Legionella and Legionnaires' Disease: 25 Years of Investigation (original) (raw)
Abstract
There is still a low level of clinical awareness regarding Legionnaires' disease 25 years after it was first detected. The causative agents, legionellae, are freshwater bacteria with a fascinating ecology. These bacteria are intracellular pathogens of freshwater protozoa and utilize a similar mechanism to infect human phagocytic cells. There have been major advances in delineating the pathogenesis of legionellae through the identification of genes which allow the organism to bypass the endocytic pathways of both protozoan and human cells. Other bacteria that may share this novel infectious process are Coxiella burnetti and Brucella spp. More than 40 species and numerous serogroups of legionellae have been identified. Most diagnostic tests are directed at the species that causes most of the reported human cases of legionellosis, L. pneumophila serogroup 1. For this reason, information on the incidence of human respiratory disease attributable to other species and serogroups of legionellae is lacking. Improvements in diagnostic tests such as the urine antigen assay have inadvertently caused a decrease in the use of culture to detect infection, resulting in incomplete surveillance for legionellosis. Large, focal outbreaks of Legionnaires' disease continue to occur worldwide, and there is a critical need for surveillance for travel-related legionellosis in the United States. There is optimism that newly developed guidelines and water treatment practices can greatly reduce the incidence of this preventable illness.
INTRODUCTION
Legionnaires' disease has a false but enduring status as an exotic plague. In reality, this disease is a common form of severe pneumonia, but these infections are infrequently diagnosed. Failure to diagnose Legionnaires' disease is largely due to a lack of clinical awareness. In addition, legionellae, the bacteria that cause this disease, are fastidious and not easily detected.
Legionellae are gram-negative bacteria found in freshwater environments. The first strains of Legionella were isolated in guinea pigs by using procedures for the isolation of Rickettsia (203). The first was isolated in 1943 by Tatlock, with another strain isolated in 1947 by Jackson et al. (268). In 1954, Drozanski isolated a bacterium that infected free-living amoebae from soil in Poland (68). This organism was classified as a species of Legionella in 1996 (143). The genus Legionella was established in 1979 after a large outbreak of pneumonia among members of the American Legion that had occurred 3 years earlier (38, 104) and was traced to a previously unrecognized bacterium, Legionella pneumophila (203). Legionellae are intracellular parasites of freshwater protozoa and use a similar mechanism to multiply within mammalian cells (91). These bacteria cause respiratory disease in humans when a susceptible host inhales aerosolized water containing the bacteria or aspirates water containing the bacteria.
Clinical Presentation
Legionellosis classically presents as two distinct clinical entities, Legionnaires' disease, a severe multisystem disease involving pneumonia (104), and Pontiac fever, a self-limited flu-like illness (114). Additionally, many persons who seroconvert to Legionella will be entirely asymptomatic (33).
It is not possible to clinically distinguish patients with Legionnaires' disease from patients with other types of pneumonia (76). Features of Legionnaires' disease include fever, nonproductive cough, headache, myalgias, rigors, dyspnea, diarrhea, and delirium (273). Although no chest X-ray pattern can separate this infection from other types of pneumonia, alveolar infiltrates are more common with Legionnaires' disease (188). The key to diagnosis is performing appropriate microbiologic testing when a patient is in a high-risk category.
There is some debate as to whether Legionnaires' disease presents as a clinical spectrum or whether the only manifestation of disease is pneumonia. In a study of persons exposed to L. pneumophila who did not develop pneumonia during a large outbreak of Legionnaires' disease in the Netherlands, there was no difference in the rate of respiratory symptoms between those who were seropositive and those who were seronegative. This would suggest that patients either develop pneumonia or are asymptomatic (33). Outbreaks of legionellosis have occurred in which some patients develop Legionnaires' disease and others Pontiac fever (21).
One species of Legionella, L. pneumophila, causes approximately 90% of all reported cases of legionellosis in the United States (199; R. E. Besser, unpublished data). This figure may be inflated because most diagnostic tests are specific for L. pneumophila. Currently, there are 48 species comprising 70 distinct serogroups in the genus Legionella (5, 23, 184, 185). Although there are now 15 serogroups of L. pneumophila, 79% of all culture-confirmed or urine antigen-confirmed cases are caused by L. pneumophila serogroup 1 (199; A. L. Benin and R. E. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 873, 2001). Approximately one-half of the 48 species of legionellae have been associated with human disease. Since all legionellae are presumed to be capable of intracellular growth in some host cell (91), it is likely that most legionellae can cause human disease under the appropriate conditions (91). Infections due to the less common strains of Legionella are infrequently reported because of their rarity and because of the lack of diagnostic reagents (90).
Studies have estimated that between 8,000 and 18,000 persons are hospitalized with legionellosis annually in the United States (200). The disease is a major concern of public health professionals and individuals involved with maintaining building water systems. Legionellosis is generally considered a preventable illness because controlling or eliminating the bacterium in certain reservoirs will prevent cases of the disease. This concept of preventable illness has resulted in a number of guidelines and new control strategies aimed at reducing the risk of legionellosis in building water systems. The factors that lead to outbreaks or cases of Legionnaires' disease are not completely understood, but certain events are considered prerequisites for infection. These include the presence of the bacterium in an aquatic environment, amplification of the bacterium to an unknown infectious dose, and transmission of the bacteria via aerosol to a human host that is susceptible to infection (103).
Microbial Ecology
Water is the major reservoir for legionellae, and the bacteria are found in freshwater environments worldwide (98). Legionellae have been detected in as many as 40% of freshwater environments by culture and in up to 80% of freshwater sites by PCR (90). A single exception to this observation is Legionella longbeachae, a frequent isolate from potting soil (257). This species is the leading cause of legionellosis in Australia and occurs in gardeners and those exposed to commercial potting soil (244). The first U.S. cases of L. longbeachae infection associated with potting soil were reported in 2000 (45).
L. pneumophila multiplies at temperatures between 25 and 42°C, with an optimal growth temperature of 35°C (159). Most cases of legionellosis can be traced to human-made aquatic environments where the water temperature is higher than ambient temperature. Thermally altered aquatic environments can shift the balance between protozoa and bacteria, resulting in rapid multiplication of legionellae, which can translate into human disease. Legionellosis is a disease that has emerged in the last half of the 20th century because of human alteration of the environment. Left in their natural state, legionellae would be an extremely rare cause of human disease, as natural freshwater environments have not been implicated as reservoirs of outbreaks of legionellosis. Some outbreaks of legionellosis have been associated with construction, and it was originally believed that the bacteria could survive and be transmitted to humans via soil. However, L. pneumophila does not survive in dry environments, and these outbreaks are more likely the result of massive descalement of plumbing systems due to changes in water pressure during construction (159, 205).
