plasmid, has enabled the engineering of plants expressing exogenous or modified genes. T-DNAs with selectable marker genes and plasmid-based origins of replication have also been very useful as an insertional mutagen. Hundreds of mutants have been characterized and the disrupted genes isolated using forward genetic strategies based on T-DNA mutagenesis (Koncz et al., 1992; Azpiroz-Leehan and Feldmann, 1997). Further modifications of T-DNA have allowed additional approaches for assessing gene function, such as enhancer and promoter trapping (Topping and Lindsey, 1997; Lindsey et al., 1998; Campisi et al., 1999) as well as activation tagging (Weigel et al., 2000).
T-DNA mutagenesis has also provided strategies for investigating gene function through genomics approaches. The most common strategy is a reverse genetics approach that utilizes PCR with oligonucleotide primers derived from a gene of interest to isolate T-DNA insertion alleles from populations of mutagenized plants (for review, see Krysan et al., 1999). Two other genomic approaches used on large collections of insertion lines include large-scale sequencing of flanking DNA from T-DNA populations, and sequence tagging projects in which mutants of a phenotypic class are identified from tagged populations and then sequences are obtained from the insertion sites from that class of mutants.
Despite its utility as an insertion mutagen and gene tag, T-DNA mutagenesis has some shortcomings. Several types of genetic anomalies have been documented in transformants of Arabidopsis following T-DNA mutagenesis. These range from base substitutions and minor additions and deletions (Negruk et al., 1996; Noguchi et al., 1999), to major chromosomal translocations (Castle et al., 1993). Truncated, tandem, and multiple T-DNA inserts are also common in T-DNA mutagenized populations (Castle et al., 1993). Several such cases have been characterized in some detail, with the hope that they can provide clues to the molecular mechanisms of T-DNA integration (e.g. Gheysen et al., 1991; De Neve et al., 1997; De Buck et al., 1999). Minor chromosomal rearrangements have also been observed at T-DNA integration sites. For example, Ohba et al. (1995) observed multiple duplications of short (<20 bp) sequences adjacent to inserts in tobacco, and Gheysen et al. (1991) identified DNA of unknown origin in T-DNA junctions analyzed by Southern blot. Two detailed studies recently documented large chromosomal rearrangements associated with T-DNA integration (Nacry et al., 1998; Laufs et al., 1999). In both of these cases, transformants contained multiple inserts, and rearrangements were apparently caused by interaction between different T-DNAs that had integrated at separate sites.
Here, we describe two examples of a novel type of T-DNA-associated inter-chromosomal rearrangement, identified in the course of both forward and reverse genetic analyses. In both cases, the transformants contain single inserts and display straightforward genetics with regard to T-DNA segregation, but sequences from loci unlinked to the insertion site flank the T-DNA LB. In one case, the T-DNA and adjacent translocated sequences have inserted into an essential gene, resulting in an embryo-defective phenotype. Our results demonstrate that major, internal chromosomal translocations may be associated with single T-DNA inserts, and that duplication of translocated sequences may accompany such rearrangements. The implications of such chromosomal rearrangements for functional genomic strategies employing T-DNA mutagenesis are discussed.
RESULTS
Molecular Characterization of the _emb_88 T-DNA Insertion
_Embryo-defective_88 (_emb_88) is a recessive T-DNA insertion mutant that displays arrested morphogenesis and embryo lethality prior to seed maturation (Meinke, 1994, 1995). As one of a large collection of Arabidopsis embryo-defective mutants generated by T-DNA insertion mutagenesis (Meinke, 1994, 1995), _emb_88 has been thoroughly characterized at the genetic level. The mutation has been mapped to 42 centimorgans (cM) on chromosome I, showing clear linkage to visible phenotypic markers near that location (Meinke 1994; Franzmann et al., 1995; linkage data available at http://mutants.lse.okstate.edu/). The _emb_88 line contains a single functional T-DNA insert that is tightly linked to the embryo-lethal phenotype, as demonstrated by cosegregation of the _emb_88 mutation with T-DNA-encoded kanamycin resistance and nopaline synthase activity (Errampalli et al., 1991; Castle et al., 1993). Genomic Southern blotting experiments with probes corresponding to the T-DNA LB, RB, and internal PBR322 sequences have confirmed the presence of a single T-DNA in _emb_88 and indicated the T-DNA had integrated at the _emb_88 locus as a simple, intact insert (Castle et al., 1993).
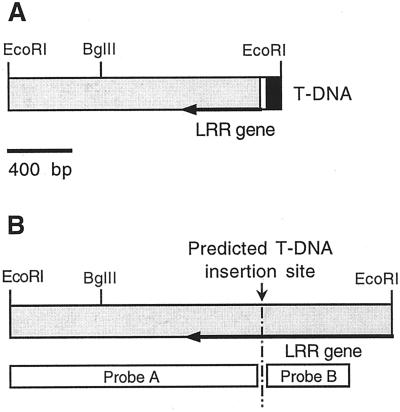
To investigate the molecular basis of the _emb_88 mutant phenotype, we characterized genomic DNA flanking the T-DNA LB, which had been isolated by plasmid rescue (Castle et al., 1993). A 1.6-kb (kbp) _Eco_RI fragment containing genomic flanking sequence was subcloned from rescued genomic DNA and sequenced. A diagram of this cloned fragment is shown in Figure 1A. The rescued fragment contained the LB of the T-DNA, 23 bases of “filler” sequence (a common feature of LB-flanking DNA; Gheysen et al., 1991; De Buck et al., 1999), the 3′ end of a gene encoding the C terminus of a novel Arabidopsis LRR protein (to be described in detail elsewhere), and 1,257 bp of unique sequence downstream of the LRR coding region. We characterized the corresponding WT allele of this DNA by isolating and sequencing three WT λ-genomic clones containing the rescued sequence. These clones all contained a 2.3-kbp _Eco_RI fragment consisting of the rescued LB sequence, plus an additional approximately 750 bp of the LRR gene (Fig. 1B). The putative T-DNA insertion site was identified within an open reading frame that constituted the 3′-most exon of this gene.
Figure 1.
Structure of T-DNA LB-flanking region from _emb_88 and of the corresponding region from wild type (WT). A, Rescued LB and plant DNA from the _emb_88 mutant. The 1,626-bp _Eco_RI fragment flanking the _emb_88 T-DNA LB consisted of 80 bp of the extreme T-DNA LB (black), 23 bp of “filler DNA” (white), and 1,523 bp of Arabidopsis genomic sequence (shaded). The genomic region contained the 3′-terminal 484 bp of a gene encoding a novel Arabidopsis Leu-rich repeat (LRR) protein (designated by arrow), and 1,039 bp of single-copy downstream extragenic sequence. B, Diagram of the corresponding genomic region from WT: a 2.3-kbp _Eco_RI fragment containing the prospective site of T-DNA insertion and the regions used as probes for the Southern blots shown in Figure 2. Probe A corresponded to the LB flanking region and probe B to the region predicted to be immediately adjacent to the T-DNA RB.
