vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain (original) (raw)
Abstract
Mutations in the homeobox gene vHnf1 are associated with human diseases MODY5 (maturity-onset diabetes of the young, type V) and familial GCKD (glomerulocystic kidney disease). In an insertional mutagenesis screen in zebrafish, we isolated mutant alleles of vhnf1. Phenotypes of these mutants include formation of kidney cysts, underdevelopment of the pancreas and the liver, and reduction in size of the otic vesicles. We show that these abnormalities arise from patterning defects during development. We further provide evidence that vhnf1 regulates the expression of key patterning genes for these organs. vhnf1 is required for the proper expression of pdx1 and shh (sonic hedgehog) in the gut endoderm, pax2 and wt1 in the pronephric primordial, and valentino (val) in the hindbrain. Complementary to the loss-of-function phenotypes, overexpression of vhnf1 induces expansion of the val expression domain in the hindbrain. We propose that vhnf1 controls development of multiple organs through regulating regional specification of organ primordia. The similarity between _vhnf1_-associated fish phenotypes and human symptoms suggests a correlation between developmental functions of vhnf1 and the molecular etiology of MODY5 and GCKD.
Keywords: vhnf1, hindbrain, liver, pancreas, kidney, regional specification
Organogenesis in vertebrates is not well understood. Zebrafish is an excellent model system for studying organogenesis. The embryo is transparent and develops ex utero, enabling easy observation of multiple internal organs during development. The conservation of organogenesis between zebrafish and mammals has already been shown for several organs, including the kidney (for review, see Drummond 2000). Furthermore, zebrafish is accessible to genetic screens. In a large insertional mutagenesis screen in the zebrafish with the goal of identifying a significant fraction of the genes required for vertebrate development (G. Golling, A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S.-Y. Lin, R.M. Nissen, M. Lee, and N. Hopkins, in prep.), we identified mutants affecting the development of internal organs. Three of these are alleles of the zebrafish homolog of the homeobox gene vHnf1 (variant Hnf1, also known as Hnf1β, LF-B3, or TCF2). Mutations in vHnf1 are associated with the human diseases MODY5 (maturity-onset diabetes of the young, type V) and familial GCKD (glomerulocystic kidney disease) (Horikawa et al. 1997; Nishigori et al. 1998; Lindner et al. 1999; Bingham et al. 2001).
MODY is a form of dominantly inherited type II diabetes mellitus characterized by pancreatic β-cell dysfunction with an onset age of 25 years or younger. Distinctive from other subtypes of MODY, MODY5 is also associated with nondiabetic, early-onset renal diseases (Horikawa et al. 1997; Nishigori et al. 1998; Lindner et al. 1999). GCKD is a kidney disease characterized by cystic dilatation of the Bowman space and the initial proximal convoluted tubule. It can occur sporadically or as an autosomal dominant inheritable disorder. Some GCKD patients display early-onset diabetes or impaired glucose tolerance (Rizzoni et al. 1982; Bingham et al. 2001). Genetic heterogeneity and hypoplastic subtypes account for part of the clinical spectrum of renal and diabetic symptoms of MODY5 and familial GCKD. Although the molecular etiology of MODY5 and GCKD is unknown, the association of vHnf1 with the renal and pancreatic symptoms of these two diseases indicates that vHnf1 is involved in the development and/or the functions of the kidney and the pancreas.
vHnf1 is highly homologous to Hnf1, which is the causal gene for another form of MODY, MODY3 (Yamagata et al. 1996). Both Hnf1 and vHnf1 are composed of an N-terminal dimerization domain, a variant homeodomain and a C-terminal transactivation domain. They can form homo- or heterodimers with each other and can recognize the same binding site (for review, see Tronche and Yaniv 1992). However, the in vivo targets of vHnf1 remain unidentified. In mouse Hnf1 and vHnf1 are expressed in overlapping yet distinctive patterns. Hnf1 is expressed in liver, kidney, intestine, pancreas, and stomach, while vHnf1 is expressed earlier in primitive endoderm, visceral endoderm, then in neural tube, gut, pancreas, kidney, and liver (Ott et al. 1991; Cereghini et al. 1992; Lazzaro et al. 1992; Barbacci et al. 1999; Coffinier et al. 1999). In both the liver and the kidney, the expression of vHnf1 precedes that of Hnf1, indicating that vHnf1 and Hnf1 may play distinct roles in development. _Hnf1_-deficient mice are afflicted with type II diabetes, renal Fanconi syndrome, hepatic dysfunctions, and hypercholesterolemia (Pontoglio et al. 1996; Shih et al. 2001). _vHnf1_-deficient mice die shortly after implantation with abnormal visceral endoderm (Barbacci et al. 1999; Coffinier et al. 1999), precluding a detailed analysis of the role of vHnf1 at later stages.
The mutant embryos of all three zebrafish vhnf1 alleles isolated in this screen can survive to day 5 of development when major organs are well-formed and functional in wild-type embryos. These mutants show underdevelopment of the liver and the pancreas, cyst formation in the pronephros, and in two strong alleles, reduction in size of the otic vesicles. Our data suggest that these abnormalities arise from patterning defects in primordia of gut, pronephros, and hindbrain. Our data also allow us to place vhnf1 in genetic networks that control the regional specification of these primordia. Analysis of the vhnf1 mutants may provide insight into mechanisms underlying _vHnf1_-associated human diseases.
Results
Isolation and characterization of the zebrafish vhnf1
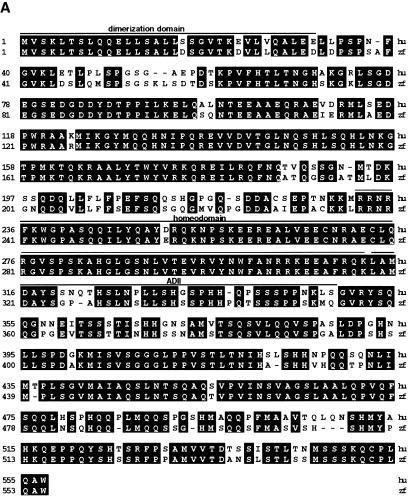
In a large-scale insertional mutagenesis screen underway in our laboratory, we isolated hi548, hi1843, and hi2169, due to cystic kidney formation in mutant embryos. Sequence analysis of junction fragments flanking proviral insertions associated with each mutant revealed that these insertions are located in a gene encoding a protein highly homologous to the human vHnf1, with an overall similarity of 83.1%, an almost identical homeodomain and a similarity of 81.1% within the transactivation domain (G. Golling, A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S.-Y. Lin, R.M. Nissen, M. Lee, and N. Hopkins, in prep.) (GenBank accession number AF430840; Fig. 1A).
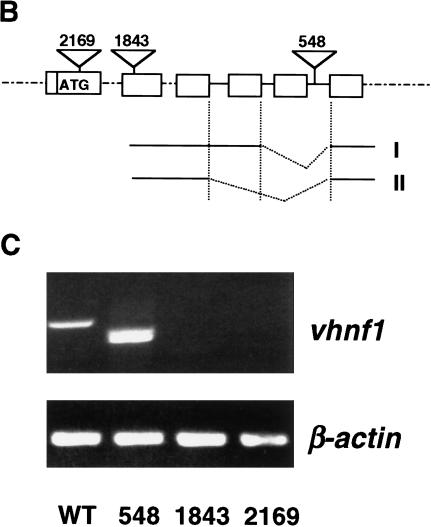
Figure 1.