Association with Amoebae
The presence of the bacteria in an aquatic environment and warm water temperature are two factors that can increase the risk of Legionnaires' disease. The third component is the presence of nutritional factors that allow the bacteria to amplify. These bacteria require a unique combination of nutrients in order to be grown in the laboratory. Initially, these unusual nutritional requirements appeared to contradict the widespread distribution of legionellae in freshwater environments. The levels of nutrients that legionellae require are rarely found in fresh water and, if present, would serve only to amplify faster-growing bacteria that would compete with the legionellae. However, these nutrients represent an intracellular environment, not soluble nutrients commonly found in fresh water.
Legionellae survive in aquatic and moist soil environments as intracellular parasites of free-living protozoa (91, 243). These bacteria have been shown to multiply in 14 species of amoebae, two species of ciliated protozoa, and one species of slime mold, while growth of legionellae in the absence of protozoa has been documented only on laboratory media (91, 123, 254). Protozoa naturally present in environments implicated as sources of Legionnaires' disease can support intracellular growth of legionellae in vitro (17). While protozoa are the natural hosts of legionellae, the infection of human phagocytic cells is opportunistic. Much of our understanding of the pathogenesis of legionellae has come from an analysis of the infection process in both protozoa and human host cells. Studies contrasting the role that virulence factors play in these two host populations allow speculation on the bacteria's transition from their obligatory relationship with protozoa to their opportunistic relationship with humans.
Association with Biofilms
Legionellae survive within biofilms in building water systems. The bacteria are more easily detected from swab samples of biofilm than from flowing water, suggesting that the majority of the legionellae are biofilm associated (235). A limited number of studies have attempted to characterize the bacteria's interaction within these complex ecosystems (236, 237, 282). These studies have evaluated the effect of temperature and surface materials on the growth of L. pneumophila as well as the effect of biocides on sessile legionellae. The use of biofilm models to evaluate biocide efficacy against L. pneumophila represents a vast improvement over previous studies, which primarily evaluated the susceptibility of agar-grown bacteria in sterile water (120, 174).
The majority of _Legionella_-biofilm studies that have been conducted employed naturally occurring microbial communities. Such studies have the advantage of representing a true and natural microbial community, but not all the organisms present have been identified and their contribution to the survival and multiplication of legionellae remains unknown. Biofilm matrices are known to provide shelter and a gradient of nutrients. The complex nutrients available with biofilms have led some researchers to propose that the biofilms support the survival and multiplication of legionellae outside a host cell (237). This concept is certainly plausible; most facultative intracellular bacteria are known to multiply extracellularly in some environments. If legionellae can multiply extracellularly within biofilms, the characterization of this phenomenon could have tremendous impact on control strategies for the prevention of legionellosis.
Investigators have attempted to detect extracellular growth of L. pneumophila by using a biofilm reactor and a defined bacterial biofilm grown on nonsupplemented potable water (211). The base biofilm was composed of Pseudomonas aeruginosa, Klebsiella pneumoniae, and the _Flavobacterium_-like organism isolated from a water sample containing legionellae. The addition of the amoeba Hartmannella vermiformis to the reactor resulted in a reproducible equilibrium between the amoeba and heterotrophic bacteria. L. pneumophila associated with and persisted in these biofilms with and without H. vermiformis. L. pneumophila cells did not appear to develop microcolonies, and growth measurement studies indicate that L. pneumophila did not multiply within this biofilm in the absence of amoebae. L. pneumophila did multiply in the biofilm and planktonic phase in the presence of H. vermiformis, and the majority of these bacteria appeared to be shed into the planktonic phase. These studies suggest that L. pneumophila may persist in biofilms in the absence of amoebae, but in the model, the amoebae were required for multiplication of the bacteria. This model biofilm was constructed of five preselected organisms and does not represent the many potentially diverse biofilms that may support the growth of legionellae in the environment. Additional studies are needed to determine if legionellae possess a means to multiply independent of a host cell within biofilms.
The control of biofilm-associated legionellae may lead to the most effective control measures to prevent legionellosis. Institutions that have experienced outbreaks of legionellosis are all too aware of how tenacious legionellae can be within building water system biofilms.
PATHOGENESIS
The Intracellular Cycle
Our understanding of the pathogenesis of Legionella spp. has grown tremendously in the past 10 years. Two major areas of accomplishment are characterization of the life cycle of legionellae and identification of virulence determinants by molecular techniques.
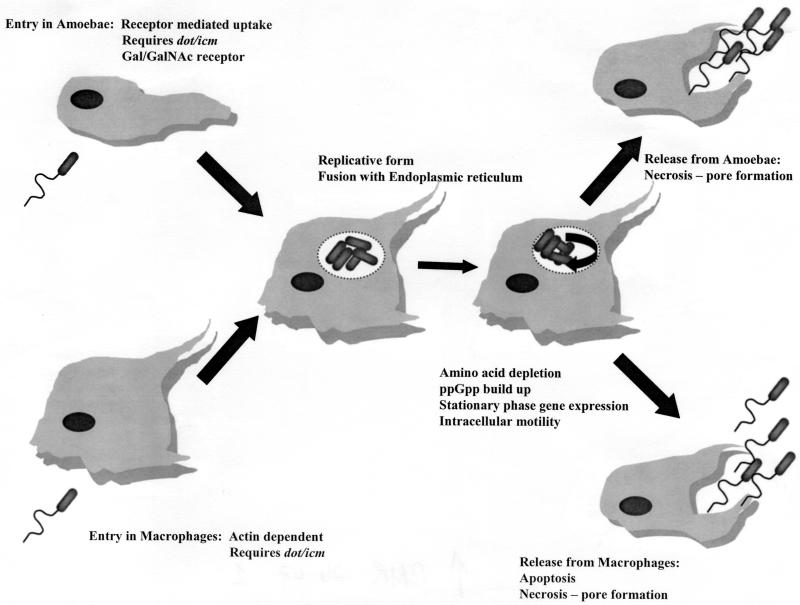
The primary feature of the pathogenesis of legionellae is their ability to multiply intracellularly. The life cycle of legionellae has been characterized in both protozoa and mammalian cells. Current understanding of this infectious cycle is outlined in Fig. 1. Studies of the infectious cycle are primarily based on microscopic observation by transmission and scanning electron microscopy and by fluorescent microscopy after labeling various bacterial and host cell components.
FIG. 1.
Life cycle of L. pneumophila in protozoa and human macrophages.
