Identification of NF-κB-Dependent Gene Networks in Respiratory Syncytial Virus-Infected Cells (original) (raw)
Abstract
Respiratory syncytial virus (RSV) is a mucosa-restricted virus that is a leading cause of epidemic respiratory tract infections in children. In epithelial cells, RSV replication activates nuclear translocation of the inducible transcription factor nuclear factor κB (NF-κB) through proteolysis of its cytoplasmic inhibitor, IκB. In spite of a putative role in mediating virus-inducible gene expression, the spectrum of NF-κB-dependent genes induced by RSV infection has not yet been determined. To address this, we developed a tightly regulated cell system expressing a nondegradable, epitope-tagged IκBα isoform (Flag-IκBα Mut) whose expression could be controlled by exogenous addition of nontoxic concentrations of doxycycline. Flag-IκBα Mut expression potently inhibited IκBα proteolysis, NF-κB binding, and NF-κB-dependent gene transcription in cells stimulated with the prototypical NF-κB-activating cytokine tumor necrosis factor alpha (TNF-α) and in response to RSV infection. High-density oligonucleotide microarrays were then used to profile constitutive and RSV-induced gene expression in the absence or presence of Flag-IκBα Mut. Comparison of these profiles revealed 380 genes whose expression was significantly changed by the dominant-negative NF-κB. Of these, 236 genes were constitutive (not RSV regulated), and surprisingly, only 144 genes were RSV regulated, representing numerically ∼10% of the total population of RSV-inducible genes at this time point. Hierarchical clustering of the 144 RSV- and Flag-IκBα Mut-regulated genes identified two discrete gene clusters. The first group had high constitutive expression, and its expression levels fell in response to RSV infection. In this group, constitutive mRNA expression was increased by Flag-IκBα Mut expression, and the RSV-induced decrease in expression was partly inhibited. In the second group, constitutive expression was very low (or undetectable) and, after RSV infection, expression levels strongly increased. In this group, NF-κB was required for RSV-inducible expression because Flag-IκBα Mut expression blocked their induction by RSV. This latter cluster includes chemokines, transcriptional regulators, intracellular proteins regulating translation and proteolysis, and secreted proteins (complement components and growth factor regulators). These data suggest that NF-κB action induces global cellular responses after viral infection.
NF-κB is a family of inducible transcription factors controlling expression of genes important for pathogen- or cytokine-induced inflammation, immune response, and cellular survival (5, 21, 37). The prototypical NF-κB complex, composed of 50-kDa NF-κB1 and 65-kDa RelA heterodimers, is regulated by its association with a family of cytoplasmic inhibitors, IκBs, whose members bind and specifically inactivate NF-κB members by masking their nuclear localization sequence, thereby preventing nuclear entry (27, 32; reviewed in reference 7).
NF-κB activation is a sequential process whose final result is to induce nuclear translocation of the inactivated cytoplasmic NF-κB complex through targeted proteolysis of the IκB inhibitors. Intracellular NF-κB-activating signals converge on the multiprotein cytoplasmic IκB kinase complex (IKK), a complex that phosphorylates IκB on two serine residues (Ser32 and Ser36) in its NH2-regulatory domain (reviewed in reference 37). Phospho-IκB is then specifically bound by the SCF-type E3 ubiquitin ligase E3RS, initiating IκB ubiquitination and proteolysis through the proteasome (10, 37, 38). A parallel pathway important in viral infection produces IκB degradation through cytoplasmic calpains (30, 34). Following IκB proteolysis, liberated NF-κB enters the nucleus to activate target gene transcription.
Because of the large number of inducible cytokine, chemokine, acute-phase reactant, and adhesion molecule genes that contain NF-κB binding sites in their promoters, NF-κB appears to play an essential role in innate immunity and the inflammatory (host) response to infectious agents (8, 11, 40, 52; reviewed in reference 56). Our laboratory has been investigating the mechanisms for chemokine expression induced by respiratory syncytial virus (RSV). RSV is a ubiquitous human respiratory tract pathogen known to produce severe lower respiratory tract infections (bronchiolitis) in infants, accounting for ∼100,000 hospitalizations in the United States annually (50; reviewed in reference 26). Human RSV infections characteristically show virus-induced damage to the epithelial surface (1, 15, 19) and a pronounced inflammatory response, consisting of a perivascular mononuclear and lymphocytic infiltrate (1, 15). Other features of immune activation that can be detected include neutrophil recruitment into the bronchoalveolar lavage (14) and eosinophilic and basophilic degranulation products in nasopharyngeal secretions whose levels correlate with disease severity (20, 31, 53).
Because RSV is a mucosally restricted virus, the airway epithelium is thought to play an important role in initiating airway inflammation. Here RSV replication induces expression and secretion of cytokines (16, 21, 44), chemokines (6, 47, 57), arachidonic acid metabolites (22), reactive oxygen species (11), and cell surface display of major histocompatibility complex (MHC) class I (36) and CD18 (46). We recently extended these observations by profiling chemokine expression patterns by using macro- and high-density oligonucleotide arrays (57). These studies suggested that RSV profoundly influenced gene expression profiles in the cells in which it replicates; focused analysis of virus-induced chemokines showed a time-dependent expression of CC (I-309, Exodus-1, TARC, RANTES, MCP-1, MDC, and MIP-1 α/β), CXC (GRO α/β/γ, ENA-78, IL-8, and I-TAC), and CX3C (fractalkine) subclasses.
Promoter analysis studies suggest that NF-κB is likely to play a central role mediating inducible cytokine and chemokine gene expression. For example, we have shown that RSV induces NF-κB translocation through a mechanism involving proteolysis of the RelA-associated inhibitor IκBα through a dual pathway involving the 26S proteasome and an as yet uncharacterized protease (34) stimulated by active viral replication (17, 34) and mediated upstream by activation of the IKK complex (25). Mutation of the NF-κB binding site or inhibition of its translocation blocks RSV-inducible interleukin-8 (IL-8) and RANTES expression (8, 12, 52). However, the spectrum of NF-κB-dependent genes expressed by replicating RSV has not been reported.
To address this, we developed a cell system expressing a nondegradable epitope-tagged IκBα (IκBα Ser32Ala/Ser36Ala, termed Flag-IκBα Mut) whose expression is strongly inhibited after addition of nontoxic concentrations of doxycycline to the culture medium. Stable clonal cell lines expressing the tetracycline-controlled transactivator and IκBα Mut were isolated and screened for doxycycline-dependent expression. Upon doxycycline withdrawal, Flag-IκBα Mut expression potently inhibited NF-κB-dependent transcription in response to the prototypical NF-κB-activating cytokine TNF-α and in response to RSV infection. High-density oligonucleotide arrays were then used to assay for profiles of gene expression in response to RSV infection in the presence of Flag-IκBα Mut versus those induced in its absence. Our data suggest a role for NF-κB in controlling constitutive gene expression and further specifically identify members of the RSV-inducible NF-κB-dependent gene network.
MATERIALS AND METHODS
RSV preparation.
The human Long strain of RSV (A2) was grown in Hep-2 cells and purified by centrifugation on discontinuous sucrose gradients (57). The virus titer of the purified RSV pools was 7.5 to 8.5 log PFU/ml, determined by methylcellulose plaque assay. No contaminating cytokines, including IL-1, TNF-α, IL-6, IL-8, granulocyte-macrophage colony-stimulating factor, and interferon, were found in these sucrose-purified viral preparations (21). Lipopolysaccharide (LPS), assayed by using the Limulus hemocyanin agglutination assay, was also not detected. Virus pools were aliquoted, quick-frozen on dry ice-alcohol, and stored at −70°C until used.
Plasmid construction.
Plasmids pUHD10-4, a plasmid containing seven copies of the tetracycline operator (tetO) upstream of the cytomegalovirus minimal promoter, and pUHD15-1 Neo, a plasmid expressing the tetracycline transactivator (tTA), were gifts of M. Gossen and H. Bujard (University of Heidelberg, Germany) (23). The doxycycline-regulated NF-κB dominant-negative expression plasmid encoding IκBα with serine residues 32 and 36 substituted for alanine (IκBα Mut) under control of tetO sequences was generated in two steps. The pUHD10-4 multiple cloning site was modified to encode a Flag epitope tag downstream of an optimized Kozak initiation site. This plasmid was constructed by inserting the double-stranded oligonucleotide produced by annealing the sequence 5′-AATT CGC CAT ATG GAT TAC AAA GAC GAT GAC GAT ACC ATG GTT AGA TCT GAT ATC G-3′ (initiation codon indicated by double underline, _Bgl_II site indicated by single underline) with 5′-GATCC GAT ATC AGA TCT AAC CAT GGT ATC GTC ATC GTC TTT GTA ATC CAT ATG GC-3′ into _Eco_RI- and _Bam_HI-restricted pUHD10-4 to generate pUHDFLAG.
The IκBα Ser32Ala/Ser36Ala mutation was produced by PCR by using mutagenic primers as previously described by us (12). The IκBα Mut cDNA was isolated containing upstream _Bam_HI and downstream _Hin_dIII (made blunt ended by Klenow polymerase) and ligated into _Bgl_II and _Bam_HI-restricted pUHDFLAG (the _Bam_HI site made blunt ended by Klenow polymerase) to generate the expression vector pUHDFLAG-IκBα Mut for use in transient transfection assays. For stable cell isolation, the Flag-IκBα Mut cDNA was excised, made blunt ended, and cloned into the _Pvu_II site of the plasmid pBI-EGFP plasmid (Clontech, Palo Alto, Calif.). pBI-EGFP-IκBα Mut expresses the enhanced green fluorescent protein (EGFP) and Flag-IκBα Mut from a bidirectional _tetO_-driven promoter for rapid visual screening of stable transfectants.
The firefly luciferase reporter genes driven by the human IL-8 (hIL-8) native promoter, −162/+44 hIL-8/LUC, and the IL-6 native promoter, −310/+25 hIL-6/LUC, have been described (8, 29). Constructs carrying three copies of the angiotensinogen RelA/NF-κB1 binding site or three copies of the IL-6 RelA/NF-κB1 site cloned upstream of an inert TATA box driving expression of firefly luciferase, (APREWT)3-p59rAT/LUC and (IL-6κBE)3-p59rAT/LUC, respectively, have been described (8, 29, 35). pTK-Hyg, encoding the hygromycin resistance marker, was obtained commercially (Clontech, Palo Alto, Calif.). Plasmids were purified by ion exchange chromatography (Qiagen, Chatsworth, Calif.) and sequenced across cloning junctions to verify authenticity prior to transfection.
Cell culture and transfection.
The human cervical epithelioid carcinoma cell line HeLa expressing tTA (HeLa Tet-Off) was constructed earlier (23). HeLa Tet-Off cells were grown in medium containing 90% Dulbecco's modified Eagle's medium, 10% heat-inactivated fetal bovine serum, 4 mM l-glutamine, 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate plus 100 μg of G418, 100 U of penicillin G sodium, and 100 μg of streptomycin sulfate per ml in a humidified atmosphere of 5% CO2.
