Null Mutation of AtCUL1 Causes Arrest in Early Embryogenesis in Arabidopsis (original) (raw)
Abstract
The SCF (for _S_KP1, C_ullin/CDC53,F_-box protein) ubiquitin ligase targets a number of cell cycle regulators, transcription factors, and other proteins for degradation in yeast and mammalian cells. Recent genetic studies demonstrate that plant F-box proteins are involved in auxin responses, jasmonate signaling, flower morphogenesis, photocontrol of circadian clocks, and leaf senescence, implying a large spectrum of functions for the SCF pathway in plant development. Here, we present a molecular and functional characterization of plant cullins. The_Arabidopsis genome contains 11 cullin-related genes. Complementation assays revealed that AtCUL1 but not AtCUL4 can functionally complement the yeast cdc53 mutant.Arabidopsis mutants containing transfer DNA (T-DNA) insertions in the AtCUL1 gene were shown to display an arrest in early embryogenesis. Consistently, both the transcript and the protein of the AtCUL1 gene were found to accumulate in embryos. The AtCUL1 protein localized mainly in the nucleus but also weakly in the cytoplasm during interphase and colocalized with the mitotic spindle in metaphase. Our results demonstrate a critical role for the SCF ubiquitin ligase in_Arabidopsis embryogenesis.
INTRODUCTION
Ubiquitin conjugation to target proteins and subsequent degradation of the target proteins by the 26S proteasome play an important role in diverse cellular processes, including cell cycle regulation, stress responses, signal transduction, metabolic regulation, and cell differentiation (for review, see Hershko and Ciechanover, 1998). Three types of enzymes are involved sequentially in the ubiquitin-conjugation pathway: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). E1 catalyzes, in an ATP-dependent reaction, the formation of a ubiquitin adenylate that is then transferred to a conserved cysteine residue within the E1, resulting in the formation of a thiolester bond between the cysteinyl sulfhydryl group of E1 and the terminal carboxyl group of ubiquitin. The activated ubiquitin is subsequently transferred onto a cysteine residue within an E2. An E3 is typically required for the final transfer of the activated ubiquitin from the E2 to the lysine residue within the target protein, resulting in an isopeptide linkage between the C terminus of ubiquitin and the ε-lysyl group of the target protein. Once a polyubiquitin chain is assembled on a substrate, the substrate is then degraded by the 26S proteasome. The 26S proteasome is composed of two large subcomplexes, the 20S proteasome and the 19S regulatory cap. The plant counterpart appears to be similar in organization and structure to animal proteasome and probably functions in an analogous manner (Parmentier et al., 1997; Fu et al., 1998).
Work in yeast and mammals indicates that the specificity of the ubiquitin pathway derives from the activity of a specific E3 or E2/E3 combination. The SCF complex is a recently identified, and currently the best characterized, E3 complex that is composed of four majors subunits: cullin (CDC53 in yeast), SKP1, RBX1/ROC1, and an F-box protein (reviewed in Krek, 1998; Patton et al., 1998a;Deshaies, 1999; Tyers and Jorgensen, 2000). Structure–function studies in yeast and mammals have demonstrated that cullin/CDC53 functions as a scaffold in assembling different subunits of the SCF complex as well as an E2 enzyme (e.g., CDC34). Different F-box proteins may be assembled onto the same core complex, forming different SCFs, which in turn catalyze the ubiquitination of different substrates .
Genetic studies have demonstrated the involvement of plant F-box proteins in a number of developmental and physiological processes (reviewed in Callis and Vierstra, 2000). The F-box protein UFO/FIM has an important role in regulating floral organ identity in_Arabidopsis_ and Antirrhinum (Ingram et al., 1997; Samach et al., 1999). The_Arabidopsis_ F-box proteins TIR1 and COI1 are essential for response to auxin and jasmonic acid, respectively (Ruegger et al., 1998; Xie et al., 1998). More recently, two closely related Arabidopsis F-box proteins, ZTL and FKF1, have been shown to be involved in the regulation of circadian rhythm (Nelson et al., 2000; Somers et al., 2000). The F-box protein EID1 is involved in phytochrome A–specific light signaling in Arabidopsis (Dieterle et al., 2001). Finally, the F-box protein ORE9 seems to play a key role in natural and hormone-induced senescence processes (Woo et al., 2001). For most of these F-box proteins, their interaction with ASK1 (the Arabidopsis SKP1-like protein) has been demonstrated by the yeast two-hybrid system and/or by immunoprecipitation assays, which implies their function through SCF complexes. The mutant ask1–1 has been shown to be defective in homologous chromosome separation in male meiosis anaphase I (Yang_et al._, 1999). Also, multiple aspects of vegetative and floral growth as well as response to auxin are affected in the_ask1–1_ mutant (Gray et al., 1999; Zhao et al., 1999).
The Arabidopsis cullin AtCUL1 has been found in a complex containing TIR1 and ASK1 or ASK2 (Gray et al., 1999). The modification of AtCUL1 by the ubiquitin-related protein RUB1 has been demonstrated, and genetic studies revealed that the enzymes responsible for this RUB1-conjugation pathway are important for auxin response (del Pozo and Estelle, 1999a; Dharmasiri and Estelle, 2002). Recently, the COP9 signalosome, first identified in Arabidopsis as a negative regulator of photomorphogenesis, has been shown to promote the removal of RUB1/NEDD8 from cullins (Lyapina et al., 2001;Schwechheimer et al., 2001; Zhou et al., 2001). Strikingly, an increase in RUB1-modified AtCUL1 by knockdown of COP9 signalosome activity has the same effect on auxin response as a decrease in the amount of modified cullin. Schwechheimer et al. (2001) suggested that the RUB1 conjugation and deconjugation cycle is important for this process. An AMP-activated protein kinase SnRK has been demonstrated to interact with the SCF complex through binding with ASK1 (Farràs et al., 2001). The function of this potential phosphorylation pathway on the SCF activity is currently unclear.
To study the role of AtCUL1 in Arabidopsis development, we identified T-DNA insertion atcul1 mutants. In yeast, the_cdc53ts_ mutants fail to enter S phase because they are unable to degrade the S phase cyclin/CDK inhibitor SIC1 (Schwob et al., 1994). The Dictyostelium culA mutants exhibit aggregation and morphogenesis defects (Mohanty et al., 2001). In nematodes, the _cul1–1_mutants show hyperplasia of blast-cell lineages (Kipreos et al., 1996). In mice, loss of the CUL1 gene arrested embryogenesis before the onset of gastrulation (Dealy et al., 1999; Wang et al., 1999). Unlike animals, plants have multicellular haploid (gametophyte) and multicellular diploid (sporophyte) stages in their life cycle. In addition, higher plants have a sedentary lifestyle; plant cells that are trapped within rigid walls divide and differentiate in place. Despite the existence of large collections of mutants that affect plant embryogenesis (Meinke, 1985), the molecular basis underlying the developmental steps leading to early embryo development remains poorly understood. In this study, we show that null mutations in AtCUL1 cause arrest before the first cell division of both embryo and endosperm cells, which originate from a double-fertilization event in which two sperm nuclei fuse with the egg cell and central cell nuclei, respectively. This work provides new insights into the role of the SCF pathway in the control of plant cell division and embryogenesis.
MATERIALS AND METHODS
Yeast Strains and Vectors
The yeast strain cdc53st and the plasmid pJS161–53 carrying the CDC53 gene were a generous gift from D. Lammer and J. Singer (Hutchinson Cancer Research Center, Washington, DC), and the vector p426TEF (Mumberg et al., 1995) from A. Camasses (Institut de Physiologie, Strasbourg, France). The cDNAs covering the entire coding region of AtCUL1, NtCUL1, and AtCUL4 were cloned into the p426TEF vector by use of_Bam_HI–_Xho_I, _Spe_I–_Xho_I, and _Eco_RI–_Sal_I restriction enzyme sites, respectively.