A series of classic experiments performed by Horwitz and associates in the 1980s outlined the basic pathway for L. pneumophila in human phagocytic cells. These studies determined that the bacteria enter the cells by coiling phagocytosis and that, once phagocytized, the bacteria reside within a unique phagosome that does not fuse with lysosomes or become highly acidic (145, 147, 148). Phagocytosis in human monocytes has been shown to be partly mediated by a three-component system composed of complement receptors CR1 and CR3, although the role of these receptors has never been determined (20, 223). Horwitz also described the interaction of the phagosome with mitochondria and ribosome-studded vesicles (144).
Rowbotham first demonstrated that L. pneumophila could infect amoebae and later characterized the life cycle of the bacterium in amoebae based entirely on observations made by light microscopy (240, 243). He noted that the bacteria would attach to extended trophozoites and enter the amoeba cell as several bacteria per vesicle. The bacteria initiated multiplication and became motile inside the host cell. Rowbotham also noted that the bacteria could either leave the host cell after lysis or be maintained within an encysted amoeba (240). Two growth phases were described for the bacterium. The multiplicative form was nonmotile and contained a rumpled wall and little or no β-hydroxybutyrate. The nonmultiplicative or infective forms were smaller, with smooth walls, motile, and contained numerous β-hydroxybutyrate inclusions.
Recently, these phases of the L. pneumophila life cycle have been characterized in greater detail, linking the expression of several traits with a particular phase of growth. Hammer and Swanson have proposed that host cell amino acid depletion causes the accumulation of 3′,5′-bispyrophosphate (ppGpp) (125). This would increase the amount of stationary-phase σ factor RpoS, resulting in the expression of stationary-phase genes. The stationary-phase proteins facilitate the infection of a new host cell and include sodium sensitivity, cytotoxicity, osmotic resistance, motility, and evasion of phagosome-lysosome fusion (263). Additional evidence for a precise cell cycle comes from the observation that expression of flagellin is coordinately regulated with the bacteria's ability to infect host cells. Pruckler et al. demonstrated a link between the loss of flagellar protein (Fla−) and loss of the ability to multiply in H. vermiformis and U937 cells (227). However, some Fla− strains retained their intracellular capability, demonstrating that the flagellar protein itself is not a virulence factor. These observations were confirmed by analysis of 30 laboratory-maintained strains of legionellae, showing that all strains possessing intact flagella were able to infect H. vermiformis, while strains that did not possess flagellar protein were unable to infect the amoebae (34).
There are striking similarities in the processes by which legionellae infect protozoa and mammalian phagocytic cells. Microscopically, the processes are virtually identical, although notable differences in the mechanism of entering and exiting the host cell do exist. The method of uptake of the bacteria has been described as coiling phagocytosis in both macrophages and amoebae (37, 146). Subsequently, studies have determined that L. pneumophila can enter host cells by conventional phagocytosis (50). The significance of coiling phagocytosis remains unclear, and this form of uptake has only been observed by transmission electron microscopy. L. pneumophila cells attach to small hair-like projections (filapodia) of the amoeba H. vermiformis (93). Filapodia are hair-like, naturally adherent structures, and the bacteria may bind to the projections by random encounter. Once attached to a Hartmannella host cell, the bacteria rapidly enter the cell by a nonphagocytic process.
Studies with the metabolic inhibitors methylamine and cytochalasin D have shown that L. pneumophila enter H. vermiformis by a form of receptor-mediated endocytosis (162). Actin polymerization, an essential component of phagocytosis, is not required for L. pneumophila to infect H. vermiformis, although polymerization is essential for the infection of human monocyte-like cells (U937). The attachment of L. pneumophila to H. vermiformis induces a time-dependent tyrosine dephosphorylation of multiple host proteins, including a 170-kDa protein homologue of the Entamoeba histolytica galactose- and _N-_acetylgalactosamine(Gal/GalNAc)-inhibitable lectin (277). This lectin is a putative receptor for the attachment of L. pneumophila to H. vermiformis.
Another difference between the uptake of L. pneumophila in human phagocytic cells and H. vermiformis is the role of host cell protein synthesis. Cycloheximide, an inhibitor of eukaryotic cell protein synthesis, inhibits the ability of L. pneumophila to enter H. vermiformis cells in a dose-dependent manner (2). L. pneumophila requires host cell protein synthesis to infect H. vermiformis but not to infect human U937 cells. The role of the proteins synthesized during bacterial cell uptake remains undefined.
Once an L. pneumophila cell has entered a host, the bacterium occupies a unique phagosome that does not follow the endosomal pathway. A number of studies have shown that the L. pneumophila phagosome does not possess markers or characteristics of conventional phagosomes. Swanson and Hammer have published an extensive review of these studies (263). Briefly, the early L. pneumophila phagosome lacks alkaline phosphatase, major histocompatibility complex (MHC) class I and II markers, transferrin receptors, Rab 7, LAMP-1, and cathepsin D. In addition, these vacuoles do not accumulate endocytic tracers such as Texas Red-ovalbumin, CM-DiI, or Alexa Fluor-streptavidin.
L. pneumophila occupies a vacuole that is independent of the endocytic pathway. Approximately 4 to 6 h after entering the host cell, the L. pneumophila phagosome is associated with ribosome-studded membranes that have been shown to be the host cell endoplasmic reticulum. Studies using antisera to detect the endoplasmic reticulum-specific protein BiP have shown that the protein colocalizes with the L. pneumophila phagosome in both protozoa and human host cells (1, 264). This capability is shared by Brucella abortus, another intracellular bacterial pathogen which multiplies within the host cell endoplasmic reticulum (60). L. pneumophila avoids lysosomes and the traditional endocytic pathway by utilizing a novel pathway into the host cell. It is not clear how much of this novel means of uptake is directed by the pathogen and what portion of this mechanism is under the control of the host cell.
The final stage of the infectious cycle is host cell death and release of the bacteria. L. pneumophila kills its host cell by either apoptosis or necrosis mediated by a pore-forming activity or both. In macrophages and alveolar epithelial cells, L. pneumophila induces apoptosis during the early stages of infection (107, 124). A second phase of necrosis induced by a pore-forming activity takes place in infected human phagocytes. In contrast, death of host amoeba cells has not been associated with apoptosis in studies utilizing Acanthamoeba castellani and Acanthamoeba polyphaga (108, 124). The pore-forming activity of L. pneumophila is required for killing and exiting A. polyphaga. These studies suggest that a different mechanism is used for killing and exiting mammalian and protozoan host cells.
Identification of Virulence Determinants by Molecular Techniques
A number of virulence factors have been described for L. pneumophila. This discussion will be limited to genes and gene products that play a role in the infection of mammalian and protozoan cells. The study of virulence factors in such diverse hosts has led to much speculation on the evolution of intracellular pathogens. This increasing body of research suggests that some intracellular pathogens of higher vertebrates acquired this ability in response to predation by protozoa (3, 91, 263). This interaction provides an excellent selective pressure for the acquisition of factors facilitating intracellular survival and, subsequently, infection.
