Premature Termination Mutations in FBN1: Distinct Effects on Differential Allelic Expression and on Protein and Clinical Phenotypes (original) (raw)
Abstract
Marfan syndrome (MFS) and other type 1 fibrillinopathies result from mutations in the FBN1 gene, which encodes the connective-tissue microfibrillar protein fibrillin 1. Attempts at correlating genotype with phenotype have suggested considerable heterogeneity. To define the subtype of fibrillinopathy caused by premature termination codon (PTC) mutations, we integrate genotype information and mRNA expression levels with clinical and biochemical phenotypes. By screening the entire FBN1 gene for mutations, we identified 34 probands with PTC mutations. With the exception of two recurrent mutations, these nonsense and frameshift mutations are unique and span the entire FBN1 gene, from IVS2 to IVS63. Allele-specific reverse-transcriptase polymerase chain reaction analyses revealed differential allelic expression in all studied samples, with variable reduction of the mutant transcript. Fibrillin protein synthesis and deposition into the extracellular matrix were studied by pulse-chase analysis of cultured fibroblasts. In the majority of PTC samples, synthesis of normal-sized fibrillin protein was ∼50% of control levels, but matrix deposition was disproportionately decreased. Probands and mutation-positive relatives were clinically evaluated by means of a standardized protocol. Only 71% (22/31) of probands and 58% (14/24) of the mutation-positive family members met current clinical diagnostic criteria for MFS. When compared with our previously reported study group of 44 individuals with FBN1 cysteine substitutions, the PTC group showed statistically significant differences in the frequency of individual signs, especially in the ocular manifestations. Whereas large-joint hypermobility was more common, lens dislocation and retinal detachment were distinctly less common in the PTC group. We conclude that PTC mutations have a major impact on the pathogenesis of type 1 fibrillinopathies and convey a distinct biochemical, clinical, and prognostic profile.
Introduction
Fibrillinopathies are characterized by defects in fibrillin proteins. Fibrillin 1 is the predominant component of extracellular matrix microfibrils (Sakai et al. 1986; Aoyama et al. 1995). The 350-kD glycoprotein fibrillin 1 is produced as a larger form (profibrillin 1), which is proteolytically processed at its unique N- and C-terminal domains before or during secretion from the cell (Reinhardt et al. 1996; Lonnqvist et al. 1998). Fibrillin 1 has a modular structure, with 47 repeats of six-cysteine epidermal-growth-factor (EGF)–like repeats, 43 of which are of the calcium-binding type (cbEGF) (Handford et al. 2000). It also contains several eight-cysteine motifs, which bear homology to motifs found in the latent transforming growth factor β binding proteins (LTBP family) (Saharinen and Keski-Oja 2000). Abnormalities in fibrillin 1 cause a spectrum of connective-tissue phenotypes, including Marfan syndrome (MFS [MIM #154700]), a pleiotropic disorder with autosomal dominant inheritance (reviewed by Robinson and Godfrey 2000). MFS affects ∼1 in 10,000 individuals and is diagnosed mainly on clinical grounds (Pyeritz 2000; Schrijver et al. 2001). Recently revised diagnostic criteria take into account skeletal, cardiovascular, and ocular manifestations; signs and symptoms in the lungs, dural sac, and skin; and family history (De Paepe et al. 1996). In addition, mutations in the FBN1 gene that are known to cause MFS can be included as major diagnostic criteria.
FBN1 (MIM 134797) is a large gene, located in chromosome band 15q21.1, that spans >235 kb (extending approximately from nt 45185000 to nt 44950000 in the UCSC Human Genome Project Working Draft sequence). FBN1 contains 65 coding exons and 3 alternatively spliced 5′ untranslated exons (Pereira et al. 1993; Biery et al. 1999). Three types of mutations, which have been identified throughout the gene, lead to a disease phenotype (Collod-Beroud et al. 1998; Robinson and Godfrey 2000). Missense mutations that substitute residues important for calcium binding and for the structural integrity of EGF-like repeats constitute 30%–40% of FBN1 mutations identified. They include substitutions of cysteine residues or any of the four amino acids that offer ligands for calcium binding (Dietz et al. 1991; Tynan et al. 1993; Halliday et al. 1999; Schrijver et al. 1999). A second category is represented by in-frame deletions due to exon-skipping/splice-site mutations (Liu et al. 1996) or genomic deletions (Kainulainen et al. 1992; Liu et al. 2001). The third mutation type, accounting for ∼25% of FBN1 mutations, consists of frameshift or nonsense mutations leading to premature termination codons (PTC) (Dietz et al. 1993_a;_ Tynan et al. 1993; Nijbroek et al. 1995; Karttunen et al. 1998; Beroud et al. 2000; Loeys et al. 2001; Pepe et al. 2001; Tiecke et al. 2001).
Genotype-phenotype correlations in the type 1 fibrillinopathies—disorders caused by FBN1 mutations—have been complicated by the large number (>250) of unique mutations reported, as well as by clinical heterogeneity among individuals with the same mutation. Some associations have emerged, however. Skipping of exons 24–32 correlates with the most severe, neonatal forms of MFS (Liu et al. 1996; Putnam et al. 1996; Booms et al. 1999). Severe manifestations are also associated with multiexon deletions (Liu et al. 2001). Mutations that change a cysteine residue or a residue that is crucial for calcium binding in one of the cbEGF-like domains usually cause classic MFS (Schrijver et al. 1999; Tiecke et al. 2001). Milder forms of type 1 fibrillinopathy are often seen in individuals who carry mutations that lead to replacement of non-cysteine, non–calcium-binding amino acids, although the degree of severity is variable (Francke et al. 1995; Furthmayr and Francke 1997; Palz et al. 2000).
In the present study, we report 19 novel premature stop-codon mutations in FBN1. Together with 13 unique PTC mutations previously reported from our laboratory (Tynan et al. 1993; Liu et al. 1997/1998) and 2 spontaneous de novo recurrent mutations, we have assembled data on 60 mutation-positive members of 16 families, with the goal of correlating the mutations with mutant transcript levels, fibrillin protein phenotypes, and clinical phenotypes. PTC mutations predict the production of truncated fibrillin 1 monomers, which can interfere with the assembly of normal monomers into extracellular microfibrils. This postulated dominant negative effect is, however, ameliorated by instability of the mutant mRNA transcripts that are preferentially degraded by the nonsense-mediated mRNA decay mechanism (Cheng and Maquat 1993; Dietz et al. 1993_a_). We observed differential expression of FBN1 mRNA in all studied skin fibroblast cultures, with drastic reduction of the mutant mRNA. In the majority of cultures, synthesis of normal-sized fibrillin protein was reduced to ⩽50% of normal, but matrix deposition was disproportionately lower. Comparison and quantitative analysis revealed significant differences between the clinical manifestations in 34 probands and 26 PTC mutation–positive relatives, on one hand, and a group of 25 probands and 19 mutation-positive relatives with FBN1 cysteine substitutions, on the other. The PTC subgroup of type 1 fibrillinopathy has more-striking skeletal features and large-joint laxity, coupled with a much lower risk of serious ocular manifestations. Ascending aortic dissections were more common in the PTC group, whereas ascending aortic aneurysms presented the prevailing indication for aortic replacement surgery in the cysteine substitution group. This is the first report of a large-scale comparison of fibrillinopathy subgroups, defined by FBN1 mutation type, that were systematically evaluated for the same clinical and biochemical features within a single MFS research center.
Subjects, Material, and Methods
Participants
Study participants were examined in the Center for Marfan Syndrome and Related Connective Tissue Disorders at Stanford Medical Center and were recruited to the study under an institutional-review-board approved protocol and informed consent. Physical examination included the parameters set forth in the Ghent Diagnostic Criteria (De Paepe et al. 1996). Patients were evaluated for dolichostenomelia by measurement of the upper-to-lower body-segment ratio (⩽0.85 in dolichostenomelia) and armspan-to-height ratio (>1.05 in dolichostenomelia). Arachnodactyly was assessed by hand measurements. Facial features, height and width of the palate, dental crowding, pectus deformities, scoliosis, contractures, hypermobility of the large and small joints, and the presence of striae atrophicae were scored by use of standardized criteria. Ophthalmology, echocardiography, cardiology, and orthopedics reports were reviewed, and patients were asked to disclose any history of hernias or pneumothorax. No imaging was performed to look for dural ectasia or protrusio acetabuli. Skin biopsies, buccal swabs (Cytobrush Plus Cell Collector; Medscand Medical AB), and peripheral blood samples were obtained. Control samples were from unaffected volunteers.
Detailed clinical information was obtained on 34 probands and 26 PTC mutation–positive family members. Two probands were lost to follow up, and four died within the 10-year study period. To compare the relative frequency of connective-tissue manifestations, the PTC group was compared with a previously published collection of individuals with cysteine substitutions in FBN1 (Schrijver et al. 1999). Ascertainment through cardiovascular surgery was similar in both groups: 10/34 (29%) for the PTC group and 5/25 (20%) for the cysteine group. The remainder were recruited from the outpatient clinic population by use of identical criteria.