Plant Materials
The Arabidopsis plants were of the Wassilewskija ecotype. Seeds were produced under greenhouse conditions.Arabidopsis and tobacco BY2 cell suspensions were maintained by weekly subculture as described by Glab et al. (1994) andNagata et al. (1992), respectively.
Antibodies
Peptides containing the N-terminal 20 amino acids of AtCUL1 were synthesized, linked to KLH carrier proteins, and used to immunize rabbits. The antiserum was immunoaffinity purified against the same peptides bound to Sepharose matrix. The affinity-purified anti-peptide antibody (@AtCUL1) was diluted 1:4000 for Western blot analysis and 1:500 for immunolocalization. Antibodies against PSTAIRE and α-tubulin were purchased from Santa Cruz Biotechnology, Santa Cruz, CA, and Amersham Pharmacia Biotech, Arlington Heights, IL, and used as recommended.
Northern and Western Analysis
Total RNAs and proteins were prepared from_Arabidopsis_ plants and suspension-cultured cells. Northern and Western blot analyses were performed as described previously (Criqui et al., 2000).
Plant Vectors
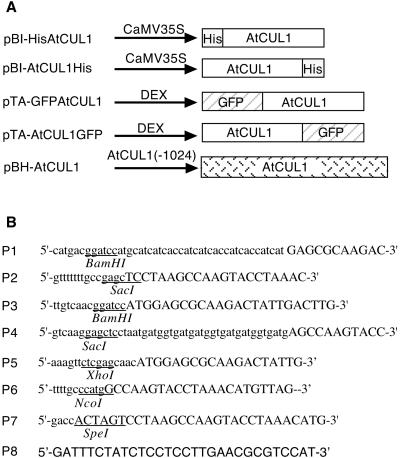
Schematic representations of different plant vectors and sequences of oligonucleotides used in PCR amplification for vector constructions are shown in Figure 4.
Figure 4.
Vectors used for plant transformation. (A) Schematic representation of the genes inserted into different vectors. Arrows with lines represent the different promoter regions and open boxes the coding sequence of different proteins. (B) Oligonucleotides used in PCR amplification for plant vector construction. Nucleotides corresponding to the AtCUL1 sequence are specified by capital letters. Restriction enzyme sites used in cloning are underlined.
An epitope of 10 histidines was fused to the N- and C-terminus of the AtCUL1 by PCR amplification of the AtCUL1 cDNA by use of oligonucleotides P1 and P2, and P3 and P4, respectively. The amplified fragments were cloned into the _Bam_HI and _Sac_I restriction sites of the binary vector pBI121.1 (Clontech, Cambridge, UK), resulting in pBI-HisAtCUL1 and pBI-AtCUL1His.
The whole coding region of the AtCUL1 cDNA was PCR-amplified by use of oligonucleotides P5 and P6 and subsequently cloned into the_Xho_I and _Nco_I sites of pSK-GFP (Criqui et al., 2000), resulting in pSKAtCUL1-GFP. Similarly, the PCR-amplified fragment of the AtCUL1 cDNA by use of oligonucleotides P3 and P7 was cloned into pKS-GFP by use of_Bam_HI and _Spe_I sites, resulting in pKS-GFPAtCUL1. After confirmation by sequencing of the AtCUL1 sequences and its in-frame fusion with green fluorescent protein (GFP), the_Xho_I-_Spe_I DNA fragments encoding the chimeric AtCUL1-GFP and GFP-AtCUL1 proteins were subcloned into the glucocorticoid-inducible vector pTA7002 (Aoyama and Chua, 1997), resulting in pTA-AtCUL1GFP and pTA-GFPAtCUL1, respectively.
The Arabidopsis BAC T10P11 containing the _AtCUL1_gene was received from the Genome Sequencing Center of the Cold Spring Harbor Laboratory. The region spining the AtCUL1 gene was PCR-amplified by use of oligonucleotides P7 and P8. The resulting PCR fragment was digested with _Eco_RI and _Spe_I and subsequently cloned into the _Eco_RI- and_Xba_I-digested pBinHyg-TX vector (Gatz, 1995), resulting in pBH-AtCUL1. Sequence analysis of the cloned fragment revealed that PCR amplification caused three T -to-C substitutions. Luckily, these substitutions were located at positions nonessential for the expression of AtCUL1.
These different plant vectors were transferred by electroporation into_Agrobacterium_, and the resulting strains were used in plant transformation.
Plant Transformation and Transgene Expression Analysis
Transgenic Arabidopsis plants were obtained by_Agrobacterium_-mediated transformation by the floral dip method (Bechtold et al., 1993; Clough and Bent, 1998). Tobacco plant transformation and the establishment of transgenic BY2 cell lines were as previously described (Shen, 2001b). The DEX induction for transgene expression and the confocal microscopy detection of GFP fluorescence were as previously described (Shen, 2001b).
Isolation of atcul1 Mutants
DNA pools of the Arabidopsis T-DNA insertion lines from the Versailles collection (Bechtold et al., 1993) were screened for T-DNA insertion in the AtCUL1 locus. Forward and reverse primers from the sequence of the AtCUL1 gene were designed for PCR screening of the DNA pools by the combination of T-DNA left and right border–specific primers. PCR products were analyzed by Southern hybridization with the AtCUL1 cDNA and the T-DNA probes. PCR fragments hybridized with both probes were further confirmed by sequencing.
Segregation Analysis
Seeds were surface-sterilized and plated onto medium supplemented with kanamycin (Km) (50 mg/L) (half-strength Murashige and Skoog salts, 1% sucrose, 0.9% agar, pH 5.7). After 2 d at 4°C, the seeds were grown under 12 h light/12 h dark cycles at 22°C. The Km phenotype (resistant or sensitive) was scored after 2 weeks.
Intact Silique Analysis and Whole-Mount Preparation of Ovules
Siliques were dissected fresh or after fixation in an ethanol/acetic acid (9:1) solution. Seeds (ovules) were removed from fixed siliques, cleared for 10 min to 2 h in Hoyer's solution (chloral hydrate/gum arabic/glycerol/water [100:7.5:5:30 g]), and imaged by use of Nomarski optics.
Immunofluorescence Staining and In Situ Hybridization
Tobacco BY2 cells as well as Arabidopsis suspension cells were fixed in 3.7% paraformaldehyde as described (Proust_et al._, 1999). Inflorescences and siliques of_Arabidopsis_ plants were fixed in 4% paraformaldehyde and embedded in paraffin wax, and 10-μm sections were prepared for immunolabeling and for in situ hybridization according to Jackson (1991). Immunolabeling was performed as described by Schmit et al. (1996). The sense and antisense AtCUL1 probes for in situ hybridization were prepared by use of the DIG RNA Labeling Kit (Roche; Catalog No. 1175025), and hybridizations were performed as described by Jackson (1991).