In 1989, the first virulence-associated gene of L. pneumophila was detected by site-specific mutagenesis. This gene, designated mip for macrophage infectivity potentiator, encodes a 24-kDa surface protein (Mip) (48). Mip is a prokaryotic homolog of the FK506-binding proteins and exhibits peptidyl-prolyl-cis/trans isomerase activity (97). Subsequently, _mip-_like genes have been detected in other species of Legionella and other bacteria (47, 231). mip is required for efficient infection of guinea pigs, mammalian phagocytic cells, and protozoa, and this was the first gene shown to be involved in the pathogenesis of both mammalian and protozoan hosts (49). The mechanism of action for Mip remains unknown.
Genes within the loci encoding the type IV secretion system of L. pneumophila were the first factors detected that were essential for infection of the host cell. These loci comprise 24 genes in two separate regions of the Legionella chromosome and have been named Dot/Icm (defective for organelle trafficking/intracellular multiplication) (28, 196). This type IV secretion system encodes factors involved in the assembly and activation of conjugal transfer of plasmid DNA (263, 281). It is believed that L. pneumophila utilizes these operons to deliver virulence factors required for entering the host cell in a manner that initiates the infectious process. It is postulated that the system delivers a protein during phagocytosis that diverts the phagosome from the endocytic pathway. The only secreted substrate that has been identified for the Dot/Icm system is DotA, a polytopic membrane protein (213). A 19-amino-acid leader peptide is removed from DotA prior to secretion.
Genes encoding the loci for type II secretion systems are required for unrestricted intracellular growth of L. pneumophila. This type II secretion system is similar to the PilBCD piliation system in Pseudomonas aeruginosa (181). These genes were detected in L. pneumophila by analysis of mutants defective in type IV pilus formation (259). The two genes that have been analyzed the most extensively are pilE (pilin protein) and pilD (prepilin peptidase) (181, 259), The pilE gene/pilin protein is not required for intracellular growth but may be involved in attachment to the host cell. The pilD gene encodes the prepilin peptidase and is essential for pilus production and type II secretion of proteins. The ability of the pilD mutant strain to multiply within U937 cells, H. vermiformis, and guinea pigs is greatly impaired (180). In addition, this mutant showed reduced secretion of enzymatic activities (11, 12). The secreted protein that facilitates intracellular growth of L. pneumophila has not been identified. Recent studies have shown that loss of type II secretion explains the intracellular defect of the pilD mutant in amoeba, while a novel _pilD_-dependent mechanism may be involved in the infection of human cells (239).
Several other loci involved in the intracellular growth of L. pneumophila have been identified. These include mak (macrophage killing), mil (macrophage-specific infectivity loci), and pmi (protozoan and macrophage infectivity) (109, 110, 245). Defects in any of these loci result in either decreased intracellular multiplication of the organism or complete abolition of intracellular growth. The mechanism of action of these genes is not known.
Additional potential virulence factors include several cytotoxins, heat shock proteins, phospholipases, lipopolysaccharides, compounds associated with acquisition of iron, and metalloproteases (263). These factors will not be addressed in this review.
DIAGNOSIS
Taxonomy
The family Legionellaceae consists of the single genus Legionella. Some investigators have proposed placing the legionellae in three separate genera: Legionella, Fluoribacter, and Tatlockia (102, 112). However, recent studies using 16S rRNA analysis confirm the family Legionellaceae as a single monophyletic subgroup within the gamma-2 subdivision of the Proteobacteria (23, 106). Within the genus Legionella, the DNA relatedness between strains of a given species is at least 70%, whereas the DNA relatedness of one species to another is less than 70% (23). This conforms to the definition for a genus and species as defined by an ad hoc committee on reconciliation of approaches to bacterial systematics (23). Phylogenetically, the nearest relative to the Legionellaceae is Coxiella burnettii, the etiologic agent of Q fever (4, 263). These organisms have similar intracellular lifestyles and may utilize common genes to infect their host.
The number of species and serogroups of legionellae continues to increase. As previously mentioned, there are currently 48 species comprising 70 distinct serogroups in the genus Legionella (Table 1). There are 15 serogroups of L. pneumophila and two each in L. bozemanii, L. longbeachae, L. feeleii, L. hackeliae, L. sainthelensi, L. spiritensis, L. erythra, and L. quinlivanii, and a single serogroup in each of the remaining species (23). Some legionellae cannot be grown on routine Legionella media and have been termed _Legionella_-like amoebal pathogens (LLAPs) (242). These organisms have been isolated and maintained by cocultivating the bacteria with their protozoan hosts. One LLAP strain was isolated from the sputum of a pneumonia patient by enrichment in amoebae and is considered a human pathogen (242). Additional LLAP strains may be human pathogens, but proving this is difficult because they cannot be detected by conventional techniques used for legionellae. Recently, three LLAP strains were named Legionella species (5).
TABLE 1.