PCR and DNA Sequencing
DNA and RNA were extracted from fresh leukocytes or cultured fibroblasts by standard procedures and from buccal epithelial cells as described by Richards et al. (1997). FBN1 mutations were initially detected by RT-PCR and SSCA, followed by sequencing (Tynan et al. 1993). For genomic analysis of the FBN1 gene, DNA was amplified with intronic primers flanking each of the 65 exons (Nijbroek et al. 1995; Liu et al. 1997/1998). If a heteroduplex pattern was detected by mutation detection enhancement (MDE) gel electrophoresis or denaturing high performance liquid chromatography (DHPLC) (Liu et al. 1997/1998), the PCR product was subsequently sequenced on an ABI 377 sequencer, as described by Schrijver et al. (1999). All FBN1 mutations were confirmed on an independently obtained sample, such as a blood or buccal smear from the proband and/or from affected relatives.
RT-PCR Analysis of Allele-Specific Transcript Levels
To compare levels of mutant relative to normal FBN1 transcripts, a combination of 32P-labeled PCR and restriction digestion was employed. Total RNA was extracted and treated with RNase-free DNase (Ambion). After incubation at 37°C for 20 min, the sample was heated to 70°C for 10 min, to destroy the DNase. Reverse transcription of total RNA was performed using Superscript II (Gibco BRL). The products of the first strand reaction were amplified using paired primers, as reported elsewhere (Tynan et al. 1993). Thermal cycling was performed as described elsewhere (Schrijver et al. 1999). After 20 or 21 cycles, one of each primer pair labeled with 32P-γATP (Amersham) was added to each reaction, followed by one more cycle and a final extension step of 10 min at 72°C. Labeled PCRs were performed with Ampli_Taq_ Gold DNA polymerase (Perkin Elmer). Cold PCRs were run in parallel but were extended to 35 cycles.
Restriction digestion was performed, on both labeled and unlabeled PCR products, with an enzyme that distinguishes the mutant from the normal allele (table 1). Unlabeled digests were electrophoresed on a 2% agarose/2% NuSieve (Bio Whittaker Molecular Probes) gel, whereas labeled digestion products were separated by electrophoresis on 8% polyacrylamide gels. Gels were dried and then autoradiographed. Quantitation of the labeled cDNA products was performed by phosphorimaging (Molecular Dynamics). The levels of intensity of digested and undigested bands were quantitated, with expression of the normal allele considered to be 100%. Genomic DNA from each proband was amplified with exon-specific primers (Nijbroek et al. 1995) and was digested as a control for digestion efficiency. The thermal cycling for genomic controls was modified to denaturation at 95°C and to 45 s for each annealing step.
Table 1.
FBN1 Mutations, Transcript Levels, and Protein Phenotypes of Probands[Note]
| Location ofTermination | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proband | ScreeningMethod | Mutation | MutationLocated at CpGHotspot | MutatedExon | Domaina | RestrictionSite Changes | Codon | Exon | Reduction ofMutantTranscriptLevelb | Fibrillin Synthesis/Matrix Depositionc(%) | BiosynthesisGroupd | Reference forMutation |
| FB1075 | DHPLC | 247+1G→A | Yes | IVS2 | Unique | Skip exon 2 | S128X | 3 | ND | ND | ND | Present study |
| FB845 | DHPLC | 526C→T | No | 5 | EGF (NCB) | _Bsr_I−, _Bfa_I+ | Q176X | 5 | ND | 44/6 | II | Liu et al. 1997/1998 |
| FB807 | DHPLC | 656delA | 6 | EGF | _Apa_I+, _Ban_II+, | I329X | 8 | ND | 39/44 | I | Present study | |
| FB860 | MDE | 745G→T | 7 | EGF (CB) | _Ase_I+, _Mse_I+ | E249X | 7 | 2 | 44/22 | II | Present study | |
| FB1305 | DHPLC | 755del8bp | 7 | EGF (CB) | _Ava_I−, _Hae_III− | N264X | 7 | 1 | 27/9 | II | Liu et al. 1997/1998 | |
| FB798 | DHPLC | 1302T→G | 10 | Proline | _Alu_I+ | Y434X | 10 | 1 | 25/12 | II | Liu et al. 1997/1998 | |
| FB1592 | DHPLC | 1642del3ins20bp | 13 | EGF (CB) | … | I578X | 14 | ND | 97/14 | IV | Present study | |
| FB969 | RT-PCR/SSCA | 1884C→A | 15 | EGF (CB) | _Hin_fI+ | C628X | 15 | 3 | 51/39 | I | Present study | |
| FB1002 | DHPLC | 1888delAAinsC | 15 | EGF (CB) | … | M717X | 17 | ND | 35/6 | II | Present study | |
| FB821 | DHPLC | 2399delC | 19 | EGF (CB) | … | L802X | 19 | 2 | 46/4 | II | Liu et al. 1997/1998 | |
| FB993 | DHPLC | 2581C→T | Yes | 21 | Hybrid | _Hph_I+ | R861X | 21 | ND | 96/63 | III | Present study |
| FB1162 | DHPLC | 2581C→T | Yes | 21 | Hybrid | _Hph_I+ | R861X | 21 | 2 | 35/13 | II | Liu et al. 1997/1998 |
| FB1103 | DHPLC | 3353delAGAG | 27 | EGF (CB) | … | L1161X | 28 | 2 | 26/12 | II | Present study | |
| FB1234 | DHPLC | 3445insC | 27 | EGF (CB) | … | E1158X | 28 | ND | 106/35 | III | Liu et al. 1997/1998 | |
| FB773 | RT-PCR/SSCA | 3464del17bp | 28 | EGF (CB) | _Alu_I−, _Dde_I+ | R1192X | 28 | 3 | 49/17 | II | Tynan et al. 1993 | |
| FB857 | MDE | 3524insA | 28 | EGF (CB) | _Bsl_I+ | R1192X | 28 | ND | 39/49 | I | Present study | |
| FB1439 | DHPLC | 3766delA | 30 | EGF (CB) | … | M1275X | 30 | ND | ND | ND | Present study | |
| FB1058 | DHPLC | 3778G→T | 30 | EGF (CB) | _Bpm_I− | E1260X | 30 | 2 | 39/10 | II | Liu et al. 1997/1998 | |
| FB751 | RT-PCR/SSCA | 4177delG | 33 | EGF (CB) | … | L1412X | 34 | ND | 44/9 | II | Present study | |
| FB1479 | DHPLC | 4429G→T | 35 | EGF (CB) | … | E1477X | 35 | ND | ND | ND | Liu et al. 1997/1998 | |
| FB997 | MDE | 4567C→T | Yes | 36 | EGF (CB) | _Taq_I− | R1523X | 36 | 2 | 42/41 | II | Present study |
| FB1186 | DHPLC | 4930C→T | Yes | 39 | EGF (CB) | _Nla_III+ | R1644X | 39 | ND | 40/22 | II | Present study |
| FB1533 | DHPLC | 5240insACACT | 42 | LTBP | … | I1892X | 46 | ND | 39/15 | II | Liu et al. 1997/1998 | |
| LCL768 | DHPLC | 5826C→A | 47 | EGF (CB) | _Hpy_CH4V− | C1942X | 47 | ND | NA | NA | Present study | |
| FB754 | DHPLC | 5863C→T | No | 47 | EGF (CB) | _Bsr_I−, _Bfa_I+ | Q1955X | 47 | ND | ND | ND | Present study |
| FB733 | MDE | 6163delG | 49 | EGF (CB) | _Tsp_5091+, _Apo_I+ | M2058X | 50 | ND | 46/29 | II | Present study | |
| FB1286 | DHPLC | 6169C→T | Yes | 50 | LTBP | … | R2057X | 50 | ND | 52/52 | I | Liu et al. 1997/1998 |
| FB1324 | DHPLC | 6185insA | 50 | LTBP | _Mse_I+ | Y2062X | 50 | 2 | 21/17 | II | Liu et al. 1997/1998 | |
| FB1220 | DHPLC | 6283insC | 50 | LTBP | … | D2104X | 50 | 1 | 47/23 | II | Present study | |
| FB939 | MDE | 6423delG | 52 | EGF (CB) | _Hpy_CH4III−, _Nsi_I+ | L2159X | 52 | 3 | 48/16 | II | Present study | |
| FB849 | DHPLC | 6513delT | 53 | EGF (CB) | … | V2184X | 53 | ND | 49/16 | II | Liu et al. 1997/1998 | |
| FB938 | MDE | 7240C→T | Yes | 58 | EGF (CB) | _Bsl_I− | R2414X | 58 | 1 | 41/21 | II | Present study |
| FB1420 | DHPLC | 7963del13bp | 63 | EGF (CB) | _Fsp_−, _Mwo_I+ | I2681X | 63 | ND | ND | ND | Present study | |
| FB1447 | DHPLC | 8051+1G→A | No | IVS63 | EGF (CB) | Skip exon 63 | V2751X | 65 | ND | 36/11 | II | Liu et al. 1997/1998 |
Allele-specific transcript levels of samples FB773, FB798, FB821, FB860, FB969, FB997, FB1058, FB1103, FB1162, and FB1220 were also assessed by RT-PCR of two FBN1 single-nucleotide polymorphisms (SNPs): C8931T, in the 3′ UTR (Tynan et al. 1993; Hewett et al. 1994), and T1875C, in exon 15 (Hayward et al. 1994). RT-PCR, _Rsa_I digestion, and electrophoresis were performed as described by Schrijver et al. (1999). The two allelic transcripts were distinguished by SSCA and silver staining, according to the protocol provided by the manufacturer (BioRad), and were subsequently quantified by scanning densitometry.