The zebrafish vhnf1 gene. (A) Alignment of amino acid sequences of human (hu) and zebrafish (zf) vHnf1 by the Jotun Hein method. Black boxes show identical residues. Human vHnf1 sequence is from NM 00458. The region C-terminal to the homeodomain is the transactivation domain. The dimerization domain, homeodomain, and the ADII region of the transactivation domain are defined as before (Rey-Campos et al. 1991; Tronche and Yaniv 1992). (B) Diagram of proviral insertion sites in vhnf1. Only relevant exons are shown. I and II show two transcripts resulted from exon-skipping events in hi548 mutant embryos. (C) Disruption of vhnf1 expression by proviral insertions. Day 3 embryos from cross of wild-type TAB fish (WT) or phenotypic embryos from heterozygous crosses of hi548 (548), hi1843 (1843), and hi2169 (2169) were analyzed by RT–PCR for vhnf1 expression. RT–PCR for the β-actin gene was performed as loading control. In lane 2 of the upper panel, two bands migrated closely to each other.
In hi548 a proviral insertion is located in an intron, 6 bp away from the 5′-side exon–intron boundary (Fig. 1B). Only two truncated vhnf1 transcripts but no detectable wild-type transcripts are produced in homozygous mutant embryos (Fig. 1C). In one aberrant form, a single exon immediately preceding the insertion site is skipped (Fig. 1B, form I). The resulting in-frame deletion (nucleotides 1503–1682 in cDNA) leads to deletion of amino acids 450 to 509, a region within the transactivation domain. In the other aberrant form, an additional 5′-side exon is skipped (Fig. 1B, form II). The resulting frameshift deletion (nucleotides 1376–1682 in cDNA) disrupts the protein sequence after amino acid 411 and leads to a premature stop codon. In hi1843 the proviral insertion is located in an exon (Fig. 1B) between nucleotides 744 and 745 (of the cDNA). In hi2169 the proviral insertion is located in the first coding exon (Fig. 1B) between nucleotides 361 and 362 (of the cDNA). vhnf1 mRNA levels are drastically reduced in both hi1843 and hi2169 (Fig. 1C).
Both hi1843 and hi2169 failed to complement hi548. Therefore, we concluded that hi548, hi1843, and hi2169 are alleles of vhnf1. As discussed below, further support that vhnf1 is the responsible gene comes from partial rescue of the mutant phenotype of hi2169 by vhnf1 mRNA injection.
In hi1843 and hi2169 mutant embryos, otic vesicles are reduced in size (see below). In contrast, hi548 mutant embryos are morphologically indistinguishable from wild type until around 50 hpf (hour postfertilization), when kidney cysts start to form. Kidney cysts become more prominent and severe edemas develop with time in all three mutants (see below). On day 5 of development, neither the liver, nor the pancreas can be detected under a dissecting-scope in all three mutants, whereas both are visible in wild-type embryos.
vhnf1 expression during development
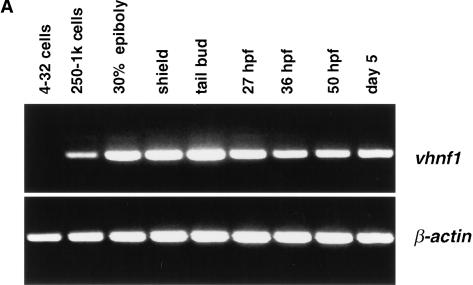
We next investigated the expression of vhnf1 during embryogenesis. Transcripts of vhnf1 are absent from early embryos but become detectable at between the 256-cell and the 1000-cell stage by RT–PCR (Fig. 2A). The zebrafish midblastula transition (MBT) begins at the 512-cell stage (Kimmel et al. 1995). Therefore, vhnf1 is an early zygotic gene. vhnf1 expression persists through at least day 5 of development, the last time point analyzed (Fig. 2A).
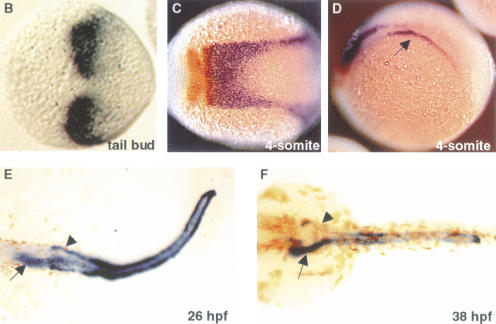
Figure 2.
vhnf1 expression during embryogenesis. (A) Time course of vhnf1 expression as indicated by RT–PCR. Samples were collected at indicated stages. (B_–_F) Whole-mount in situ hybridization shown in dorsal views, except for D. Anterior is to the left. (B) vhnf1 expression in tailbud-stage embryos. (C,D) vhnf1 (blue) expression in 4-somite-stage embryos. Embryos were double stained with a krox20 (red) probe. D is in a lateral view to show the intermediate mesoderm (arrow). (E,F) vhnf1 expression in the gut (arrow), the pronephric tubules, and ducts (arrow head) in embryos at 26 hpf (hour postfertilization; E) and 38 hpf (F).
In situ hybridization experiments revealed dynamic expression patterns of vhnf1 during development. Specific vhnf1 expression domains first emerge at the end of gastrulation, as two patches in the presumptive hindbrain (Fig. 2B). During the early segmentation period, these two patches appear to have fused at the dorsal midline. This expression domain has a sharp anterior border, extends caudally into the presumptive spinal cord, and tapers off gradually (Fig. 2C). By the 8-somite stage, the expression in the hindbrain has already regressed (data not shown).
At the 4-somite stage, vhnf1 expression also appears in the intermediate mesoderm (Fig. 2D). This expression domain similarly has a sharp anterior border and a more diffuse posterior border. The paired-box-containing gene pax2 is expressed in the intermediate mesoderm that is destined to become the pronephric tubule and ducts (Drummond et al. 1998; Serluca and Fishman 2001). Double in situ hybridization with pax2 and vhnf1 probes indicated that the vhnf1 expression domain in the intermediate mesoderm colocalized with that of pax2 (data not shown). vhnf1 expression in the kidney field persists through the pharyngula period. From the early pharyngula period, vhnf1 also starts to be expressed in the presumptive foregut endoderm and weakly in the hindgut endoderm (Fig. 2E). At 38 hpf vhnf1 is strongly expressed in the foregut and the entire length of the pronephric tubules and ducts (Fig. 2F).