To establish stable cell lines in which IκBα Mut is regulated by exogenous doxycycline, HeLa Tet-Off cells were cotransfected with pBI-EGFP-IκBα Mut and pTK-Hyg plasmids by using Lipofectamine (Life Technologies, Inc.). At 24 h after transfection, cells were changed into medium containing 250 μg of hygromycin B and 2 μg of doxycycline per ml. Hygromycin-resistant clones were isolated and screened after doxycycline withdrawal by culture in tetracycline-free fetal bovine serum (Clontech) for 7 days. Only strongly fluorescent clones were selected further. These clones were further screened for IκBα Mut expression by Western blotting (with either anti-ΙκBα or anti-Flag antibodies) or by inhibition of TNF-α-stimulated NF-κB binding in electrophoretic mobility shift assays. Only the clones whose expression of IκBα Mut was tightly regulated by doxycycline were selected for further experiments.
For TNF-α stimulation, recombinant TNF-α (rTNF-α) was added directly to the culture medium for the indicated times (25 ng/ml, final concentration). For RSV infection, freshly isolated cells were split into two cultures; one group was maintained in doxycycline, and the other was maintained without for 7 days, a time at which IκBα Mut expression was maximal. Cells were then changed into Dulbecco's modified Eagle's medium containing 2% (vol/vol) fetal bovine serum. Cell monolayers were infected with sucrose cushion-purified RSV at a multiplicity of infection of 1.
For transient transfections, cells freshly plated into triplicate 60-mm plates were transfected by using Lipofectamine (Life Technologies, Inc.) with 8 μg of indicated luciferase reporter plasmid and 3 μg of the constitutive alkaline phosphatase internal control plasmid pSV2PAP (35). Cells were cultured for an additional 40 h and, where indicated, stimulated with rTNF-α for the indicated times. For reporter assays, cells were harvested 6 h after rTNF-α stimulation, cytoplasmic lysates were prepared, and luciferase activity was measured (35). As an internal control for transfection efficiency, alkaline phosphatase activity was measured in 50 μg of cell lysate as described previously (35). Fold induction of reporter activity was calculated by dividing the mean normalized luciferase activity from three treated cultures by the mean normalized luciferase activity from three untreated cultures.
Preparation of subcellular extracts.
Cells were washed two times with phosphate-buffered saline and disrupted by hypotonic nonionic detergent lysis by addition of 2 cell volumes of buffer A (50 mM HEPES [pH 7.4], 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol plus 0.1 mg of phenylmethylsulfonyl fluoride, 1 μg of pepstatin A, 1 μg of leupeptin, 10 μg of soybean trypsin inhibitor, and 10 μg of aprotinin per ml, and 0.5% IGEPAL CA-630 [(octylphenoxy)polyethoxyethanol]). After 10 min on ice, the nuclei were pelleted by centrifugation at 4,000 × g for 4 min at 4°C (the supernatant constituting the cytoplasmic extract). The nuclear pellets were further purified by centrifugation through a discontinuous sucrose cushion by resuspending in buffer B (1.7 M sucrose, 50 mM HEPES [pH 7.4], 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol plus 0.1 mg of phenylmethylsulfonyl fluoride, 1 μg of pepstatin A, 1 μg of leupeptin, 10 μg of soybean trypsin inhibitor, and 10 μg of aprotinin per ml), and centrifuging at 12,000 × g for 10 min at 4°C.
Proteins in the pelleted nuclei were extracted by incubating in buffer C (10% glycerol, 50 mM HEPES [pH 7.4], 400 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol plus 0.1 mg of phenylmethylsulfonyl fluoride, 1 μg of pepstatin A, 1 μg of leupeptin, 10 μg of soybean trypsin inhibitor, and 10 μg of aprotinin per ml) with frequent vortexing for 30 min at 4°C. After centrifugation at 12,000 × g for 30 min at 4°C, the supernatant was frozen at −70°C. With this technique, the nuclear fractions are free of cytoplasmic contamination and the cytoplasmic fractions lack nuclear markers (9). Both cytoplasmic and nuclear extracts were normalized for protein concentration by using bovine serum albumin as a standard (Bio-Rad, Hercules, Calif.).
Western immunoblot.
For Western immunoblot, a constant amount of cytoplasmic or nuclear extracts (200 to 300 μg as indicated) was boiled in Laemmli buffer, fractionated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE), and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) in 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS)-methanol (35). Membranes were blocked in 5% milk-Tris-buffered saline (TBS)-0.1% Tween for 1 h and immunoblotted with either anti-Flag M2 monoclonal antibody-peroxidase conjugate (Sigma Chemical, no. A8592) or affinity-purified rabbit polyclonal antibody anti-ΙκBα (Santa Cruz Biotechnology, Santa Cruz, Calif.) for 1 h at 4°C. Membranes were washed four times in TBS-0.1% Tween 20. Immune complexes were detected by reaction in the enhanced chemiluminescence assay (ECL; Amersham) according to the manufacturer's recommendations. Protein loading controls were performed by probing the blot with monoclonal antibody to β-actin (9).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (9) with minor modifications. Nuclear extracts (15 μg) were incubated with 40,000 cpm of 32P-labeled APRE WT duplex oligonucleotide probe and 2 μg of poly(dA-dT) in a buffer containing 8% glycerol, 100 mM NaCl, 5 mM MgCl2, 5 mM dithiothreitol, and 0.1 μg of phenylmethylsulfonyl fluoride per ml in a final volume of 20 μl for 20 min at room temperature. The complexes were fractionated on 6% or 7% native polyacrylamide gels run in 1× TBE buffer (89 mM Tris, 89 mM boric acid, and 2.0 mM EDTA), dried, and exposed to Kodak X-AR film at 70°C. Competition was performed by the addition of a 100-fold molar excess of nonradioactive double-stranded oligonucleotide competitor at the time of addition of radioactive probe. The sequences of the APRE double-stranded oligonucleotides are shown below:
APRE WT: GATCCACCACAGTTGGGATTTCCCAACCTGACCAGTGGTGTCAACCCTAAAGGGTTGGACTGGTCTAGAPRE M6: GATCCACCACATGTTGGATTTCCGATACTGACCAGTGGTGTACAACCTAAAGGCTATGACTGGTCTAG
Antibody supershift assays were performed by adding to the binding reaction 1 μl of affinity-purified polyclonal antibodies and incubating for 1 h on ice. All of the antibodies used in these assays were obtained commercially (Santa Cruz Biotechnology).
RNase protection assay.
Steady-state mRNA levels in RSV-infected cells were analyzed by RNase protection assay by using the RiboQuant multiprobe kit (PharMingen, San Diego, Calif.) following the manufacturer's recommendations. In brief, 32P-labeled specific antisense RNA probes were synthesized by in vitro transcription from multiple DNA template sets. An equal amount of total RNA (15 μg per sample) was hybridized overnight to the 32P-labeled RNA probes and digested with RNases A and T1. The protected cRNA probes were precipitated with ethanol and resolved on QuickPoint precast denaturing polyacrylamide gels (Novex, San Diego, Calif.). The gel was dried and then exposed to X-ray film. The abundance of each mRNA transcript was quantified on a PhosphorImaging screen and normalized to housekeeping L32 transcripts in the same reaction.
Northern blot.
Total cellular RNA was extracted by acid guanidium-phenol extraction (Tri Reagent; Sigma). RNA (20 μg) was denatured, fractionated by electrophoresis on a 1.2% agarose-formaldehyde gel, capillary transferred to a nitrocellulose membrane (Zeta-Probe GT; Bio-Rad), and prehybridized as described previously (8). The 1.1-kb RSV N cDNA probe was produced by PCR by using poly(A)-primed cDNA as a template from RSV-infected A549 cells by using the upstream primer 5′-CAAATGGATCCATGGCTCTTAGCAAAGTCAAG-3′ and the downstream primer 5′-TTCCCGGTTCAAAGCTCTACATCATTATC-3′. The membrane was hybridized with 1 × 106 to 2 × 106 cpm of 32P-RSV-N cDNA probe per ml at 60°C overnight in 5% sodium dodecyl sulfate (SDS) hybridization buffer. The membrane was washed with a buffer containing 5% SDS and 1× saline-sodium citrate (0.15 M NaCl and 0.015 M sodium citrate) for 20 min at room temperature, followed by 30 min at 60°C. Internal control hybridization was performed with 18S RNA. The membrane was exposed to XAR film (Kodak) for 24 to 48 h at 70°C and quantified by exposure to PhosphorImager cassette.
Measurement of apoptotic index.
DNA fragmentation was assayed by using a commercially available enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). This assay detects small nucleosomal fragments in the cytoplasmic fractions of affected cells that arise during apoptosis but not as a result of necrosis. After the cells were infected with RSV for the indicated time period, cells were harvested by trypsinization, and 5 × 105 cells were lysed for 30 min at room temperature and cleared of cellular debris for 20 min at 200 × g. The resulting cytoplasmic fraction was used as a source of nucleosomal fragments. Following incubation of this supernatant with capture antibodies and detection reagent, the absorbance was measured at 405 nm by Multiskan (model MCC/340; Titertek Instruments, Inc., Irvine, Calif.) and compared with substrate solution as a blank. The apoptotic index was calculated according to the manufacturer's instructions by dividing the absorbance of stimulated cells by the absorbance of control cells.
Oligonucleotide probe-based microarrays.
Hu95A GeneChip (Affymetrix Inc., Santa Clara, Calif.) containing 12,626 sequenced human genes was used as previously described (57). Briefly, first-strand cDNA synthesis was performed by using total RNA (10 to 25 μg), a T7-(dT)24 oligomer [5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′] and SuperScript II reverse transcriptase (Life Technologies). The T7 promoter introduced during first-strand cDNA synthesis is then used to direct the synthesis of cRNA by using bacteriophage T7 RNA polymerase. The biotin-labeled target RNAs are frag-mented to a mean size of 200 bases and initially hybridized to a test array containing a set of probes representing genes that are commonly expressed in the majority of cells (actin, glyceraldehyde-3-phosphate dehydrogenase [GAPDH], transferrin receptor, transcription factor ISGF-3, 18S RNA, 28S RNA, and alu) to confirm their successful labeling.
Hybridization was performed at 45°C for 16 h in hybridization buffer (0.1 M morpholineethanesulfonic acid [MES, pH 6.6], 1 M NaCl, 0.02 M EDTA, and 0.01% Tween 20). Four prokaryotic genes (bioB, bioC, and bioD from the Escherichia coli biotin synthesis pathway and cre, the recombinase gene from bacteriophage P1) were added to the hybridization cocktail as internal controls. Arrays were washed by using both nonstringent (1 M NaCl, 25°C) and stringent (1 M NaCl, 50°C) conditions prior to staining with phycoerythrin-streptavidin (10 μg/ml final). Gene Chip arrays were scanned by using a Gene Array Scanner (Hewlett Packard) and analyzed by using the Gene Chip Analysis Suite 3.3 software (Affymetrix Inc.). For each gene, 16 to 20 probe pairs were immobilized as ∼25-mer oligonucleotides that hybridized throughout the mRNA; each probe pair was represented as a perfect match oligonucleotide and a mismatch oligonucleotide as a hybridization control.