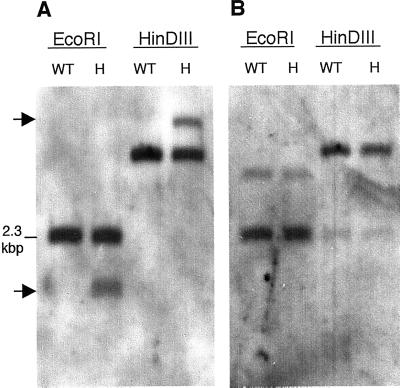
The presence of T-DNA LB sequence in the rescued 1.6-kbp _Eco_RI fragment indicated that this DNA lies adjacent to the _emb_88 T-DNA insert. To further confirm this, we carried out genomic Southern blots on DNA isolated from _emb_88 heterozygotes and from WT Arabidopsis, using the rescued LB-flanking DNA as a probe. The rescued LB sequence hybridized to a single band in WT DNA (Fig. 2A). As expected, the size of this band in _Eco_RI-digested DNA corresponded to that of the 2.3-kbp _Eco_RI fragment present in the WT genomic clones. In DNA isolated from _emb_88 heterozygotes, the rescued LB-flanking region probe detected the WT band plus a polymorphic band representing the mutant allele (Fig. 2A). In heterozygote DNA digested with _Eco_RI, the size of the polymorphic fragment corresponded to the size of the _Eco_RI fragment rescued from the mutant line (approximately 1.6 kbp). These data, together with the previous results establishing the presence of a single T-DNA insert in _emb_88, established that the cloned genomic flanking DNA was that disrupted by the _emb_88 T-DNA insert.
Figure 2.
Southern-blot analysis of genomic DNA from _emb_88 heterozygotes and WT. A, LB-flanking probe detected a T-DNA-associated polymorphism in _emb_88 heterozygotes. Genomic DNA isolated from _emb_88 heterozygotes (H) or WT Arabidopsis (WT) was digested with _Eco_RI or HinDIII, resolved by agarose gel electrophoresis, Southern blotted, and probed with the 1.6-kbp _Eco_RI fragment rescued from the T-DNA LB of the _emb_88 mutant. Arrows indicate positions of bands corresponding to the disrupted allele, in lanes containing DNA from heterozygotes. B, Sequences on the prospective T-DNA RB were not disrupted in _emb_88 heterozygotes. A genomic Southern blot identical to that shown in A was probed with a 250-bp genomic fragment that lies on the “right” side of the prospective T-DNA insertion site, immediately adjacent to the rescued LB sequence in WT DNA (“probe B,” in Fig. 1B). This blot was probed at medium stringency in an effort to detect a disrupted allele in heterozygotes (see “Materials and Methods”). High-resolution scans of autoradiographs are shown.
If rescued LB-flanking DNA represented one portion of a gene disrupted by simple T-DNA insertion, then the region on the other side of the putative T-DNA insertion site should also be disrupted in _emb_88 heterozygotes. To test this, we probed genomic Southern blots with the WT LRR gene sequence predicted to be on the immediate right flank of the putative T-DNA insertion site (see Fig. 1B). Surprisingly, this sequence appeared to be intact in DNA from _emb_88 heterozygotes, showing no evidence of adjacent T-DNA insertion (Fig. 2B). This indicated that the predicted 5′ region of the LRR gene was actually not present on the T-DNA RB; apparently the _emb_88 T-DNA did not incorporate as a straightforward insertion event. As more information from the Arabidopsis genome project became available in GenBank, further sequence characterization of the LB-flanking regions also yielded unexpected results. Database searches indicated that the rescued LB-flanking sequences, including the LRR gene and downstream extragenic sequence, aligned unambiguously with P1 clone MJJ3 from chromosome V (Sato et al., 1997). Sequence identity was >99.9% over 1,523 bp, with only a single base mismatch, which we attributed to the ecotype difference between our Wassilewskija-2 (Ws-2)-derived gene and the Columbia (Co)-derived MJJ3 sequence. Furthermore, our rescued LB and WT clones hybridized at high stringency to bacteria artificial chromosomes (BACs) that map to chromosome V (data not shown). The rescued DNA and the corresponding WT clones showed no comparable alignment to any other Arabidopsis sequences present in GenBank, a result consistent with our genomic Southern blots, which indicated that this sequence was present in single copy in the WT genome (Fig. 2A). Thus, despite the thoroughly established linkage of the T-DNA to the _emb_88 phenotype, and the mapping of the _emb_88 mutation to chromosome I, the DNA adjacent to the LB of the _emb_88 T-DNA appeared to originate from chromosome V.
The sequence alignments and genomic Southern blots suggested a scenario in which a portion of chromosome V had inserted on the T-DNA LB at the e_mb_88 locus in chromosome I. To investigate this possibility, we mapped the _emb_88 T-DNA insert using molecular markers. _Emb_88 heterozygotes (ecotype Ws-2) were crossed to plants of the Landsberg erecta (Ler) ecotype. F1 progeny segregating for the _emb_88 mutation were identified by their kanamycin resistance, and their identity was confirmed by scoring for the characteristic emb phenotype (production of 25% defective seeds following self-fertilization). Viable F2 progeny from these F1 heterozygotes were then analyzed for segregation of simple sequence length polymorphism (SSLP) markers AthS0392 and nga158. These markers were chosen due to their proximity to regions of interest on chromosomes I and V: AthS0392 is located on chromosome I within approximately 3 cM of the genetically mapped _emb_88 locus, whereas nga158 is located on chromosome V only approximately 40 kbp (<1 cM) downstream of the LRR gene identified in our cloned LB sequence (Bell and Ecker, 1994; Sato et al., 1997). We made specific predictions regarding the segregation of each these markers in kanamycin-resistant and kanamycin-sensitive F2 progeny, then determined the SSLP genotypes of F2 seedlings germinated on media containing kanamycin. Predictions and results of SSLP analysis are summarized in Table I. Consistent with previous genetic analysis of _emb_88 (Castle et al., 1993; Franzmann et al., 1995), these experiments demonstrated that the _emb_88 phenotype and T-DNA insertion map to chromosome I near the AthS0392 SSLP marker. However, the T-DNA also appeared linked to a copy of the nga158 marker because all kanamycin-resistant F2 plants also contained a Ws-2 allele of this SSLP. Thus, a large (≥40 kbp) fragment of Ws chromosome V, containing the nga158 marker, is linked to the T-DNA in _emb_88 chromosome I.
Table I.