vhnf1 regulates the patterning of the gut endoderm
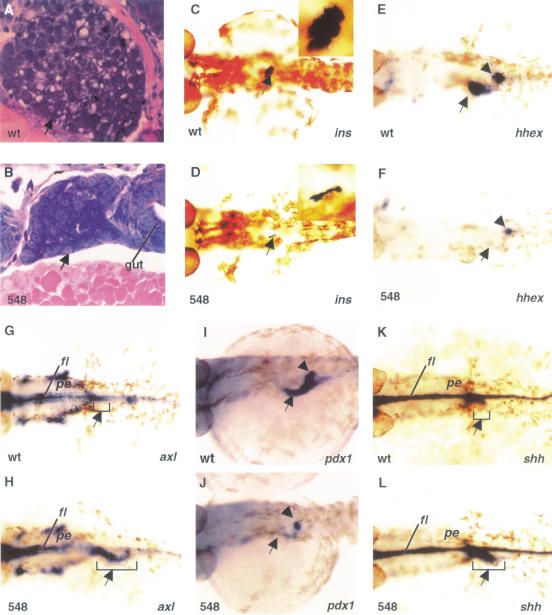
Several digestive organs are affected by vhnf1 inactivation. On histological sections of wild-type embryos at 72 hpf, the liver is well-formed, with differentiated hepatocytes showing typical vacuolar structures of stored substances (Fig. 3A). The pancreas has formed as well with a single islet surrounded by exocrine cells (data not shown). In sections of hi548 mutant embryos, the liver is underdeveloped and the hepatocytes appear undifferentiated (Fig. 3B). In addition, we were unable to detect a pancreas. To confirm the underdevelopment of the pancreas, we performed whole-mount in situ hybridization for the insulin gene. In wild-type embryos, a single islet reacted strongly with the insulin probe, whereas only a vestigial signal was detected in hi548 mutant embryos (Fig. 3C,D). The insulin signal was completely absent in hi2169 mutant embryos (data not shown). These data suggest that vhnf1 is required for the development of the liver and the pancreas.
Figure 3.
Effects of vhnf1 inactivation on gut development. Anterior is to the left. (A,B) Longitudinal sections comparing the liver (arrow) in embryos at 72 hpf (hour postfertilization). (C_–_L) Whole-mount in situ hybridization in dorsal views. (C,D) insulin expression (arrow) in 48-hpf embryos. Insets show islets at higher magnifications. (E,F) hhex expression in the presumptive liver primordium (arrow) and the pancreatic bud (arrowhead) of 30-hpf embryos. (G,H) axl expression in the anterior foregut (arrow) of embryos at 26-hpf. axl is also present in more anterior endoderm (pe). (I,J) pdx1 expression in the duodenal endoderm (arrow) and the pancreatic bud (arrowhead) of 36-hpf embryos. (K,L) shh expression in the foregut (arrow) of 30-hpf embryos. Wild-type embryo, wt; hi548 mutant embryo, 548; floor plate, fl; pharyngeal endoderm, pe.
Specific regions of the gut endoderm give rise to gut-associated digestive organs, including the liver and the pancreas. The liver derives from the anterior duodenal endoderm (posterior foregut), whereas the pancreas buds from the posterior duodenal endoderm. Because vhnf1 is expressed in the gut endoderm before visible organogenesis of the liver and the pancreas, we investigated whether it is required for the expression of several genes that mark specific regions of the gut endoderm.
The homeobox gene Hex is a marker for liver development. It is expressed in the liver primordium of mouse and chick embryos and is required for the liver organogenesis of mouse (Yatskievych et al. 1999; Martinez Barbera et al. 2000). hhex, the zebrafish homolog of Hex, is expressed in the liver primordium (Fig. 3E; Liao et al. 2000). In hi548 mutant embryos, this hhex expression is reduced (Fig. 3E,F). The underdevelopment of the liver in vhnf1 mutants may indicate that hhex is also required for the liver development of zebrafish and is downstream of vhnf1.
The absence of hhex expression could indicate a patterning defect of the gut endoderm. Alternatively, the formation of the gut endoderm itself could have been disrupted. To distinguish these two possibilities, we examined the expression of hnf3β/axl, which encodes a winged-helix transcription factor. In mouse, Hnf3β/Axl is expressed in the gut endoderm and is required for foregut morphogenesis (Ang et al. 1993; Krauss et al. 1993; Weinstein et al. 1994; Strahle et al. 1996). In wild-type zebrafish embryos at 26 hpf, axl expression in the gut endoderm is restricted to the anterior foregut (Fig. 3G). In hi548 mutant embryos, this expression is expanded into the posterior foregut (duodenum; Fig. 3H). The caudal expansion of the axl expression is complementary to the reduction of the hhex expression in the anterior duodenum. Together, these data indicate that regional specification of the foregut endoderm is disrupted in vhnf1 mutants.
One important aspect of regional specification of the foregut endoderm is the restricted expression of Pdx1 and Shh. The ParaHox homeoprotein Pdx1 and the intercellular signal transducer Shh are among the earliest markers of pancreas development. In amniotes, Pdx1 and Shh are expressed in complementary domains of the gut endoderm. Pdx1 is expressed in the duodenal endoderm, whereas Shh is expressed in the gut endoderm, with the exception of the pancreatic endoderm (Guz et al. 1995; Offield et al. 1996). The proper expression of Pdx1 and Shh is essential for pancreas development. Recent studies suggest that Shh and Pdx1 can repress each other's expression and may form a regulatory loop in the gut endoderm of chick embryos (Apelqvist et al. 1997; Hebrok et al. 1998; Kim and Melton 1998; Grapin-Botton et al. 2001). In zebrafish embryos, pdx1 is expressed in the presumptive duodenal endoderm (Fig. 3I; Argenton et al. 1999), whereas shh is expressed in the rostral tip of the foregut but is absent from the presumptive duodenal endoderm (Fig. 3K; Roy et al. 2001). The pdx1 expression is greatly reduced in hi548 mutant embryos (Fig. 3J) and is nearly eliminated in hi2169 mutants (data not shown). On the contrary, in mutant embryos, shh expression in the anterior foregut is expanded caudally (Fig. 3L). Our results suggest that vhnf1 may regulate regional specification of the gut endoderm through the shh_–_pdx1 network.