Legionella species and serogroupsa
| Species | No. of serogroups | No. associated with disease |
|---|---|---|
| 1. L. pneumophila | 15 | 15 |
| 2. L. bozemanii | 2 | 2 |
| 3. L. dumoffii | 1 | 1 |
| 4. L. micdadei | 1 | 1 |
| 5. L. longbeachae | 2 | 2 |
| 6. L. jordanis | 1 | 1 |
| 7. L. wadsworthii | 1 | 1 |
| 8. L. hackeliae | 2 | 2 |
| 9. L. feeleii | 2 | 2 |
| 10. L. maceachernii | 1 | 1 |
| 11. L. birminghamensis | 1 | 1 |
| 12. L. cincinnatiensis | 1 | 1 |
| 13. L. gormanii | 1 | 1 |
| 14. L. sainthelensi | 2 | 2 |
| 15. L. tucsonensis | 1 | 1 |
| 16. L. anisa | 1 | 1 |
| 17. L. lansingensis | 1 | 1 |
| 18. L. erythra | 2 | 1b |
| 19. L. parisiensis | 1 | 1 |
| 20. L. oakridgensis | 1 | 1 |
| 21. L. spiritensis | 1 | 0 |
| 22. L. jamestowniensis | 1 | 0 |
| 23. L. santicrucis | 1 | 0 |
| 24. L. cherrii | 1 | 0 |
| 25. L. steigerwaltii | 1 | 0 |
| 26. L. rubrilucens | 1 | 0 |
| 27. L. israelensis | 1 | 0 |
| 28. L. quinlivanii | 2 | 0 |
| 29. L. brunensis | 1 | 0 |
| 30. L. moravica | 1 | 0 |
| 31. L. gratiana | 1 | 0 |
| 32. L. adelaidensis | 1 | 0 |
| 33. L. fairfieldensis | 1 | 0 |
| 34. L. shakespearei | 1 | 0 |
| 35. L. waltersii | 1 | 0 |
| 36. L. genomospecies | 1 | 0 |
| 37. L. quateirensis | 1 | 0 |
| 38. L. worsleiensis | 1 | 0 |
| 39. L. geestiana | 1 | 0 |
| 40. L. natarum | 1 | 0 |
| 41. L. londoniensis | 1 | 0 |
| 42. L. taurinensis | 1 | 0 |
| 43. L. lytica | 1 | 0 |
| 44. L. drozanskii | 1 | 0 |
| 45. L. rowbothamii | 1 | 0 |
| 46. L. fallonii | 1 | 0 |
| 47. L. gresilensis | 1 | 0 |
| 48. L. beliardensis | 1 | 0 |
Legionellae are identified by growth on buffered charcoal yeast extract (BCYE), appropriate colonial morphology, and a requirement for the amino acid l-cysteine (excepting L. oakridgensis and L. spiritensis) (92). Isolates that react with specific antisera against known Legionella species are confirmed legionellae. When such isolates fail to react with specific antisera to all known Legionella species, they must be evaluated as potential new species within this genus. Species within the Legionellaceae can be distinguished by biochemical analysis, fatty acid profiles, protein banding patterns, serology, and nucleic acid analysis (23).
Biochemical data for legionellae other than L. pneumophila are limited. Legionellae are gram-negative, catalase-positive, motile rods with polar or lateral flagella (23). Most species produce beta-lactamase and liquefy gelatin. The oxidase reaction is variable, and reactions for nitrate reduction, urease, and carbohydrate utilization are negative. Amino acids are the carbon source for legionellae (224). Strains belonging to all serogroups of L. pneumophila except serogroups 4 and 15 strongly hydrolyze hippurate (133). Several laboratories have described methods for identifying putative Legionella isolates to the genus level and in some cases to the species level by using phenotypic characteristics (101, 129, 278).
All legionellae contain large amounts of branched-chain cellular fatty acids and contain ubiquinones with side chains of 9 to 14 isoprene units (208). The use of fatty acid and ubiquinone profiles allows all members of the Legionellaceae to be assigned to the genus. Although not all strains can be reliably identified to the species level, the narrowing of strains to groups facilitates further serologic testing and confirms additional tests.
Legionellae have been characterized based on analysis of soluble peptides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and protein profiling of native proteins by PAGE of soluble cytoplasmic protein banding patterns (89, 178). This method was shown to be complementary to analysis by fluorescent antibody, fatty acid, and ubiquinone analysis for identification of Legionella spp. prior to DNA-DNA hybridization.
Identification of Legionella species by serologic methods is the most frequently used technique. Antisera produced in rabbits have been prepared against all species and serogroups of Legionella and have been used in the Centers for Disease Control and Prevention (CDC) laboratory to identify most Legionella strains in a slide agglutination test (23, 270). It should be noted that these sera have been found to cross-react with new serogroups and species not previously identified. These antisera are not available commercially, and only a very limited number of laboratories have antisera to all species. Several commercial suppliers have produced antisera to a limited number of Legionella species and serogroups for use in identifying cultures by either direct fluorescent antibody or latex agglutination. Cross-reactions with non-Legionella bacteria have been reported for antisera produced against several Legionella species and serogroups (25, 80, 154). Monoclonal antibodies against genus-wide antigens have been produced to facilitate identification of isolates. Several investigators have produced monoclonal antibodies to the Legionella heat shock protein with various ranges of specificity (246, 258). Helbig et al. produced monoclonal antibodies against the Mip protein which reacted with 82 Legionella strains representing 34 species tested (138). A Legionella genus-wide antibody against flagella was produced by Bornstein et al. (32). No single antiserum is routinely used to identify all legionellae.
DNA-DNA hybridization is the definitive procedure for establishing the identity of a strain of Legionella or a new species. This procedure requires that DNA from the test strain be hybridized with DNA from all known species of Legionella. Therefore, only a very limited number of specialty laboratories are able to perform this procedure. Sequence analysis of specific genes has been employed for taxonomic analysis of legionellae. Analysis of 16S rRNA genes led to the designation of Legionella within the gamma-2 subdivision of the class Proteobacteria and has been used to show the phylogenetic relatedness of new species of this genus (106). Recently, a sequence-based classification scheme has been developed for legionellae which targets the mip gene (232). This procedure can unambiguously discriminate among the 39 species of Legionella tested. Many researchers are convinced that all taxonomic analysis will eventually become sequence-based in the near future.
Culture of Legionella
L. pneumophila was first isolated by using Mueller-Hinton agar supplemented with hemoglobin and IsoVitaleX (MH-IH) (88). The essential component in hemoglobin was found to be a soluble form of iron, and l-cysteine is the essential amino acid provided by the IsoVitaleX. These refinements led to the development of Feeley-Gorman agar, which provides better recovery of the organism from tissue (88). Later, starch was replaced with charcoal to detoxify the medium and the amino acid source was changed to yeast extract, resulting in charcoal yeast extract agar (87). Charcoal yeast extract agar is the base form for most media used to grow legionellae. The medium used for the culture of legionellae has been improved several times, eventually resulting in the medium currently used, buffered charcoal-yeast extract (BCYE) agar enriched with α-ketoglutarate with and without selective agents added (71, 73, 221). Culture requires the use of selective and nonselective media. These media can be prepared with or without indicator dyes, which impart a color specific for certain species of Legionella (279). Although the majority of Legionella spp. grow readily on BCYE agar, some require supplementation with bovine serum albumin to enhance growth (207).
Culture diagnosis remains the gold standard for diagnosis of legionellosis and is the most specific diagnostic procedure. Based upon culture of serologically positive (fourfold rise) patients, the sensitivity is near 60% and the specificity is near 100% (74, 192). However, a number of factors limit the sensitivity of culture. First, laboratories experienced in the isolation of legionellae are more likely to recover the organism. A survey by the College of American Pathologists indicated that as many as two-thirds of clinical microbiology laboratories in the United States are unable to grow a pure and heavy culture of L. pneumophila (76). Moreover, to improve the specificity of the diagnosis of pneumonia caused by pyogenic bacteria, hospital laboratories commonly reject sputum specimens containing many squamous epithelial cells or few polymorphonuclear leukocytes. However, some patients with Legionnaires' disease produce little or nonpurulent sputum, and sputum specimens that would be routinely discarded may contain culturable legionellae (150). The bacteria survive poorly in respiratory secretions, and immediate culture of these specimens is critical (R. F. Benson, unpublished data). The reported sensitivity of culture isolation from respiratory secretions ranges widely, from 20 to 80%, reflecting the perishable nature of these specimens and the expertise required (238).