SSLP mapping of the emb88 T-DNA: linkage to markers from chromosomes I and Va
| SSLP (Position)b | Segregation Predictions | Observed | Interpretation |
|---|---|---|---|
| AthS0392 (45 cM; I) | ∼25% (∼9/37) of KanR plants should be Ws/Ws for this marker, if it is unlinked to the T-DNAc | 1/37 Ws/Ws among KanR plants | The T-DNA is closely linked to this marker in Ws chromosome I. |
| ∼25% (∼8/30) of KanS plants should be Ws/Ws for this marker, if it is unlinked to the T-DNAc | 0/30 Ws/Ws among KanS plants | The T-DNA is closely linked to this marker in Ws chromosome I. | |
| <3% of viable F2 plants should be Ws/Ws for this marker, if it is closely linked to the T-DNAd | 1/67 Ws/Ws among entire F2 population | The lethal _emb_88 mutation and the T-DNA are closely linked to this marker in Ws chromosome I. | |
| All KanS plants should be Ler/Ler for this marker, if it is closely linked to the T-DNAe | 30/30 Ler/Ler among KanS plants | The T-DNA is closely linked to this marker in Ws chromosome I. | |
| nga158 (18 cM; V) | If this marker is unlinked to the T-DNA, ∼25% (9/36) of KanR plants should be Ler/Ler (lack the Ws allele)c | 0/36 Ler/Ler among KanR plants | A copy of the nga158 marker from Ws chromosome V is closely linked to the T-DNA |
| Unless a copy of nga158 segregates independently of the T-DNA, 100% of KanR plants should contain the Ler alleled | 14/36 Ws/Ws (lack Ler allele) among KanR plants | A copy of nga158 segregates independently of the T-DNA; the T-DNA is not linked to this marker in Ws chromosome V. |
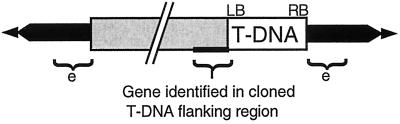
Although a copy of Ws nga158 was linked to the _emb_88 T-DNA, it was clear that an additional copy of this marker was also segregating independently of the lethal T-DNA insertion because a large number of viable kanamycin-resistant F2 seedlings (_emb_88 heterozygotes) lacked the Ler allele of this marker. Such nga158Ws/nga158Ws, _emb_88/_EMB_88 seedlings could not have been obtained in this F2 population unless a copy of the nga158 marker was present in a location unlinked to the lethal _emb_88 mutation. Thus, these _emb_88 heterozygotes were homozygous for the Ws-2 allele of nga158 at its native location on chromosome V. Taken together, these results can be best explained by a model in which the T-DNA, along with a duplicated fragment of chromosome V, has inserted into the _emb_88 locus on chromosome I, causing an embryo-lethal mutation in a yet unidentified gene. It is difficult to conceive of a straightforward alternative model that can account for our sequence, hybridization, and genetic results. This model is illustrated in Figure 5.
Figure 5.
General model for T-DNA-associated rearrangements in _emb_88 and _LRRPK_-TKO. In each case, T-DNA (white) has inserted into a target locus (black) along with LB-flanking sequences originating from a different chromosome (shown in gray). Sequences from this latter chromosome are not associated with the T-DNA RB. e, Presumed location of the gene responsible for the embryo-defective phenotype in the _emb_88 line. Sizes of the translocated LB-flanking regions are ≥than 40 kbp for _emb_88, and at least 1,020 bp for the _LRRPK_-TKO line.
Analysis of a Putative T-DNA Knockout Allele of the Light-Regulated Receptor Protein Kinase (LRRPK) Gene
LRRPK was originally identified as a light-regulated mRNA based on decreases in abundance after light pulses (Deeken and Kaldenhoff, 1997). Sequence analysis revealed LRRPK encoded a protein with a putative signal peptide, a predicted transmembrane domain, and an intracellular kinase domain, indicating LRRPK is a member of the large receptor-like kinase gene family in Arabidopsis. The predicted protein sequence also contained extracellular LRRs, which are a common feature of plant receptor-like kinases (Becraft, 1998). The LRRPK gene had been mapped using a recombinant inbred approach (http://www.Arabidopsis.org/cgi-bin/maps/RIintromap) to position 79.4 on chromosome IV, and this location has been confirmed by the genome sequencing project (http://www.Arabidopsis.org/cgi-bin/maps/Seqtable.pl?chr = 4).
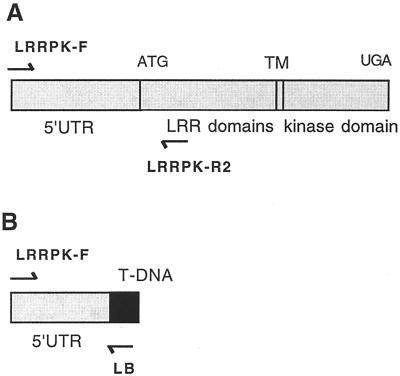
Using a PCR-based reverse genetics approach to identify insertions in specific genes by screening large populations of T-DNA-mutagenized lines (Krysan et al., 1996), we identified a strain of plants with a T-DNA insertion in LRRPK. This line is now referred to as _LRRPK_-translocated knockout (TKO). The PCR product containing the LRRPK/T-DNA junction fragment from this line consisted of 102 bp of T-DNA LB sequence and 1,020 bases of flanking plant genomic DNA (see Fig. 3A). The T-DNA insertion was located 134 bp upstream of the start codon in the 5′-untranslated region (UTR) of LRRPK, and would be predicted to result in a complete loss-of-function (null) allele (see Fig. 3B). Sequence alignment of the entire 1,020 bp of genomic flanking DNA with the WT LRRPK sequence in the database revealed no differences, despite their origin from different Arabidopsis ecotypes (C24 for the LRRPK sequence, Deeken and Kaldenhoff, 1997; Ws-2 for the PCR insertion, data not shown). The complete identity over the entire length of the non-coding sequence and adjacent extragenic region confirmed that the insertion is located within the LRRPK gene because potential LRRPK homologs would not be expected to be identical in promoter and 5′-UTR sequences. BLAST analysis confirmed the sequence similarity to the LRRPK annotation and in addition identified a match to the BACs F6G3 and F27B13. These BACs are located in the middle of chromosome IV in a position corresponding to the previously determined location of LRRPK (http://www.Arabidopsis.org/cgi-bin/maps/Seqtable.pl?chr = 4; http://www.Arabidopsis.org/cgi-bin/maps/Riintromap). Together, these data established that the line obtained in reverse genetic screens contained a T-DNA-disrupted allele of LRRPK.
Figure 3.
LRRPK gene structure and identification of a T-DNA insert adjacent to LRRPK sequences. A, Diagram showing the structure of WT LRRPK gene (Deeken and Kaldenhoff, 1997). Positions of PCR primers used in subsequent experiments are shown. B, Map of the PCR fragment obtained from the prospective LRRPK knockout line, _LRRPK_-TKO. The 1,122-bp product contained 102 bp of the T-DNA LB (black), adjacent to 1,020 bp of the LRRPK gene, containing 5′-untranslated region (UTR; 458 bp) and upstream extragenic sequences.