Fibrillin 1 Pulse-Chase Analysis
Fibrillin protein was metabolically labeled with 35S-cysteine (“pulse”) for 30 min and either (a) harvested immediately or (b) harvested after 8-h or 20-h incubations with unlabeled cysteine (“chase”). The quantitative assay of fibrillin in the soluble intracellular and insoluble extracellular fractions was performed as described elsewhere (Brenn et al. 1996; Schrijver et al. 1999). After protein separation on SDS-PAGE, the gels were dried and exposed to a phosphor screen. Protein bands were examined on a phosphorimager (Molecular Dynamics). Fibrillin 1 was identified in its characteristic position and was subsequently quantified (Aoyama et al. 1994; Brenn et al. 1996). Samples were assigned to biosynthetic groups I–IV, as described by Aoyama et al. (1994). In that original article, we also described a group V that is indistinguishable from the normal control; none of the samples with PTC mutations were assigned to group V.
Results
PTC Mutations in the FBN1 Gene
Thirty-four unrelated probands with mutations leading to PTCs were identified by RT-PCR/single-strand conformation analysis of overlapping segments of the FBN1 coding region or by amplification of all 65 FBN1 exons followed by MDE or denaturing HPLC heteroduplex screening (table 1). On the genomic level, the mutations were precisely defined by sequencing of PCR products directly or after subcloning. Thirteen mutations discovered by DHPLC have been described elsewhere, in a methods article that did not include any clinical data (Liu et al. 1997/1998). Twenty PTC mutations are reported in the present study for the first time. The mutations span the FBN1 gene from IVS2 to IVS63, with a rather even distribution. They affect all domain types of the fibrillin 1 protein except for the unique region located at the carboxyl terminus (table 1). All of the 34 mutations are unique, except for one recurrence (2581C→T) in our sample and one recurrence of a previously reported 247+1G→A mutation that causes skipping of exon 2 (Dietz et al. 1993_a;_ Halliday et al. 1999; Guo et al. 2001; Loeys et al. 2001). Both recurring mutations involve C→T transitions on the sense or antisense strand at a CpG dinucleotide. Of the 13 unique nonsense mutations due to single-nucleotide substitutions that change an amino acid codon to a stop codon, 5 are CGA (R) to TGA (X) mutations at CpG hotspots (table 1). The 20 frameshift mutations include 7 single-nucleotide deletions, 4 single-nucleotide insertions, 4 multinucleotide deletions, 2 complex insertion/deletions, and 1 five-nucleotide tandem duplication. In addition, two splice-site mutations (247+1G→A and 8051+1G→A) result in skipping of exons 2 and 63, respectively. These are two of the three out-of-frame exons in the FBN1 gene and, therefore, deletion of these exons leads to frameshifts (fig. 1).
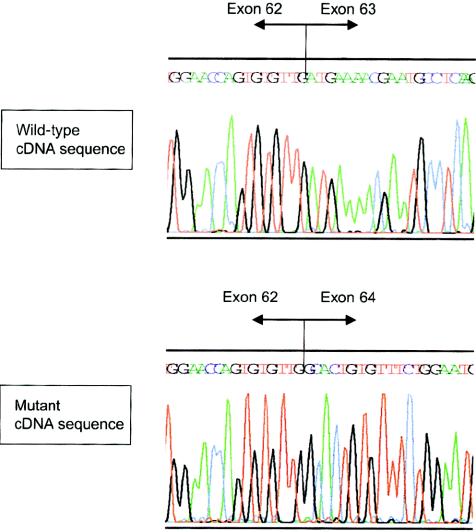
Figure 1.
Effect of splice-site mutation 8051+1G→A on the FBN1 transcript. Direct sequencing of RT-PCR products from fibroblast of a normal control (top) and proband FB1477 with 8051+1G→A (bottom) reveals skipping of exon 63. This event leads to deletion of the two most C-terminal cbEGF-like domains and to a frameshift, resulting in premature termination of translation at amino acid position 2751 in exon 65 in the unique C-terminal region.
Assessment of Allele-Specific FBN1 Transcript Levels
Since relative levels of mutant RNA were estimated by three different methods and on more than one occasion, the combined results were placed into categories defined by brackets (table 1). Representative samples in which the FBN1 mutation either generated a new restriction site or abolished one (FB860, FB969, FB939, FB938, FB1305, FB1162, FB1058, and FB1324) were evaluated by restriction digestion of radioactively labeled and unlabeled RT-PCR products from cultured skin fibroblasts. Labeled and unlabeled PCR products of the corresponding genomic regions were used as controls. In all cases studied, the mutant transcripts were reduced; the degree of reduction was expressed as nondetectable/severely reduced (<10%) (level 3), markedly reduced (10%–29%) (level 2), and slightly reduced (30%–50%) (level 1) (table 1 and fig. 2).
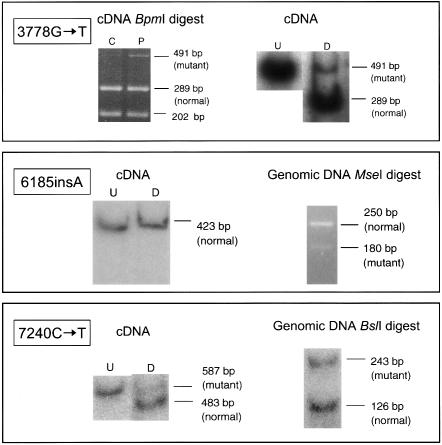
Figure 2.
Detection of allele-specific transcripts. Top, Mutation 3778G→T abolishes a _Bpm_I site. Top left, Unlabeled RT-PCR of a region spanning nts 3506–3996 of the FBN1 cDNA sequence, digested with _Bpm_I. Digestion products from both the proband (P) and a normal control (C) are shown. Top right, 32P-labeled RT-PCR and digestion with _Bpm_I of proband cDNA. Both digested (D) and undigested (U) products are shown. Middle, Mutation 6185insA generates new _Mse_I site. Middle left, 32P-labeled RT-PCR and digestion with _Mse_I. Undigested (U) and digested (D) samples from proband are shown. Middle right, Genomic PCR of exon 50 from proband and digestion with _Mse_I. Bottom, Mutation 7240C→T abolishes _Bsl_I site. Bottom left, 32P labeled RT-PCR and digestion with _Bsl_I. Undigested (U) and digested (D) samples from proband are shown. Bottom right, 32P labeled genomic PCR of exon 58 from proband and digestion with _Bsl_I.
In FB1058, the 3778G→T (E1260X) nonsense mutation in exon 30 destroys a _Bpm_I site. Fragments representative of both the normal (289 bp and 202 bp) and mutant alleles (491 bp) were observed (fig. 2, top). A normal control RNA sample was reverse transcribed, amplified, and digested in parallel. Although the 491-bp full-length PCR amplicon was still faintly visible, it was much less intense, when visualized on an agarose gel, than the corresponding band in the FB1058 sample. Labeled RT-PCR with subsequent _Bpm_I digestion resulted in an abundance of the 289-bp fragment and a disproportionately small amount of the mutant 491-bp fragment upon phosphorimaging. _Bpm_I digestion of labeled genomic PCR products resulted in bands representative of both mutant (491 bp) and normal (289 bp + 202 bp) alleles. These were present at an ∼1:1 ratio (data not shown).
FB1324 is heterozygous for the frameshift mutation 6185insA in exon 50 (table 1 and fig. 2, middle). _Mse_I digestion of unlabeled and radioactively labeled RT-PCR products allowed the visualization of only the original 423-bp PCR amplicon, reflecting the normal allele. The 345-bp band expected to be generated from the mutant allele was not detectable. Digestion of unlabeled as well as 32P-labeled genomic PCR amplicons produced a 250-bp band (normal allele) and a 180-bp fragment (mutant allele). Thus, failure to detect the mutant allele in cDNA was not a result of enzyme inefficiency. In the genomic digest, however, the mutant 180-bp fragment is considerably weaker than the 250-bp undigested normal allele, probably because of formation of heteroduplex molecules that are resistant to _Mse_I digestion. Therefore, a small amount of mutant cDNA would not have been detected in this assay.
Unlabeled and labeled cDNA PCR products from FB938, known to carry a 7240C→T (R2414X) nonsense mutation in exon 58, were digested with _Bsl_I (fig. 2, bottom). The restriction site is only present in the normal sequence. After agarose gel electrophoresis, bands of both the normal (483 bp) and mutant (587 bp) transcripts were identified. cDNA from a normal control was also amplified and fully digested with _Bsl_I. Phosphorimaging quantitation of the relative intensities of the bands representing the normal versus the mutant transcript in FB938 indicated mutant allele expression at 36%–45% of the normal allele. This may be an overestimate, however, because heteroduplex molecules would be included in the “mutant” undigested band.
To obtain an independent estimate of allele-specific transcript levels, samples FB773, FB798, FB821, FB860, FB969, FB997, FB1058, FB1103, FB1162, and FB1220 were also studied by RT-PCR of two regions with common FBN1 SNPs. C8931T, in the 3′ UTR, has a heterozygote frequency of 47% (Tynan et al. 1993; Hewett et al. 1994) and T1875C, in exon 15, has a heterozygosity level of 30% (Hayward et al. 1994). Nine studied samples were heterozygous for the _Rsa_I polymorphism in the 3′ UTR, and seven were heterozygous for the SNP in exon 15. Representative results for the 3′ UTR SNP are shown in figure 3. The level of unequal expression was designated by one of three degrees of severity defined above (i.e., level 3 = only one allele detected; level 2 = marked difference; and level 1 = slight difference) (table 1).