vhnf1 regulates the patterning of the pronephric primordia
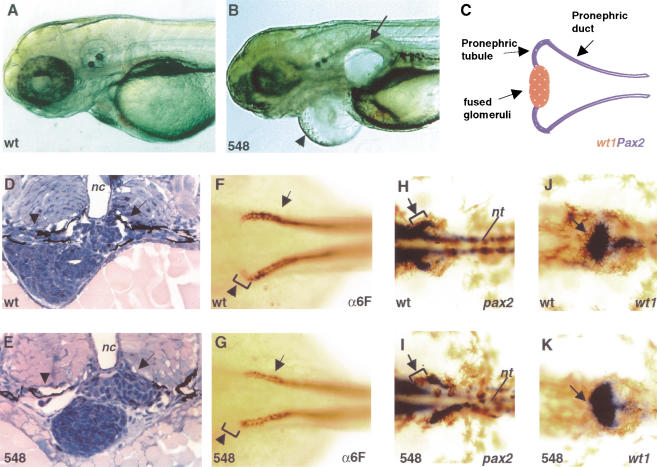
The most striking phenotype of our insertional alleles is the formation of kidney cysts. In all three alleles, kidney cysts start to form at around 50 hpf (data not shown) and severe edemas develop subsequently (Fig. 4A,B). The pronephros is the functional kidney of the zebrafish embryo and larva. It contains a pair of fused glomeruli, which lie ventral to the notochord and dorsal aorta. The glomeruli are connected laterally via a pair of pronephric tubules to the pronephric ducts, which run along both sides of the fish and eventually fuse at the midline and exit the embryo at the anus (Fig. 4C; Kimmel et al. 1995; Drummond et al. 1998). Tissue cross-sections show that at 54 hpf, the fused glomeruli are present in hi548 mutant embryos (Fig. 4D,E). However, the pronephric tubules are disrupted (Fig. 4D,E). Immuno-staining revealed even earlier defects. In 38-hpf embryos, the α6F antibody (Developmental Studies Hybridoma Bank), which recognizes a subunit of the chick Na+/K+ ATPase, reacts strongly with the entire pronephric duct and stains the forming pronephric tubules at the tip of the ducts. At this stage, hi548 embryos are morphologically indistinguishable from wild-type animals. However, in about a quarter of the embryos from a heterozygous cross, the α6F staining in the tubules is absent (Fig. 4F,G). Subsequent genotyping experiments verified that these embryos were homozyous for the hi548 proviral insertion. This result suggests that the pronephric tubules are the primary sites affected by _vhnf1_-deficiency prior to cyst formation in the kidney.
Figure 4.
Effects of vhnf1 inactivation on pronephros development. Anterior is to the left, except for D and E. (A,B) Embryos raised in fish water containing PTU (1-phenyl-2-thiourea; Westerfield 1993) on day 4 of development showing kidney cyst (arrow) and pericardiac edema (arrowhead) in lateral views. (C) Schematic diagram of the zebrafish pronephros in a dorsal view. Posterior parts of the ducts are not shown. (D,E) Cross-sections comparing the proneprhic tubules (arrowheads) and glomeruli (arrows) in embryos at 54 hpf (hour postfertilization). (F,G) Whole-mount antibody staining with α6F on 38 hpf embryos in dorsal views. Pronephric tubules (arrowheads) started to form at the tip of pronephric ducts (arrows). (H_–_K) Whole-mount in situ hybridization on 38-hpf embryos in dorsal views. (H,I) pax2.1 expression in pronephric tubules (arrows) and ducts. (J,K) wt1 expression in the future glomeruli (arrows). Wild-type embryo, wt; hi548 mutant embryo, 548; notochord, nc; neural tube, nt.
This early defect prompted us to investigate the expression of genes involved in the patterning of the pronephric primordia. The paired-domain protein Pax2 and the zinc-finger containing Wt1 are two transcription factors critical during renal development. In mouse both Pax2 and Wt1 are expressed in kidney anlagen and are required for kidney development (Pritchard-Jones et al. 1990; Armstrong et al. 1993; Kreidberg et al. 1993; Rackley et al. 1993; Rothenpieler and Dressler 1993; Torres et al. 1995). In zebrafish embryos at 38 hpf, pax2.1 is expressed in the pronephric tubules and the anterior pronephric ducts (Fig. 4H; Krauss et al. 1991; Majumdar et al. 2000). Meanwhile, wt1 is expressed in the future glomerulus (Fig. 4J; Majumdar et al. 2000; Serluca and Fishman 2001), complementary to pax2 expression. In hi548 mutant embryos, the pax2 expression in the tubules is absent (Fig. 4I). Complementarily, the wt1 expression in the future glomerulus is expanded mediolaterally to regions corresponding to the future pronephric tubules in wild-type embryos (Fig. 4J,K). These data suggest that this region has adopted molecular attributes of the future glomeruli. We also examined pax2.1 and wt1 expression in hi2169 embryos and observed similar changes (data not shown). Therefore, inactivation of vhnf1 leads to transformations at the molecular level in kidney development.
vhnf1 regulates the patterning of the hindbrain
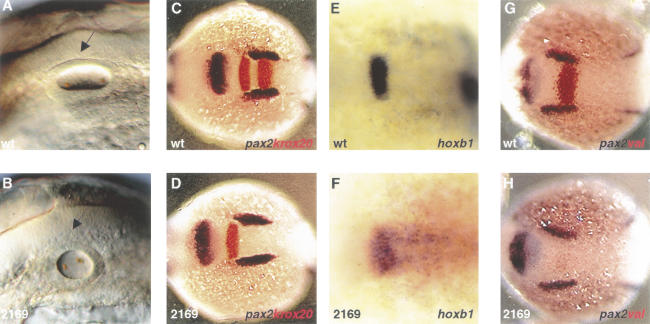
The strong alleles hi1843 and hi2169 revealed additional functions of vhnf1. In mutant embryos of these two alleles, otic vesicles are reduced in size (Fig. 5A,B). During development, vhnf1 expression is detected in the hindbrain but not in the otic vesicles (Fig. 2C). The hindbrain is involved in the induction of the otic vesicles (Phillips et al. 2001); thus the reduction in size of the otic vesicles may result from patterning defects in the hindbrain. To test this hypothesis, we first investigated the hindbrain expression of vhnf1 in detail. The hindbrain is subdivided transiently into a series of rhombomeres along the anterior–posterior (AP) axis. The presumptive rhombomeres 3 and 5 (r3 and r5) are marked by the expression of krox20, which encodes a zinc-finger transcription factor (Gilardi et al. 1991; Oxtoby and Jowett 1993). Double in situ hybridization with krox20 and vhnf1 probes revealed that the anterior border of vhnf1 expression coincided with that of r5 (Fig. 2C). We next tested whether the r5 expression of krox20 was affected by vhnf1 inactivation. In hi2169 mutant embryos, the r5 krox20 expression is diminished, whereas the r3 expression remains (Fig. 5C,D). Immediately anterior to r5, r4 is marked by hoxb1 expression (Prince et al. 1998). Complementary to the loss of the r5 krox20 expression, the r4 expression of hoxb1 expands caudally in hi2169 mutant embryos (Fig. 5E,F), suggesting that r5 takes on molecular attributes of r4 as a result of vhnf1 inactivation.
Figure 5.
Hindbrain patterning is affected by vhnf1 inactivation. Anterior is to the left. (A,B) Otic vesicles (arrows) in embryos at the 25-somite stage in lateral views. (C_–_H) Whole-mount in situ hybridization in dorsal views. In C, D, G, and H embryos were double stained with a pax2.1 probe (blue) to show relative positions. (C, D) krox20 (red) expression in 8-somite stage embryos. (E,F) hoxb1 expression in 4-somite stage embryos. (G,H) valentino expression (red) in embryos at the 4-somite stage. Wild-type embryo, wt; hi2169 mutant embryo, 2169.
vhnf1 could regulate the expression of krox20 and hoxb1 directly, or it could do so through other factors. valentino (val) is the zebrafish homolog of the mouse bZIP transcription factor Kreisler (Kr) and is expressed in r5 and r6 (Moens et al. 1998). In val mutants, the otic vesicles are reduced in size, the expression of the hoxb1 gene is expanded posteriorly, and the r5 krox20 expression is diminished (Prince et al. 1998), reminiscent of what is observed in hi2169 mutants. We, therefore, investigated the hindbrain expression of val in hi2169 embryos. In mutant embryos, the val expression in both r5 and r6 is nearly eliminated (Fig. 5G,H). In addition, we observed touch insensitivity and an ectopic cluster of cells that resemble the interhyal cartilage at the tip of the first ceratobranchial (data not shown), just as in val mutants (Moens et al. 1998). The similarity between the phenotypes of hi2169 and val mutants and the requirement of vhnf1 for val expression support the hypothesis that vhnf1 acts upstream of val to regulate hindbrain patterning.