Legionellae can be isolated from a number of specimens, including blood, lung tissue, lung biopsy specimens, respiratory secretions (sputum, bronchial alveolar lavage [BAL], and bronchial aspirates), and stool (74, 241). Respiratory secretions are considered the specimen of choice (74). In a recent review, Maiwald et al. suggested that sputum was less suitable than other respiratory secretions, especially in early disease, when few patients have a productive cough (192). When culturing sputum, it is best to pretreat the sample by either acidification or heat (40, 81). Legionella has also been cultured from a number of extrapulmonary sites such as bone marrow, prosthetic heart valves, and sternal wounds (67, 75). Several guidelines give detailed methods for the isolation of Legionella spp. from clinical samples (67, 260, 280, 285).
DFA Detection of Legionella
Microscopic examination of specimens using direct fluorescent antibody (DFA) staining was the first method used to detect legionellae in lung tissue (from autopsy or biopsy specimens) and respiratory secretions. Legionellae can be detected in respiratory secretions by DFA for several days after the start of antimicrobial therapy. DFA staining has also been used for serologic identification of Legionella isolates, as mentioned previously. A limited number of fluorescein-conjugated polyclonal sera are available for L. pneumophila serogroups and non-L. pneumophila species. The specificity and sensitivity of these reagents have not been evaluated, and cross-reactions with non_-Legionella_ bacteria have been reported (25, 79, 154, 219). It has been recommended that polyclonal antisera to non-L. pneumophila species not be used for routine testing of clinical samples unless there is a high suspicion of disease due to non-L. pneumophila species or in epidemic investigations (192). The sensitivity of DFA testing of respiratory secretions for the diagnosis of Legionnaires' disease has ranged from 25 to 75%, and the specificity is >95% (74). While DFA provides a rapid method of identifying Legionella species, immunofluorescent microscopy is technically demanding and should be performed only by laboratory personnel experienced in the procedure. The specificity of polyvalent DFA reagents is probably lower than that of monoclonal reagents, leading some clinical microbiologists to recommend against routine use of polyvalent reagents (76).
A commercially available monoclonal antibody (Genetic Systems, Seattle, Wash.) which reacts with an outer membrane protein and detects all serogroups of L. pneumophila can be used to detect Legionella in clinical samples (117). This reagent is highly specific for L. pneumophila strains, making it possible to confirm isolates belonging to this species. It should be noted that a cross-reaction with Bacillus cereus spores from a strain isolated from water has been reported (99). However, this organism is easy to differentiate from Legionella in pure culture. The reagent's utility in detecting Legionella in clinical specimens has been evaluated and found to be comparable with polyvalent antisera (77). Genus-specific monoclonal antibodies have been produced to detect either a heat shock protein or the MIP protein, but these have not been shown to be useful for detection of Legionella in clinical samples (138, 139, 246, 258).
Serologic Diagnosis
A number of serologic tests have been developed to detect antibodies to Legionella spp. The use of the indirect immunofluorescence assay (IFA) was used to detect antibodies in patients from the Philadelphia outbreak and was instrumental in determining the cause of the illnesses. Enzyme immunoassay (EIA) and microagglutination have also been used to detect antibodies to the Legionnaires' disease bacterium (84, 85, 165). An indirect hemagglutination assay has been used for diagnosis of Legionnaires' disease due to L. pneumophila serogroups 1 to 4 (293). A counterimmunoelectrophoresis test and latex agglutination have been used to diagnose Legionnaires' disease (140-142). In addition to the IFA test, a rapid microagglutination test is used extensively in Europe for diagnosis (130).
The rapid microagglutination test has some advantages over immunofluorescence, such as the ease of testing a large number of samples and the early appearance of agglutinating antibodies. The specificity is in the range of 97 to 99%, and the sensitivity is 80% (128). Cross-reactions due to antibodies to Pseudomonas aeruginosa in cystic fibrosis patients have been reported as well as cross-reactions between Legionella and Campylobacter spp. (35, 54). Klein reported cross-reactions in sera with elevated titers to Pseudomonas pseudomallei (164).
Of all these tests, the IFA has been evaluated extensively and is available commercially. The IFA originally developed by CDC used a progression of antigen preparations from live bacteria (L. pneumophila serogroup 1, Philadelphia 1) grown in infected hen yolk sacs to ether-killed antigen grown on enriched Mueller-Hinton agar and finally to a heat-killed antigen suspension grown on various laboratory media (203, 289). Validation of this heat-killed antigen with epidemic sera yielded a sensitivity of 78 to 91% and a specificity of 99% (287). Subsequent CDC studies demonstrated that a heat-killed antigen grown on BCYE medium gave the same level of sensitivity and specificity as formalin-killed antigen from the same medium if a higher cutoff value for positive results was used (286).
Additional factors to consider in the serologic diagnosis of Legionnaires' disease are the use of an anti-human immunoglobulin which recognizes immunoglobulin G (IgG), IgM, and IgA (15, 288). These antibody responses may be serogroup specific or may react with an antigen common to L. pneumophila; therefore, it is not possible to reliably determine the serogroup or species causing infection (290). Early studies recommended the use of polyvalent antigen pools to screen sera and subsequent testing with the monovalent components (83, 290). Recent experience by several laboratories has indicated that 25% of seroconversions with polyvalent antigens could not be confirmed when retested with the monovalent antigen (72). Current recommendations suggest the use of L. pneumophila serogroup 1 antigen only and the avoidance of polyvalent antigen pools. It is extremely difficult to make a diagnosis based on results of IFA tests with polyvalent pools.
The IFA test used in Europe is slightly different from the test described above. The strain of L. pneumophila serogroup 1 used is Pontiac 1, which is grown in embryonated hens' eggs and formalin killed (127). Harrison et al. evaluated both the IFA and the rapid microagglutination test mentioned earlier for the diagnosis of Legionnaires' disease (126). The sensitivity of both assays was 80%, and additionally, 40% of patients had suggestive titers within the first week of hospital admission. Evaluation of the specificity of the IFA with sera from patients with Mycoplasma pneumoniae, Chlamydia psittaci, Coxiella burnetti, influenza A virus, and adenovirus showed no cross-reactions with the formalin yolk sac antigen (269).