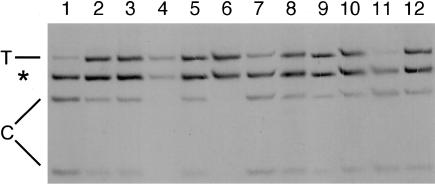
Figure 3.
Comparison of transcript levels for FBN1 alleles distinguished by the C6236T _Rsa_I polymorphism in the 3′ UTR. All samples are from heterozygotes. RT-PCR products from fibroblast RNA samples were digested with _Rsa_I and electrophoresed on an SSCA gel under denaturing conditions. The band labeled “T” represents the uncut T allele, the two bands labeled “C” represent the cut C allele, and the band marked by an asterisk (*) is constant. Samples 3 and 12 are normal controls. Unequal transcript levels are evident in four samples with PTC mutations. Lane 1, FB 1058 (E1260X); lane 4, FB 1103 (L1161X); lane 6, FB773 (R1192X); and lane 11, FB997 (R1523X). Samples 1 and 11 reveal predominant transcripts with the C allele, whereas in samples 4 and 6 only the T allele is detectable. In contrast, the sample in lane 2 is from FB774, with a genomic deletion of exons 42 and 43 (Liu et al. 2001), and the one in lane 8 is from FB808, with the cysteine substitution C637R (Schrijver et al. 1999). Both of these samples, as well as the ones in lanes 5, 9, and 10, generated the normal pattern of fragments, indicating equal transcripts of both alleles. Lane 7 shows slight skewing towards the C allele. For samples 5, 7, 9, and 10, no FBN1 mutations have been identified as yet.
Of the 14 samples with PTC mutations, 8 were informative with two of the three assays used, 5 were informative with one, and 1 was informative with all three. In all 14 samples, the mutant transcript levels were reduced compared with levels in the normal allele, or, in phase-unknown cases, one of the transcript levels was lower than the other. The combined results were expressed as one of three categories of reduction (table 1).
Pulse-Chase Studies of Fibrillin Protein
Pulse-chase analysis, a quantitative assay for monitoring protein synthesis in the cell and deposition of newly synthesized protein in the extracellular connective tissue, enables subdivision of patients with type 1 fibrillinopathies into four different pathologic phenotypic groups (see table 1 of Aoyama et al. 1994). The assay was performed on 28 fibroblast cultures with PTC mutations (table 1 and fig. 4). The vast majority (25/28) of these samples were assigned to protein group I or II, both of which are characterized by reduced synthesis of normal fibrillin molecules (<70% of normal control values). This result is consistent with the presence of a single wild-type FBN1 allele in these cells. In the 21 group II samples, there was disproportionate reduction of extracellular matrix deposition (<35% of control values) of the newly synthesized full-length fibrillin molecules. The four samples in protein group I (characterized by reduced fibrillin synthesis and proportionate reduction of extracellular matrix deposition at 35%–70% of control values) carry mutations that lead to PTC in exons 8, 15, 28, and 50. Mutant transcript levels, studied in only one of the four group I samples (FB969), were undetectable.
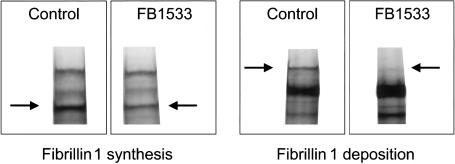
Figure 4.
Reduced fibrillin 1 synthesis and deposition in a proband with a premature termination mutation in the FBN1 gene. Radioactively labeled fibrillin (arrows) is visualized after pulse-chase analysis of cultured skin fibroblasts from a normal control subject and FB1533 (5240insACACT, leading to a PTC in exon 46). After a 30-min pulse with 35S-cysteine, FB1533 demonstrates reduced synthesis of normal-sized fibrillin (34% of the normal control sample). Protein deposition is assessed after a 20-h “chase” period in which labeled fibrillin is secreted from the cells and accumulates in the extracellular matrix. Matrix deposition is reduced in FB1533. At 20% of the normal control level, it is disproportionately low, suggesting a dominant-negative mechanism. These results place FB1533 into protein group II (Aoyama et al. 1994).
In this study, which was performed blindly, only 3/28 samples showed apparently normal levels of fibrillin synthesis: two in group III (with moderately reduced deposition) and one in group IV (with severely reduced deposition). Normal fibrillin protein synthesis has also been reported for the frameshift mutation 4365delCT (Halliday et al. 1999). It is possible that skipping of the PTC-containing exon has generated an apparently normal-sized mutant protein or that the normal FBN1 allele is somehow upregulated in these cases. Remarkably, none of our samples were found to have normal fibrillin synthesis and normal deposition (i.e., group V), which supports the sensitivity of the pulse-chase assay for the detection of molecular pathology in type 1 fibrillinopathies, as well as the specificity of the group assignments (table 1) (Brenn et al. 1996).
PTC-Associated Clinical Phenotypes
Detailed clinical information on 34 probands and 26 mutation-positive family members is listed in table 2. Of the individuals with a PTC mutation, dilatation of the ascending aorta was present in 44/57 individuals (77%) and required surgical aortic root replacement in 26/60 individuals (43%). The average age at cardiovascular surgery was 37 years, with a range of 22–59 years. Mitral-valve prolapse (MVP), in 38/57 individuals (67%), and mitral regurgitation (MR), in 35/53 individuals (66%), were also common findings.
Table 2.
Phenotypic Details of Probands and Relatives with Premature Termination Mutations
| PTC Location | Clinical Statusb | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Skeletalc | Oculard | Cardiovasculare | Otherf | ||||||||||||||||||||||
| Subjecta(Age inYears) | Mutated Exon | Stop Exon | DCS | PE/PC | SC | H/NP | DC | CTR | HSJ | HLJ | WS | TS | ARD | Myopia | EL | RD | AAA | AD | ARR | MVP | MR | PMT | Hernia | Striae | Age at Death(years) |
| 1075 (32) | IVS2 | del 2 | − | C | + | + | + | − | − | + | − | − | + | S | − | − | 32 | + | 32 | − | + | − | − | + | |
| 1075-2 (41) | IVS2 | del 2 | ? | C | + | − | − | − | − | ? | − | − | + | S | − | − | + | − | 32 | ? | + | + | B | + | |
| 845 (35) | 5 | 5 | + | E | S, SU | + | + | − | + | + | + | + | + | + | − | − | 26 | + | 27 | + | + | + | B | + | |
| 807 (50) | 6 | 8 | + | C | + | + | + | + | + | + | + | + | + | + | − | − | 38 | − | 49 | − | + | − | U | + | |
| 860 (67) | 7 | 7 | + | C | + | + | + | + | − | − | − | − | + | + | − | − | 67 | − | 67 | + | + | − | BR | − | |
| 860-01 (34) | 7 | 7 | + | − | + | + | + | + | + | + | + | + | + | + | B | − | 33 | − | − | + | + | − | U | + | |
| 1305 (37) | 7 | 7 | ? | ? | ? | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | 27 | + | 28 | − | + | + | ? | ? | |
| 798 (59) | 10 | 10 | ? | C | + | + | ? | ? | ? | ? | ? | ? | + | − | − | − | 30 | + | 30 | + | + | − | − | + | |
| 798-01 (29) | 10 | 10 | − | C | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | + | |
| 1592 (45) | 13 | 14 | ? | C | + | + | − | + | + | + | − | + | ? | − | − | − | 35 | + | 42 | + | + | − | − | + | |
| 969 (57) | 15 | 15 | ? | E | + | + | + | + | + | + | − | − | + | S | − | − | ? | − | − | + | + | − | U | + | |
| 969-01 (33) | 15 | 15 | + | − | + | + | + | − | − | + | + | − | + | S | − | − | + | + | 31 | − | + | − | H | + | |
| 969-02 (23) | 15 | 15 | − | − | − | + | + | − | − | + | − | + | − | − | − | − | 22 | + | 22 | − | + | − | − | + | |
| 1002 (37) | 15 | 17 | − | − | − | − | − | − | + | + | − | − | + | + | − | − | + | + | 29 | − | − | − | − | + | |
| 1002-01 (14) | 15 | 17 | + | C | + | + | + | + | + | + | − | − | + | + | B | + | 7 | − | − | − | − | − | − | − | |
| 821 (43) | 19 | 19 | − | − | + | + | + | − | + | + | − | − | + | + | B | − | 42 | − | − | S | + | − | IN | + | |
| 993 (49) | 21 | 21 | + | C | + | + | + | + | + | + | + | − | + | S | B | + | 49 | − | 49 | − | + | + | U | + | |