Partial rescue of hi2169 phenotype
The early requirement of vhnf1 activity for the hindbrain val expression provided us an avenue to test whether vhnf1 can rescue our mutants. We injected vhnf1 mRNA into hi2169 embryos and performed in situ hybridization for the val gene. All _vhnf1_-injected embryos were positive for val expression, including embryos homozygous for the hi2169 insertion (14 out of 42 and 7 out of 27 tested embryos were homozygous for hi2169 in two independent experiments). As controls, all _vhnf1_−/− embryos were negative for val expression when injected with LacZ mRNA, verifying that mutations in vhnf1 cause the phenotypes observed in our mutants. The restoration of val expression by vhnf1 mRNA also supports the hypothesis that vhnf1 is upstream of val.
Effects of vhnf1 overexpression
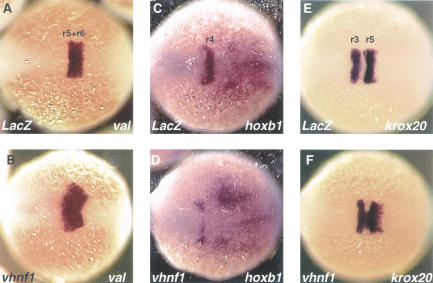
All the above data support the hypothesis that vhnf1 regulates the regional specification of multiple-organ primordia. We tested this hypothesis further in the hindbrain. If vhnf1 is a key regional specification factor of the hindbrain, overexpression of it may be able to confer attributes of rhombomeres that normally express vhnf1 to other regions. We overexpressed vhnf1 by injecting mRNA into embryos at the 1- to 4-cell stage. In vhnf1 mRNA injected embryos, val expression is expanded anteriorly with a diffuse border (Fig. 6A,B). Complementarily, the r4 hoxb1 expression is regressed rostrally, with only a residual signal left in the most anterior part of r4 (Fig. 6C,D). In some embryos, the entire r4 hoxb1 expression is eliminated (data not shown). Consistent with the expansion of the val expression, the r5 krox20 expression is expanded, whereas the r3 krox20 is not affected (Fig. 6E,F). As a control, LacZ mRNA injection did not change the expression of val, hoxb1, or krox20 (Fig. 6A,C,E). Interestingly, ectopic val and krox20 expression can only be detected in regions neighboring their normal expression domains, suggesting combinatorial regulation of val and krox20 expression by vhnf1 and other factors. In summary, as indicated by the expansion of val and krox20 expression and the absence of hoxb1 expression, r4 takes on molecular attributes of r5 as a result of vhnf1 overexpression.
Figure 6.
Effects of vhnf1 overexpression on hindbrain patterning. All show whole-mount in situ hybridization on embryos at the 4-somite stage in dorsal views. Anterior is to the left. (A,B) val expression. (C,D) hoxb1 expression. (E,F) krox20 expression. LacZ,embryo injected with LacZ mRNA; vhnf1, embryo injected with vhnf1 mRNA.
Discussion
Zebrafish vhnf1 gene is the causal gene for hi548, hi1843, and hi2169
Zebrafish mutants hi548, hi1843, and hi2169 belong to a single complementation group and are each linked to a proviral insertion located in a gene that is highly homologous to the human and mouse vHnf1 gene. The homology between the fish protein and the human vHnf1 encompasses the entire sequence, including the transactivation domain, which defines the difference between Hnf1 and vHnf1. In addition, the fish gene is expressed in a similar spatial and temporal pattern as the mouse vHnf1 gene, indicating that it is the fish vHnf1 homolog. Injection of vhnf1 mRNA can rescue val expression in hi2169 mutant embryos, providing further evidence that mutations in vhnf1 are responsible for all three mutants.
In hi1843 and hi2169 mutant embryos, vhnf1 expression is below the level of detection, suggesting that they are null or strong hypomorphic alleles. In hi548 mutant embryos, altered messages are produced. Hi548 mutant embryos display similar kidney, liver, and pancreas phenotypes as hi1843 and hi2169 mutants but lack the hindbrain phenotype. In addition, the hi548 mutation has milder effects on the expression of several downstream genes, such as pdx1 and the insulin gene. Together, these data suggest that hi548 is hypomorphic to hi1843 and hi2169.
vhnf1 regulates regional specification of the gut, kidney, and hindbrain primordia
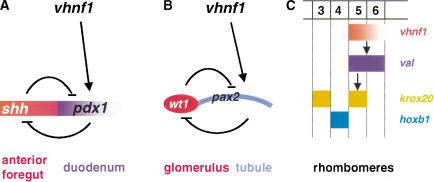
In this study, we showed that vhnf1 plays similar roles in the development of multiple organs. Mutations in vhnf1 lead to transformation of the gene expression profile along the AP axis in the gut endoderm and the hindbrain primordium and mediolaterally in the pronephric primordium. In addition, misexpression of vhnf1 can confer molecular characteristics of regions that normally express vhnf1 to regions more rostral in the hindbrain. These data suggest that vhnf1 is a key regulator of regional specification during the development of these organs (Fig. 7).
Figure 7.
Models for vhnf1 functions in the development of the gut endoderm, the pronephros, and the hindbrain. (A) vhnf1 activates the pdx1 expression and subsequently regulates the regional specification of the foregut endoderm via the _shh_-pdx1 network. (B) vhnf1 regulates regional specification of the pronephric primordia through the pax2–wt1 network. (C) vhnf1, during hindbrain development, establishes the anterior border of the val expression, subsequently activates the r5 krox20 expression, and maintains the posterior border of the r4 hoxb1 expression.
In all vertebrates, gut-associated organs start to form by swelling and budding of specific regions of the gut endoderm. Before the onset of visible organogenesis, the expression of several genes is already restricted (for review, see Grapin-Botton and Melton 2000). The expression of Shh and Pdx1 in the gut endoderm is under strict temporal and spatial control and this regulation is essential for pancreas development in amniotes (Offield et al. 1996; Apelqvist et al. 1997; Hebrok et al. 1998; Grapin-Botton et al. 2001). How regional specification is achieved for the seemingly uniform sheet of endoderm cells is a long-standing question. Our results show that vhnf1 is required for the expression of pdx1 in the duodenal endoderm and the repression of shh expression in the same region. Intriguingly, Hnf1/vHnf1-consensus-binding sites can be found within the conserved region of the mammalian Pdx1 enhancer (Ben-Shushan et al. 2001), suggesting that Hnf1 or vHnf1 may regulate Pdx1 expression directly. We propose that inactivation of vhnf1 causes imbalance of the pdx1_–_shh network, which in turn leads to underdevelopment of the liver and the pancreas (Fig. 7A). The expression domain of vhnf1 extends beyond that of pdx1, indicating that other factors are involved as well. Multiple approaches will provide insight to the complex regulation of regional specification of the gut endoderm, including cloning of responsible genes for additional zebrafish gut mutants.