The average intensity of each probe cell was calculated after subtraction of the local background (the lowest 2% intensity of each sector; each probe cell is divided into 16 sectors). The normalized average intensity value is used to determine the number of positive and negative probe pairs. Based on the positive/negative ratio, positive fraction and log average ratio of the perfect match to mismatch, the absolute call [e.g., the gene is detected (present) or not (absent)] is determined (57, 58). Finally the average difference is determined by calculating the difference intensity between the perfect match and mismatch of every probe pair and averaging the differences over the entire probe set.
Data analysis.
Three independent experiments were performed identically, including control without doxycycline treatment, control with doxycycline treatment, RSV 12-h infection without doxycycline treatment, and RSV 12-h infection with doxycycline treatment. For comparison of the fluorescent intensity (average difference) values among multiple experiments, the average difference values for each “experimental” GeneChip were scaled to that of the “base” GeneChip. This was done first by calculating the “2% trimmed mean” (a measurement of global signal intensity) for each GeneChip. The trimmed mean is obtained by calculating the mean signal intensity of the chip after discarding the top and bottom 2% average difference values (representing the outliers). Next, scaling was performed by multiplying all the average differences in the “experimental” array by a scaling factor defined as the ratio of the base trimmed mean to the experimental trimmed mean (the base array was defined to be the control plus doxycycline GeneChip).
Because both RSV and doxycycline can be considered experimental treatments, the scaled average difference values were then subjected to a two-way analysis of variance (ANOVA; Splus 6; Insightful Inc.) with replications to determine which genes were significantly influenced by either the RSV or doxycycline treatment. Genes (probe sets) with a P value [Pr(F)] at the <0.05 confidence level as a result of either treatment were deemed highly significant and were selected for further analysis. Agglomerative hierarchical clustering by using the unweighted pair-group method with arithmetic mean (43) was performed on the indicated genes (Spotfire Array Explorer, v. 3.01; Spotfire Inc., Cambridge, Mass.). The data are graphically presented as heat maps in which fluorescence intensity is represented by a color gradient. For the heat maps shown, green represents the minimum average difference value (5 scaled units), black represents the mid average difference value (5,000 scaled units), and red represents the maximum average difference value (10,000 scaled units). Investigators may obtain the primary data at our website http://bioinfo.utmb.edu/Brasier_Lab/.
RESULTS
To identify the gene network under control of the NF-κB transcription factor in the setting of RSV replication, we examined methods for expressing dominant negative inhibitors that would be suitable for high-density microarray analysis. Although others have had success with transducing cells with replication-deficient adenovirus encoding the nonproteolyzed IκBα Mut (52), we sought alternative approaches as adenovirus infection may have significant confounding effects on host cell gene expression or unrecognized interactions with RSV replication or signaling. Additionally, isolating a cell line constitutively expressing an NF-κB inhibitor would be problematic as we and others have shown that NF-κB is required for cell survival (4, 41).
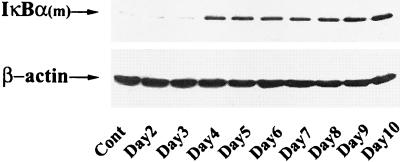
For these reasons, we employed a tightly regulated system in which expression of the nonproteolyzable IκBα Mut could be controlled by exogenously added tetracycline (doxycycline). Clonal isolates of Flag epitope-tagged IκBα Mut under tetO control selected by hygromycin resistance in HeLa cells containing the tetracycline transactivator were selected and characterized. To determine the kinetics of IκBα Mut expression, cells were withdrawn from doxycycline for various times from 0 to 7 days, and cytoplasmic lysates were prepared. Figure 1 shows a Western immunoblot of IκBα Mut expression detected by probing with monoclonal antibody to the Flag epitope. Undetectable in control cells, steady-state levels of the specific ∼42-kDa Flag-IκBα Mut protein plateaued 4 days after doxycycline withdrawal, and its expression remained constant for the duration of the experiment. These data suggested that Flag-IκBα Mut expression is tightly controlled and would be suitable for further analysis.
FIG. 1.
Time-course of Flag-IκBα Mut expression after doxycycline deprivation. IκBα Mut-expressing cells isolated in the presence of doxycycline (2 μg/ml in growth medium) were transferred to doxycycline-free medium. At the indicated times, cytoplasmic extracts were prepared, and 150 μg was fractionated on SDS-10% PAGE and subjected to Western immunoblot by using anti-Flag M2 monoclonal antibody-peroxidase conjugate. After Flag detection, the immunoblot was reprobed for β-actin as a loading control.
Flag-IκBα Mut is a potent inhibitor of cytokine-inducible NF-κB activation.
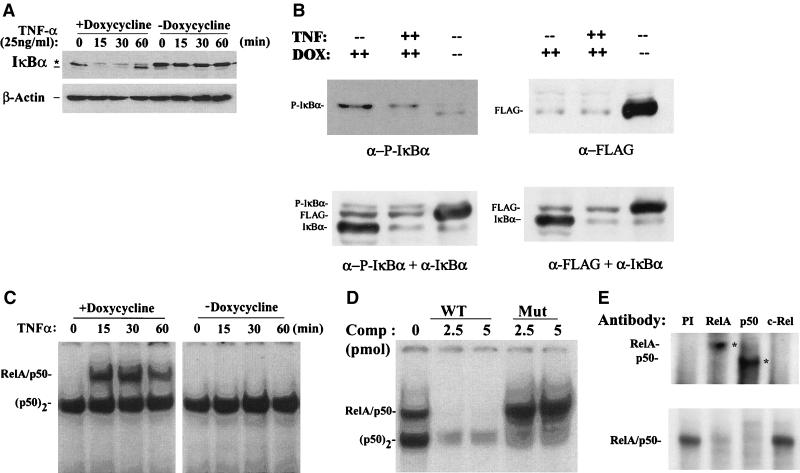
To determine whether a sufficient level of Flag-IκBα Mut was produced to inhibit NF-κB activation, freshly isolated cells were divided and cultured for 7 days in the absence or presence of doxycycline (to allow maximal accumulation of Flag-IκBα Mut). Individual plates were then briefly stimulated with rTNF-α (25 ng/ml), a rapid and potent inducer of IκBα proteolysis and NF-κB binding (8). Western immunoblots with polyclonal antibody to IκBα detected the relative abundance of both endogenous and mutant IκBα (Fig. 2A). In cells maintained in the presence of doxycycline, endogenous ∼39-kDa IκBα was detected, and in this experiment, no appreciable levels of the ∼42-kDa IκBα Mut were detected. Within 15 min of stimulation, IκBα was completely proteolyzed; concomitantly we noted the appearance of a slower migrating IκBα-specific band.
FIG. 2.
Effect of doxycycline withdrawal on rTNF-α-induced IκBα proteolysis and NF-κB binding. (A) Time course of IκBα degradation after rTNF-α stimulation. Flag-IκBα Mut-expressing cells were cultured with or without doxycycline for 7 days. The cells were then treated without (lane 0) or with rTNF-α (25 ng/ml) for the indicated times (minutes). Top panel, cytoplasmic extracts of the cells were prepared and subjected to Western immunoblot with anti-IκBα antibody. ∗, migration of phospho-IκBα and Flag-IκBα Mut; ---, migration of endogenous IκBα. Bottom panel, similar protein loading was confirmed by stripping the blot and reprobing with anti-β-actin. (B) Analysis of IκBα isoforms in doxycycline-treated cells. Cytoplasmic lysates corresponding to lanes 1, 2, and 5 from panel A were fractionated and probed with anti-phospho-specific IκBα and anti-Flag alone and in combination with anti-IκBα. Although the Flag-IκBα Mut protein migrated slower than the native IκBα, it was clearly separable from phospho-IκBα and represents the slower migrating band observed in A. (C) Time course of NF-κB binding. Sucrose cushion-purified nuclear extracts were prepared from the same experiment and subjected to EMSA by using 32P-labeled APRE WT duplex oligonucleotide probe. The complexes were fractionated on 6% native polyacrylamide gels; an autoradiographic exposure is shown. The relative migration of the RelA/NF-κB1 (p50) heterodimer and the p50 homodimer (p50)2 complexes are indicated (34). (D) Competition assay in EMSA. Nuclear extracts from rTNF-α-stimulated cells (15 min, cultured in the presence of doxycycline) were subjected to competition analysis by the addition of a molar excess of nonradioactive double-stranded oligonucleotide competitor at the time of addition of radioactive probe. Competitors used were APRE WT and APRE M2 (Mut). (E) Antibody supershift in EMSA. Nuclear extracts from rTNF-α-stimulated cells (15 min, plus doxycycline) were mixed with 1 μl of affinity-purified polyclonal antibodies for 1 h at 4°C prior to EMSA analysis. PI, preimmune IgG. Bottom panel, brief autoradiographic exposure of the native NF-κB binding complexes. Top panel, longer exposure to detect the supershifted complexes (indicated by asterisk). Addition of either anti-RelA or anti-NF-κB1(p50) antibodies completely disrupted the RelA/NF-κB1(p50) complex.
After 60 min of stimulation, IκBα was resynthesized, due to the NF-κB activation of the endogenous IκBα promoter in a process termed the NF-κB-IκB autoregulatory feedback loop (28). Conversely, in unstimulated cells cultured in the absence of doxycycline, the level of Flag-IκBα Mut expression exceeded that of endogenous IκB and the level of endogenous IκBα was apparently reduced (compare lane 5 with lane 1, Fig. 2A). After rTNF-α stimulation, the Flag-IκBα Mut was not significantly proteolyzed, although the endogenous IκBα disappeared.
Additional experiments were performed to identify the slower migrating IκBα-specific band in the doxycycline-treated cells (Fig. 2B). Cytoplasmic extracts from cells cultured in the presence of doxycycline in the absence or presence of rTNF-α were cofractionated with cytoplasmic extracts of cells cultured in the absence of doxycycline (as a source of the Flag-IκBα Mut antigen). These blots were probed with antibodies directed to phospho-specific IκBα and Flag alone and in combination with anti-IκBα. We observed that only a small fraction of the total IκBα pool whose pseudo-steady-state levels did not appreciably change after rTNF-α treatment was phosphorylated (Fig. 2B, lower left panel). Also, we noted that the phospho-IκBα migrated at a distinct location from the slower migrating Flag-IκBα Mut band detected with the anti-Flag and anti-IκBα antibodies in the doxycycline-treated cells (Fig. 2B).
We interpret these data to mean that a small amount of Flag-IκBα Mut can be detected in cells cultured in the presence of doxycycline. More importantly, the data in Fig. 2A indicate that Flag-IκBα Mut was tightly regulated, producing steady-state levels that exceeded endogenous IκBα after doxycycline withdrawal. Finally, the observations in doxycycline-deprived cells that the abundance of endogenous IκBα was reduced in unstimulated cells and was not resynthesized 60 min after stimulation suggested that Flag-IκBα Mut disrupted NF-κB-dependent target gene activation.
We next analyzed the effect of Flag-IκBα Mut expression on inducible NF-κB binding. For this, nuclear extracts from the experiment in Fig. 2A were subjected to EMSA. As we have described before, rTNF-α is a potent and rapid inducer of nuclear translocation and DNA binding of the NF-κB heterodimer RelA/NF-κB1(p50), whose characteristic migration could be distinguished from constitutive NF-κB isoforms by nondenaturing electrophoresis (8, 21, 34). The kinetics of RelA/NF-κB1 DNA binding exactly parallels the pattern of IκBα degradation in cells cultured in the presence of doxycycline (Fig. 2C). In contrast, no inducible RelA/NF-κB1 complexes could be detected in nuclear extracts from cells cultured in the absence of doxycycline (Fig. 2C).