| 1162 (58) | 21 | 21 | − | C | + | + | + | + | + | + | + | + | + | + | − | − | 51 | + | 51 | − | − | − | − | + | |
| 1162-01 (34) | 21 | 21 | + | − | S | + | + | + | + | + | + | + | − | + | B | − | − | − | − | + | − | − | − | + | |
| 1162-02 (30) | 21 | 21 | + | − | + | − | − | − | − | + | − | + | ? | + | − | − | − | − | − | + | − | − | − | + | |
| 1103 (43) | 27 | 28 | + | E | S, SU | − | − | − | + | + | + | + | + | S | − | − | 31 | + | 31 | + | ? | − | UM | + | 45 |
| 1234 (34) | 27 | 28 | − | EC | + | + | − | + | + | + | + | + | + | − | U | + | − | − | − | + | + | + | H | + | |
| 1234-11 (56) | 27 | 28 | ? | E | + | + | + | − | + | + | − | − | + | + | − | − | − | − | − | + | ? | − | H | + | |
| 1234-2 (29) | 27 | 28 | ? | − | − | + | + | + | + | + | + | + | + | + | B | − | − | − | − | + | + | − | − | + | |
| 857 (23) | 28 | 28 | + | E | + | + | − | + | + | + | + | + | + | + | − | − | 17 | − | − | + | + | + | − | + | |
| 773 (55) | 28 | 28 | ? | E | − | + | ? | − | ? | ? | ? | ? | + | − | − | − | 45 | − | 45 | + | + | + | − | + | 55 |
| 773-02 (25) | 28 | 28 | − | C | + | + | − | + | + | + | − | − | + | + | B | − | + | − | − | + | − | − | − | − | |
| 773-002 (6) | 28 | 28 | + | C | − | + | + | − | + | + | + | + | + | + | B | − | 6 | − | − | + | + | − | − | − | |
| 1439 (38) | 30 | 30 | − | C | − | + | + | + | + | − | + | + | + | − | − | − | 34 | + | 34 | − | − | − | − | + | |
| 1439-01 (7) | 30 | 30 | − | − | − | + | − | + | + | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | |
| 1058 (13) | 30 | 30 | + | + | S, SU | + | + | + | + | + | + | + | + | + | − | − | − | − | − | + | + | − | − | − | |
| 751 (62) | 33 | 34 | + | − | S | + | + | − | + | + | + | + | + | + | − | − | 54 | − | 59 | − | + | − | IN | + | |
| 751-02 (40) | 33 | 34 | ? | E | S, SU | + | + | − | + | + | + | + | + | − | − | − | − | − | − | + | − | − | − | + | |
| 751-04 (38) | 33 | 34 | ? | E | + | + | + | − | + | + | ? | ? | + | − | U | − | + | − | − | + | − | − | − | + | |
| 751-001 (8) | 33 | 34 | ? | − | + | + | + | − | + | − | ? | ? | + | − | − | − | − | − | − | − | − | − | − | − | |
| 1479 (36) | 35 | 35 | ? | C | − | + | + | ? | + | ? | + | ? | + | + | U | − | − | − | − | − | + | − | − | + | |
| 997 (49) | 36 | 36 | + | C | S | + | + | − | + | + | + | + | + | + | − | − | 47 | − | 48 | + | + | − | − | + | 49 |
| 1186 (38) | 39 | 39 | ? | E | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | 37 | + | 37 | − | − | ? | ? | ? | |
| 1533 (38) | 42 | 46 | ? | C | − | − | + | − | + | ? | ? | ? | + | S | B | − | 26 | − | 37 | + | − | − | − | + | |
| 1533-3 (34) | 42 | 46 | ? | C | − | + | + | + | + | ? | ? | ? | + | S | B | − | 22 | − | − | + | − | − | − | + | |
| 754 (33) | 47 | 47 | ? | C | ? | + | ? | ? | ? | ? | ? | ? | + | ? | ? | ? | 31 | − | 31 | ? | ? | ? | ? | ? | |
| 768 (24) | 47 | 47 | − | E | S | + | + | + | + | + | + | + | + | + | − | − | 7 | − | − | + | + | − | − | + | |
| 768-2 (22) | 47 | 47 | − | C | + | + | + | + | + | + | + | + | − | − | − | − | 5 | − | − | + | + | − | B | + | |
| 768-3 (20) | 47 | 47 | − | EC | + | + | + | + | + | + | + | + | + | + | − | − | 3 | − | − | + | + | − | − | + | |
| 768-4 (18) | 47 | 47 | − | C | + | + | + | + | + | + | + | + | − | + | − | − | 8 | − | − | + | + | − | − | + | |
| 733 (33) | 49 | 50 | − | − | + | + | − | + | + | − | + | + | + | − | B | − | 25 | − | 30 | + | + | − | − | + | |
| 1286 (33) | 50 | 50 | − | C | + | + | − | + | + | + | + | + | + | − | − | − | 27 | + | 33 | − | + | − | − | + | |
| 1286-8 (46) | 50 | 50 | ? | C | − | + | − | − | + | + | − | − | + | − | − | − | ? | − | − | ? | ? | − | − | + | |
| 1286-12 (55) | 50 | 50 | + | C | + | + | − | + | + | + | − | + | + | − | − | − | 54 | − | − | + | ? | − | − | − | |
| 1324 (42) | 50 | 50 | + | EC, SU | − | + | + | + | + | + | − | − | + | + | B | − | 32 | + | 37 | + | − | − | B | + | |
| 1324-01 (16) | 50 | 50 | + | − | − | + | + | − | + | + | − | − | + | + | − | − | − | − | − | + | + | − | − | + | |
| 1220 (22) | 50 | 52 | + | EC | + | + | + | − | + | + | + | + | + | + | − | − | 21 | − | 22 | − | + | + | + | + | |
| 939 (64) | 52 | 52 | − | − | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | H | + | |
| 939-01 (45) | 52 | 52 | + | C | S | + | + | + | + | − | + | + | + | + | B | − | 11 | − | − | + | − | − | − | + | |
| 939-001 (14) | 52 | 52 | + | E | + | + | + | + | + | − | + | + | + | − | − | − | 8 | − | − | + | − | − | − | − | |
| 849 (44) | 53 | 53 | − | E | − | + | + | + | + | + | + | − | − | − | − | − | 37 | − | − | + | + | − | − | + | |
| 849-01 (19) | 53 | 53 | − | − | S | + | + | + | + | + | + | + | − | − | B | − | 14 | − | − | + | + | − | − | + | |
| 938 (22) | 58 | 58 | C | S | + | + | + | + | ? | + | + | + | + | − | − | 22 | + | 22 | + | ? | − | − | + | 29 | |
| 1420 (24) | 63 | 63 | − | E | + | + | − | − | + | + | + | + | + | S | B | − | 17 | − | − | + | + | − | − | + | |
| 1447 (9) | IVS63 | 65 | − | E | − | + | + | − | + | + | + | + | + | + | − | − | − | − | − | + | + | − | − | − |
Ocular-system manifestations were distinctly less frequent. Lens dislocation, or ectopia lentis (EL), was diagnosed in only 18/57 individuals who had received dilated eye examinations (32%). EL was bilateral in 15 and unilateral in 3 individuals. Of 10 families, 9 were discordant for the presence of EL. Myopia was present in 38/57 individuals (67%). Retinal detachment (RD) was rare, having occurred in only 3/57 patients (5%).
Skeletal features were pronounced, often with excessive height measurements. The vast majority of study participants had pectus deformities, scoliosis, high and narrow palate, joint hypermobility, and arachnodactyly. Wrist and thumb signs were present in 29 subjects, out of 50 who were evaluated for both features (58%). Although skeletal features were common, only five patients underwent surgical treatment, one for severe pectus deformity and four for scoliosis.
Cutaneous striae atrophicae (not associated with weight change or pregnancy) were present in 47/57 (82%); inguinal, incisional, or umbilical hernias were present in 17/57 (30%); and spontaneous pneumothorax was reported in 8/58 (14%) of study subjects.
Genotype-Phenotype Correlations: Comparing PTC to Cysteine Substitution Mutations
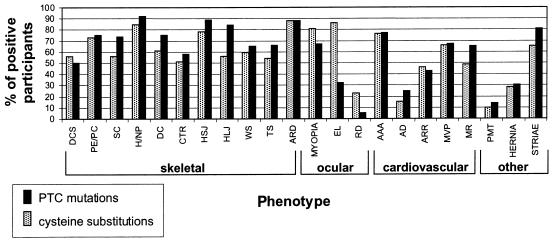
The phenotypic manifestations of the 60 study subjects with PTC mutations were compared to those of the 44 previously reported individuals with FBN1 cysteine substitutions (Schrijver et al. 1999), by χ2 analysis with 1 df. These are the largest mutation-specific groups of type 1 fibrillinopathies for which detailed phenotypic data are available. Distinct differences between the two groups were observed (fig. 5).
Figure 5.
Frequency of connective-tissue clinical features in study participants with PTC mutations, compared with those with cysteine substitutions (Schrijver et al. 1999). The percentage of all participants who are positive for the studied skeletal, ocular, cardiovascular, and other phenotypes in each of the two groups is demonstrated along the _Y_-axis. Individual manifestations are listed along the _X_-axis. Blackened columns represent PTC mutations, and shaded columns reflect cysteine substitutions. DCS = dolichostenomelia; PE/PC = pectus excavatum/pectus carinatum; SC = scoliosis; H/NP = high/narrow palate; DC = dental crowding; CTR = contractures; HSJ = hypermobile small joints; HLJ = hypermobile large joints; WS = wrist sign; TS = thumb sign; ARD = arachnodactyly; EL = ectopia lentis; RD = retinal detachment; AAA = ascending aortic aneurysm; AD = aortic dissection; ARR = aortic root replacement; MVP = mitral valve prolapse; MR = mitral regurgitation; and PMT = pneumothorax. Differences for HLJ, EL, and RD were statistically significant.