Patients with nonlegionellosis pneumonia and bacteremia have been found to have false-positive titers to Legionella spp. These infections have included Bacteroides fragilis, C. psittaci, and Pseudomonas aeruginosa in cystic fibrosis patients, as well as mycobacteria, mycoplasmas, campylobacters, Citrobacter freundii, Pseudomonas pseudomallei, Haemophilus influenzae, Coxiella burnetti, Rickettsia typhi, and Proteus vulgaris OX19 (31, 36, 54, 69, 80, 118, 119, 164, 198, 204, 212, 218, 288). Cross-reactions occur more frequently with non-L. pneumophila species.
Despite the obvious utility of serologic diagnosis, these tests have a number of important limitations. Even in cases of culture-confirmed Legionnaires' disease, a fourfold rise in antibody by IFA can be documented for only 70 to 80% of patients, and seroconversion following legionellosis may not occur for up to 2 months after illness onset (72). Past case definitions for Legionnaires' disease included a presumptive diagnosis based on an elevated titer of ≥1:256. Recent studies of patients hospitalized with community-acquired pneumonia showed that a single titer of ≥1:256 did not distinguish between Legionnaires' disease and pneumonia due to other agents. Only 10% of 68 patients with legionellosis confirmed by culture or a fourfold rise in antibody had acute-phase titers of ≥256, compared with 6% of 636 patients with pneumonia caused by other agents (226).
Various studies have demonstrated the presence of L. pneumophila serogroup-specific antigens, _L. pneumophila_-specific antigens, and _Legionellaceae-_specific antigens (14, 52, 53). Several investigators have attempted to identify specific antigens that could be used to improve the specificity of antibody determinations. Sampson et al. (247), using immunoblot analysis, demonstrated antibody response to major protein antigens of L. pneumophila SG 1. Further studies with a purified 60-kDa protein antigen demonstrated the presence of _Legionella_-specific and nonspecific epitopes (225). This protein was later shown to be a heat shock protein which is a cross-reactive common antigen. The use of flagella in an EIA test was useful only for L. pneumophila serogroup 1 (216). Studies have been performed with whole bacteria (18), heat-killed soluble antigens (248), and sonicated bacteria (15). A lipopolysaccharide fraction for detecting IgG and IgM was the most specific, followed by an IgA assay with a sonicated antigen, which was the most sensitive (15).
Recently, two enzyme-linked immunosorbent assay (ELISA) test kits have been introduced. The Wampole Laboratories (Cranbury, N.J.) kit utilizes a heat-inactivated, solubilized pool of L. pneumophila serogroups 1 to 6 and detects IgG and IgA. The Zeus Scientific, Inc. (Raritan, N.J.) kit uses a formalin-inactivated, sonicated suspension of L. pneumophila serogroups 1 to 6 and detects IgG, IgM, and IgA. This test is also sold by Sigma Diagnostics (St. Louis, Mo.). These test systems have not been independently evaluated.
It is important to use reagents that detect all major immunoglobulin classes (IgG, IgM, and IgA) to increase test sensitivity (288). However, commercial kits for measuring antibody response do not use a secondary antibody proven to react with IgM and IgA (72). Studies have shown that many patients produce primarily IgM antibodies and that these are useful for early diagnosis of Legionnaires' disease (59, 214, 294). However, IgM may be present later in some confirmed cases, limiting the usefulness of the assay for early diagnosis in all patients. In a 2-year follow-up study of IgM in 35 patients, 13 had seroconversion to IgM within 7 days. Seven patients had titers of >64 at 2 years after the disease. The authors stated that persisting titers made it impossible to determine a reliable value of a single titer (157).
Serologic diagnosis of Legionnaires' disease was once the most frequently used diagnostic test. The need for testing of paired serum samples collected 3 to 6 weeks apart has diminished the use of serology, and urine antigen detection is now the most frequently used diagnostic test (A. L. Benin and R. E. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 873, 2001). Perhaps the development of sensitive and specific assays for detecting IgM will increase the usefulness of serologic testing.
Urine Antigen Detection
Urinary antigen testing has led to the recognition of outbreaks of Legionnaires' disease and allowed a rapid public health response (96, 100, 179). In addition, urine antigen testing permits early diagnosis and initiation of appropriate antibiotic therapy (158). The capture antibody used in the majority of these assays is considered to be specific for L. pneumophila serogroup 1. Therefore, even though the majority of human disease is associated with L. pneumophila serogroup 1, total dependence on this diagnostic assay may miss as many as 40% of cases of legionellosis. Legionella antigen present in urine specimens appears to degrade upon prolonged storage (234). There is a single report of a false-positive urinary antigen assay on a specimen obtained from a renal transplant patient who had received rabbit serum with antibodies to thymocytes (57).
Changes in the use of diagnostic tests for Legionnaires' disease may have had an impact on the number of reported cases, the reported mortality, and the distribution of serotypes. In the United States, cases diagnosed by culture have declined, while cases diagnosed by urine antigen have increased. Since urine antigen testing allows earlier diagnosis, this may have allowed more appropriate antimicrobial therapy and improved the outcome of Legionnaires' disease. However, since urine antigen testing primarily detects serogroup 1 infections, cases of Legionnaires' disease caused by other serogroups and species may have been missed (A. L. Benin and R. E. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 873, 2001).
Shortly after the outbreak of Legionnaires' disease at the American Legion Convention in Philadelphia, two investigators reported the ability to detect antigen in the urine of serologically confirmed Legionnaires' disease patients by ELISA (27, 271). Both studies tested a small number of patients but suggested the utility of a rapid technique for diagnosing Legionnaires' disease by using a clinical sample that is easily obtained. These studies were followed by experiments with a radioimmunoassay to detect antigen in nine patients confirmed by culture, direct fluorescent antibody, or serologic testing of specimens collected on or before erythromycin therapy. The sensitivity was 100%, and urine samples from 245 controls were negative, for a specificity of 100% (168). The antigen detected is a component of the lipopolysaccharide portion of the Legionella cell wall and is heat stable (168, 292). Antigens are generally detectable in urine within a few days of illness onset and can remain so for several weeks after initiation of appropriate antimicrobial therapy (167). Studies have shown that antigen is excreted as soon as 3 days after the onset of symptoms and can persist for >300 days (167).
A study relating the onset of symptoms and antigen detection indicated that antigen was detected in 88% of patients tested during days 1 to 3, 80% tested during days 4 to 7, 89% tested during days 8 to 14, and 100% tested after day 14 (167). In another study, multiple urine samples from 23 patients were analyzed after initiation of therapy to determine how long antigen continued to be excreted. In some patients, antigen was no longer detected within 4 days of therapy, but it persisted for >300 days in one patient. The duration of antigen excretion has implications for the diagnostic relevance of antigen detection in patients with recurrent pneumonia in the months following illness. Also, concentration of urine specimens can lead to an increased sensitivity for the urine antigen assay, although specificity is not affected (64, 166).