RESULTS
Arabidopsis Contains Multiple Putative Cullins
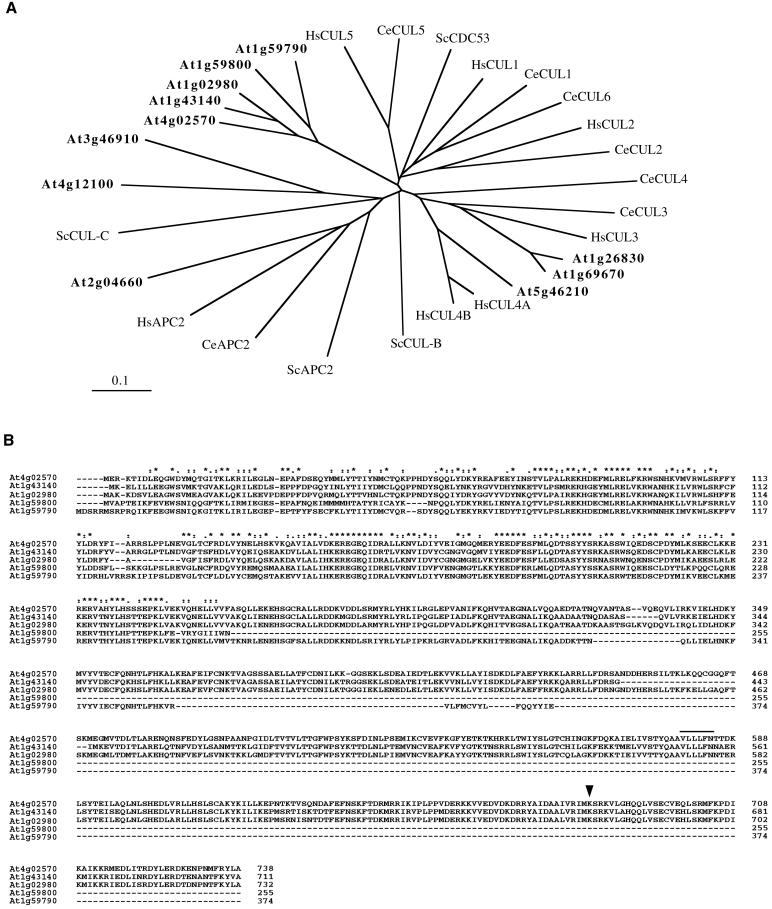
The cullin family encompasses at least six genes in humans and in_Caenorhabditis elegans_. The genome sequence of_Arabidopsis_ has been determined (_Arabidopsis_genome initiative, 2000). A sequence similarity search reveals that the Arabidopsis genome contains 11 cullin-related genes that together with other eukaryotic members can be classified into distinct, distantly related groups (Figure1A). The APC2 group member contains a cullin homology region but is a subunit of the anaphase-promoting complex (APC), an E3 enzyme responsible for ubiquitination of mitotic regulators (for review, see Zachariae and Nasmyth, 1999). The APC-mediated destruction box pathway also seems to be conserved in plants (Genschik et al., 1998; Criqui et al., 2000). The ubiquitin-ligase complexes containing CUL3, CUL4, CUL5, and CeCUL6 are poorly characterized, although CUL3 has been demonstrated to be involved in the degradation of cyclin E (Singer et al., 1999). The HsCUL2 protein functions in a ubiquitin-ligase complex containing the VHL tumor suppressor protein elongin-B (a ubiquitin-like protein), RBX1/ROC1, and elongin-C (a SKP1 functional homologue) that recruits a SOCS-box containing protein (reviewed in Tyers and Jorgensen, 2000; Ivan and Kaelin, 2001). Also, CeCUL2 is not functionally redundant with CeCUL1 (Feng et al., 1999). The mammalian and nematode CUL1 show the highest homology with ScCDC53 and form SCF complexes with similar partners: SKP1, ROC1/RBX1, and an F-box protein (reviewed in Krek, 1998; Deshaies, 1999; Tyers and Jorgensen, 2000). The AtCUL1 (At4g02570) protein, together with four other_Arabidopsis_ proteins (At1g43140, At1g02980, At1g59800, and At1g59790), are the closest orthologues of ScCDC53, but their sequence does not allow assignment to either the CUL1 or CUL2 group (Figure 1A). Among these Arabidopsis proteins, only AtCUL1 has been demonstrated to be expressed (del Pozo and Estelle, 1999b; Gray_et al._, 1999; Farràs et al., 2000); whether the other proteins are also expressed is currently unknown. In addition, two of them (At1g59800 and At1g59790) contain the conserved N-terminal region (Figure 1B), which is involved in the interaction with SKP1 (Patton et al., 1998b; Wu et al., 2000), but do not contain the conserved C-terminal region, which is required for interaction with RBX1/ROC1 and RUB1/NEDD8 modification (Furukawa et al., 2000; Wu et al., 2000).
Figure 1.
Sequence analysis of Arabidopsis_cullin-related proteins. (A) Phylogenetic tree of the_Arabidopsis proteins (bold letters), together with cullins and APC2 of Saccharomyces cerevisiae, C. elegans, and Homo sapiens, was established by use of ClustalW and TreeViewPPC programs. DDBJ/EMBL/GenBank accession numbers: Q12018 for ScCDC53, P53202 for ScCUL-B, NP_012488for ScCUL-C, NP_013228 for ScAPC2, Q17389 for CeCUL-1,Q17390 for CeCUL-2, Q17391 for CeCUL-3, Q17392 for CeCUL-4, Q23639 for CeCUL-5, Q21346 for CeCUL-6, AAF99984 for CeAPC2,NP_003583 for HsCUL1, NP_003582 for HsCUL2,NP_003581 for HsCUL3, NP_003580 for HsCUL4A,AAK16812 for HsCUL4B, AAK07472 for HsCUL5, NP_037498 for HsAPC2, AAK76704 for At4g02570 (AtCUL1, this work),NP_175007 for At1g43140, NP_171797 for At1g02980, NP_176189 for At1g59800, NP_176188for At1g59790, NP_174005 for At1g26830,NP_177125 for At1g69670, AJ318018 for At5g46210 (AtCUL4, this work), NP_178543 for At2g04660,NP_192947 for At4g12100, and NP_190275 for At3g46910. (B) Sequence alignment of AtCUL1 (At4g02570) and its most closely related Arabidopsis proteins was performed by use of ClustalX. Numbers refer to amino acid positions in the corresponding proteins. Consensus symbols on top of the alignment: * for the identical or conserved residues in all sequences; : and . for the conserved and semiconserved substitutions, respectively. The RBX1/ROC1 binding domain and the RUB1/NEDD8 conjugation site are indicated by a line and an arrow, respectively.
AtCUL1 but Not AtCUL4 Complements the Yeast_cdc53_ts Mutant Phenotype
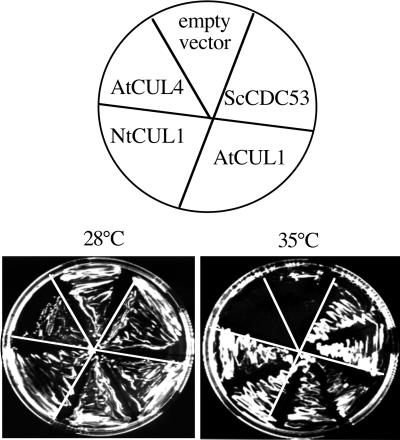
To test whether plant cullins can functionally replace ScCDC53 to form active SCF complexes in yeast, we introduced the plant cullin cDNAs under the control of the TEF promoter (Mumberg et al., 1995) into the yeast cdc53ts mutant strain, carrying a temperature-sensitive mutation in the Sc_CDC53_ gene. Transformants that expressed ScCDC53 (positive control) or AtCUL1 were able to grow at a restrictive temperature (35°C), whereas the negative control containing the empty vector was not (Figure 2). The tobacco orthologue of AtCUL1, the NtCUL1, was also able to complement the yeast mutant. AtCUL4 (At5g46210, Figure 1A), however, failed to complement, which in addition inhibited yeast growth even at a permissive temperature (28°C).
Figure 2.
Complementation of the yeast cdc53_mutant by plant cullins. The yeast temperature-sensitive mutant_cdc53 ts was transformed with the empty vector or the vectors expressing CDC53, AtCUL1, NtCUL1, and AtCUL4, respectively. Individual transformants were plated on selective media and grown either at permissive (28°C) or at restrictive (35°C) temperature. Photographs were taken after 4 d.