Confirmation of the identity of the inducible RelA/NF-κB1 complexes was determined in competition analysis in EMSA (Fig. 2D). Here, the inclusion of unlabeled wild-type probe strongly competed for inducible RelA/NF-κB1 DNA binding, whereas the mutant did not. The constitutive NF-κB1 homodimer (p50)2 competes with lower affinity, being partly inhibited with the mutant probe (9). Supershift analysis in EMSA was further used to identify the presence of RelA in the rTNF-α-inducible NF-κB binding complex. Addition of anti-RelA or anti-p50 completely disrupted the RelA/p50 complex and produced a corresponding supershift (Fig. 2E). These data are consistent with our earlier demonstrations that the predominant rTNF-α-inducible NF-κB complex is composed of RelA/NF-κB1 in epithelial cells (8, 21). Together, these data indicate that Flag-IκBα Mut completely blocks inducible RelA/NF-κB1 binding in response to the potent cytokine TNF-α.
Flag-IκBα Mut is a potent inhibitor of RSV-inducible NF-κB activation.
Although the Ser32Ala/Ser36Ala site mutation is a potent inhibitor of cytokine-inducible IκBα proteolysis through the IKK-proteasome pathway, we have observed significant differences between RSV- and cytokine-inducible IκBα proteolysis. For example, the kinetics of RSV-inducible IκBα degradation is distinctly slower than that of rTNF-α (being detectable after 12 to 24 h) and is only weakly blocked by pretreatment with specific proteasome inhibitors, and no significant IκBα resynthesis occurs (34). Consequently, we determined whether Flag-IκBα Mut would interfere with RSV-inducible NF-κB activation.
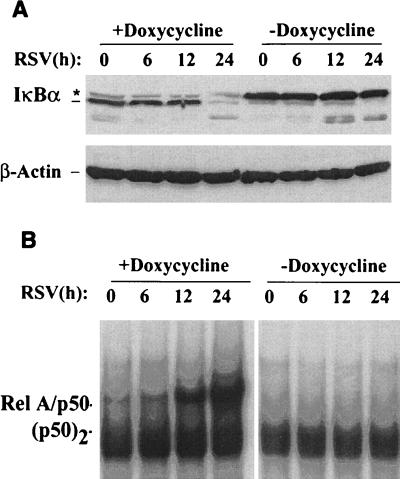
Freshly isolated Flag-IκBα Mut-expressing cells were divided and cultured for 7 days in the absence or presence of doxycycline. Individual plates were then infected with sucrose cushion-purified RSV (multiplicity of infection, 1), after which cytoplasmic and nuclear extracts were prepared. Western immunoblots of the cytoplasmic extracts for IκBα detected a fourfold greater relative abundance of endogenous IκBα relative to that of the Flag-IκBα Mut (Fig. 3A). After RSV infection, a significant amount of IκBα proteolysis was detected 24 h later, a time course consistent with our earlier descriptions (34). In cells maintained in the presence of doxycycline, like that seen in Fig. 2, loss of endogenous ∼39-kDa IκBα was noted, and no significant degradation of the Flag-IκBα Mut occurred at 24 h. These data suggest that RSV degrades Flag-IκBα Mut less efficiently than the endogenous form.
FIG. 3.
Effect of Flag-IκBα Mut expression on RSV-induced IκBα proteolysis and NF-κB binding. (A) Western immunoblot for cytoplasmic IκBα levels. Flag-IκBα Mut-expressing cells were cultured in the presence or absence of doxycycline for 7 days. The cells were infected without (lane 0) or with purified RSV (multiplicity of infection of 1) for the indicated times (hours) prior to harvest. Cytoplasmic extracts of the cells were analyzed by Western immunoblot for IκBα by using affinity-purified anti-IκBα antibodies. ∗, migration of phospho-IκBα and Flag-IκBα Mut; ---, migration of endogenous IκBα. Bottom panel, similar protein loading was confirmed by stripping the blot and reprobing with anti-β-actin. (B) Time course of NF-κB binding. Sucrose cushion-purified nuclear extracts were subjected to EMSA as in Fig. 2B except twice as much nuclear protein was used to detect the relatively weaker induction of DNA binding. The positions of the RelA/NF-κB1 (p50) heterodimer and the p50 homodimer (p50)2 complexes are indicated (34). The RSV-inducible complexes have composition and binding specificity identical to those of the rTNF-α-inducible ones (34).
Nuclear extracts were subjected to EMSA to detect the effect of Flag-IκBα Mut expression on RSV-inducible NF-κB binding. In cells cultured in the presence of doxycycline, significant RelA/NF-κB1 DNA binding was observed at 12 h and continued to increase in abundance 24 h after RSV infection (Fig. 3B). Conversely, RelA/NF-κB1 DNA binding was completely abolished at all points in the cells cultured in the absence of doxycycline (Fig. 3B). These data suggest that Flag-IκBα Mut is also a potent inhibitor of RSV-inducible RelA/NF-κB1 binding.
Flag-IκBα Mut blocks NF-κB-dependent promoter transactivation.
We next determined whether Flag-IκBα Mut expression was sufficient to interfere with NF-κB-dependent transcription. For this, several strategies were employed. We first determined whether Flag-IκBα Mut expression interfered with inducible activity of a purely NF-κB-dependent reporter gene. Multimeric RelA/NF-κB1 binding sites from either the angiotensinogen (AGT) gene or the IL-6 gene cloned upstream of the inert rAGT minimal promoter driving firefly luciferase [termed (APREWT)3-p59rAT/LUC and (IL-6κBE)3-p59rAT/LUC, respectively] (29, 35) were transfected into Flag-IκBα Mut-expressing cells maintained in the absence or presence of doxycycline.
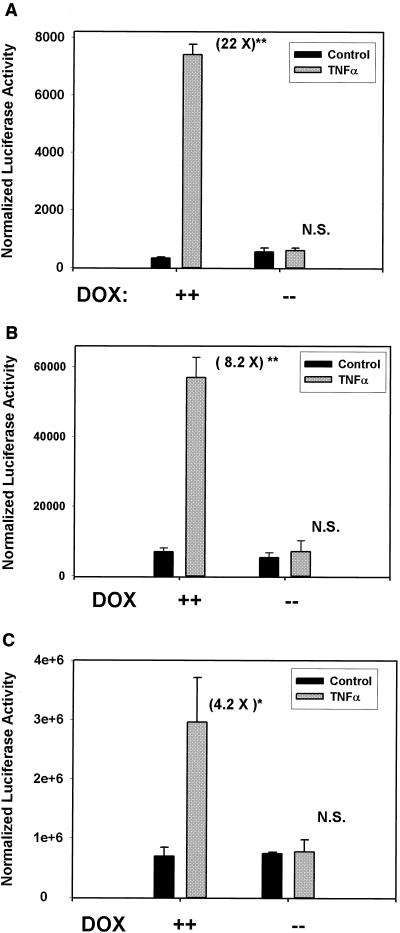
Figure 4A shows the results of a representative transfection of (APREWT)3-p59rAT/LUC in the absence or presence of acute stimulation with rTNF-α. In the presence of doxycycline, rTNF-α induced a 22-fold increase in normalized luciferase reporter activity (P < 0.05, n = 3 independent experiments); however, in the absence of doxycycline, no statistically significant induction could be recorded (a 1.1-fold increase). Similar results were obtained for the (IL-6κBE)3-p59rAT/LUC reporter activity, in which an 8.2-fold increase was observed relative to the control in the presence of doxycycline and a 1.3-fold increase was observed in its absence (Fig. 4B).
FIG. 4.
Effect of Flag-IκBα Mut expression on NF-κB-dependent transcription. (A) Flag-IκBα Mut-expressing cells were transiently transfected with (APREWT)3-p59rAT/LUC reporter vector and the alkaline phosphatase-internal control plasmid pSV2PAP in the absence or presence of doxycycline as indicated. Cells were cultured for an additional 40 h after transfection and stimulated with rTNF-α (25 ng/ml, 6 h). Normalized luciferase activity is presented for a representative transfection. The number in parentheses corresponds to the fold induction of reporter activity from the TNF stimulation relative to unstimulated cells. ∗∗, P < 0.001, Student's t test, compared to control, n = 3 independent experiments. (B) Experiment as in A in which (IL-6κBE)3-p59rAT/LUC reporter was used. (C) The cells were transfected with the native −162/+44 hIL-8/LUC. Experiment and data analysis are as in A. ∗, P < 0.05, Student's t test, compared to control.
Because (APREWT)3-p59rAT/LUC and (IL-6κBE)3-p59rAT/LUC are artificial constructs taken from genes not normally expressed by HeLa cells, we determined if Flag-IκBα Mut expression would block transcriptional activation of a native chemokine gene by testing its effect on the IL-8 promoter, a highly RSV-inducible gene in epithelial cells. We selected this target because we have previously demonstrated that rTNF-α activates hIL-8 transcription in a manner requiring RelA/NF-κB1 binding to a site in its proximal promoter (8). Flag-IκBα Mut-expressing cells maintained in the absence or presence of doxycycline were transfected with −162/+44 hIL-8/LUC, and reporter activity was determined after rTNF-α stimulation. Flag-IκBα Mut expression completely inhibited reporter activity driven by −162/+44 hIL-8/LUC (Fig. 4C).
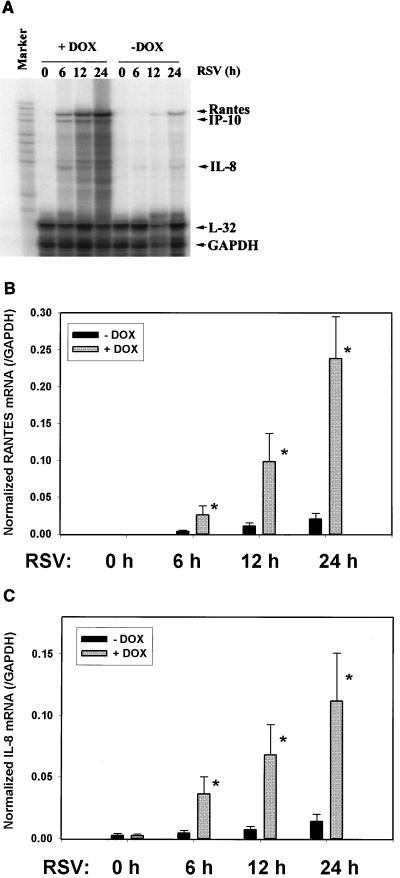
To ensure that Flag-IκBα Mut expression would interfere with endogenous gene expression of NF-κB-dependent promoters in the setting of RSV infection, we performed multiprobe RNase protection assays in Flag-IκBα Mut-expressing cells maintained in the absence or presence of doxycycline. In cells not expressing Flag-IκBα Mut, RSV infection induced RANTES, IP-10 (interferon-inducible protein of 10 kDa), and IL-8 expression in a time-dependent fashion (Fig. 5A). By contrast, in cells expressing Flag-IκBα Mut, RANTES, IP-10, and IL-8 were significantly reduced after RSV infection.