The _LRRPK_-TKO strain then was characterized by determining the number of independent T-DNA insertions, and by looking for phenotypes associated with the insertion. To determine the number of T-DNA inserts, LRRPK_-TKO plants were backcrossed to Ws-2 WT. F1 heterozygotes containing the LRRPK/T-DNA junction were identified by PCR, selfed, and resulting F2 progeny were germinated on kanamycin. A ratio of 223:67 kanamycin-resistant:-sensitive plants was observed, consistent with the 3:1 ratio expected for a single T-DNA insert. It is important that all kanamycin-resistant F2 plants contained the LRRPK/T-DNA junction when tested with PCR (24 of 24 plants), and kanamycin-sensitive plants did not contain the junction (12 of 12 plants). These results indicated that the T-DNA in LRRPK_-TKO plants segregates as a single functional insert that cosegregates with the T-DNA/LRRPK junction fragment identified by PCR. We then scored the progeny of a selfed _LRRPK_-TKO parental plant for kanamycin resistance. All progeny (28 of 28) were kanamycin resistant, indicating that the line was homozygous for the T-DNA insertion. There was no obvious phenotype in the homozygous _LRRPK-_TKO strain, or among any of the F2 plants from the backcross.
To further characterize the _LRRPK_-TKO strain, we mapped the site of the T-DNA insertion. _LRRPK_-TKO was crossed to WT of the Columbia ecotype, and the segregation of SSLP and cleaved amplified polymorphic sequence (CAPS) markers in F2 progeny was determined. F2 plants lacking the T-DNA were predicted to be predominantly homozygous for the Columbia allele of markers linked to the site of T-DNA insertion. F2 plants that lacked the _LRRPK/_T-DNA junction fragment were identified by PCR, and the segregation of markers dimorphic between Ws-2 and Columbia was analyzed. The results are summarized in Table II. Surprisingly, the T-DNA insertion was tightly linked to two markers on chromosome V, and was unlinked to the chromosome IV markers near the previously determined location of the LRRPK gene (Table II). As a control, the segregation ratios for markers 20 to 30 cM from the map position of LRRPK on chromosome IV and from the insertion site on chromosme V were determined. These results demonstrated that segregation for other chromosome IV and V markers is normal in the _LRRPK_-TKO line (Table II). Thus, sequences flanking the T-DNA in _LRRPK_-TKO represented a fragment of the LRRPK gene that had translocated to chromosome V. Translocated sequences were restricted to the T-DNA LB because, in results similar to those observed for _emb_88 in Figure 2A, Southern blots indicated that LRRPK sequences were not present adjacent to the T-DNA RB (data not shown). Furthermore, we were consistently unable to PCR amplify an RB-flanking fragment using an RB-specific primer and gene-specific primers from the 3′ region of LRRPK predicted to flank the T-DNA RB. Taken together, our results suggest that _LRRPK_-TKO contains a single functional T-DNA insertion in chromosome V, which is flanked on the LB by a translocated fragment of the LRRPK gene from chromosome IV. This scenario is similar to that determined for _emb_88, and is illustrated in Figure 5.
Table II.
Linkage of the T-DNA-tagged allele of LRRPK to chromosome V
| Marker (Map Position) | Segregation Prediction Co/Totala | Observed Co/Totalb |
|---|---|---|
| Chromosome IV | ||
| 4F16G20 (73.8) | 21 /22c | 13 /22 |
| LRRPK (79.0) | ||
| nga1139 (83.4) | 21 /22c | 11 /22 |
| DHS1 (108.5) | 15 /22c | 10 /22 |
| Chromosome V | ||
| nga139 (50.4) | 11 /22d | 15 /22 |
| so191 (77.3) | 11 /22d | 21 /22 |
| nga129 (105.4) | 11 /22d | 20 /22 |
We further characterized _LRRPK_-TKO to determine if the WT gene coexisted with the translocated mutant allele in this putative knockout line. Based on the sequence and mapping results presented above, two types of events might have led to the genomic rearrangements in the _LRRPK_-TKO line. The first was a unilateral translocation, in which sequence from chromosome IV inserted with the T-DNA into chromosome V, leaving a corresponding deletion at the LRRPK locus in chromosome IV. The second possibility was that the translocated portion of the LRRPK gene was duplicated as well as translocated, such that an intact copy of LRRPK remains in chromosome IV. This second model predicts that plants homozygous for the LRRPK/T-DNA junction would also contain the WT gene. We tested this prediction by using LRRPK forward and reverse gene-specific primers to detect WT LRRPK in DNA from _LRRPK_-TKO homozygotes. As shown in Figure 4, the WT gene was present in a representative _LRRPK_-TKO homozygote. Identical results were obtained with all other homozygotes tested among the F2 (n = 28). In a similar manner, when a homozygous _LRRPK_-TKO parental was backcrossed to WT, no homozygotes lacking the WT gene could be identified (n = 50). Thus, despite the identification of a prospective T-DNA knockout allele, it appears that the translocated LRRPK sequences flanking the T-DNA have been duplicated, such that the WT gene is still present at its original location in the prospective knockout line.
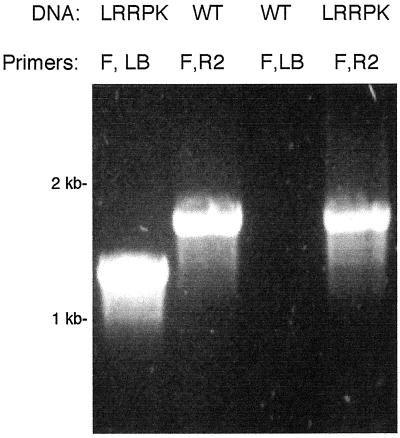
Figure 4.
PCR detection of the WT LRRPK allele in _LRRP_K/T-DNA homozygotes. Gene-specific and T-DNA-specific primers were used to detect the presence of the WT and mutant LRRPK alleles in genomic DNA from a representative _LRRPK_-TKO homozygote, and from WT controls. Genomic DNA template (DNA) and primer combinations (Primers) used in each reaction are labeled above each lane. Primers: F, LRRPK-specific forward (5′) primer; R2, LRRPK-specific reverse (3′) primer; LB, T-DNA specific LB primer. Primer positions are indicated on LRRPK gene diagrams in Figure 3. Lanes, in order shown: 1, control amplification of the LRRPK/T-DNA junction product from a representative _LRRPK_-TKO homozygote; 2, control amplification of the WT LRRPK gene from WT genomic DNA; 3, negative control, showing lack of the LRRPK/T-DNA junction product in WT DNA; 4, amplification of the WT LRRPK PCR product from the LRRPK/T-DNA homozygote.
DISCUSSION
We have described T-DNA-associated interchromosomal rearrangements identified in the course of both forward and reverse genetic investigations in Arabidopsis. In both cases, molecular mapping, along with analysis of the mutated gene flanking the T-DNA, revealed that flanking DNA had originated from a locus unrelated to the site of T-DNA insertion. WT copies of these translocated flanking DNA sequences also remained intact at their native locations.