Expression of AtCUL1 in _Arabidopsis_Plants and Suspension Cells
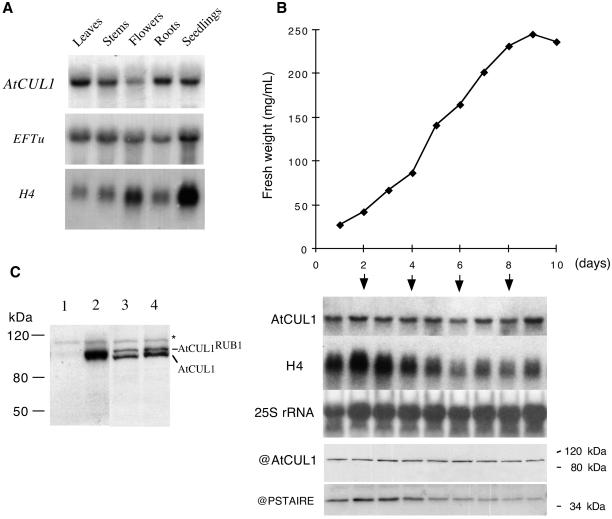
RNA blot analysis shows that AtCUL1 encodes a single transcript of ∼2.5 kb, which is present in different organs of_Arabidopsis_ plants (Figure3A). Whereas the histone H4 transcripts show higher levels in organs containing actively dividing cells (such as flowers and young seedlings), the AtCUL1 did not exhibit such specificity of expression, suggesting that the AtCUL1_gene is not cell cycle regulated. In agreement with this assumption, the AtCUL1 transcript was found at a relatively constant level during different growth phases of suspension-cultured cells (Figure 3B). Also, in the synchronized tobacco BY2 cells, the_NtCUL1 transcript was constantly present during different phases of the cell cycle (data not shown).
Figure 3.
Northern and Western blot analyses of_AtCUL1_ expression. (A) Total RNA was isolated from different organs of Arabidopsis plants, and Northern analysis was performed by successive hybridizations with different probes, as indicated. EFTu: elongation factor EF-1α; H4: histone H4. (B) Samples were taken at different days of subculture from an_Arabidopsis_ cell suspension culture and used for fresh weight measurement and for RNA and protein analysis. Northern analysis was performed by successive hybridizations with the indicated probes. Western blots were performed with the antibodies against AtCUL1 (@AtCUL1) and the conserved CDK kinase motif PSTAIRE (@PSTAIRE). (C) Total proteins prepared from tobacco BY2 cells (lane 1), transgenic BY2 cells expressing 10×his-tagged AtCUL1 (lane 2), and 2-week-old_Arabidopsis_ seedling of wild-type Wassilewskija genotype (lane 3) and Km-resistant_atcul1–1_ +/− (see Table 1) genotype (lane 4) were Western blotted with @AtCUL1. The asterisk indicates an aspecific protein band, which cannot be competed by the AtCUL1 peptide (data not shown).
Antibodies directed against the N-terminal 20-amino-acid peptide of AtCUL1 were produced in rabbits and affinity purified against the antigen. Western blot analysis revealed that the antibodies specifically recognized AtCUL1 expressed in transgenic tobacco BY2 cells but not the endogenous tobacco cullins (Figure 3C). As described previously (del Pozo and Estelle, 1999b; Gray et al., 1999), two predominant bands that migrate close together were detected in the total protein extract from Arabidopsis seedlings (Figure3C). They correspond to unconjugated and RUB1-conjugated isoforms of AtCUL1 (del Pozo and Estelle, 1999b). Interestingly, in both transgenic tobacco BY2 cells expressing AtCUL1 (Figure 3C, lane 2) and_Arabidopsis_ suspension-cultured cells (Figure 3B), only the unconjugated isoform was observed. Like its transcript, the AtCUL1 protein was present at a relatively constant level during different growth phases of suspension-cultured cells (Figure 3B).
AtCUL1 Is Localized in Nucleus, Cytoplasm, and Metaphase Spindles
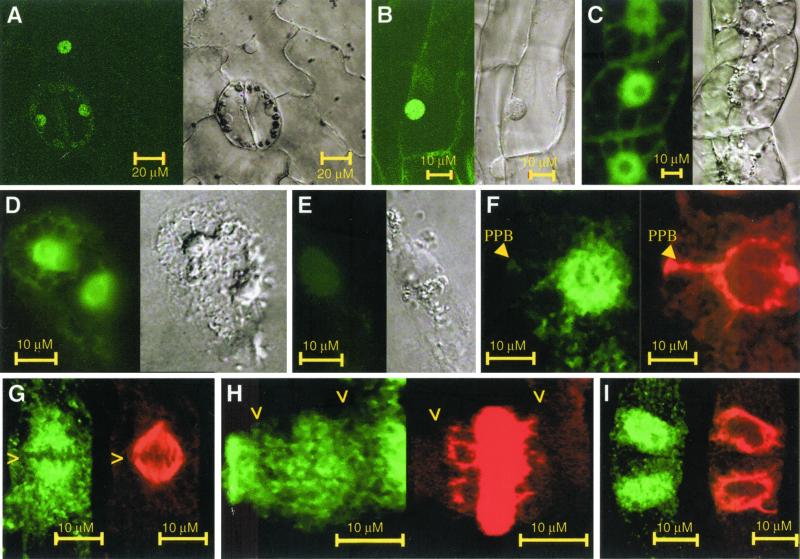
To study the localization of AtCUL1, both GFP and immunolocalization technologies were used. Transgenic tobacco BY2 cell lines expressing the 10×his-tagged AtCUL1 or the GFP-fused AtCUL1 (Figure 4) as well as transgenic tobacco plants expressing the GFP-fused AtCUL1 were generated. In interphase cells, GFP-AtCUL1 as well as AtCUL1-GFP were localized mainly to nucleus and weakly to cytoplasm in transgenic BY2 cells as well as in transgenic plants (Figure 5, A–C). Immunolocalization in Arabidopsis cells confirms this pattern of AtCUL1 localization (Figure 5D). Control immunolocalization experiments using the anti- AtCUL1 preimmune serum, either with_Arabidopsis_ (data not shown) or with tobacco BY2 cells constitutively expressing AtCUL1 (Figure 5E), did not show any detectable staining above background levels. In late G2 phase, a microtubule array called the preprophase band defines the future division plane of the plant cell. At this stage, AtCUL1 was still detected mainly in the nucleus and barely on the preprophase band (Figure 5F). Colocalization of AtCUL1 with mitotic spindle was observed at metaphase (Figure 5G). At telophase, AtCUL1 weakly colocalized with the phragmoplast (Figure 5H). On entrance into interphase, AtCUL1 localized primarily to the newly formed nucleus (Figure 5I).
Figure 5.
Subcellular localization of AtCUL1. (A) Leaf epidermal pavement and stomata cells of a transgenic tobacco plant expression GFP-AtCUL1. (B) Root cortex cells of a transgenic tobacco plant expression GFP-AtCUL1. (C) Transgenic TBY2 cells expressing GFP-AtCUL1. (D) Arabidopsis suspension culture cells showing immunofluorescence after incubation with @AtCUL1 as primary antibody and the Alexa 488-conjugated goat anti-rabbit IgG (Molecular Probes) as secondary antibody. (E) Transgenic TBY2 cells expressing 10×His-tagged AtCUL1 under the control of CaMV 35S promoter were used for immunolocalization with rabbit @AtCUL1 preimmune serum. (F to I) Transgenic TBY2 cells expressing 10×His-tagged AtCUL1 used for coimmunolocalization studies using as primary antibodies the rabbit @AtCUL1 and the mouse @α-tubulin, together with their corresponding secondary antibodies (the Alexa 488–conjugated goat anti-rabbit IgG and the Alexa 568–conjugated goat antimouse IgG, respectively). The fluorescence of Alexa 488 (green, representing AtCUL1) and Alexa 568 (red, representing α-tubulin) was visualized in preprophase (F), metaphase (G), telophase (H), and early interphase (I) cells. > indicates the chromosome position. PPB, preprophase band.