FIG. 5.
Effect of Flag-IκBα Mut expression on RSV-inducible chemokine expression. Flag-IκBα Mut-expressing cells were cultured with or without doxycycline for 7 days. Cells were infected with purified RSV (multiplicity of infection, 1) for indicated times (in hours) prior to harvest of total cellular RNA. mRNA abundance was determined in each sample by multiprobe RNase protection assay. Shown is a representative autoradiographic exposure after denaturing gel electrophoresis. Left lane is undigested input probe (marker). Locations of protected fragments for RANTES, IP-10, IL-8, and the two housekeeping genes L32 and GAPDH are indicated at right. (B) Time course of normalized RANTES mRNA abundance (relative to GAPDH) was determined by exposure of the gel to a PhosphorImager cassette and plotted as a function of the duration of RSV infection (in hours). Errors are standard deviations from three independent experiments. ∗, P < 0.05, Student's t test, compared to no doxycycline treatment for each time point. (C) Time course of normalized IL-8 mRNA abundance (relative to GAPDH) was determined as in B.
Normalized values for RANTES and IL-8 were determined in three independent experiments and plotted in Fig. 5B and C. At all times after 6 h of RSV infection, Flag-IκBα Mut expression induced a statistically significant reduction in chemokine expression. We noted that the Flag-IκBα Mut-mediated inhibition of IL-8 mRNA expression exactly paralleled the behavior of the native IL-8 promoter in transient transfection assays (Fig. 4C). These data suggest that the cell system expressing Flag-IκBα Mut was indeed tightly regulated and could be used to block RSV-inducible NF-κB-dependent expression of gene networks.
Flag-IκBα Mut expression does not interfere with RSV transcription or cellular viability.
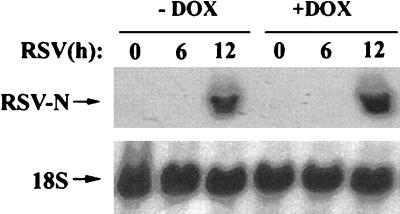
We and others have previously shown that RSV transcription/replication is required for IκB proteolysis (34), NF-κB binding (17, 34), and IL-8 (21) or RANTES (12) expression. To ensure that Flag-IκBα Mut expression does not alter RSV transcription, Northern blots for RSV nucleocapsid (N) were performed in cells treated with or without doxycycline. In this experiment, total cellular RNA was assayed for N expression in RSV-infected cells 0 to 12 h after RSV infection [a time during which the first round of viral transcription occurs (39)]. As shown in Fig. 6, RSV N expression was strongly detected 12 h after RSV infection irrespective of whether cells were treated with or without doxycycline. These data suggest that Flag-IκBα Mut expression does not interfere with transcription of this cytoplasmic paramyxovirus. Finally, changes in the apoptotic index for the Flag-IκBα Mut-expressing cells were compared for the first 24 h of infection. No significant differences were observed for doxycycline-treated or untreated cells relative to controls (data not shown). These data indicate that, at least for these short-term experiments, Flag-IκBα Mut did not artifactually influence the RSV life cycle or alter cellular viability.
FIG. 6.
Effect of Flag-IκBα Mut expression on RSV transcription. Flag-IκBα Mut-expressing cells were cultured with or without doxycycline for 7 days and infected with purified RSV (multiplicity of infection, 1) for the indicated times (in hours). Top, total cellular RNA was extracted, and abundance of RSV N transcript was determined by Northern blot (shown is a representative autoradiographic exposure). Bottom, the blot was reprobed with 18S RNA as an RNA recovery marker.
Identification of NF-κB-dependent gene networks by high-density microarrays.
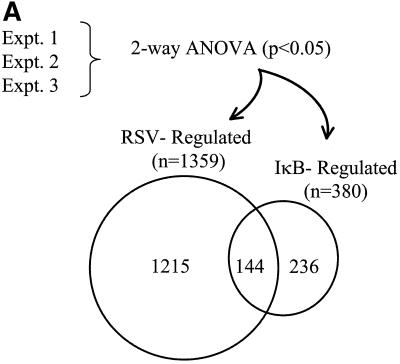
To identify NF-κB-dependent genes, Flag-IκBα Mut-expressing cells were cultured in the presence or absence of doxycycline for 7 days prior to infection with RSV. We harvested RNA from the cells after 12 h, a time at which NF-κB activation (see Fig. 3) and chemokine expression (Fig. 5) were easily detectable, and profiled gene expression changes by using high-density oligonucleotide arrays containing ∼12,696 sequenced human genes (Affymetrix Hu95A GeneChip). Two-way ANOVA was used to identify genes whose expression levels were statistically significantly altered by the treatment. RSV infection significantly changed the abundance of 1,359 mRNAs present on the GeneChip (Fig. 7A) . Conversely, Flag-IκBα Mut expression produced significant changes in gene expression profiles for a significantly smaller number of genes (a total of 380). Comparison of the two groups found only 144 genes common to the two treatments (Fig. 7A).
FIG. 7.
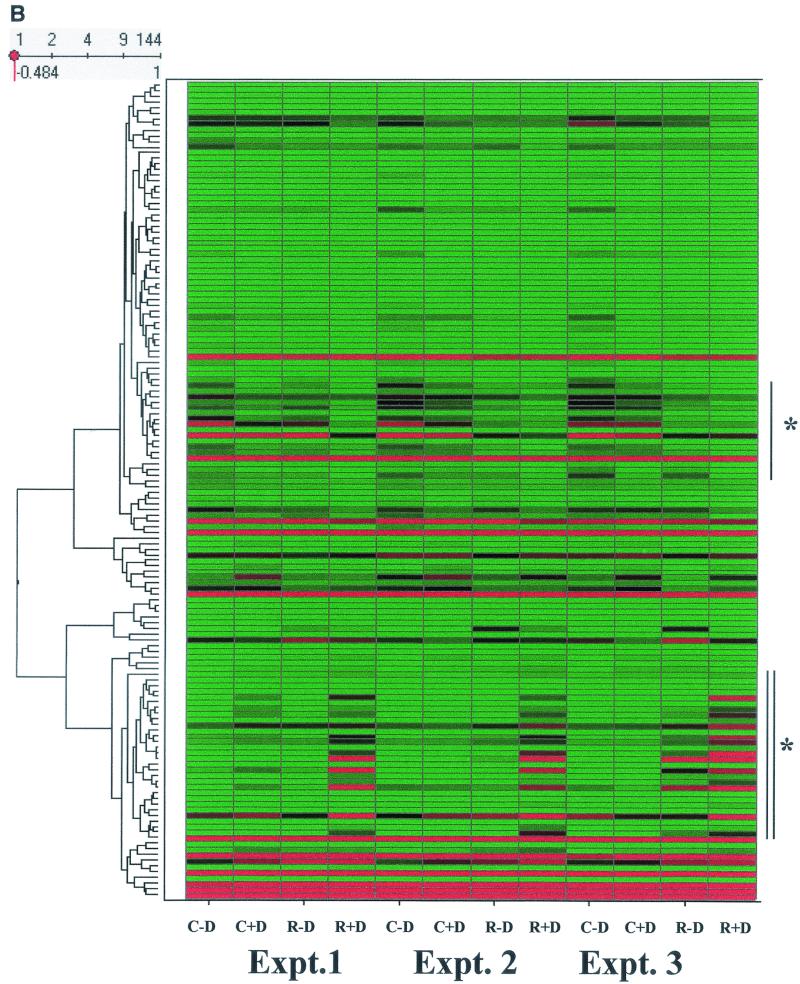
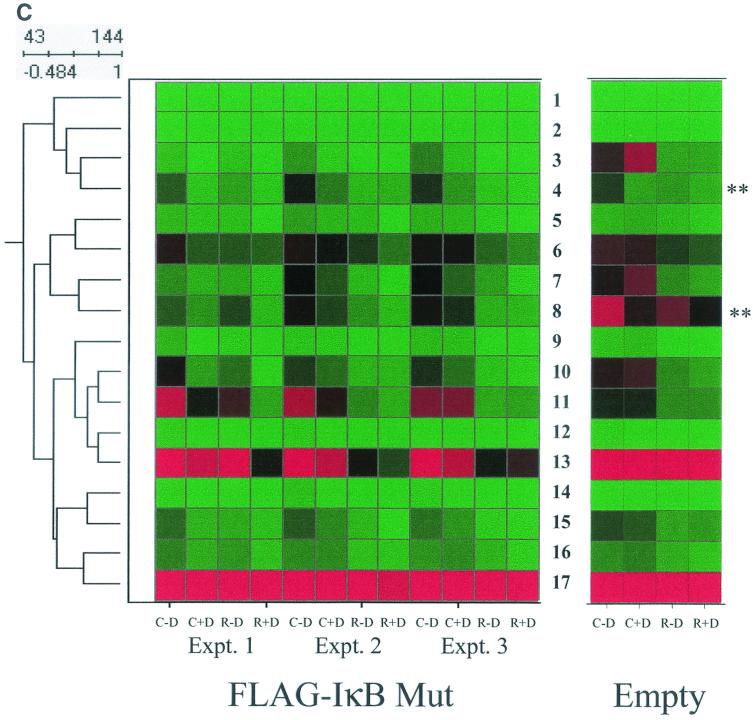
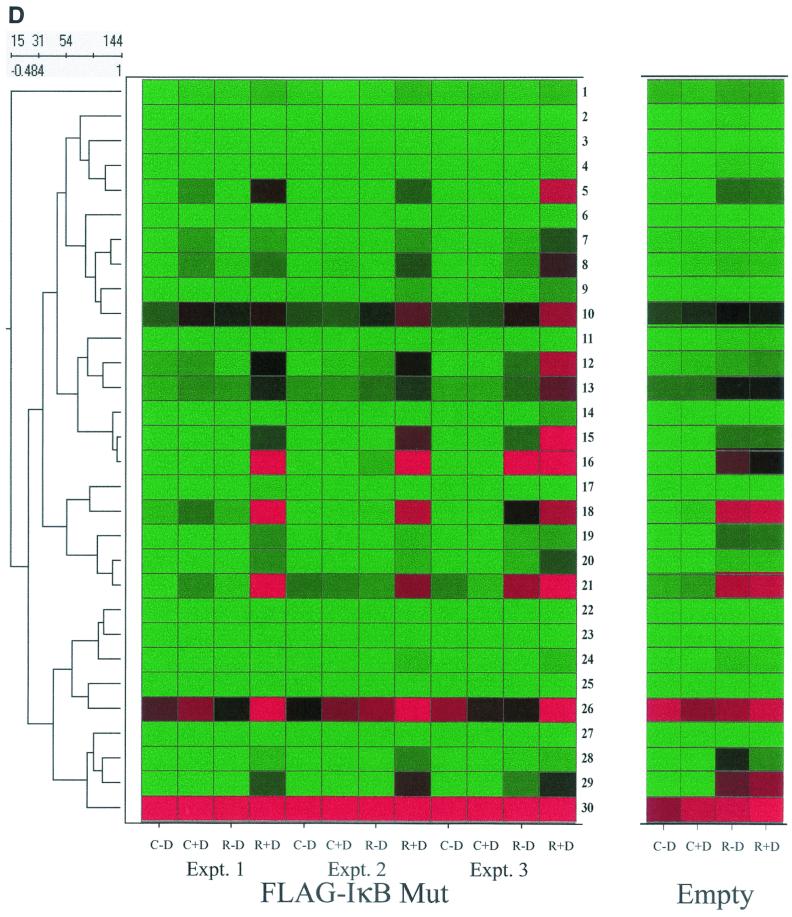
NF-κB-dependent gene networks identified by high-density microarrays. (A) Analysis of RSV- and doxycycline-regulated genes. Two-way ANOVA was used to analyze the data set from three independent array experiments corresponding to control (no doxycycline treatment), control (with doxycycline treatment), RSV 12-h infection (no doxycycline treatment), and RSV 12-h infection (with doxycycline treatment) and genes with a P value [Pr(F)] of <0.05 as a result of either treatment were compared. Shown is a Venn diagram of genes common to both datasets. (B) Clustering and heat map analysis of the RSV- and Flag-IκBα Mut-regulated data set. Agglomerative hierarchical clustering was performed by using the unweighted pair-group method with arithmetic mean technique (see text). A heat map for each gene for the three independent experimental data points is shown at right. For each experiment, the treatment conditions are given as control (C), doxycycline treated (D), and RSV infected (R). Green represents the minimum value of 5 scaled fluorescence intensity units, black represents the middle value of 5,000 scaled units, and red represents the maximum value of 10,000 scaled units. Top left is the number of nodes at each point on the horizontal scale. Single vertical bar is a subnode further analyzed in C; the double vertical is further analyzed in D. (C) Clustering and heat map analysis of the genes whose constitutive expression is influenced by Flag-IκBα Mut. Left, an enlargement of the dendrogram and heat map from B is shown; right, data from identically treated cells expressing empty vector. Gene 1 is growth hormone (GenBank accession no. J03071), 2 is protein tyrosine phosphatase (X54131), 3 is unknown (AL050002), 4 is serine/threonine kinase (AB018324), 5 is RAB4 (M28211), 6 is pp21 (M99701), 7 is frizzled (L37882), 8 is RNP A0 (U23803), 9 is unknown (AF035292), 10 is enolase (X56832), 11 is α1 type XI collagen (J04177), 12 is selenium donor (U34044), 13 is α1 type XVI collagen (M92642), 14 is Rar (U05227), 15 is microophthalmia-associated transcription factor (AB006909), 16 is UFD2 (AF043117), and 17 is FIP-1 (U41654). ∗∗ indicates genes influenced by doxycycline treatment alone. (D) Clustering and heat map analysis of genes upregulated by RSV and inhibited by Flag-IκBα Mut. Left, an enlargement of the dendrogram and heat map from B is shown; right are array data from identically treated cells expressing empty vector. Details of the genes numbered to the right of the Flag-IκB Mut heat map are provided in Table 1.