The rearrangements characterized in _emb_88 and _LRRPK_-TKO have additional features in common. In both cases, translocated sequences are adjacent to the T-DNA LB, and there is no evidence that related translocated sequences are also present on the RB. Although we did not directly characterize RB flanking sequences from either of these lines, Southern-blot analysis of _emb_88 clearly indicated that the 5′ portion of the disrupted LRR gene predicted to be on the T-DNA right flank was not adjacent to the T-DNA insertion (Fig. 2B). Similar results were obtained with _LRRPK_-TKO, in which the LRRPK 3′ region, which was predicted to flank the RB, was found to be undisrupted by the T-DNA insert. Thus, at the level of resolution detection provided by hybridization, translocated sequences appear to be restricted to the T-DNA LB. We cannot rule out that tiny, undetectable fragments of translocated sequence may be present on the T-DNA RB in our mutants. Rescue of RB-flanking DNA and detailed sequence characterization would be required to determine RB flanking structure at this resolution.
Another common feature of _emb_88 and _LRRPK_-TKO is that translocated sequences adjacent to the T-DNA appear to have been duplicated. We demonstrated this directly with _LRRPK_-TKO: all plants homozygous for the T-DNA-disrupted form of LRRPK also contained the intact, WT allele. Similar direct demonstration could not be carried out with _emb_88 because T-DNA homozygotes are early embryo lethals and cannot be analyzed at the molecular level. However, it is likely that the large translocated region from chromosome V also remains intact at its native location in this mutant line because _emb_88 does not segregate for any defects other than _emb_88 itself, which is in chromosome I. A chromosome V-linked phenotype (most likely recessive lethality) would be expected to segregate in this line if >40 kbp of this chromosome had been lost from its original location upon translocation. Also, such a large deletion would likely result in distorted segregation patterns in the _emb_88 line. We cannot rule out that, for both _emb_88 and _LRRPK_-TKO, deletions corresponding to the translocated sequences could have been present in the initially transformed cells; chromosomes containing these large deletions could have segregated away in subsequent generations prior to our analyses. Because T-DNA mutagenized populations had been propagated for several generations prior to this study, this possibility cannot be experimentally addressed. From a practical perspective, the significant point is that major, stably inherited unilateral translocations are present in publicly available T-DNA mutant populations.
The duplication/translocations we report are clearly a consequence of T-DNA transformation, and are not the result of T-DNA insertion adjacent to preexisting evolutionary duplications. Genomic Southern blots suggested that LB-flanking sequences were present in single copy in WT (e.g. see Fig. 2A). Furthermore, sequences of our flanking DNAs allowed unambiguous identification of their chromosomal origins: We observed >99.9% sequence identity between our flanking regions and unique Arabidopsis sequences in GenBank. It is important that most of this sequence was from extragenic DNA, which would be predicted to diverge rapidly, even if it represented a relatively “recent” duplication specific to the Ws-2 ecotype. The best characterized ecotype-specific evolutionary duplication in Arabidopsis, the Ws PAI 4 gene, exhibits a level of divergence an order of magnitude higher than we observed in our LB-flanking regions (Bender and Fink, 1995), even within the gene. Our mapping results provide the most direct evidence that the duplications we identified were T-DNA induced: the nga158 SSLP marker was duplicated in _emb_88. Nga158 is an established SSLP marker that segregates as a single locus in crosses between Ws and other ecotypes (Bell and Ecker, 1994; N. Forsthoefel and D.M. Vernon, unpublished data). Thus, the presence of this marker near the site of T-DNA insertion in chromosome I was not due to preexisting evolutionary duplication in Ws, but rather to the T-DNA mutagenesis itself.
The model we present in Figure 5 is a general model that illustrates the translocations we have identified in _emb_88 and _LRRPK_-TKO, and underscores the common features of the T-DNA insertions in these two lines. The exact sizes of the translocated regions in these mutants are not known and would be difficult to determine. For _emb_88, this region must be ≥40 kbp, because a copy of the Ws-2 nga158 marker is linked to the T-DNA insert. It is known from published chromosome V sequence that this marker resides approximately 40 kbp downstream of the LRR gene adjacent to the T-DNA LB (Sato et al., 1997; see Fig. 1A). The size of the rearrangement in _LRRPK_-TKO is less well defined, but direct sequencing of the LB-flanking region indicated that it exceeds 1,020 bp (see “Results”). Estimates of translocation size in these lines could perhaps be refined to some degree by more exhaustive mapping, were more tightly linked markers available, but such an effort would still not provide information at the nucleotide level, and thus would not likely alter the fundamental features of our proposed model.
Novel Features of the emb88 and LRRPK-TKO Rearrangements
A wide range of chromosomal defects have been observed in populations of Arabidopsis subjected to T-DNA mutagenesis. Genetic defects characterized from such populations range from small additions and deletions of a few bases (Negruk et al., 1996; Noguchi et al., 1999) to modest rearrangements and major chromosomal translocations (Gheysen et al., 1991; Castle et al., 1993). In two well-characterized cases, large chromosomal translocations have been found in conjunction with multiple T-DNA inserts (Nacry et al., 1998; Laufs et al., 1999). In both of these cases (an inversion and a reciprocal translocation/inversion), rearrangements apparently occurred during integration, due to interaction between T-DNAs that ultimately inserted at separate loci. The rearrangements we describe here differ in several respects from these previously characterized major rearrangements. First, the rearrangements in _emb_88 and _LRRPK_- TKO are associated with a single functional T-DNA insert rather than with multiple T-DNAs inserted at different loci. Second, the _emb_88 and _LRRPK_-TKO rearrangements appear to involve unilateral transfer of sequence from one chromosome to another, rather than reciprocal translocation. Third, the translocations in our lines appear to be internal, T-DNA-associated insertions, rather than terminal exchanges originating at chromosomal breakpoints, or intrachromosomal inversions. And finally, as discussed above, the translocations we characterized appear to have been accompanied by duplication, such that WT copies of the rearranged genes remain intact at their native loci.
Potential Mechanisms for Observed Rearrangements
The origins of the T-DNA associated rearrangements in _emb_88 and _LRRPK_-TKO are not clear. One unlikely possibility is that they arose from recombination between unlinked T-DNAs, as has previously been suggested for some rearrangements in lines containing multiple T-DNA inserts (Nacry et al., 1998; Laufs et al., 1999). We cannot entirely rule out this scenario because additional T-DNAs, if they were unlinked to the inserts we analyzed, could have segregated out of our lines immediately following mutagenesis. However, even if they had existed, it is not clear how such additional inserts could have contributed to the unilateral translocations we observed in our lines.