Mutants of AtCUL1 Show Reduced Inheritance in the Gametophyte and Embryonic Lethality
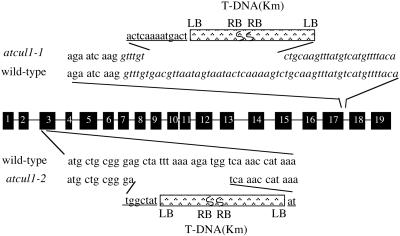
Two T-DNA insertion lines, atcul1–1 and_atcul1–2_, were identified by PCR screening of a total of 40,000 independent transgenic lines of the Versailles T-DNA collection. In atcul1–1, two copies of T-DNA were inserted head-to-head in the intron between the 17th and 18th exons (Figure6). The insertion also caused a deletion of 25 nucleotides at the 5′ end of the intron and an addition of 13 nucleotides of unknown origin at the 5′ end junction between_AtCUL1_ and the T-DNA. In atcul1–2, two copies of T-DNA were inserted head-to-head in the third exon, which also resulted in a small deletion of 16 nucleotides of the exon and the addition of 7 and 2 nucleotides at the 5′ and 3′ end junctions between_AtCUL1_ and the T-DNA, respectively (Figure 6).
Figure 6.
Schematic representation of the T-DNA insertions in the AtCUL1 gene. The comparison between the genomic and cDNA sequences of the AtCUL1 revealed that the coding region of the gene consists of 19 exons (black boxes) separated by 18 introns. The junctions between AtCUL1 and T-DNA are detailed by the representation of the AtCUL1 exon sequence in triple-nucleotide codon format, intron in italics, and those of unknown origin (corresponding neither to _AtCUL1_nor to T-DNA) in underlined letters. LB and RB indicate the orientation of the left and right borders of the T-DNA, respectively.
Heterozygous plants of the atcul1–1 line appeared normal in morphology. A decreased level of AtCUL1 was barely evident in these heterozygous plants (Figure 3C). The T-DNA inserted in the_atcul1_ mutants contains the chimeric nptII gene that confers Km resistance (Figure 6). Segregation tests for Km resistance on seeds produced by self-pollination of more than 60 individual atcul1–1 plants revealed that homozygous_atcul1–1_ plants could not be obtained. In addition, as shown in Table 1, the ratio of Km-resistant-to-Km-sensitive in self-progeny of individual heterozygous plants was significantly lower than the expected ratio of 3:1. The atcul1–2 line behaved very similarly to_atcul1–1_ (Table 1). To determine the inheritance of the_atcul1_ mutations in the male and female gametophytes, reciprocal backcrosses of heterozygous mutant plants with the wild-type plants were performed. Genetic analysis of Km resistance in the F1 progeny revealed that the inheritance of both _atcul1–1_and atcul1–2 mutations was reduced through both male and female gametes (Table 1). Together, these genetic studies reveal that mutations in the AtCUL1 gene affect the development, viability, or function of both male and female gametophytes and that homozygous atcul1 mutant embryos are aborted before seed production.
Table 1.
F1 mutant progeny
| Genotype | Progeny | Hypothesis | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of plants Km-resistant (R) | No. of plants Km-sensitive (S) | R/S | df | χ2 | ||||
| R/S = 1 | R/S = 2 | R/S = 3 | 1< R/Sa <2 | |||||
| atcul1-1+/− selfing | 14183 | 8082 | 1.75 | 64 | 1304.95 | 166.48 | 1572.8 | 36.23b |
| atcul1-2+/− selfing | 3187 | 2100 | 1.52 | 18 | 228.67 | 102.86 | 617.91 | 13.47b |
| atcul1-1+/− ×atcul1-2+/− | 118 | 69 | 1.71 | 2 | 12.83 | 1.07 | 14.12 | 0.02b |
| atcul1-2+/− ×atcul1-1+/− | 71 | 56 | 1.27 | 2 | 1.77 | 6.61 | 24.69 | 0.29b |
| atcul1-1+/− × wild-type | 279 | 356 | 0.78 | 4 | 9.69 | 148.03 | 327.26 | |
| wild-type ×atcul1-1+/− | 235 | 348 | 0.67 | 4 | 21.51 | 183.21 | 372.85 | |
| atcul1-2+/− × wild-type | 67 | 144 | 0.47 | 2 | 28.09 | 115.73 | 210.46 | |
| wild-type ×atcul1-2+/− | 135 | 149 | 0.91 | 2 | 0.69 | 46.77 | 114.25 | |
| atcul1-1+/− (pAtCUL1) | 3216 | 1102 | 2.92 | 12 | 1034.95 | 118.37 | 1.68b | |
| atcul1-2+/− (pAtCUL1) | 745 | 244 | 3.05 | 4 | 252.78 | 33.39 | 0.06b |
Mutations in AtCUL1 Are Responsible for the Phenotype
To confirm that atcul1–1 and atcul1–2 are alleles, crosses between plants that were heterozygous for the two mutations were performed. The resulting F1 progeny exhibited a ratio of Km-resistant-to-Km-sensitive similar to that of the self-progeny of either mutant (Table 1). PCR-amplification analysis revealed that the Km-resistant plants produced from the crosses were either_atcul1–1_ or atcul1–2 genotype but never both (data not shown). These results indicate that atcul1–1 and_atcul1–2_ are allelic mutations responsible for the mutant phenotype.
To further confirm that the mutation of AtCUL1 is responsible for the phenotype, genetic complementation was carried out. The first construct used carried the AtCUL1 cDNA under the control of the CaMV 35S promoter and failed to rescue the mutant phenotype of atcul1–1 (data not shown). A genomic fragment spanning from the −1024 base pairs upstream of the ATG to the stop codon of the AtCUL1 gene was subsequently cloned into a vector carrying the hpt gene, which confers hygromycin (Hyg) resistance (pBH-AtCUL1, Figure 4). Hyg-resistant plants were obtained from transformation of heterozygous mutant plants, and their self-progeny were scored for Km resistance. Of six independent transformants of atcul1–1 that produced Km-resistant progeny, three were found to display a ratio of Km-resistant-to-Km-sensitive of ∼3:1 (one of them is shown in Table1), as expected for rescue by the transgene. One transformant obtained on atcul1–2 exhibited the rescued segregation phenotype as well (Table 1).
Mutants of AtCUL1 Are Arrested Before the First Cell Divisions after Fertilization during Embryogenesis
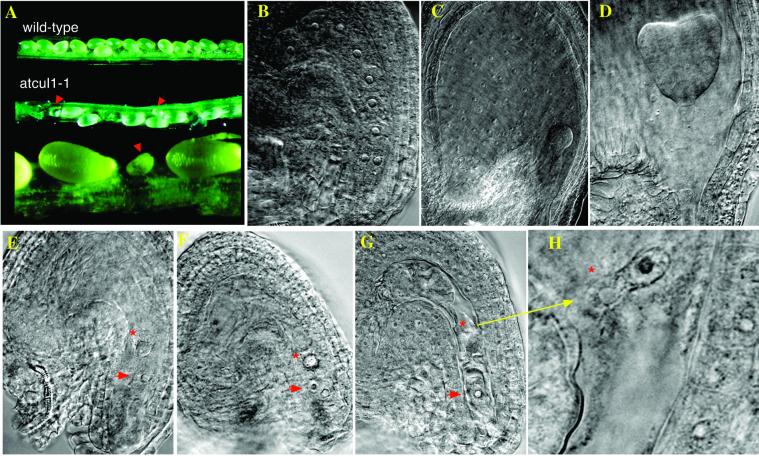
More than 50 siliques (with a total of more than 2000 ovules) on heterozygous atcul1–1 plants were dissected after self-pollination. Among these, ∼27% of embryos failed to develop (indicated by arrows), whereas the others developed normally into mature seeds (Figure 7A). A similar embryo-lethal phenotype was observed in the atcul1–2 line and also in two other atcul1 mutant alleles (H. Hellmann and M. Estelle, unpublished observations) recovered from the Wisconsin collection of T-DNA insertion lines. Differential interference contrast imaging of cleared ovules revealed that whereas the normal ovules contained zygotes that develop through the characteristic preglobular (Figure 7B), globular (Figure 7C), and heart (Figure 7D) stages into mature embryos, the arrested ovules do not contain developed zygotes (Figure 7, E–G). In addition, mutant ovules are missing the endosperm cells, which divide earlier and fill the space around the zygotes. The most advanced stage of an arrested ovule contained one zygote and one endosperm cell with either one or two nuclei (Figure 7, F and G) that were not properly separated from each other (Figure 7H).