Most notably, EL was present in 36/42 (86%) informative individuals with cysteine substitutions but in only 18/57 (32%) of those with PTC mutations (P<.001). RD was also much more prevalent in the cysteine substitution group, at 9/39 (23%), versus 3/57 (5%) in the PTC group (P<.01). For all ocular findings, the fraction of positives were very similar when probands and relatives were considered separately. The observed differences cannot be accounted for by differences in age. Skeletal system manifestations, in contrast, were more pronounced in patients with PTC mutations. Large-joint hypermobility was present in 42/50 (84%) individuals in the group with PTC mutations and in 19/34 (56%) individuals in the cysteine substitution group (P<.01). Scoliosis was associated with 42/57 (74%) of all PTC mutations and with 20/36 (56%) of cysteine substitutions, but this difference was not significant (_P_=.07). Aortic dissections appeared to be more common in the premature termination group, occurring in 15/60 (25%) patients, compared with 6/40 (15%) individuals in the cysteine substitution group (P value not significant). As expected, aortic dissection in both groups was more frequent in probands: it was present in 38% of probands with PTC versus 8% of mutation-positive relatives, and in 23% of probands with cysteine substitution and 6% of relatives.
In the cysteine substitution group, 6/19 (32%) ascending aortic replacements were performed for dissection and the remainder for aortic dilatation, whereas in the PTC group dissection was the predominant indication (15/26 [58%]; _P_=.08). In addition to aortic dissection, MR was also more common in the PTC group, with 35/54 (65%) affected, compared with 18/37 (49%) in the cysteine substitution group (_P_=.12; difference not significant). Spontaneous pneumothorax occurred in 8/58 (14%) individuals in the PTC group and in 4/39 (10%) individuals in the cysteine substitution group. Both incidence figures are higher than in a clinically ascertained heterogeneous group of patients with MFS, in which only 11/249 (4.4%) had spontaneous pneumothorax (Hall et al. 1984). Our detailed presentation of the clinical data for mutation-specific subgroups should enable their inclusion in future meta-analyses of larger patient cohorts.
Discussion
FBN1 mutations have been associated with a broad spectrum of phenotypes, ranging from single connective-tissue manifestations, such as isolated EL (Kainulainen et al. 1994; Lonnqvist et al. 1994), to lethal neonatal MFS (Milewicz and Duvic 1994; Putnam et al. 1996). In this article on PTC mutations and in our previous work on in-frame internal deletions (Liu et al. 1996, 1997, 2001) and cysteine substitutions (Schrijver et al. 1999), we have presented extensive studies integrating data on mutation, transcript, and protein and clinical phenotypes. Distinct mutation-type–specific profiles have been established at the transcript and protein level. It now appears that it may also be possible to subdivide MFS and other type 1 fibrillinopathies clinically, depending on the type of FBN1 mutation present. Although no specific manifestation or set of features are diagnostic for a particular subtype, the relative risk for specific organ involvement shows some statistically significant differences.
Here, we report the detection of 34 FBN1 nonsense or frameshift (PTC) mutations, 32 of which are unique and 2 of which are recurrent. The 34 probands and 26 mutation-positive family members were studied to assess the clinical and biochemical characteristics of this mutation category. Phenotypes in our study participants showed a range of features. On clinical grounds alone, 22/31 (71%) of probands for whom complete clinical data were available met the revised diagnostic criteria for MFS (De Paepe et al. 1996), when the FBN1 mutation was not included as a criterion. If imaging studies to look for dural ectasia and protrusio acetabuli had been performed, the number of participants meeting clinical criteria for MFS might have been higher (De Paepe et al. 1996; Rose et al. 2000). Age at ascertainment is clearly important. With the median proband age of 38 years, only two of the nine informative probands who did not meet criteria were children who may not yet express diagnostic cardiovascular manifestations.
To assess the effects of the various PTC mutations on mutant mRNA stability, we performed FBN1 mRNA quantitation. Eight samples were studied by RT-PCR and mutation-specific restriction digestion of the amplicon after a low number of PCR cycles (Schrijver et al. 1999). In all individuals studied by this method, mutant transcript levels were decreased, which suggests that mutant mRNA is preferentially degraded. In addition, two FBN1 SNPs were analyzed in 10 individuals, and, in each case, transcript levels of one allele were reduced. Since the alleles were distinguished by SNPs unrelated to the mutation, however, the mutant allele could only be inferred.
Cellular recognition and degradation of mRNA that contains premature termination codons is a process whereby potentially harmful effects of truncated proteins may be limited (Culbertson 1999; Frischmeyer and Dietz 1999). This RNA surveillance or nonsense-mediated mRNA decay (NMD) pathway controls the effects of transcription errors, aberrant pre-mRNA splicing, and nonproductive rearrangements in the immune system. In human genetic diseases, NMD directly impacts the pathological effects of PTC mutations. PTC mutations make up ∼25% of recognized spontaneous disease-causing mutations in hundreds of human genes. The effectiveness of NMD on PTC-containing transcripts is highly relevant to the postulated mechanism of disease. Rapid decay of mutant transcripts leads to a loss-of-function phenotype, as first described in recessive β-thalassemia (Baserga and Benz 1992), while synthesis of truncated proteins may have a dominant negative effect—for example, in the adenomatous polyposis coli gene (APC [MIM 175100]), leading to dominant familial polyposis coli. On the other hand, a truncated protein may restore partial function and, therefore, the reduction of NMD may have a beneficial effect, as, for example, in CFTR. Regarding the mechanism of NMD, recent studies support a direct connection between RNA splicing and NMD (Le Hir et al. 2001). An intron introduced within the 3′ UTR at a certain distance downstream of the termination codon will target a normal mRNA for NMD (Thermann et al. 1998). Introns are not absolutely required for occurrence of decay, however, as was demonstrated in the HEXA gene. Thus, sequences in cis may also play a role (Rajavel and Neufeld 2001). Cytoplasmic translation is a necessary component of the NMD pathway (Thermann et al. 1998).
For FBN1 mutations, preferential degradation of transcripts containing premature stop codons has been described, with mutant transcript levels ranging from 2%–28%, compared with the normal transcript (Dietz et al. 1993_a,_ 1993_b;_ Aoyama et al. 1994; Karttunen et al. 1998; Halliday et al. 1999). Normal transcript levels have only rarely been associated with a PTC mutation; examples include a nonsense mutation in exon 24 (Karttunen et al. 1998) and a frameshift in exon 65, the 3′-most exon (Nijbroek et al. 1995). The latter finding is expected, since PTCs in the final exon do not interfere with mRNA stability. Early studies had suggested that low mutant transcript levels are associated with a milder phenotype (Dietz et al. 1993_a;_ Aoyama et al. 1994). More recently, however, a 7% level of mutant transcript and near absence of immunohistological staining of fibrillin 1 in cultured fibroblasts were reported for an individual with an inherited R529X mutation who had severe aortic and skeletal manifestations (Montgomery et al. 1998). A reported individual with severe MFS and an R1541X mutation had mutant mRNA of 2% compared with the transcript of the normal allele (Hewett et al. 1994; Halliday et al. 1999). These observations throw doubt on the hypothesis that mutant mRNA levels in cultured cells are the major determinants of the clinical and biochemical consequences of FBN1 PTC mutations.
In pulse-chase protein studies, null mutations, or total lack of mutant transcripts, would lead to reduced fibrillin synthesis and deposition of only normal fibrillin 1, albeit in reduced amounts. This scenario is compatible with protein group I, characterized by fibrillin synthesis at 50% of control values (±2 SD; range 35%–70%) and proportional reduction of deposition (Aoyama et al. 1994, 1995). This protein phenotype was identified in only 4 of the 28 probands we studied. The majority of fibrillin 1 truncation mutations in the present study were classified as protein phenotype II, with fibrillin synthesis at 50% of control values (±2 SD; range 35%–70%) and severely reduced deposition (<2 SD of control values; <35%). Attempts to establish a correlation between mutant mRNA (MT) levels and protein groups in the 14 samples studied for both parameters revealed no significant results. The 13 samples in protein group II were associated with MT levels from (1) mild (_n_=4), to (2) intermediate (_n_=7), and (3) severe reduction (_n_=2). The one group I sample was associated with level 3, as expected.
The mechanism by which PTC mutations exert their effect on extracellular fibrillin monomers and their incorporation into microfibrils is not entirely clear. The existence of putative truncated proteins has been difficult to prove. Because of NMD of mutant transcripts, they are expected to be made at low abundance. Although western blotting and immunoprecipitation experiments have failed, we have previously published evidence of a putative truncated protein of ∼60 kD isolated from the medium of a group II fibroblast culture after enrichment on an immunoaffinity column (see fig. 6 of Aoyama et al. 1994). We subsequently identified a FBN1 mutation, 4177delG in exon 33, leading to a stop codon in exon 34 after a stretch of missense codons, in this sample (FB751; table 1). Although this mutation would predict a truncated protein of ∼150 kD, we found a ∼60-kD product that might reflect degradation, either during the isolation procedure or because of protein instability and intra- or extracellular proteolysis. This finding points to yet another variable—namely, that the location of the stop codon may not be indicative of the ultimate size of the mutant product and, hence, of its putative biological activity.