Results from this study indicate a high level of conservation between pancreatic development in zebrafish and higher vertebrates. shh expression is repressed in the pancreatic endoderm of wild-type zebrafish embryos, just as in amniotes (Fig. 3K; Roy et al. 2001). In addition, we observed expansion of shh expression into the pancreatic endoderm, reduction of pdx1 expression in this region, and underdevelopment of the pancreas in vhnf1 mutants. Together, these data suggest that the repression of shh in the duodenal endoderm is important for the pancreas specification of zebrafish as well. A recent study showed that the specification of the zebrafish pancreas requires the activities of hedgehog (Hh) proteins and that overexpression of shh can induce ectopic pdx1 expression (Roy et al. 2001). These results seem to indicate a contrasting role of shh in pancreas development of zebrafish to that in amniotes (Roy et al. 2001), and seem to contradict our hypothesis. However, it is possible that shh plays different roles in distinct steps during regional specification of the gut endoderm and that different stages of gut specification may be targeted in different experiments. Targeted expression of shh in the pancreatic endoderm of zebrafish embryos at later stages may help to clarify the degree of conservation between the pancreatic development in zebrafish and amniotes.
Glomeruli, tubules, and ducts arises de novo from the intermediate mesoderm to form the pronephos of zebrafish (Drummond et al. 1998; Serluca and Fishman 2001). Proper regional specification has to be achieved for the cells in the intermediate mesoderm to adopt distinct fates. Our results show that in vhnf1 mutants, α6f signal in the future tubules is lost while wt1 expression is expanded into the same region, indicating a molecular transformation from the tubular to the glomerular characteristics. pax2 seems to contribute to cell fate choices between tubular and glomerular cells (Majumdar et al. 2000). We show that the vhnf1 expression and the pax2 expression in the intermediate mesoderm colocalize and that vhnf1 is required for the pax2 expression in the future tubules. Furthermore, a consensus site for vHnf1 can be found in the upstream region of mammalian Pax2, although the significance of this site is unknown. We propose that vhnf1 activates the expression of pax2, which in turn regulates regional specification of the pronephric primordia (Fig. 7B). It has been shown that pax2 and wt1 can repress each other's expression under certain conditions (for review, see Dressler 1995). Thus vhnf1 may inhibit wt1 expression indirectly through pax2 (Fig. 7B). In noi, a zebrafish pax2 mutant, α6F staining in both the pronephric tubules and the anterior ducts is lost (Majumdar et al. 2000). In contrast, in vhnf1 mutants, only staining in the tubules is affected (Fig. 4F,G). That vhnf1 inactivation only affects pax2 expression in the tubules (Fig. 4H,I) may explain this difference and may indicate that other factors are involved in establishing the posterior border of pax2 expression.
Similarly, the vertebrate hindbrain is specified through multiple steps. Although Val/Kr is one of the earliest factors known to regulate hindbrain patterning, the restricted pattern of val expression indicates that the onset of the regional specification of the hindbrain primordium is an earlier event. We found that the anterior expression border of vhnf1 overlaps with that of val. Inactivation of vhnf1 leads to strikingly similar molecular consequences, as well as morphological phenotypes in the hindbrain, the otic vesicle, and the head skeleton as inactivation of val. Furthermore, val expression is lost in vhnf1 mutants and conversely vhnf1 overexpression can induce the expression of val. Taken together, these data suggest that vhnf1 regulates hindbrain patterning by activating val expression (Fig. 7C). val is probably a direct target of vhnf1, because consensus binding sites for vHnf1 can be found within the upstream region of mammalian Val/Kr, although whether these sites are developmentally relevant is unknown. The vhnf1 expression domain extends more posteriorly than that of val, suggesting that vhnf1 is involved in setting the anterior boundary and that other factors are involved in specifying the posterior border of val expression. The _vhnf1_-induced anterior expansion of val expression is consistent with this hypothesis. The spatially restricted expression of vhnf1 in the hindbrain arises almost simultaneously with val. What establishes the expression domain of vhnf1 is a question that requires further investigation.
Probable association of the developmental functions of vhnf1 with human diseases
In both human and zebrafish, the kidney and the pancreas are two of the main organs affected by mutations in vhnf1. In addition, retarded motor development was noted for one MODY5 patient (Lindner et al. 1999). Therefore, there is a correlation between _vhnf1_-associated fish phenotypes and human symptoms. Our mutants could serve as model systems to study MODY5 and GCKD.
Mutations in Pdx1 are associated with MODY4 (Stoffers et al. 1997). Our results suggest that vhnf1 functions upstream of pdx1 to regulate pancreatic development, raising the possibility that other components of this pathway might contribute to pancreatic diseases as well. We also show that vhnf1 functions in the same genetic network as pax2 and wt1 to regulate patterning of the pronephros. Mutations in both Pax2 and Wt1 are associated with human syndromes that have renal symptoms (Burgin et al. 1990; Pelletier et al. 1991; Eccles and Schimmenti 1999; Patek et al. 1999). Therefore multiple genes in this pathway are associated with human kidney diseases. Dissecting this network may provide new avenues for identifying novel genetic components of renal diseases.
In transfection experiments, vHnf1 can regulate the expression of several genes directly involved in adult organ functions, such as the insulin gene in the pancreas (Okita et al. 1999) and the albumin gene in the liver (Rey-Campos et al. 1991). It is possible that MODY5 and GCKD arise from defects in vHnf1 functions that are unrelated to its role in organogenesis. However, pancreatic tissue and kidney tissue undergo constant renewal, presumably from stem cells or progenitor cells. It is tantalizing to postulate that the replenishing processes of adult tissues might utilize similar pathways as the developmental processes. Furthermore, the onset of both GCKD and MODY5 is early in life. The majority of familial GCKD cases are observed in infants. The earliest detection of renal defects in MODY5 patients is at 27 wk of gestation (Nishigori et al. 1998). The early-onset ages raise the possibility that MODY5 and familial GCKD are developmental diseases. Studying vhnf1 functions in early development may provide insight for mechanisms of MODY and GCKD.
Materials and methods
Cloning of vhnf1
The cloning procedures for our insertional mutants will be described elsewhere (G. Golling, A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S.-Y. Lin, R.M. Nissen, M. Lee, and N. Hopkins, in prep.). For pcs2(+)–vhnf1, the coding region of vhnf1 was amplified with primers 5′-TGGAATTCATTTAG ATGTTTGCAACCATGG-3′ and 5′-AGCTCGAGGTTGCCA TGGATATAGGTGCGATGAAG-3′ from cDNA, digested with _Eco_RI and _Xho_I, and subcloned into the same sites of pcs2(+).