Figure 7.
Phenotypes of atcul1 mutants. (A) Open siliques from self-pollinated wild-type and_atcul1–1_ mutant plants. Arrows indicate the developmental arrested siblings. (B–H) Differential interference contrast images of cleared ovules from self-pollinated_atcul1–1_ mutant plants. (B–D) Embryogenesis of developing siblings (containing wild-type or heterozygous zygotes and endosperm) at preglobular (B), globular (C), and heart (D) stages. (E–H) Ovules of arrested siblings from siliques of globular to early heart age. The arrested zygote (arrow) and endosperm (asterisk) cells contain 1 or 2 nuclei each.
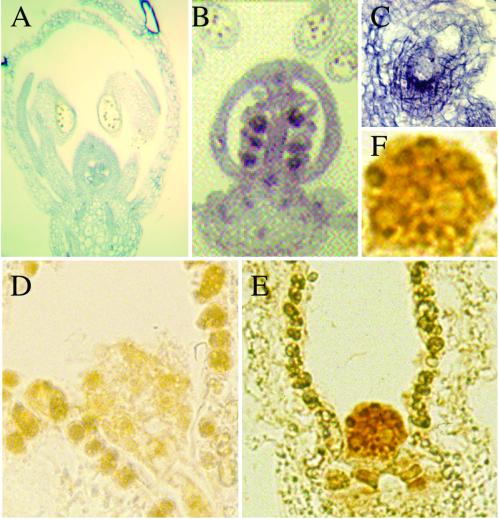
The expression of the AtCUL1 during embryogenesis was further analyzed. In situ hybridization showed that the AtCUL1 antisense probe detected a strong signal in the embryo (Figure 8, B and C) but not the negative control sense probe (Figure 8A). Immunolocalization using the AtCUL1 antibody detected a strong staining in embryos, particularly in the nucleus (Figure 8, E and F). Such staining was not detected in the negative control by use of the preimmune serum (Figure 8D). These results together demonstrated high expression levels of_AtCUL1_ transcript and protein in embryos, which further underscores the important function of AtCUL1 during embryogenesis.
Figure 8.
Expression of AtCUL1 in embryos. (A) Negative control of in situ hybridization with_AtCUL1_ sense probe in a flower section. (B) A flower section probed with the AtCUL1 antisense probe showing strong staining in ovules of a silique. (C) Close-up of an ovule from (B) showing staining of the embryo. (D) Negative control of immunostaining with the preimmune serum of an ovule containing globular embryo. (E) Immunostaining of an ovule containing globular embryo with @AtCUL1 antibodies, showing strong staining of the embryo. (F) Close-up of the stained embryo from (F) showing strong staining in the nuclei.
DISCUSSION
Arabidopsis Contains Functionally Distinct Cullins
All of the SCF subunits, including the cullins, are highly conserved from yeast to mammals, suggesting a common biochemical mechanism of protein ubiquitination. The Arabidopsis genome sequence reveals that plants contain a large number of homologues of SCF components. Whereas only a single SKP1 protein has been identified so far in humans, there are 18 SKP1 orthologues present in the_Arabidopsis_ genome. The F-box proteins function in substrate recognition and are the most diverse and specific components of the SCF complex. The Arabidopsis genome contains more than 300 F-box–containing proteins (del Pozo and Estelle, 2000; Xiao and Jang, 2000; Andrade et al., 2001; our unpublished data). Cullins also belong to gene families, and in Arabidopsis, there are 11 members. This large number of different proteins underscores the potential importance of SCF complexes and regulated protein degradation in various cellular processes in plants.
Yeast complementation tests demonstrated that two distantly related members of the Arabidopsis cullin family, AtCUL1 and AtCUL4, have distinct functions in yeast. The AtCUL1 group consists of four additional proteins in Arabidopsis. Two of them (At1g59800 and At1g59790) seem unlikely to exhibit the full function of cullins, because they lack a conserved C-terminal region demonstrated to be required for interaction with RBX1/ROC1 (Furukawa et al., 2000; Wu et al., 2000). The other two (At1g43140, At1g02980), however, show high similarities to AtCUL1 and thus might act to some extent in the same or overlapping regulatory pathway. The fact that AtCUL1 mutants display an arrest in embryogenesis indicates that AtCUL1 is not redundant with any of the other_Arabidopsis_ cullin homologues, at least during embryogenesis. There are at least two possible explanations for this. First, different cullins could be required nonredundantly in plant development. In support of this, the close homologues of C. elegans cullins CeCUL1 and CeCUL2 are not functionally redundant (Feng et al., 1999). Second, in contrast to the broad expression of the AtCUL1 in plants, some cullins might be stage- or tissue-specific variants. Consistent with this, no ESTs corresponding to At.g43140 or At.g02980 have been identified thus far. It will be important to examine mutants in different_Arabidopsis_ cullin genes to determine the scope of cullin-dependent regulation in plant development.
Subcellular Localization of AtCUL1
Cellular localization studies lend further support to the broad distribution and potential function of AtCUL1. In transgenic tobacco plants expressing GFP-tagged AtCUL1, GFP fluorescence detected in living cells was distributed mainly in the nucleus but also weakly in the cytoplasm (see Figure 5). Similar results were obtained by immunofluorescence (Farràs et al., 2001; this work). In mammalian cells, the CUL1 protein is found in the cytoplasm, in the nucleus, and associated with centrosomes (Freed et al., 1999). Biochemical purification of the centrosome demonstrated that the CUL1 is present in NEDD8-conjugated form. Western blot analysis revealed the presence of the RUB1-conjugated AtCUL1 in_Arabidopsis_ plants (del Pozo and Estelle, 1999b; this work). However, plant cells are known to lack distinct centrosomes. Instead, the microtubule nucleation sites are located on the periphery of the nucleus (reviewed in Canaday et al., 2000). At present, it is not clear whether AtCUL1 is colocalized with the nucleation sites. In suspension-cultured cells, AtCUL1 was present mainly in the unconjugated form. In these cells, both GFP labeling and immunofluorescence staining revealed a pattern of AtCUL1 localization similar to that in plant tissues. During mitosis, the patterns of AtCUL1 localization were also quite similar to those demonstrated in_Arabidopsis_ plants (Farràs et al., 2001). Therefore, our results do not provide evidence that RUB1-conjugation influences the pattern of AtCUL1 localization. Furthermore, AtCUL1 colocalized with the mitotic spindle. Such colocalization is not reported in other organisms. ASK1 has also been demonstrated to colocalize with the mitotic spindle (Farràs et al., 2001). Together, these results suggest that the AtCUL1-based SCF complexes regulate mitotic processes.
AtCUL1 Is Essential for Embryogenesis
Arabidopsis embryogenesis is initiated upon the delivery of two sperm nuclei to the ovules, one fusing with the nucleus of the egg cell and the other with the central cell nuclei (Berger, 1999; Harada, 1999). The fertilized egg cell is the true zygote, whereas the fertilized central cell divides and forms the endosperm. Development of the embryo is initiated by an asymmetric division of the zygote, producing cells with different fates. The apical cell goes on to produce the embryo proper, and the basal cell generates the hypophysis and the suspensor, a transient organ that plays structural and physiological roles in embryo development. Although large collections of mutants that affect Arabidopsis embryogenesis are available, the atcul1 mutants are the first to be characterized at the molecular level that block the earliest divisions in the developing embryo. Unlike medea mutants, which display zygote arrest without affecting endosperm development (Grossniklaus et al., 1998), the atcul1 mutants are defective in both zygote and endosperm development.