Among 41 probands with PTC mutations reported by others, there are two recurrent and 35 unique mutations, a total of 14 nonsense and 23 frameshift mutations (Dietz et al. 1993_a;_ Nijbroek et al. 1995; Wang et al. 1997; Montgomery et al. 1998; Halliday et al. 1999; Guo et al. 2001; Loeys et al. 2001; Pepe et al. 2001; Tiecke et al. 2001). We included only 19 true PTC mutations from the study by Loeys et al. (2001), excluding those cases listed under “frameshift” that have in-frame deletions or insertions or that lead to skipping of in-frame exons. It is remarkable that the ratio of nonsense to frameshift mutations is very consistent, with 14/23 in the literature and 13/20 in our cases. The most common recurrent PTC mutation is 247+1G→A in the exon 2 donor splice site. It represents a C→T transition at a CpG mutational hotspot on the antisense strand. As a consequence of this mutation, exon 2 is skipped, which causes a frameshift and PTC. Of the five unrelated individuals with this mutation, the first one (MS8) reported (Dietz et al. 1993_a_) had classic and severe manifestations at age 29 years. Three subsequent individuals with the same mutation had no EL but had major aortic and minor skeletal involvement at ages 39, 41, and 52 (Halliday et al. 1999; Guo et al. 2001; Loeys et al. 2001). We assume that the individual listed as “del ex 2” by Loeys et al. (2001) had this same mutation, because the methods used would not have detected a genomic deletion of this exon. In our proband FB1075 and in her sibling, the same 247+1G→A mutation was identified. Both individuals lack EL and have relatively mild skeletal symptoms (table 2). The amount of fibroblast cDNA that was amplified from the mutant allele, as compared with the normal allele, was 16% for MS8 (Dietz et al. 1993_a_) and 9.9% for the 41-year-old patient reported by Guo et al. (2001). Pulse-chase analysis of proband MS8 revealed fibrillin synthesis of 50.7% and matrix deposition of 25.1% (Aoyama et al. 1994). In the patient reported by Guo et al. (2001), pulse-chase analysis revealed synthesis of 41% and deposition of <5% of that in normal controls. Without quantitation, Halliday et al. (1999) reported reduced synthesis and reduced deposition in all four individuals with PTC studied by pulse-chase analysis, including the 247+1G→A mutation carrier.
Comparison of our clinical data (table 2) with the that from 41 patients with FBN1 PTC mutations reported in the literature is limited to features that have been consistently reported. Our most striking finding—that is, that EL is present in only 32% (18/57) of subjects—is corroborated by the frequency of 15/41 (37%) reported in the literature, when we interpret “major criterion in the ocular system” (Loeys et al. 2001) as indicating the presence of EL. Although major criteria in the cardiovascular system were reported in 37/41 (90%) individuals, we recorded ascending aortic dilation and/or dissection in only 44/57 (77%) of participants who were evaluated for both features. This difference may be explained by most published cases being in probands, whereas 43% of our study population were secondarily ascertained mutation-positive relatives. When we consider only the probands, the frequency is 28/34 (82%).
In an effort to define mutation-type–specific clinical subgroups, we compared extensive clinical data on 60 study participants (table 2) to a large group of subjects with type 1 fibrillinopathy due to cysteine substitutions (Schrijver et al. 1999). The PTC-associated phenotype emerges as a distinguishable clinical entity. Whereas large-joint hypermobility (P<.01), scoliosis, and aortic dissection were more common in the PTC population, EL (P<.001) and RD (P<.01) were strikingly less frequent than in the cysteine-substitution group.
Comparison of the protein phenotypes demonstrates that the preponderance (75%) of probands with PTC mutations have markedly reduced extracellular fibrillin deposition with reduced synthesis. In contrast, 76% of individuals with cysteine substitutions had normal levels of fibrillin synthesis and markedly reduced matrix deposition. In this group, newly synthesized fibrillin may be sequestered intracellularly, since secretion was delayed (Schrijver et al. 1999). These comparisons represent a first step in understanding the differential molecular pathology of type 1 fibrillinopathies.
In conclusion, our study provides the first comprehensive genotype-phenotype correlation in individuals with PTC mutations in FBN1. The new pathogenic insights about this mutation category will allow more-precise assessment of the clinical prognosis and potentially will lead to tailored patient management. For example, PTC mutation carriers may not meet clinical criteria of MFS, and the absence of EL in the majority of patients with PTC may delay diagnosis. Yet, these patients are at high risk for aortic dissection and mitral valve insufficiency. They must be monitored closely for these life-threatening cardiovascular complications. The risk for EL is distinctly lower than the 60% usually quoted for patients with MFS (GeneTests Web site). This risk figure was derived by studying heterogeneous populations with MFS, merging higher-risk groups (with FBN1 cysteine substitutions and in-frame exon skipping mutations or deletions) with the lower-risk PTC group. Therefore, FBN1 mutation testing and/or fibrillin protein phenotyping performed in persons suspected of having a type 1 fibrillinopathy may assist in assignment to a mutation-type–specific subgroup for individualized prognosis and management.
Acknowledgments
We thank D. C. Miller, for samples; C. Qian, K. Bogard, A. Roxas, and V. Meyers, for technical support; C. Gasner, J. Mendoza, and the physicians and genetic counselors associated with the Stanford Center for Marfan Syndrome and Related Connective Tissue Disorders, for help in collecting samples and clinical data; and L. Lazzeroni, for statistical analysis. This research was supported by the Howard Hughes Medical Institute (support to I.S., W.L., R.O., and U.F.), National Institutes of Health grant R01 GM28428 (to P.O.), the Deutsche Forschungsgemeinschaft (support to T.B.), the Kyle Mann Research Fund (support to U.F. and H.F.), and the National Marfan Foundation (support to H.F.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GeneTests, http://www.genetests.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MFS [MIM 154700], FBN [MIM 1134743], and APC [MIM 175100])
- UCSC Human Genome Project Working Draft, http://genome.cse.ucsc.edu/
References
- Aoyama T, Francke U, Dietz HC, Furthmayr H (1994) Quantitative differences in biosynthesis and extracellular deposition of fibrillin in cultured fibroblasts distinguish five groups of Marfan syndrome patients and suggest distinct pathogenetic mechanisms. J Clin Invest 94:130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Francke U, Gasner C, Furthmayr H (1995) Fibrillin abnormalities and prognosis in Marfan syndrome and related disorders. Am J Med Genet 58:169–176 [DOI] [PubMed] [Google Scholar]
- Baserga SJ, Benz EJ Jr (1992) Beta-globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc Natl Acad Sci USA 89:2935–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroud C, Collod-Beroud G, Boileau C, Soussi T, Junien C (2000) UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum Mutat 15:86–94 [DOI] [PubMed] [Google Scholar]
- Biery NJ, Eldadah ZA, Moore CS, Stetten G, Spencer F, Dietz HC (1999) Revised genomic organization of FBN1 and significance for regulated gene expression. Genomics 56:70–77 [DOI] [PubMed] [Google Scholar]
- Booms P, Cisler J, Mathews KR, Godfrey M, Tiecke F, Kaufmann UC, Vetter U, Hagemeier C, Robinson PN (1999) Novel exon skipping mutation in the fibrillin-1 gene: two “hot spots” for the neonatal Marfan syndrome. Clin Genet 55:110–117 [DOI] [PubMed] [Google Scholar]
- Brenn T, Aoyama T, Francke U, Furthmayr H (1996) Dermal fibroblast culture as a model for studies of fibrillin assembly and pathogenetic mechanisms: defects in distinct groups of individuals with Marfan's syndrome. Lab Invest 75:389–402 [PubMed] [Google Scholar]
- Cheng J, Maquat LE (1993) Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol 13:1892–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collod-Beroud G, Beroud C, Ades L, Black C, Boxer M, Brock DJH, Holman KJ, de Paepe A, Francke UF, Grau U, Hayward C, Klein H-G, Liu W, Nuytinck L, Peltonen L, Alvarez Perz AB, Rantamaki T, Junien C, Boileau C (1998) Marfan database (third edition): new mutations and new routines for the software. Nucleic Acids Res 26:229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet 15:74–80 [DOI] [PubMed] [Google Scholar]
- De Paepe A, Devereux RB, Dietz HC, Hennekam RCM, Pyeritz RE (1996) Revised diagnostic criteria for the Marfan syndrome. Am J Med Genet 62:417–426 [DOI] [PubMed] [Google Scholar]
- Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, et al (1991) Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 362:337–339 [DOI] [PubMed] [Google Scholar]