RT–PCR
Embryos were collected according to the staging method described by Kimmel et al. (1995). RNA was extracted with the trizol reagent (GIBCO BRL) following the manufacturer's instructions. First-strand cDNA was generated with the superscript II RT–PCR system (GIBCO BRL) and subsequently amplified with PCR. The primers used for vhnf1 in Figure 1 were 5′-CACAATACCGCATTTCCCTCTCTAG-3′ and 5′-GTCAC TAAATTGGGCGCCATGTTGATCA-3′. The primers used for vhnf1 in Figure 2 were 5′-AGGTCTTCCTCCAGTAAGCAC CCTGACC-3′ and 5′-GGCTGTTGGCGTCTGTCACCAC CAT-3′. The primers used for β_-actin_ controls were 5′-CAT CAGCATGGCTTCTGCTCTGTATGG-3′ and 5′-GACTTGT CAGTGTACAGAGACACCCTG-3′.
In situ hybridization and histological analysis
Whole-mount in situ hybridization was performed as described with minor modifications (Westerfield 1993). For double in situ, RNA probes were labeled with digoxygenin and fluorescein, respectively, and incubated with embryos together. Embryos were then incubated with one of the alkaline phosphatase-coupled antibodies (anti-digoxygenin or anti-fluorescein). Blue color was first developed with NBT/BCIP as the chromogenic substrate. Alkaline phosphatase activity was removed by incubation with 0.1 M glycine/HCl at pH2.2 (Hauptmann and Gerster 1994) at room temperature for 30 min before incubation with the second antibody. Red color was developed with INT/BCIP (Roche Molecular Biochemicals). Embryos older than 24 hpf were bleached with 10% hydrogen peroxide in methanol at room temperature overnight, cleared with benzyl benzoate, and flattened with coverslips for photography.
Antibody staining and histological sections were carried out as described by Drummond et al. (1998). Briefly, α6F from DSHB was used at a concentration of 1:5. Horseradish-peroxidase coupled anti-mouse IgG (Vector Laboratories) was used at 1:500, and DAB was used as the chromogenic substrate (Vector Laboratories). For sections, embryos were fixed in formalin and embedded in JB-4 resin (Polysciences) following manufacturer's instructions and cut at 4 μm. Slides were then stained with methylene blue/azureII/basic fuchsin (Humphrey and Pittman 1974).
Genotyping
Individual embryo was digested in 25 μL lysis buffer (50 mM KCl, 10 mM Tris, 0.01% Gelatin, NP-40, 0.45% Tween-20, 5 mM EDTA, 200 μg/mL Proteinase K) at 55°C for 2 h. Proteinase K was deactivated by incubation at 94°C for 15 min; 1 μL lysate was used for each PCR reaction. Three primers were used in a single reaction. One was a viral-specific primer, the other two were a pair of genomic-specific primers flanking the proviral insertion. The presence of a proviral insertion would disrupt the amplification between the genomic pair. A fragment between the viral primer and one of the genomic primers would amplify instead. For hi548, the genomic-specific primer pair were 5′-GGCTGTTGGCGTCTGTCACCACCAT-3′ and 5′-CATCAC AAGCACAGACGGTGCCTGTC-3′. The viral primer was 5′-GCTAGCTTGCCAAACCTACAGGT-3′. For hi2169 the genomic primers were 5′-CACAATACCGCATTTCCCTCTCTAG 3′ and 5′-TCCGGATAACTTCCCTTTACTGTG-3′. The viral primer was 5′-CTGTTCCATCTGTTCCTGAC-3′.
Embryo injections
Full-length, capped mRNA was generated in vitro with the SP6 RNA polymerase from pcs2-vhnf1 and pcs2-LacZ DNA template and purified with G-50 spin columns for RNA (Roche Molecular Biochemicals). mRNA was injected into 1- to 4-cell-stage embryos at 25 ng/μL. For the rescuing experiment, mRNA was injected into hi2169 embryos. In situ hybridization for val was performed and embryos were subsequently genotyped by PCR. For the overexpression experiment, mRNA was injected into wild-type embryos instead. In situ hybridization for val, hoxb1, and krox20 was performed on injected embryos at the 4-somite stage.
Acknowledgments
We thank A. Amsterdam, S. Burgess, G. Golling, S.-Y. Lin, R.M. Nissen, and S. Zhang for critical reading of the manuscript; A. Amsterdam and G. Golling for help with cloning and sequence analysis; K. Artzt and M.A. Haldi for help on phenotypic analysis; W. Chen, I.A. Drummond, J. Moss, L.G. Moss, B. Riley, H. Sive, E. Wiellette, and C.V. Wright for helpful discussions; and S. Farrington and the technical staff of the Hopkins lab for superb support. We also thank W. Chen and S. Burgess for plasmids and probes, the Developmental Studies Hybridoma Bank (maintained by the Department of Biological Sciences, The University of Iowa) for the α6F antibody, and A.M. Caron of the MIT CCR Core Histology Facility for technical assistance on histological sections. We apologize for citing reviews instead of original papers due to space limitation. This work was supported by grants from Amgen, NIH, and NCRR.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nhopkins@mit.edu; FAX 617-258-0258.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad946701.