To further analyze the RSV- and Flag-IκBα Mut-regulated gene sets and determine if there were distinct gene expression profiles within it, hierarchical clustering was performed (57). In this technique, each gene expression profile is grouped with its nearest neighbor and the mathematical proximity of this gene expression profile is indicated by the height of a common line that connects the two nodes. Figure 7B shows the hierarchical clustering analysis for the 144 RSV- and Flag-IκBα Mut-regulated genes adjacent to the primary data visually represented by a heat map. From visual inspection of the dendrogram, the gene expression patterns were divided into two large clusters: the top cluster contained a group of genes whose constitutive (unstimulated) expression was high and fell with RSV infection (this group is indicated by the single vertical line), and the bottom cluster contained a group of genes whose constitutive expression was low and was strongly increased by RSV infection (indicated by the vertical double line).
The subnode within the top cluster (indicated by single vertical line, Fig. 7B) was expanded for detailed inspection (Fig. 7C). Inspection of gene 11, α1-collagen, can be used to illustrate the regulation pattern of this group. For all three experiments in the Flag-IκB Mut-expressing cells, the fluorescence intensity measurements of the control plus doxycycline values are in the mid (black) to high (red) range. After RSV infection, the values fall dramatically to the low (green) range (compare intensity for control plus doxycycline versus RSV plus doxycycline). In cells cultured in the absence of doxycycline, the signal intensity in two of the three experiments is increased (for each experiment, compare intensity of control plus doxycycline versus control minus doxycycline). Finally, the reduction in expression after RSV infection is reduced (compare RSV plus doxycycline versus RSV minus doxycycline).
To control for nonspecific effects of doxycycline, parallel arrays were performed in empty-vector-transfected cells, and the data were retrieved for side-by-side comparison (Fig. 7C). Genes represented by the numbers 4 (a putative serine-threonine kinase) and 8 (RNP A0) are artifactually influenced by doxycycline, as seen by the changes in their expression with doxycycline treatment in the empty-vector-transfected array. However, for the other genes, doxycycline alone does not significantly influence their expression. This expression profile suggests a role for NF-κB in controlling constitutive gene expression and perhaps in mediating RSV-induced downregulation of promoter activity.
The subnode corresponding to the bottom cluster (indicated by double vertical line, Fig. 7B) was also expanded for inspection with control data (Fig. 7D). As illustrated by gene numbers 5 (complement B), 7 and 8 (both NF-κB2), 10 (5-aminolevulinate synthase), 12 (insulinlike growth factor-binding protein 6 [IGF-BP6]), 13 (human GCN1), 14, 15, and 16 (all RANTES), 18 (IRF-1), 21 (STAT1), 26 (B94), and 29 (IL-8), similar patterns of expression are also seen. For these genes, constitutive expression is at or close to background (green). After RSV infection, the gene expression levels are dramatically upregulated (compare intensity of control plus doxycycline versus RSV plus doxycycline). In cells expressing the Flag-IκBα Mut, the constitutive levels fall to background (control plus doxycycline versus control minus doxycycline), and the genes are significantly inhibited in their response to RSV (compare control minus doxycycline versus RSV minus doxycycline). The behavior of these genes was not significantly influenced by the effect of doxycycline alone (Fig. 7D, right panel), excluding a nonspecific artifact of doxycycline treatment. Finally, we emphasize that the identification of RANTES and IL-8, well-characterized NF-κB-dependent genes, further validates the array data (compare with the RNase protection assay in Fig. 5). Together, these data suggest that NF-κB plays a multifaceted role in controlling constitutive gene expression, involvement in RSV-mediated downregulation of genes, as well as its role in mediating upregulation of a network of RSV-inducible genes.
DISCUSSION
Because of its almost ubiquitous activation in response to pathogen exposure, the NF-κB signaling pathway is frequently modified by viral agents (reviewed in reference 33). Depending on the viral agent, NF-κB may serve to enhance viral replication by binding to NF-κB sites in the long terminal repeat (LTR) of retroviruses, such as the human immunodeficiency virus LTR, evade or manipulate the host immune response, influence cellular proliferation, or control host cell apoptosis (reviewed in references 3 and 33). In the case of paramyxovirus infection, particularly RSV, NF-κB has no apparent role in viral transcription, and its activation appears to be important to the host as an activator of the innate immune and inflammatory response.
A number of highly inducible genes encoding cytokine, chemokine, acute-phase reactant, and adhesion molecules that contain NF-κB binding sites in their proximal promoters in a certain number of cases are induced by RSV (8, 11, 40, 52; reviewed in reference 56). However, whether NF-κB is truly a master regulator of gene expression and which genes are controlled by this transcription factor have not been subjected to direct experimental investigation.
The requirement for NF-κB in controlling cellular survival and its important role in immune development have made genetic knock-out approaches to this question problematic. Here we address the role of NF-κB by using a tightly regulated system of homogeneous cells in which the expression of a dominant negative inhibitor of NF-κB translocation could be controlled by exogenous administration of nontoxic concentrations of doxycycline. By multiple independent criteria, including the expressed IκB molecule's resistance to degradation, inhibition of NF-κB binding and complete blockade of NF-κB-dependent reporter gene expression, we have successfully isolated a cell line in which an empirical genomic approach can be applied to begin to understand the complex role of NF-κB in constitutive and virus-inducible gene transcription.
A surprising finding from our study is the relatively small number of genes that were sensitive to Flag-IκBα Mut expression. Of the 1,359 RSV-inducible genes, only ∼10% (144 genes) were shown to be NF-κB dependent. We recognize that our analysis may miss genes which have redundant activation mechanisms (e.g., those activated by both NF-κB and/or JNK-AP-1, CREB/ATF, NF-IL-6, IRF-3, or other pathways that we and others have shown to be activated in the context of RSV infection), in which significant induction may still occur in the absence of translocated NF-κB. The activating signals controlling expression of the remaining 90% of the RSV-inducible genes in this system will require further investigation. Additionally, it is important to emphasize that our analysis is dependent on whether a given gene is induced by virus in order for us to identify it as an NF-κB target, so this analysis underrepresents the entire spectrum of NF-κB-dependent target genes.
Moreover, the cutoff used to determine statistical significance could omit genes that are NF-κB dependent. For example, we note that IP-10, a gene whose expression was consistently inhibited in the RNase protection assay (Fig. 5A), did not meet our statistical cutoff, with a Pr(F) of 0.07 in the ANOVA. Clearly these observations will have to be extended for other NF-κB-activating stimuli and into other cell types. Nevertheless, our observations suggest that not only does NF-κB play a role in virus-dependent gene activation, it also plays a role in constitutive and virus-dependent inhibition of distinct genetic elements.
NF-κB is important in RSV-inducible gene expression for a number of genes with apparently diverse functions, including chemokines, transcription factors (IRF/STATs and NF-κB/IκB members), proteins controlling translation and proteolysis, secreted proteins, cytoskeletal elements, and signaling proteins, in those whose function is known (Table 1). The chemokine group contains the CXC chemokines GROα and IL-8 and the CC chemokine RANTES. IL-8 contains a potent NF-κB binding site located between nucleotides −80 and −71 upstream of the cap site (8) whose mutation completely blocks RSV-inducible IL-8 transcription (21). Similarly, RANTES has two such NF-κB binding sites (located between nucleotides −71 and −39 upstream of the cap site) which bind members of the RelA/NF-κB1 family (12). We (12) and others (52) have shown that IκB Mut expression blocks virus-inducible RANTES expression.
TABLE 1.