A more likely explanation for our rearrangements is that they resulted from a single T-DNA that underwent aborted insertion at one site, accompanied by successful integration at another, incorporating LB-flanking plant DNA from the initial failed insertion. T-DNA insertion apparently aborts frequently during transformation: Minor additions, deletions, and substitutions unaccompanied by T-DNA inserts are common in T-DNA-mutagenized populations, and are thought to be a consequence of failed integration and perhaps subsequent repair (Castle et al., 1993; Negruk et al., 1996; Tinland, 1996; Nacry et al., 1998). One proposed model of T-DNA integration suggests that, at least in some integration events, the LB inserts at a target site before the RB (Tinland, 1996; Laufs et al., 1999). Interruption of such “LB first” integration could explain one aspect of the rearrangements found in the _emb_88 and _LRRPK_-TKO lines: the association of translocated DNA with only the T-DNA LB. If T-DNA integration was aborted following LB insertion but prior to RB insertion, and the T-DNA subsequently re-inserted with flanking DNA into a different site, any flanking sequences that were translocated would be expected to be found only on the LB. This is what we observed. It should be noted, however, that the LB first model is not universally accepted (De Buck et al., 1999). And even within the context of this model, it is not clear why LB integration aborted in the first place, or how adjacent DNA could have been translocated.
Studying T-DNA integration through the structure of insertions after the fact has always posed a problem, and it would be difficult to deduce the mechanisms by which rearrangements arose in _emb_88 and _LRRPK_-TKO, even with the most detailed analysis of insertion structure. Nevertheless, it is tempting to speculate about how the duplication/translocations we observed arose. We suggest two possible scenarios. One possibility is that the LBs and RBs of a single T-DNA inserted simultaneously into nicked sites in separate chromosomes, resulting in a transient “T-DNA bridge” between two distinct chromosomes. Resolution of this aberrant structure could have resulted in the transfer of the T-DNA to the site of RB insertion, with concomitant transfer of portions of the LB-associated chromosome attached to the transferred LB. The chromosome of initial LB insertion then could have been restored through cellular DNA repair mechanisms, thus resulting in duplication of translocated sequences and maintenance of WT alleles at their original location. The second possibility is that ongoing DNA repair in the first target chromosome interfered with integration and somehow resulted in duplication of LB-flanking sequences, which were then somehow transferred with the T-DNA to a second, unlinked, target site. Chromosomal regions undergoing DNA repair are thought to be hotspots for T-DNA insertion (Tinland, 1996; De Buck et al., 1999). Double-strand repair processes may play an important role in normal T-DNA insertion (Salomon and Puchta, 1998; De Buck et al., 1999). Thus, DNA repair processes may have been active in T-DNA-associated regions in our lines; had these processes gone awry they could have interfered with initial integration and contributed to the rearrangements we observed. Finally, we cannot rule out that each individual rearrangement might have arisen through a different mechanism. Perhaps due to the complexity of the T-DNA integration, there is a certain potential for sloppiness inherent in the process. Given the wide diversity of chromosomal defects associated with T-DNA transformation (ranging from single-base mutations to large chromosomal translocations), it seems that the T-DNA transformation process can go awry in any number of ways (Castle et al., 1993; Negruk et al., 1996; Nacry et al., 1998; Laufs et al., 1999). In the absence of a universally accepted model even for normal T-DNA integration, the cause(s) of aberrant T-DNA-associated rearrangements will remain unclear.
Implications for Genomics
Regardless of the exact mechanism of the translocations in _emb_88 and _LRRPK_-TKO, the results presented here demonstrate that significant chromosomal rearrangements may be present in T-DNA-transformed lines containing single inserts with straightforward genetics. The _emb_88 and LRRPK-TKO rearrangements are not likely to be isolated flukes: Similar rearrangements were present in both these lines, which represent two of eight T-DNA insertion lines characterized in detail by our laboratories thus far. Rearrangements such as those we describe can only be detected through detailed characterization—either by sequencing of flanking DNA from both T-DNA borders, or by the combination of sequencing and T-DNA mapping we employed. Therefore, these sorts of rearrangements have likely been overlooked in prior estimates of translocations in T-DNA-mutagenized populations. Taken together, these findings suggest that duplication/translocations may be somewhat more common than has been assumed in populations of T-DNA-transformed Arabidopsis.
Our findings have implications for two types of genomic research currently being employed with Arabidopsis: (a) reverse genetic analysis of T-DNA knockout lines, which relies on detection of T-DNA-disrupted alleles by PCR; and (b) the generation of databases containing genomic sequence tags associated with specific phenotype classes. For reverse genetic analysis, two scenarios can be envisioned by which duplication/rearrangements could interfere with characterization of T-DNA knockout lines. In one scenario, resembling what occurred in _LRRPK_-TKO, duplications might prevent the expression of a phenotype that would otherwise result from a T-DNA insertion in a gene of interest. Under such a scenario, a line containing a disrupted allele of a gene of interest would be identified by PCR, but, because the T-DNA-flanking sequence had been duplicated, an intact WT allele would also be present and no information on gene function could be deduced. A second way that duplication/rearrangements could interfere with reverse genetic analysis is by causing defective phenotypes: A T-DNA and adjacent flanking sequences containing the gene of interest could be inserted into a second locus, causing an insertion mutation. In this case, PCR screening for a knockout allele would identify a line containing the disrupted gene of interest, but the observed phenotype would not actually be due to the mutation in the gene of interest. This scenario is analogous to what was found during forward genetic analysis of _emb_88: A gene adjacent to the T-DNA was actually on a translocated genomic fragment that was not related to the mutant phenotype.
In a similar manner, duplication/translocations can complicate the interpretation of databases containing genomic sequence tags associated with specific phenotypes. Such databases can presumably provide a resource for forward genetic analyses of a given phenotype, and provide a broad, “genome-wide” view of the types of genes involved in a biological process. However, our results suggest that a significant percentage of the sequence tags in such collections may not represent true knockouts, but rather may actually represent rearranged sequences unrelated to the phenotype of interest. Thus, information on gene function based solely on such sequence tags from a single T-DNA border should be viewed with caution; it is essential to obtain sequence information from both sides of a T-DNA insert before inferring a biological role for a tagged sequence in such databases.
The general phenomenon of rearrangements in T-DNA-mutagenized Arabidopsis populations has been acknowledged before. Krysan et al. (1999) termed such events “knockworsts” and suggested some measures by which they could be identified. Their suggestions included first identifying mutant lines with phenotypes, then obtaining genomic flanking sequence information from both T-DNA borders, identifying more than one mutant allele of a gene of interest, and/or carrying out molecular complementation of the mutant phenotype. The results we present here indicate that rearrangements in T-DNA lines may be more common, and often harder to identify, than previously imagined. They also underscore the need for cautious interpretation and thorough genetic analysis of insertion lines, including prospective knockout lines that do not exhibit phenotypes. We show that a lack of phenotype hypothetically can be due to duplication of the locus of interest. Furthermore, it is likely that many single-knockout mutants will not display obvious phenotypes, due to genetic redundancy. Determination of gene function in Arabidopsis will therefore require double- and multiple-mutant analyses; obtaining KO lines with true insertions—phenotypes or not—is an essential first step for this.