References
- Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- Argenton F, Zecchin E, Bortolussi M. Early appearance of pancreatic hormone-expressing cells in the zebrafish embryo. Mech Dev. 1999;87:217–221. doi: 10.1016/s0925-4773(99)00151-3. [DOI] [PubMed] [Google Scholar]
- Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JB. The expression of the Wilms' tumour gene, WT1, in the developing mammalian embryo. Mech Dev. 1993;40:85–97. doi: 10.1016/0925-4773(93)90090-k. [DOI] [PubMed] [Google Scholar]
- Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- Ben-Shushan E, Marshak S, Shoshkes M, Cerasi E, Melloul D. A pancreatic beta-cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta ), HNF-1alpha, and SPs transcription factors. J Biol Chem. 2001;276:17533–17540. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- Bingham C, Bulman MP, Ellard S, Allen LI, Lipkin GW, Hoff WG, Woolf AS, Rizzoni G, Novelli G, Nicholls AJ, et al. Mutations in the hepatocyte nuclear factor-1β gene are associated with familial hypoplastic glomerulocystic kidney disease. Am J Hum Genet. 2001;68:219–224. doi: 10.1086/316945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin AB, Parodos K, Lane DJ, Pace NR. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- Cereghini S, Ott MO, Power S, Maury M. Expression patterns of vHNF1 and HNF1 homeoproteins in early postimplantation embryos suggest distinct and sequential developmental roles. Development. 1992;116:783–797. doi: 10.1242/dev.116.3.783. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Thepot D, Babinet C, Yaniv M, Barra J. Essential role for the homeoprotein vHNF1/HNF1beta in visceral endoderm differentiation. Development. 1999;126:4785–4794. doi: 10.1242/dev.126.21.4785. [DOI] [PubMed] [Google Scholar]
- Dressler GR. Transcription factors in renal development: The WT1 and Pax-2 story. Semin Nephrol. 1995;15:263–271. [PubMed] [Google Scholar]
- Drummond IA. The zebrafish pronephros: A genetic system for studies of kidney development. Pediatr Nephrol. 2000;14:428–435. doi: 10.1007/s004670050788. [DOI] [PubMed] [Google Scholar]
- Drummond IA, Majumdar A, Hentschel H, Elger M, Solnica-Krezel L, Schier AF, Neuhauss SC, Stemple DL, Zwartkruis F, Rangini Z, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- Eccles MR, Schimmenti LA. Renal-coloboma syndrome: A multi-system developmental disorder caused by PAX2 mutations. Clin Genet. 1999;56:1–9. doi: 10.1034/j.1399-0004.1999.560101.x. [DOI] [PubMed] [Google Scholar]
- Gilardi P, Schneider-Maunoury S, Charnay P. Krox-20: A candidate gene for the regulation of pattern formation in the hindbrain. Biochimie. 1991;73:85–91. doi: 10.1016/0300-9084(91)90079-g. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Melton DA. Endoderm development: from patterning to organogenesis. Trends Genet. 2000;16:124–130. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Majithia AR, Melton DA. Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes & Dev. 2001;15:444–454. doi: 10.1101/gad.846001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes & Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- Humphrey CD, Pittman FE. A simple methylene blue-azure II-basic fuchsin stain for epoxy-embedded tissue sections. Stain Technology. 1974;49:9–14. doi: 10.3109/10520297409116929. [DOI] [PubMed] [Google Scholar]
- Kim SK, Melton DA. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc Natl Acad Sci. 1998;95:13036–13041. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmer SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113:1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Lazzaro D, De Simone V, De Magistris L, Lehtonen E, Cortese R. LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development. 1992;114:469–479. doi: 10.1242/dev.114.2.469. [DOI] [PubMed] [Google Scholar]
- Liao W, Ho CY, Yan YL, Postlethwait J, Stainier DY. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development. 2000;127:4303–4313. doi: 10.1242/dev.127.20.4303. [DOI] [PubMed] [Google Scholar]
- Lindner TH, Njolstad PR, Horikawa Y, Bostad L, Bell GI, Sovik O. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet. 1999;8:2001–2008. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Lun K, Brand M, Drummond IA. Zebrafish no isthmus reveals a role for pax2.1 in tubule differentiation and patterning events in the pronephric primordia. Development. 2000;127:2089–2098. doi: 10.1242/dev.127.10.2089. [DOI] [PubMed] [Google Scholar]
- Martinez Barbera JP, Clements M, Thomas P, Rodriguez T, Meloy D, Kioussis D, Beddington RS. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127:2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- Moens CB, Cordes SP, Giorgianni MW, Barsh GS, Kimmel CB. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- Nishigori H, Yamada S, Kohama T, Tomura H, Sho K, Horikawa Y, Bell GI, Takeuchi T, Takeda J. Frameshift mutation, A263fsinsGG, in the hepatocyte nuclear factor-1beta gene associated with diabetes and renal dysfunction. Diabetes. 1998;47:1354–1355. doi: 10.2337/diab.47.8.1354. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Okita K, Yang Q, Yamagata K, Hangenfeldt KA, Miyagawa J, Kajimoto Y, Nakajima H, Namba M, Wollheim CB, Hanafusa T, et al. Human insulin gene is a target gene of hepatocyte nuclear factor-1alpha (HNF-1alpha) and HNF-1beta. Biochem Biophys Res Commun. 1999;263:566–569. doi: 10.1006/bbrc.1999.1412. [DOI] [PubMed] [Google Scholar]
- Ott MO, Rey-Campos J, Cereghini S, Yaniv M. vHNF1 is expressed in epithelial cells of distinct embryonic origin during development and precedes HNF1 expression. Mech Dev. 1991;36:47–58. doi: 10.1016/0925-4773(91)90071-d. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patek CE, Little MH, Fleming S, Miles C, Charlieu JP, Clarke AR, Miyagawa K, Christie S, Doig J, Harrison DJ, et al. A zinc finger truncation of murine WT1 results in the characteristic urogenital abnormalities of Denys-Drash syndrome. Proc Natl Acad Sci. 1999;96:2931–2936. doi: 10.1073/pnas.96.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Bruening W, Li FP, Haber DA, Glaser T, Housman DE. WT1 mutations contribute to abnormal genital system development and hereditary Wilms' tumour. Nature. 1991;353:431–434. doi: 10.1038/353431a0. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–365. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: Expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Pritchard-Jones K, Fleming S, Davidson D, Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J, Housman D, et al. The candidate Wilms' tumour gene is involved in genitourinary development. Nature. 1990;346:194–197. doi: 10.1038/346194a0. [DOI] [PubMed] [Google Scholar]
- Rackley RR, Flenniken AM, Kuriyan NP, Kessler PM, Stoler MH, Williams BR. Expression of the Wilms' tumor suppressor gene WT1 during mouse embryogenesis. Cell Growth Differ. 1993;4:1023–1031. [PubMed] [Google Scholar]
- Rey-Campos J, Chouard T, Yaniv M, Cereghini S. vHNF1 is a homeoprotein that activates transcription and forms heterodimers with HNF1. EMBO J. 1991;10:1445–1457. doi: 10.1002/j.1460-2075.1991.tb07665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoni G, Loirat C, Levy M, Milanesi C, Zachello G, Mathieu H. Familial hypoplastic glomerulocystic kidney. A new entity? Clin Nephrol. 1982;18:263–268. [PubMed] [Google Scholar]
- Rothenpieler UW, Dressler GR. Pax-2 is required for mesenchyme-to-epithelium conversion during kidney development. Development. 1993;119:711–720. doi: 10.1242/dev.119.3.711. [DOI] [PubMed] [Google Scholar]
- Roy S, Qiao T, Wolff C, Ingham PW. Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr Biol. 2001;11:1358–1363. doi: 10.1016/s0960-9822(01)00402-x. [DOI] [PubMed] [Google Scholar]
- Serluca FC, Fishman MC. Pre-pattern in the pronephric kidney field of zebrafish. Development. 2001;128:2233–2241. doi: 10.1242/dev.128.12.2233. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, et al. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- Strahle U, Blader P, Ingham PW. Expression of axial and sonic hedgehog in wildtype and midline defective zebrafish embryos. Int J Dev Biol. 1996;40:929–940. [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- Tronche F, Yaniv M. HNF1, a homeoprotein member of the hepatic transcription regulatory network. Bioessays. 1992;14:579–587. doi: 10.1002/bies.950140902. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish book: A guide for the laboratory use of zebrafish (brachydanio rerio). Eugene, OR: The University of Oregon Press; 1993. [Google Scholar]
- Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox RD, Lathrop GM, Boriraj VV, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- Yatskievych TA, Pascoe S, Antin PB. Expression of the homebox gene Hex during early stages of chick embryo development. Mech Dev. 1999;80:107–109. doi: 10.1016/s0925-4773(98)00204-4. [DOI] [PubMed] [Google Scholar]