In C. elegans, maternal contributions of CUL1 from heterozygous parents suffice for complete development of_cul1−/−_ mutants into sterile adults that exhibit the remarkable propensity to undergo extra rounds of cell division in all tissues (Kipreos et al., 1996). Mouse cul1 mutants show embryo arrest before the onset of gastrulation (Dealy et al., 1999; Wang et al., 1999). Arabidopsis atcul1 mutants exhibited a much earlier and stricter arrest of embryogenesis, before the first cell divisions after fertilization. This difference may reflect different contributions of CUL1 from heterozygous maternal tissues (mRNA or protein) to zygote development and/or different requirements for protein degradation during embryogenesis in each organism. Consistent with its essential function, the AtCUL1 gene is highly expressed, at both transcript and protein level, in the embryos.
Mutations in genes encoding F-box proteins as well as ASK1 have been identified previously in Arabidopsis. These mutants differ from atcul1 in that they are viable as homozygotes. The homozygous ask1–1 mutant is male sterile, indicating a more strict requirement for ASK1 in male gametogenesis (Yang et al., 1999). Genetic analysis demonstrated that the heterozygous_atcul1_ mutant plants produced functional_atcul1−_ pollen and ovules, and microscopic examination revealed that both male and female gametogenesis are morphologically normal (data not shown). This makes AtCUL1 functionally distinct from genes specifically required for gametophyte biogenesis (Yang and Sundaresan, 2000). Nevertheless, a reduced inheritance in both male and female_atcul1_− gametophytes was observed, suggesting that the gene does have a function in the gametophyte. The severe phenotype of the atcul1 mutants compared with the previously characterized mutants in the other SCF components strongly suggests that AtCUL1 forms multiple SCF complexes with different ASKs and F-box proteins, which are ultimately required for plant cell division and embryogenesis. At present, only the SCFTIR1 complex containing the AtCUL1, ASK1, and TIR1 (Gray et al., 1999) and SCFCOI1containing the AtCUL1, ASK1, and COI1 (D. Xie, personal communication) have been demonstrated. Other SCF complexes remain to be biochemically characterized.
SCF Pathway, Cell Division, and Embryogenesis
The best characterized plant SCF complex is SCFTIR1, which is involved in auxin signaling (del Pozo and Estelle, 1999a; Gray et al., 1999; Dharmasiri and Estelle, 2002). It is well known that auxin plays a crucial role in cell division and embryogenesis (Harada, 1999; Chen et al., 2001). Targets of SCFTIR1 include at least some of the large family of transcriptional regulators, the Aux/IAA proteins, involved in auxin response (Gray et al., 2001). The important function of SCFTIR1 in plant development is further evidenced by the finding that the COP9 signalosome, involved in photomorphogenesis, regulates the RUB1 conjugation of AtCUL1 and consequently the SCFTIR1-mediated auxin response (Schwechheimer_et al._, 2001). However, because of the absence of embryo arrest phenotype in the loss-of-function mutant tir1, the SCFTIR1 pathway alone does not suffice to explain the phenotype of the atcul1 mutants. It is likely that several SCF complexes are defective in_atcul1_−/− cells and that the accumulation of multiple misregulated target proteins is responsible for the embryo arrest phenotype of the atcul1 mutants.
In view of the emerging roles of SCF pathways in many cellular processes, numerous substrates are likely to accumulate in cul1−/− cells, one or more of which may account for the embryogenesis arrest. The SCF pathway plays an essential role in the cell cycle control. In yeast, SCF complexes function in both G1/S and G2/M transitions.cdc53 mutants are defective for the G1-to-S phase transition, because of accumulation of the CKI SIC1, whose degradation depends on the SCFCDC4 complex (Schwob et al., 1994). In cells in which SIC1 has been deleted, _cdc53_mutants undergo a block at G2-to-M transition, because of accumulation of the CDK-inhibitory kinase Wee1 (a negative regulator of G2/M transition), whose degradation depends on the SCFMet30 complex (Kaiser et al., 1998; Michael and Newport, 1998). Known substrates of SCF complexes in mammals include cell cycle regulators, such as the CKI (p27KIP1), G1-type cyclins (cycD and cycE), and the transcription factor E2F, as well as the signaling protein IκBα (reviewed in Krek, 1998; Deshaies, 1999;Maniatis, 1999; Tyers and Jorgensen, 2000). Although the G1-type cyclin E accumulates in the arrested mice cul1−/− embryos, it seems unlikely that this accumulation should be the cause of arrest, because cells can tolerate high levels of cyclin E expression (Wang et al., 1999). Thus, the reason for the developmental arrest of cul1−/− embryos in mice remains unknown.
Although homologues of cyclin E have not been identified in plants, orthologues of Wee1, CKI, cyclin D, and E2F are found in plants (Shen, 2001a). The plant D-type cyclins contain the conserved PEST motif, suggesting that they are degraded through the SCF complex, similar to mammalian cyclin D. Recent transgenic studies reveal that ectopic expression of cyclin D and CKI dramatically affects plant development (Riou-Khamlichi et al., 1999; Cockcroft et al., 2000; Wang et al., 2000; De Veylder et al., 2001). It is reasonable to speculate that accumulation of such cell cycle regulators in _atcul1_−/− cells might profoundly affect cell division. In addition, colocalization of AtCUL1 with the mitotic spindle suggests that SCF complexes might be involved in the control of chromosome segregation. Consistent with a role of the ubiquitin pathway in cell cycle control, several other mutants in genes encoding a proteasome subunit and ubiquitin-specific proteases are also embryo-lethal or affect cell divisions in Arabidopsis(Doelling et al., 2001; Smalle et al., 2002;Tzafrir et al., 2002).
ACKNOWLEDGMENTS
We thank M.-C. Criqui for help in microscopy analysis, A. Camasses for yeast complementation, and P. Hammann for DNA sequencing. L.L. acknowledges M. Caboche for his continuous support. The InterInstitut confocal microscopy plate-form was cofinanced by the CNRS, the Université Louis Pasteur, the Region Alsace, and the Association pour la Recherche sur le Cancer (ARC). H.H. is supported by the Deutsche Forschungsgemeinschaft (HE3224/1–1), E.L. by Action Concertée Incitative “Jeune Chercheurs” (ACI), and A.D. by the Program de Recherche Avancées de Coopération Franco-Chinoise (PRA BT98–06). Research in the laboratory of M.E. is supported by grants from the National Institutes of Health (43644). This work was partially supported by the French plant genomic program “Génoplante.”