We offer the following suggestions to detect translocations and ensure that putative T-DNA insertion lines merit further study. These methods are rapid and simple, and offer an alternative to obtaining flanking sequence from both T-DNA borders (which is not always a routine endeavor). In addition to the standard determination of T-DNA insert number and segregation, genetic characterization of putative knockout mutants should also include: (a) mapping of the T-DNA insert to confirm linkage to the known map position of the gene of interest, and (b) PCR analysis of T-DNA homozygotes with gene-specific primers flanking the gene of interest, to determine whether intact, WT copies of the gene are present in putative homozygous mutants. It is especially important that putative knockout lines that do not display any phenotype be characterized by these methods, to ensure that the lack of phenotype is not simply due to T-DNA-induced sequence duplication.
MATERIALS AND METHODS
Plant Material
Plants used for genetic crosses, DNA isolation, or seed collection were seeded in soil, chilled at 4°C for 2 to 3 d, and grown to maturity at 22°C under 16-h-light/8-h-dark cycles in a controlled climate growth chamber. To identify seedlings containing T-DNA, seeds were sterilized and germinated in culture in Petri dishes containing 50 μg mL−1 kanamycin as previously described (Errampalli et al., 1991). Seedlings germinated in culture that were chosen for further use in genetic crosses, SSLP mapping, or seed collection were transplanted to soil after 7 to 10 d in culture and grown to maturity under the conditions described above_._
Genetic Analysis and T-DNA Linkage
For _emb_88, T-DNA copy number, insert structure, and cosegregation with the mutant phenotype have been published previously (Errampalli et al., 1991; Castle et al., 1993). In addition, _emb_88 has been genetically mapped to approximately 42 cM on chromosome I (Franzmann et al., 1995; data can be viewed at http://mutant.lse.okstate.edu/).
For LRRPK, T-DNA insert number was determined by crossing the _LRRPK_-TKO strain to plants of the Ws-2 ecotype. Seeds from the F2 of this cross were tested for kanamycin resistance as previously described (Errampalli et al., 1991). Linkage of the insert to the kanamycin resistance gene was demonstrated by isolating DNA (Krysan et al., 1996) from 24 individual kanamycin resistant plants from the F2 of this cross, and demonstrating that each of these contained the T-DNA insert in the LRRPK gene by PCR.
Isolation and Characterization of T-DNA- Flanking Regions
For molecular characterization of _emb_88, genomic sequences flanking the T-DNA LB were isolated by plasmid rescue (Castle et al., 1993). A 1.6-kbp _Eco_RI fragment containing the extreme T-DNA LB and adjacent genomic DNA was subcloned into Bluescript KS+ (Strategene, La Jolla, CA) for sequencing and probe synthesis. Corresponding WT clones containing this LB-flanking sequence were obtained by screening a λgem11 WT genomic library (Castle and Meinke, 1994) using random-primed digoxygenin (DIG)-labeled probes (Roche Biochemicals, Indianapolis) derived from gel-purified LB _Eco_RI fragment. Three independent overlapping WT clones were obtained; each contained an identical 2.3-kbp _Eco_RI fragment that included the LB-flanking genomic sequence. This WT _Eco_RI fragment was subcloned into Bluescript KS+ for sequence characterization and probe synthesis. Sequencing of LB flanking DNA and WT clones was carried out by the Washington State University sequencing facility (Pullman), the Arizona Research Laboratory (University of Arizona, Tucson), and the Oklahoma State University DNA Core facility (Stillwater). Genomic sequence was analyzed and native chromosomal location was determined by BLAST searches against the GenBank sequence database (Altschul et al., 1997). Linkage of cloned genomic fragments to chromosome V was further confirmed by hybridization of the cloned flanking DNA fragment to a BAC filter provided by the Arabidopsis Biological Resource Center (Ohio State University, Columbus).
The reverse genetics method described in Krysan et al. (1996) was used to isolate the insertion in the LRRPK gene. Gene-specific primers were designed from the LRRPK sequence deposited in GenBank (accession nos. X83614 and X97774, Deeken and Kaldenhoff, 1997) using the Oligo 4.0 program. The two primers used for the reverse genetics screening were LRRPK-F1 (1,291) (TTAGACATAACGCCGACGCAGCAAAAGTA) and LRRPK-R1 (5,680) (CATCACTCTTCTTCGTTCATTTGCCGTTGC). LRRPK-R2 (2,694) (AGAGACTATGCTATGACTTGTTCCAGACT) was designed for the purpose of more efficiently amplifying the WT allele than LRRPK-R1. The number in parentheses refers to the base pair number in accession no. X97774, which contains genomic sequence. Plant DNA isolation, PCR, and gel elelectropheresis were performed as described in Krysan et al. (1996). To confirm that the DNA sequence of the insertion matched the LRRPK gene, the PCR product synthesized using the LRRPK-F1 primer and the LB primer was cloned into a TA plasmid (Invitrogen, Carlsbad, CA) and sequenced at the Arizona Research Laboratory (University of Arizona, Tucson).
Genomic DNA Isolation and Southern-Blot Analysis
Arabidopsis genomic DNA isolation was carried out using the procedures described by Castle et al. (1993) or Krysan et al. (1996). For genomic blots, DNA was subject to restriction digestion, agarose gel electrophoresis, and blotting according to standard procedures (Sambrook et al., 1989). DIG-labeled probes were synthesized with a DIG-labeling kit from Roche Biochemicals, either by random priming or by PCR in the presence of DIG deoxy-uridine triphosphate, according to the manufacturer's instructions.
For Southern-blot analysis of _emb_88, the LB-specific probe was generated by random-priming of the gel-purified 1.6-kbp LB _Eco_RI fragment subcloned from rescued LB-flanking DNA (see above and Fig. 1). The probe specific for the prospective RB-flanking DNA was generated by PCR using cloned 2.3-kbp WT _Eco_RI fragments (see above) as template. Primers defining a 250-bp region adjacent to the LB-flanking sequence, on the opposite side of the prospective site of T-DNA insertion, were designed from WT sequence and used for probe synthesis: 5′-GGATCCCAACAAAATCAG-3′ and 5′-GCTCAAGTCGAGTTCTTGG-3′. Blots were probed, processed, and visualized according to DIG-labeling kit instructions from Roche Biochemicals. Genomic Southern blots were hybridized at 65°C and washed at high stringency (68°C in 0.1× SSC). When no T-DNA-associated polymorphism was detected using the RB-specific probe, blots were reprobed and washed at lower stringency (50°; 0.5× SSC).
Molecular Mapping
The SSLP mapping technique has been described by Bell and Ecker (1994). For mapping the _emb_88 T-DNA, markers that map close to loci of interest on chromosomes I and V, and which exhibit dimorphism between ecotypes Ws-2 and Ler, were chosen: nga158 (18 cM, chromosome V; http://Arabidopsis.org/) and AthS0392 (44.5 cM, chromosome I; Bell and Ecker, 1994). Because _emb_88 homozygotes die prior to germination, heterozygotes (Ws-2 ecotype) were used for crosses to Ler. Kanamycin-resistant F1 seedlings were selected, transferred to soil, grown to maturity, and self-fertilized. Segregation for the _emb_88 mutation was confirmed in these plants by screening siliques for the presence of 25% defective seeds. F2 progeny from the _emb_88 (Ws-2) × Ler crosses were germinated on media containing kanamycin, and segregation of SSLP markers among kanamycin-resistant and kanamycin-sensitive individuals was determined using the PCR protocol described by Bell and Ecker (1994).