- Dietz HC, McIntosh I, Sakai LY, Corson GM, Chalberg SC, Pyeritz RE, Francomano CA (1993_a_) Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics 17:468–475 [DOI] [PubMed] [Google Scholar]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ Jr, Pyeritz RE, Cutting GR (1993_b_) The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259:680–683 [DOI] [PubMed] [Google Scholar]
- Francke U, Berg MA, Tynan K, Brenn T, Liu W, Aoyama T, Gasner C, Miller DC, Furthmayr H (1995) A Gly1127Ser mutation in an EGF-like domain of the fibrillin-1 gene is a risk factor for ascending aortic aneurysm and dissection. Am J Hum Genet 56:1287–1296 [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8:1893–1900 [DOI] [PubMed] [Google Scholar]
- Furthmayr H, Francke U (1997) Ascending aortic aneurysm with or without features of Marfan Syndrome and other fibrillinopathies: new insights. Semin Thorac Cardiovasc Surg 9:191–205 [PubMed] [Google Scholar]
- Guo D, Tan FK, Cantu A, Plon SE, Milewicz DM (2001) FBN1 exon 2 splicing error in a patient with Marfan syndrome. Am J Med Genet 101:130–134 [DOI] [PubMed] [Google Scholar]
- Hall J, Pyeritz J, Dudgeon D, Haller JJ (1984) Pneumothorax in the Marfan syndrome: prevalence and therapy. Ann Thorac Surg 37:500–504 [DOI] [PubMed] [Google Scholar]
- Halliday D, Hutchinson S, Kettle S, Firth H, Wordsworth P, Handford PA (1999) Molecular analysis of eight mutations in FBN1. Hum Genet 105:587–597 [DOI] [PubMed] [Google Scholar]
- Handford PA, Downing AK, Reinhardt DP, Sakai LY (2000) Fibrillin: from domain structure to supramolecular assembly. Matrix Biol 19:457–470 [DOI] [PubMed] [Google Scholar]
- Hayward C, Rae AL, Porteous EM, Logie LJ, Brock DJH (1994) Two novel mutations and a neutral polymorphism in EGF-like domains of the fibrillin gene (FBN1): SSCP screening of exons 15–21 in Marfan syndrome patients. Hum Mol Genet 3:373–375 [DOI] [PubMed] [Google Scholar]
- Hewett D, Lynch J, Child A, Firth H, Sykes B (1994) Differential allelic expression of a Fibrillin gene (FBN1) in patients with Marfan syndrome. Am J Hum Genet 55:447–452 [PMC free article] [PubMed] [Google Scholar]
- Kainulainen K, Karttunen L, Puhakka L, Sakai L, Peltonen L (1994) Mutations in the fibrillin gene responsible for dominant ectopia lentis and neonatal Marfan syndrome. Nat Genet 6:64–69 [DOI] [PubMed] [Google Scholar]
- Kainulainen K, Sakai LY, Child A, Pope FM, Puhakka L, Ryhanen L, Palotie A, Kaitila I, Peltonen L (1992) Two mutations in Marfan syndrome resulting in truncated fibrillin polypeptides. Proc Natl Acad Sci USA 89:5917–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karttunen L, Ukkonen T, Kainulainen K, Syvanen AC, Peltonen L (1998) Two novel fibrillin-1 mutations resulting in premature termination codons but in different mutant transcript levels and clinical phenotypes. Hum Mutat 1998 Suppl 1:S34–S37 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001) The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. Embo J 20:4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Oefner PJ, Qian C, Odom RS, Francke U (1997/1998) Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorders. Genet Test 1:237–242 [DOI] [PubMed] [Google Scholar]
- Liu W, Qian C, Comeau K, Brenn T, Furthmayr H, Francke U (1996) Mutant fibrillin-1 monomers lacking EGF-like domains disrupt microfibril assembly and cause severe Marfan syndrome. Hum Mol Genet 5:1581–1587 [DOI] [PubMed] [Google Scholar]
- Liu W, Qian C, Francke U (1997) Silent mutation induces exon skipping of fibrillin-1 gene in Marfan syndrome. Nat Genet 16:328–329 [DOI] [PubMed] [Google Scholar]
- Liu W, Schrijver I, Brenn T, Furthmayr H, Francke U (2001) Multi-exon deletions of the FBN1 gene in Marfan syndrome. BMC Med Genet 2:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeys B, Nuytinck L, Delvaux I, De Bie S, De Paepe A (2001) Genotype and phenotype analysis of 171 patients referred for molecular study of the fibrillin-1 gene FBN1 because of suspected Marfan syndrome. Arch Intern Med 161:2447–2454 [DOI] [PubMed] [Google Scholar]
- Lonnqvist L, Child A, Kainulainen K, Davidson R, Puhakka L, Peltonen L (1994) A novel mutation of the fibrillin gene causing ectopia lentis. Genomics 19:573–576 [DOI] [PubMed] [Google Scholar]
- Lonnqvist L, Reinhardt D, Sakai L, Peltonen L (1998) Evidence for furin-type activity-mediated C-terminal processing of profibrillin-1 and interference in the processing by certain mutations. Hum Mol Genet 7:2039–2044 [DOI] [PubMed] [Google Scholar]
- Milewicz DM, Duvic M (1994) Severe neonatal Marfan syndrome resulting from a de novo 3-bp insertion into the fibrillin gene on chromosome 15. Am J Hum Genet 54:447–453 [PMC free article] [PubMed] [Google Scholar]
- Montgomery RA, Geraghty MT, Bull E, Gelb BD, Johnson M, McIntosh I, Francomano CA, Dietz HC (1998) Multiple molecular mechanisms underlying subdiagnostic variants of Marfan syndrome. Am J Hum Genet 63:1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijbroek G, Sood S, McIntosh I, Francomano CA, Bull E, Pereira L, Ramirez F, Pyeritz RE, Dietz HC (1995) Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am J Hum Genet 57:8–21 [PMC free article] [PubMed] [Google Scholar]
- Palz M, Tiecke F, Booms P, Goldner B, Rosenberg T, Fuchs J, Skovby F, Schumacher H, Kaufmann UC, von Kodolitsch Y, Nienaber CA, Leitner C, Katzke S, Vetter B, Hagemeier C, Robinson PN (2000) Clustering of mutations associated with mild Marfan-like phenotypes in the 3′ region of FBN1 suggests a potential genotype-phenotype correlation. Am J Med Genet 91:212–221 [DOI] [PubMed] [Google Scholar]
- Pepe G, Giusti B, Evangelisti L, Porciani MC, Brunelli T, Giurlani L, Attanasio M, Fattori R, Bagni C, Comeglio P, Abbate R, Gensini GF (2001) Fibrillin-1 (FBN1) gene frameshift mutations in Marfan patients: genotype-phenotype correlation. Clin Genet 59:444–450 [DOI] [PubMed] [Google Scholar]
- Pereira L, D'Alessio M, Ramirez F, Lynch JR, Sykes B, Pangilinan T, Bonadio J (1993) Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum Molec Genet 2:961–968 [DOI] [PubMed] [Google Scholar]
- Putnam E, Cho M, Zinn A, Towbin J, Byers P, Milewicz D (1996) Delineation of the Marfan phenotype associated with mutations in exons 23-32 of the FBN1 gene. Am J Med Genet 62:233–242 [DOI] [PubMed] [Google Scholar]
- Pyeritz RE (2000) The Marfan syndrome. Annu Rev Med 51:481–510 [DOI] [PubMed] [Google Scholar]
- Rajavel KS, Neufeld EF (2001) Nonsense-mediated decay of human HEXA mRNA. Mol Cell Biol 21:5512–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt DP, Keene DR, Corson GM, Poschl E, Bachinger HP, Gambee JE, Sakai LY (1996) Fibrillin-1: organization in microfibrils and structural properties. J Mol Biol 258:104–116 [DOI] [PubMed] [Google Scholar]
- Richards CS, Ward PA, Roa BB, Friedman LC, Boyd AA, Kuenzli G, Dunn JK, Plon SE (1997) Screening for 185delAG in the Ashkenazim. Am J Hum Genet 60:1085–1098 [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Godfrey M (2000) The molecular genetics of Marfan syndrome and related microfibrillopathies. J Med Genet 37:9–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PS, Levy HP, Ahn NU, Sponseller PD, Magyari T, Davis J, Francomano CA (2000) A comparison of the Berlin and Ghent nosologies and the influence of dural ectasia in the diagnosis of Marfan syndrome. Genet Med 2:278–282 [DOI] [PubMed] [Google Scholar]
- Saharinen J, Keski-Oja J (2000) Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol Biol Cell 11:2691–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai LY, Keene DR, Engvall E (1986) Fibrillin, a new 350-kD glycoprotein is a component of extracellular microfibrils. J Cell Biol 103:2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijver I, Alcorn DM, Francke U (2001) Marfan syndrome. In: Cassidy SB, Allanson JE (eds) Management of genetic syndromes. Wiley-Liss, New York, pp 207–228 [Google Scholar]
- Schrijver I, Liu W, Brenn T, Furthmayr H, Francke U (1999) Cysteine substitutions in epidermal growth factor–like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am J Hum Genet 65:1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. Embo J 17:3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiecke F, Katzke S, Booms P, Robinson PN, Neumann L, Godfrey M, Mathews KR, Scheuner M, Hinkel GK, Brenner RE, Hovels-Gurich HH, Hagemeier C, Fuchs J, Skovby F, Rosenberg T (2001) Classic, atypically severe and neonatal Marfan syndrome: twelve mutations and genotype-phenotype correlations in FBN1 exons 24–40. Eur J Hum Genet 9:13–21 [DOI] [PubMed] [Google Scholar]
- Tynan K, Comeau K, Pearson M, Wilgenbus P, Levitt D, Gasner C, Berg MA, Miller DC, Francke U (1993) Mutation screening of complete fibrillin-1 coding sequence: report of five new mutations, including two in 8-cysteine domains. Hum Mol Genet 2:1813–1821 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang JY, Cisler J, Imaizumi K, Burton BK, Jones MC, Lamberti JJ, Godfrey M (1997) Three novel fibrillin mutations in exons 25 and 27: classic versus neonatal Marfan syndrome. Hum Mutat 9:359–362 [DOI] [PubMed] [Google Scholar]