Abbreviations used:
APC
anaphase-promoting complex
E1
ubiquitin-activating enzyme
E2
ubiquitin-conjugating enzyme
E3
ubiquitin ligase
GFP
green fluorescent protein
Hyg
hygromycin
Km
kanamycin
SCF
_S_KP1, _C_ullin/CDC53,_F_-box protein
T-DNA
transfer DNA
Footnotes
REFERENCES
- Andrade MA, Gonzalez-Guzman M, Serrano R, Rodriguez PL. A combination of the F-box motif and kelch repeats defines a large Arabidopsisfamily of F-box proteins. Plant Mol Biol. 2001;46:603–614. doi: 10.1023/a:1010650809272. [DOI] [PubMed] [Google Scholar]
- Arabidopsis genome initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Berger F. Endosperm development. Curr Opin Plant Biol. 1999;2:28–32. doi: 10.1016/s1369-5266(99)80006-5. [DOI] [PubMed] [Google Scholar]
- Callis J, Vierstra RD. Protein degradation in signaling. Curr Opin Plant Biol. 2000;3:381–386. doi: 10.1016/s1369-5266(00)00100-x. [DOI] [PubMed] [Google Scholar]
- Canaday J, Stoppin-Mellet V, Mutterer J, Lambert AM, Schmit AC. Higher plant cells: gamma-tubulin and microtubule nucleation in the absence of centrosomes. Microsc Res Tech. 2000;49:487–495. doi: 10.1002/(SICI)1097-0029(20000601)49:5<487::AID-JEMT11>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Chen J-G, Ullah H, Young JC, Sussman MR, Jones AM. ABP1 is required for organized cell elongation and division in Arabidopsisembryogenesis. Genes Dev. 2001;15:902–911. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BGW, Healy JM, Murray JAH. Cyclin D control growth rate in plants. Nature. 2000;405:575–579. doi: 10.1038/35014621. [DOI] [PubMed] [Google Scholar]
- Criqui MC, Parmentier Y, Derevier A, Shen W-H, Dong A, Genschik P. Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 2000;24:763–773. doi: 10.1111/j.1365-313x.2000.t01-1-.x. [DOI] [PubMed] [Google Scholar]
- Dealy MJ, Nguyen KV, Lo J, Gstaiger M, Krek W, Elson D, Arbeit J, Kipreos ET, Johnson RS. Loss of CUL1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. Function of the ubiquitin–proteosome pathway in auxin response. Trends Plant Sci. 1999a;4:107–112. doi: 10.1016/s1360-1385(99)01382-5. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. The Arabidopsiscullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA. 1999b;96:15342–15347. doi: 10.1073/pnas.96.26.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol. 2000;44:123–128. doi: 10.1023/a:1006413007456. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and cullin/ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1667. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri S, Estelle M. The role of regulated protein degradation in auxin response. Plant Mol Biol. 2002;49:401–409. [PubMed] [Google Scholar]
- Dieterle M, Zhou Y-C, Schäfer E, Funk M, Kretsch T. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 2001;15:939–944. doi: 10.1101/gad.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 2001;27:393–405. doi: 10.1046/j.1365-313x.2001.01106.x. [DOI] [PubMed] [Google Scholar]
- Farràs R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C. SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001;20:2742–2756. doi: 10.1093/emboj/20.11.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- Freed E, Lacey KR, Huie P, Lyapina SA, Deshaies RJ, Stearns T, Jackson PK. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 1999;13:2243–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics. 1998;149:677–692. doi: 10.1093/genetics/149.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol Cell Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz C. Novel inducible/repressible gene expression systems. Methods Cell Biol. 1995;50:411–424. doi: 10.1016/s0091-679x(08)61047-x. [DOI] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. Cell cycle-dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell. 1998;10:2063–2075. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glab N, Labidi B, Qin L-X, Trehin C, Bergounioux C, Meijer L. Olomoucine, an inhibitor of the cdc2/cdk2 kinases activity, blocks plant cells at the G1 to S and G2 to M cell cycle transitions. FEBS Lett. 1994;353:207–211. doi: 10.1016/0014-5793(94)01035-8. [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada J-P, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb-group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Harada JJ. Signaling in plant embryogenesis. Curr Opin Plant Biol. 1999;2:23–27. doi: 10.1016/s1369-5266(99)80005-3. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES. Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J. 1997;16:6521–6534. doi: 10.1093/emboj/16.21.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kaelin WG. The von Hippel-Lindau tumor suppressor protein. Curr Opin Genet Dev. 2001;11:27–34. doi: 10.1016/s0959-437x(00)00152-0. [DOI] [PubMed] [Google Scholar]
- Jackson DP. In-situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPherson M, editors. Molecular Plant Pathology: A Practical Approach. Oxford, UK: Oxford University Press; 1991. pp. 63–174. [Google Scholar]
- Jefferson R, Kavanagh T, Bevan MW. GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Sia RA, Bardes EG, Lew DJ, Reed SI. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitor kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E, Lander L, Wing J, He W, Hedgecock E. cul-1 is required for cell cycle exit in C. elegansand identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Krek W. Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Maniatis T. A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- Meinke DW. Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Theor Appl Genet. 1985;69:543–552. doi: 10.1007/BF00251102. [DOI] [PubMed] [Google Scholar]
- Michael WM, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- Mohanty S, Lee S, Yadava N, Dealy MJ, Johnson RS, Firtel RA. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 2001;15:1435–1448. doi: 10.1101/gad.871101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cells in the biology of higher plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Parmentier Y, Bouchez D, Fleck J, Genschik P. The 20S proteasome gene family in Arabidopsis thaliana. FEBS Lett. 1997;416:281–285. doi: 10.1016/s0014-5793(97)01228-3. [DOI] [PubMed] [Google Scholar]
- Patton E, Willems A, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998a;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Patton E, Willems A, Sa D, Kuras L, Thomas D, Craig K, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998b;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust J, Houlne G, Schantz ML, Shen W-H, Schantz R. Regulation of biosynthesis and cellular localization of Sp32 annexins in tobacco BY2 cells. Plant Mol Biol. 1999;39:361–372. doi: 10.1023/a:1006199814795. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH. Cytokinin activation of Arabidopsiscell division through a D-type cyclin. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsisfunctions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thalianais an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 1999;20:433–445. doi: 10.1046/j.1365-313x.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- Schmit A-C, Endlé M-C, Lambert A-M. The perinuclear microtubule-organizing center and the synaptonemal complex of higher plants share a common antigen: its putative transfer and role in meiotic chromosomal ordering. Chromosoma. 1996;104:405–413. doi: 10.1007/BF00352264. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Shen W-H. The plant cell cycle: G1/S regulation. Euphytica. 2001a;118:223–232. [Google Scholar]
- Shen W-H. NtSET1, a member of a newly identified subgroup of SET-domain-containing proteins, is chromatin-associated and its ectopic overexpression inhibits tobacco plant growth. Plant J. 2001b;28:371–383. doi: 10.1046/j.1365-313x.2001.01135.x. [DOI] [PubMed] [Google Scholar]
- Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD. Cytokinin growth responses in Arabidopsisinvolve the 26S proteasome subunit RPN12. Plant Cell. 2002;14:17–32. doi: 10.1105/tpc.010381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Tzafrir I, McElver JA, Liu C-M, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW. Diversity of TITAN functions in Arabidopsisseed development. Plant Physiol. 2002;128:38–51. [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou YM, Gilmer S, Whitwill S, Fowke LC. Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 2000;24:613–623. doi: 10.1046/j.1365-313x.2000.00899.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Penfold S, Tang X, Hattori N, Riley P, Harper JW, Cross JC, Tyers M. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr Biol. 1999;9:1191–1194. doi: 10.1016/S0960-9822(00)80024-X. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park J-H, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Fuchs SY, Chen A, Tan P, Gomet C, Ronai Z, Pan Z-Q. The SCFHOS/β-TRCP-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformation. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- Xiao W, Jang J-C. F-box proteins in Arabidopsis. Trends Plant Sci. 2000;5:454–457. doi: 10.1016/s1360-1385(00)01769-6. [DOI] [PubMed] [Google Scholar]
- Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsisgene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yang M, Hu Y, Lodhi M, McCombie R, Ma H. The ArabidopsisSKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc Natl Acad Sci USA. 1999;96:11416–11421. doi: 10.1073/pnas.96.20.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-C, Sundaresan V. Genetic of gametophyte biogenesis in Arabidopsis. Curr Opin Plant Biol. 2000;3:53–57. doi: 10.1016/s1369-5266(99)00037-0. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zhao D, Yang M, Solava J, Ma H. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev Genet. 1999;25:209–223. doi: 10.1002/(SICI)1520-6408(1999)25:3<209::AID-DVG4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Zhou C, Seibert V, Geyer R, Rhee E, Lyapina S, Cope G, Deshaies RJ, Wolf DA. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2:7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]