For mapping the insertion in LRRPK, plants containing the T-DNA insertion of the Ws-2 ecotype were crossed to WT plants of the Columbia ecotype to generate a mapping population. F2 progeny lacking the T-DNA/LRRPK junction fragment were predicted to contain predominantly the Columbia alleles of markers linked to the site of T-DNA insertion. DNA was isolated from individual F2 plants and these plants were first tested to see if they contained the T-DNA LB-LRRPK junction fragment. DNA from those plants that did not contain the insertion was tested for whether they contained Ws-2 or Columbia polymorphisms using either SSLP (Bell and Ecker, 1994) or CAPS (Konieczny and Ausubel, 1993) markers. Linkage to chromosome IV was tested by using the SSLP markers 4F16G20 (located near 73.8 cM, http://www.Arabidopsis.org/cgi-bin/maps/Seqtable.pl?chr = 4, using primers 4F16G20F-ggttcagtgttactatgtaccaag and 4F16G20R-gagaccagtggcctaagtagat; F. Tax and S. Choe, unpublished data), nga1139 (83.4 cM, Bell and Ecker, 1994), and the CAPS marker DHS1 (108.5 cM, Konieczny and Ausubel, 1993). Linkage to chromosome V was tested using the SSLP markers nga139 (located at 50.4 cM, Bell and Ecker, 1994), S0191 (1 BAC from gln1, 77.3 cM), and nga129 (105.4 cM; Bell and Ecker, 1994; recombinant inbred map, http://www.Arabidopsis.org/cgi-bin/maps/RIintromap).
ACKNOWLEDGMENTS
The authors are grateful to Nancy Forsthoefel (Whitman College) and Heng Yuan and Robert Schmitz (University of Arizona) for technical assistance. The _emb_88 mutant was originally identified in David Meinke's lab at Oklahoma State University. Reverse genetic analysis of LRRPK was initiated in Michael Sussman's lab (University of Wisconsin, Madison). We thank Drs. Richard Jorgensen and Sunghwa Choe of the University of Arizona (Tucson) for comments on the manuscript.
Footnotes
1
This work was supported by the National Science Foundation (award IBN–960–4344 to D.M.V.) and by the U.S. Department of Agriculture (award no. 97353044708 to F.E.T.).
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Becraft P. Receptor kinases in plant development. Trends Plant Sci. 1998;3:384–388. [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. doi: 10.1046/j.1365-313x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton T, Franzmann L, Yoon E, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Castle LA, Meinke DW. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A. The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 1999;20:295–304. doi: 10.1046/j.1365-313x.1999.t01-1-00602.x. [DOI] [PubMed] [Google Scholar]
- Deeken R, Kaldenhoff R. Light-repressible receptor protein kinase: a novel light-regulated gene from Arabidopsis thaliana. Planta. 1997;202:479–486. doi: 10.1007/s004250050152. [DOI] [PubMed] [Google Scholar]
- De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A. T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from cointegration of separate T-DNAs. Plant J. 1997;11:15–29. doi: 10.1046/j.1365-313x.1997.11010015.x. [DOI] [PubMed] [Google Scholar]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansoen K, Schnall J, Feldmann K, Meinke DW. Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell. 1991;3:149–157. doi: 10.1105/tpc.3.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann L, Yoon E, Meinke DW. Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J. 1995;7:341–350. [Google Scholar]
- Gheysen G, Villarroel R, Van Montagu M. Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- Gordon MP. Discovery of the T-DNA of Agrobacterium tumefaciens. In: Kung S, Yang S, editors. Discoveries in Plant Biology. Vol. 1. Singapore: World Scientific; 1998. pp. 111–115. [Google Scholar]
- Koncz C, Németh K, Rédei GP, Schell J. T-DNA insertional mutagenesis in Arabidopsis. Plant Mol Biol. 1992;20:963–976. doi: 10.1007/BF00027166. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Autran D, Traas J. A chromosomal paracentric inversion associated with T-DNA integration in Arabidopsis. Plant J. 1999;18:131–139. doi: 10.1046/j.1365-313x.1999.00436.x. [DOI] [PubMed] [Google Scholar]
- Lindsey K, Topping JF, Muskett PR, Wei W, Horne KL. Dissecting embryonic and seedling morphogenesis in Arabidopsis by promoter trap insertional mutagenesis. Symp Soc Exp Biol. 1998;51:1–10. [PubMed] [Google Scholar]
- Meinke DW. Seed development in Arabidopsis thaliana. In: Meyerowitz E, Somerville C, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1994. pp. 253–295. [Google Scholar]
- Meinke DW. Molecular genetics of plant embryogenesis. Ann Rev Plant Physiol Plant Mol Biol. 1995;46:369–394. [Google Scholar]
- Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics. 1998;149:641–650. doi: 10.1093/genetics/149.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negruk V, Eisner G, Lemieux B. Addition-deletion mutations in transgenic Arabidopsis thaliana generated by the seed co-cultivation method. Genome. 1996;39:1117–1122. doi: 10.1139/g96-140. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Yoshioka Y, Machida C, Machida Y. DNA rearrangements associated with the integration of T-DNA in tobacco: an example for multiple duplications of DNA around the integration target. Plant J. 1995;7:157–164. doi: 10.1046/j.1365-313x.1995.07010157.x. [DOI] [PubMed] [Google Scholar]
- Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- Sato S, Kotani H, Hakamura Y, Kaneko T, Asamizu E, Fukami M, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5: I. Sequence features of the 1.6 Mb regions coverd by twenty physically assigned P1 clones. DNA Res. 1997;4:215–230. doi: 10.1093/dnares/4.3.215. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, English JJ, Carroll BJ, Bennetzen JL, Harrison K, Burbidge A, Bishop GJ, Jones JD. Analysis of the chromosomal distribution of transposon-carrying T-DNAs in tomato using the inverse polymerase chain reaction. Mol Gen Genet. 1994;242:573–585. doi: 10.1007/BF00285281. [DOI] [PubMed] [Google Scholar]
- Tinland B. The integration of T-DNA into plant genomes. Trends Plant Sci. 1996;1:178–183. [Google Scholar]
- Topping JF, Lindsey K. Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell. 1997;9:1713–1725. doi: 10.1105/tpc.9.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Larebeke N, Genetello C, Schell J, Schilperoort RA, Hermans AK, Van Montagu M, Hernalsteens JP. Acquisition of tumor-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature. 1975;255:742–743. doi: 10.1038/255742a0. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn J, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1014. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P, Tempe J, Schell J. Transfer and function of T-DNA genes from Agrobacterium Ti and Ri plasmids in plants. Cell. 1989;56:193–201. doi: 10.1016/0092-8674(89)90892-1. [DOI] [PubMed] [Google Scholar]