Genes in the RSV-regulated chemokine subnodea
| Function | Name | Gene no. | GenBank accession no. | Locus | Avg fold change | Comparisonb | |
|---|---|---|---|---|---|---|---|
| A549 | SAE | ||||||
| Chemokines | GROα | 17 | X54489 | 4q21 | 5.5 | I | NC |
| IL-8 | 28 | M17017 | 4q13-q21 | 7.4 | I | I | |
| IL-8 | 29 | M28130 | 4q13-q21 | 22.7 | I | I | |
| RANTES | 14 | M21121 | 17q11.2-q12 | 5.5 | I | NC | |
| RANTES | 15 | M21121 | 17q11.2-q12 | 30.6 | I | I | |
| RANTES | 16 | M21121 | 17q11.2-q12 | 53 | I | I | |
| Interferon signaling | IRF-1 | 18 | M87503 | 14q11.2 | 10.5 | I | NC |
| IRF-7B | 19 | U53831 | 11p15.5. | 3.5 | I | I | |
| STAT1 | 21 | M97936 | 2q32.2 | 4.6 | I | I | |
| Metabolism | Cholesterol 25-hydroxylase | 20 | AF059214 | 10q23 | 10 | I | NC |
| Cytochrome P-450 | 23 | X55764 | 8q21 | 3.3 | NC | NC | |
| NF-_κ_B signaling | IAP-1 | 2 | U45878 | 11q22 | 1.8 | I | I |
| NF-κB2 | 7 | X61498 | 10q24 | 4 | I | I | |
| NF-κB2 | 8 | S76638 | 4.3 | I | I | ||
| NF-κB2 | 9 | S76638 | 6.5 | I | I | ||
| IκBɛ | 24 | U91616 | 6pter-p24.1 | 3.5 | I | I | |
| Protein synthesis/turnover | E2 Ubiquitin conjugating | 11 | AA883502 | 11q12 | 2.3 | I | I |
| GCN1 | 13 | D50919 | 9p24.1-q22.33 | 2.1 | I | I | |
| Secreted proteins | Complement B | 5 | L15702 | 6p21.3 | 16 | I | I |
| IGF-BP6 | 12 | M62402 | 12q13 | 4.9 | I | NC | |
| Structural | Membrane protein | 1 | AB015633 | 1 | 1.4 | D | NC |
| Nuclear phosphoprotein | 4 | L22342 | 2q37.1 | 3 | I | I | |
| Tyrosine kinase | Tyrosine phosphatase | 3 | M33684 | 20q13.1-q13.2 | 0.5 | I | NC |
| Other | 5-Aminolevulinate synthase | 10 | Y00451 | 3p21.1 | 1.6 | I | I |
| IL-15Rα | 6 | AF035279 | 10p15-p14 | 3.6 | I | I | |
| GAGE-2c | 22 | U19143 | Xp11.4-p11.2 | 1.8 | NC | NC | |
| Cadherin-like 22 | 25 | AF035300 | 20 | 1.3 | I | NC | |
| B94 | 26 | M92357 | 14q32 | 1.7 | I | I | |
| Unknown | 27 | AF070549 | 7 | 2.5 | I | NC | |
| Unknown | 30 | W27517 | 11 | 1.1 | NC | NC |
We suggest that the identification of IL-8 and RANTES by our genomic analysis is a highly significant confirmation of our approach and results. The mechanism for induction of GROα by RSV infection has not been reported; however, GROα contains a high-affinity NF-κB binding site in its proximal promoter, an element that mediates its cytokine-induced activation (55). In a separate study we used high-density oligonucleotide arrays to profile the temporal changes in RSV-inducible chemokine expression patterns (57). Using similar informatics analyses (clustering), IL-8, RANTES, and GROα were identified as members of one of three distinct chemokine families based solely on their gene expression profiles. That these chemokines are all related by being NF-κB dependent may be one explanation for their common expression pattern.
The NF-κB-dependent gene list contains important members of two transcription factor families. Interferon response factors (IRF) 1 and 7B are members of a family of virus-inducible transcription factors that mediate inducible gene expression (reviewed in reference 51). Although the mechanism for IRF-7B upregulation is unknown, the IRF-1 promoter contains an NF-κB binding element that mediates IRF-1 activation in response to cytokine stimulation (49). STAT1, to our knowledge, is not known to be NF-κB dependent. However, we suggest that the STAT/IRF family members are downstream of NF-κB in viral infection, suggesting an important mechanism for how NF-κB plays an important role in innate immunity by facilitating signaling through the interferon pathway.
Of the NF-κB members identified in this study, the inhibitor of apoptosis (IAP-1), an upstream regulator of caspase activation that is associated with the death domain-containing receptors and may be one mechanism through which NF-κB exerts its antiapoptotic effect, is known to be transcriptionally regulated by NF-κB (13). NF-κB2 is an NF-κB1 homolog encoded by a large 105-kDa precursor that must be processed into its 50-kDa binding form; although regulation of NF-κB2 expression has not been studied, we suggest that there may be NF-κB binding sites in its proximal promoter.
A well-established NF-κB-dependent target is the IκB inhibitors themselves. Activation of the IκB members is an autoregulatory feedback loop in which NF-κB induces the synthesis of its own inhibitor to terminate its action. Previously we showed that the NF-κB-IκBα autoregulatory loop was impaired in RSV-infected cells; this finding explains why IκBα was not identified in the present analysis (34). IκBε, in contrast to IκBα, is strongly upregulated by RSV infection and suggests stimulus-specific differences in the NF-κB-dependent expression control of individual IκB subunits. Notwithstanding, the induction of IκBε may compensate for a relative deficiency in IκBα; the existence of multiple independent NF-κB-IκB and NF-κB-BCL-3 inhibitory loops suggests that unregulated NF-κB activation is highly deleterious (9, 28, 34).
Although there is strong bias towards a role of NF-κB in mediating chemokine and acute-phase reactant gene expression, we have identified a number of other highly NF-κB-dependent genes which do not easily fit into these categories (Table 1). The functional consequences of enhanced expression of 5′-aminolevulinate synthase, a rate-limiting enzyme in heme biosynthesis, are unknown to us but suggest a role for NF-κB in virus-regulated heme metabolism in nonerythroid cells. Conversely, the NF-κB dependence of the E2 ubiquitin-conjugating enzyme suggests that NF-κB activation may have an important role in determining cellular capacity to break down proteins regulated through the ubiquitin-proteasome pathway, a process important in cell surface presentation of viral antigens in the context of MHC class I molecules.
We were surprised to identify the human homolog of the Saccharomyces cerevisiae GCN1 gene, whose protein product controls translational efficiency through modifying upstream activation of the eIF2 protein kinase (42). Gene expression and regulation studies of human GCN1 in the setting of viral infection have not been reported to our knowledge, although viral infections are known to profoundly influence translational regulation. Complement factor B, a hepatic acute-phase response factor important in the alternative complement pathway, is well known to be NF-κB inducible (45); however, viral induction of the alternative complement pathway and its role in response to infection have not been investigated. The induction of IGF-BP6 suggests that virus-infected cells exert paracrine control on the mitogenic actions of insulin-like growth factor II (18). Perhaps IGF-BP6 expression is beneficial to prevent local cellular proliferation in the presence of an infecting viral agent.
Upregulation of the α subunit of the IL-15 receptor may suggest that virus-infected cells have distinct signaling phenotypes to cytokines as a result of NF-κB action. This will require further investigation. Finally, B94 is a protein of unknown function, perhaps important in angiogenesis, that was identified by differential display to be a highly cytokine- and LPS-inducible transcript (54). We suggest that B94 is also a virus-inducible transcript in epithelial cells through an NF-κB-dependent mechanism.
Recent work suggests that NF-κB has genomic actions even in the absence of exogenous stimuli. This “constitutive” NF-κB activation may be important to inhibit apoptosis (reviewed in reference 4). For example, NF-κB appears to be required to maintain low levels of the Bcl-2 protein A1 at levels to prevent loss of mitochondrial transmembrane potential and apoptosis in macrophages (48). Alternatively, constitutive NF-κB activity may be important in cellular immortalization (2). Although our experimental design cannot distinguish these or other potential roles for constitutive NF-κB activity, we interpret our data to mean that constitutive NF-κB appears to downregulate expression of the collagen genes and others. The mechanism (transcriptional or posttranscriptional) by which NF-κB influences the abundance of these genes will require further investigation. In this regard, we note that a recent study has implicated NF-κB in the posttranscriptional control of MyoD mRNA abundance (24); perhaps collagen is regulated in a similar way.
In this report, we have identified 380 genes which are controlled by NF-κB by using microarray analysis of a tetracycline-regulated expression system controlling expression of the nonproteolyzable IκBα inhibitor. These data suggest that NF-κB regulates expression of distinct genetic networks of constitutive genes whose expression is further inhibited by viral infection as well as controlling a distinct subset of those that are virus inducible. Moreover, these data suggest that NF-κB is an upstream regulator of RSV-inducible gene expression through controlling expression of the proteins involved in interferon signaling, STAT/IRF, perhaps providing insights into mechanisms for how NF-κB controls the innate immune response.
Acknowledgments
We thank M. Gossen and H. Bujard (University of Heidelberg) for tTA plasmids and the UTMB Genomics Core Laboratory (T. Wood, Director) for performing the arrays.
This project was supported by grant R21 AI48163 from the NIAID and in part by grants R01 AI40218 (to A.R.B.) and AI 15939 (to R.P.G.), Child Health and Human Development (R30HD 27841), and P30 ES06676 from the NIEHS (to R. S. Lloyd, UTMB).
REFERENCES
- 1.Aherne, W. T., T. Bird, S. D. B. Court, P. S. Gardner, and J. McQuillin. 1970. Pathological changes in virus infections of the lower respiratory tract in children. J. Clin. Pathol. 23**:**7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsura, M., F. Mercurio, A. L. Oliver, S. S. Thorgeirsson, and G. E. Sonenshein. 2000. Role of the IκB kinase complex in oncogenic Ras- and Raf-mediated transformation of rat liver epithelial cells. Mol. Cell. Biol. 20**:**5381-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, A. S. J. 2001. The transcription factor NF-κB and human disease. J. Clin. Investig. 107**:**3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkett, M., and T. Gilmore. 1999. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18**:**6910-6924. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, P. J., and M. Karin. 1997. Nuclear factor-κB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336**:**1066-1071. [DOI] [PubMed] [Google Scholar]
- 6.Becker, S., W. Reed, F. W. Henderson, and T. L. Noah. 1997. RSV infection of human airway epithelial cells causes production of the β-chemokine RANTES. Am. J. Physiol. 272**:**L512-L520. [DOI] [PubMed] [Google Scholar]
- 7.Beg, A. A., and A. S. J. Baldwin. 1993. The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 7**:**2064-2070. [DOI] [PubMed] [Google Scholar]
- 8.Brasier, A. R., M. Jamaluddin, A. Casola, W. Duan, Q. Shen, and R. Garofalo. 1998. A promoter recruitment mechanism for TNF-α-induced IL-8 transcription in type II pulmonary epithelial cells: dependence on nuclear abundance of RelA, NF-κB1 and c-Rel transcription factors. J. Biol. Chem. 273**:**3551-3561. [DOI] [PubMed] [Google Scholar]
- 9.Brasier, A. R., M. Lu, T. Hai, Y. Lu, and I. Boldogh. 2001. NF-κB inducible BCL-3 expression is an autoregulatory loop controlling nuclear p50/NF-κB1 residence. J. Biol. Chem. 276**:**32080-32093. [DOI] [PubMed] [Google Scholar]
- 10.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of IκB-alpha proteolysis by site-specific, signal-induced phosphorylation. Science 267**:**1485-1488. [DOI] [PubMed] [Google Scholar]
- 11.Casola, A., N. Burger, T. Liu, M. Jamaluddin, A. R. Brasier, and R. P. Garofalo. 2001. Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with RSV. J. Biol. Chem. 276**:**19715-19722. [DOI] [PubMed] [Google Scholar]
- 12.Casola, A., R. P. Garofalo, H. Haeberle, T. Elliott, M. Jamaluddin, and A. R. Brasier. 2000. Multiple inducible cis elements control RANTES promoter activation in alveolar epithelial cells infected with respiratory syncytial virus. J. Virol. 75**:**6428-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erl, W., G. Hansson, R. de Martin, G. Draude, K. Weber, and C. Weber. 1999. NF-κB regulates induction of apoptosis and inhibitor of apoptosis protein-1 expression in vascular smooth muscle cells. Circ. Res. 84**:**668-677. [DOI] [PubMed] [Google Scholar]
- 14.Everard, M. L., A. Swarbrick, M. Wrightham, J. McIntyre, C. Dunkley, P. D. James, H. F. Sewell, and A. D. Milner. 1994. Analysis of cells obtained by bronchial lavage of infants with respiratory syncytial virus infection. Arch. Dis. Child. 71**:**428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferris, J. A., W. A. Aherne, and W. S. Locke. 1973. Sudden and unexpected deaths to infants: histology and virology. Br. Med. J. 2**:**439-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiedler, M. A., K. Wernke-Dollries, and J. M. Stark. 1995. Respiratory syncytial virus increases IL-8 gene expression and protein release in A549 cells. Am. J. Physiol. 269**:**L865-L872. [DOI] [PubMed] [Google Scholar]
- 17.Fiedler, M. A., K. Wernke-Dollries, and J. M. Stark. 1996. Inhibition of viral replication reverses respiratory syncytial virus-induced NF-κB activation and interleukin-8 gene expression in A549 cells. J. Virol. 70**:**9079-9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabbitas, B., and E. Canalis. 1997. Growth factor regulation of insulin-like growth factor binding protein-6 expression in osteoblasts. J. Cell. Biochem. 66**:**77-86. [DOI] [PubMed] [Google Scholar]
- 19.Gardner, P. S., J. McQuillin, and S. D. Court. 1970. Speculation on pathogenesis of death from respiratory syncytial virus infection. Br. Med. J. 1**:**327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garofalo, R., J. L. L. Kimpen, R. C. Welliver, and P. L. Ogra. 1992. Eosinophil degranulation in the respiratory tract during naturally acquired respiratory syncytial virus infection. J. Pediatr. 120**:**28-32. [DOI] [PubMed] [Google Scholar]
- 21.Garofalo, R., M. Sabry, M. Jamaluddin, R. K. Yu, A. Casola, P. L. Ogra, and A. R. Brasier. 1996. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 70**:**8773-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garofalo, R., R. C. Welliver, and P. L. Ogra. 1991. Concentrations of LTB4, LTC4, LTD4 and LTE4 in bronchiolitis due to respiratory syncytial virus. Pediatr. Allergy Immunol. 2**:**30-37. [Google Scholar]
- 23.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89**:**5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttridge, D. C., M. W. Mayo, L. V. Madrid, C. Y. Wang, and A. S. Baldwin, Jr. 2000. NF-κB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science 289**:**2363-2366. [DOI] [PubMed] [Google Scholar]
- 25.Haeberle, H., A. Casola, Z. Gatalica, H.-J. Dieterich, P. B. Ernst, A. R. Brasier, and R. Garofalo. 2000. Respiratory syncytial virus-inducible activation of the transcription factor NF-κB mediates pulmonary inflammation. Pediatr. Res. 47**:**17A.10625078 [Google Scholar]
- 26.Hall, C. B., and C. A. McCarthy. 1995. Respiratory syncytial virus, p. 1501-1519. In G. L. Mandel, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingston, New York, N.Y.
- 27.Han, Y., and A. R. Brasier. 1997. Mechanism for biphasic RelA:NF-κB1 nuclear translocation in tumor necrosis factor α-stimulated hepatocytes. J. Biol. Chem. 272**:**9823-9830. [DOI] [PubMed] [Google Scholar]
- 28.Han, Y., T. Meng, N. R. Murray, A. P. Fields, and A. R. Brasier. 1999. IL-1 induced NF-κB-IκBa autoregulatory feedback loop in hepatocytes: a role for PKCα in posttranscriptional regulation of ΙκBα resynthesis. J. Biol. Chem. 274**:**939-947. [DOI] [PubMed] [Google Scholar]
- 29.Han, Y., M. S. Runge, and A. R. Brasier. 1999. Angiotensin II induces IL-6 transcription in vascular smooth muscle cells through pleiotropic activation of NF-κB transcription factors. Circ. Res. 84**:**695-703. [DOI] [PubMed] [Google Scholar]
- 30.Han, Y., S. A. Weinman, S. Boldogh, and A. R. Brasier. 1999. TNF-α-inducible IκBα proteolysis and NF-κB activation mediated by cytosolic m-calpain. J. Biol. Chem. 274**:**787-794. [DOI] [PubMed] [Google Scholar]
- 31.Harrison, A. M., C. A. Bonville, H. F. Rosenberg, and J. B. Domachowske. 1999. Respiratory syncytial virus-induced chemokine expression in the lower airways: eosinophil recruitment and degranulation. Am. J. Respir. Crit. Care Med. 159**:**1918-1924. [DOI] [PubMed] [Google Scholar]
- 32.Henkel, T., T. Machleidt, I. Alkalay, M. Kronke, Y. Ben-Neriah, and P. A. Baeuerle. 1993. Rapid proteolysis of IκB-alpha is necessary for activation of transcription factor NF-κB. Nature 365**:**182-185. [DOI] [PubMed] [Google Scholar]
- 33.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral appropriation of the NF-κB pathway. J. Clin. Investig. 107**:**143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamaluddin, M., A. Casola, R. P. Garofalo, Y. Han, T. Elliott, P. L. Ogra, and A. R. Brasier. 1998. The major component of ΙκBα proteolysis occurs independently of the proteasome pathway in respiratory syncytial virus-infected pulmonary epithelial cells. J. Virol. 72**:**4849-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jamaluddin, M., T. Meng, J. Sun, I. Boldogh, Y. Han, and A. R. Brasier. 2000. Angiotensin II induces nuclear factor (NF)-κB1 isoforms to bind the angiotensinogen gene acute-phase response element: a stimulus-specific pathway for NF-κB activation. Mol. Endocrinol. 14**:**99-113. [DOI] [PubMed] [Google Scholar]
- 36.Jamaluddin, M., S. Wang, R. Garofalo, T. Elliott, A. Casola, S. Baron, and A. R. Brasier. 2001. IFNβ mediates coordinate expression of antigen-processing genes in RSV infected pulmonary epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 280**:**L248-L257. [DOI] [PubMed] [Google Scholar]
- 37.Karin, M. 1999. The beginning of the end: IκB kinase (IKK) and NF-κB activation. J. Biol. Chem. 274**:**27339-27342. [DOI] [PubMed] [Google Scholar]
- 38.Karin, M., and Y. Ben Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18**:**621-663. [DOI] [PubMed] [Google Scholar]
- 39.Levine, S., and R. Hamilton. 1969. Kinetics of the respiratory syncytial virus growth cycle in HeLa cells. Arch. Ges. Virusforsch. 28**:**122-132. [DOI] [PubMed] [Google Scholar]
- 40.Li, J., and A. R. Brasier. 1996. Angiotensinogen gene activation by AII is mediated by the RelA (NF-κB p65) transcription factor: one mechanism for the renin angiotensin system (RAS) positive feedback loop in hepatocytes. Mol. Endocrinol. 10**:**252-264. [DOI] [PubMed] [Google Scholar]
- 41.Lu, Y., L. Jamieson, A. R. Brasier, and A. P. Fields. 2001. NF-κB/RelA transactivation is required for atypical protein kinase Ciota-mediated cell survival. Oncogene 20**:**4777-4792. [DOI] [PubMed] [Google Scholar]
- 42.Marton, M. J., C. R. Vazques de Aldana, H. Qium, K. Chakraburtty, and A. G. Hinnebusch. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosome in activation of eIF2α kinase GCN2. Mol. Cell. Biol. 17**:**4474-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirkin, B. 1996. Mathematical classification and clusterin: nonconvex optimization and its applications. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 44.Noah, T. L., and S. Becker. 1993. Respiratory syncytial virus-induced cytokine production by a human bronchial epithelial cell line. Am. J. Physiol. 265**:**L472-L478. [DOI] [PubMed] [Google Scholar]
- 45.Nonaka, M., and Z. M. Huang. 1990. Interleukin-1-mediated enhancement of mouse factor B gene expression via NF-κB-like hepatoma nuclear factor. Mol. Cell. Biol. 10**:**6283-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olszewska-Pazdrak, B., K. Pazdrak, P. L. Ogra, and R. Garofalo. 1998. Respiratory syncytial virus-infected pulmonary epithelial cells induce eosinophil degranulation by a CD18-mediated mechanism. J. Immunol. 160**:**4889-4895. [PubMed] [Google Scholar]
- 47.Olszewska-Pazdrak, B., A. Casola, T. Saito, R. Alam, S. E. Crowe, F. Mei, P. L. Ogra, and R. P. Garofalo. 1998. Cell-specific expression of RANTES, MCP-1, and MIP-1α by lower airway epithelial cells and eosinophils infected with respiratory syncytial virus. J. Virol. 72**:**4756-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagliari, L. J., H. Perlman, H. Liu, and R. M. Pope. 2000. Macrophages require constitutive NF-κB activation to maintain A1 expression and mitochondrial homeostasis. Mol. Cell. Biol. 20**:**8855-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pine, R. 1997. Convergence of TNF-α and IFNγ signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/κB promoter element. Nucleic Acids Res. 25**:**4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shay, D. K., R. C. Holman, R. D. Newman, L. L. Liu, J. W. Stout, and L. J. Anderson. 1999. Bronchiolitis-associated hospitalizations among US children, 1980-1996. JAMA 282**:**1440-1446. [DOI] [PubMed] [Google Scholar]
- 51.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19**:**623-655. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, L. H., J. S. Friedland, M. Sharland, and S. Becker. 1998. Respiratory syncytial virus-induced RANTES production from human bronchial epithelial cells is dependent on nuclear factor-κB nuclear binding and is inhibited by adenovirus-mediated expression of inhibitor of κb alpha. J. Immunol. 161**:**1007-1016. [PubMed] [Google Scholar]
- 53.Welliver, R. C., D. T. Wong, M. Sun, E. Middleton, Jr., R. S. Vaughan, and P. L. Ogra. 1981. The development of respiratory syncytial virus-specific IGE and the release of histamine in nasopharyngeal secretions after infection. N. Engl. J. Med. 305**:**841-846. [DOI] [PubMed] [Google Scholar]
- 54.Wolf, F. W., V. Sarma, M. Seldin, S. Drake, S. J. Suchard, H. Shao, K. O'Shea, and V. Dixit. 1994. B94, a primary response gene inducible by tumor necrosis factor-alpha, is expressed in developing hematopoietic tissues and the sperm acrosome. J. Biol. Chem. 269**:**3633-3640. [PubMed] [Google Scholar]
- 55.Wood, L. D., and A. Richmond. 1995. Constitutive and cytokine-induced expression of the melanoma growth stimulatory activity/GRO alpha gene requires both NF-κB and novel constitutive factors. J. Biol. Chem. 270**:**30619-30626. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, G., and S. Ghosh. 2001. Toll-like receptor-mediated NF-κB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Investig. 107**:**13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75**:**9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, H., J.-P. Cong, G. Mamtora, T. Gingera, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95**:**14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]