These early studies on antigen detection in legionellosis patients led to the development of a commercial radioimmunoassay (RIA) (Du Pont, Wilmington, Del.) for urine specimens, later manufactured and distributed by Binax, Inc. (Portland, Maine). An evaluation of the urinary antigen testing by RIA for the diagnosis of L. pneumophila serogroup 1 infection indicated a sensitivity of 93% and specificity of 100% (6). In studies comparing urinary antigen testing, DFA, and culture, the authors concluded that urinary antigen detection was the most useful test (230). Several studies assessing the overall sensitivity (all Legionella infections, not just L. pneumophila serogroup 1) of the Binax Equate Legionella Urinary Antigen kit determined this to range from 53 to 56% (29, 226). A major drawback of urinary antigen testing with the RIA is the difficulty involved in the handling and disposal of radioisotopes required to perform RIA. For this reason, the RIA test was replaced by an ELISA in the mid-1980s (Binax, Inc.). Resulting comparisons of these two assays showed that the ELISA format was positive for 67 to 88% of specimens that were positive with the RIA kit (65, 122, 160). Another study showed that the sensitivity of the EIA was significantly greater than that of the RIA, and if the cutoff was lowered to ≥2.5, the sensitivity increased to 90%, while the specificity remained 100% (46).
Currently, there are three commercially available ELISA urinary antigen tests: Wampole (formerly distributed by Binax, Inc.), Cranbury, N.J.; Trinity Biotech, Bray, Ireland; and Biotest AG, Dreieich, Germany. Two of these tests (Wampole and Trinity Biotech) are intended to be specific for L. pneumophila serogroup 1 and are not likely to detect the 20 to 30% of Legionnaires' disease cases that are caused by other serogroups and species (199). The Biotest ELISA is intended to detect antigen of other L. pneumophila serogroups and Legionella species. Domínguez et al. (63) compared the Binax (Wampole) and Biotest EIA kits with concentrated and nonconcentrated urine samples. There were no significant differences in sensitivity between the two tests, and concentrating urine samples improved the sensitivity of both assays. Both tests detected antigens from 14 serogroups of L. pneumophila and L. bozemanii, and both tests were negative for L. longbeachae serogroup 1.
The most recent method for detecting antigenuria is the immunochromatographic (ICT) membrane assay. The ICT assay is similar to a home pregnancy test and is commercially available (NOW Legionella urinary antigen test; Binax, Inc.). The test is simple to perform and does not require special laboratory equipment, and results can be obtained within 15 min. A comparison of the Binax NOW with the Binax EIA showed 98% overall agreement between the two tests and a specificity of 100% (62). The authors suggested using concentrated urine samples to increase test sensitivity. Another study determined that the sensitivity of the ICT assay for L. pneumophila serogroup 1 was 80% and the specificity was 97% (135).
There is still a need to develop antigen capture assays that can diagnose infections with all species and serogroups of Legionella. Development of a genus-wide urinary antigen test appears feasible and would provide a distinct diagnostic advantage (265, 266). Tang and Toma (266) developed a broad-spectrum ELISA that could detect soluble antigens from numerous L. pneumophila serogroups and species by using rabbit antisera from a combination of intradermal, intramuscular, and intravenous inoculations over a period of 8 months. The spectrum of serogroups and species detected with the broad-spectrum ELISA has since been reported to include L. pneumophila serogroup 12, L. sainthelensi, L. bozemanii, L. cincinnatiensis, L. hackeliae, L. maceachernii, and L. parisiensis (24, 111, 265, 267).
Another recent review of the impact of urine antigen testing on diagnosis and therapy and a more thorough review of urine antigen testing was written by Kashuba and Ballow (158).
Detection of Legionella Nucleic Acid
The first assay designed to detect the DNA of L. pneumophila was a radiolabeled ribosomal probe specific for all strains of Legionella spp. (Gen-Probe, San Diego, Calif.). Researchers reported varying sensitivity and specificity for this assay (61, 222, 291). The use of the probe at one hospital resulted in 13 false-positive cases, with no symptoms consistent with Legionnaires' disease in 13 of 23 patients (175). The assay was removed from the market soon after this pseudo-outbreak.
PCR represents one of the few diagnostic tests with the potential to detect infections caused by all of the known species of Legionella. Since most rapid tests only detect infections due to L. pneumophila serogroup 1, an accurate PCR test would greatly enhance the ability to diagnose these infections. Various PCR tests that have been developed for legionellae target either random DNA sequences for L. pneumophila (256), the 5S rRNA gene (187, 189), the 16S rRNA gene (156, 183, 206), or the mip gene (82, 189, 229). A summary of DNA targets for various PCR assays is presented in Table 2. The most widely used test was a commercially produced kit designed to detect legionellae in the environment (EnviroAmp kit; Perkin-Elmer, Inc., Norwalk, Conn.). This test was removed from the market in 1997. It simultaneously amplified two targets, a 5S rRNA target specific for the Legionella genus and a portion of the mip gene specific for L. pneumophila. The kit was designed for testing environmental samples, and the manufacturers did not attempt to have it approved for clinical use. Nevertheless, several researchers successfully used these kits to detect legionella DNA in human specimens (161, 201, 284).
TABLE 2.
Detection of Legionella nucleic acid by gene probe and PCR in clinical samples
| Clinical sample tested | Gene target(s) | Results | Reference |
|---|---|---|---|
| Frozen respiratory secretions | rRNA (Gen-Probe) | 112 culture-positive, 63 positive; 230 culture-negative, 228 negative | 78 |
| Prospective respiratory secretions | rRNA (Gen-Probe) | 427 samples; compared to DFA, 63% sensitive, 95% specific | 222 |
| Respiratory secretions | rRNA (Gen-Probe) | 167 patients; 31-67% sensitive, 99% specific | 94 |
| BAL | mip gene | 68 samples, 15 positive | 151 |
| BAL | 5S rRNA, mip gene | 52 samples, 3 positive (EnviroAmp kit) | 161 |
| Respiratory secretions | 5S rRNA, mip gene | 31 samples, 12 positive (EnviroAmp kit) | 201 |
| BAL | 16S rRNA gene | 51 samples, 7 positive | 183 |
| Serum | mip gene | 5 patients, acute- and convalescent-phase sera positive | 182 |
| Urine | 5S rRNA gene | 21 pneumonia patients, 11 positive by PCR, 9 confirmed | 193 |
| BAL | 16S rRNA | 256 samples, 14 PCR positive | 156 |
| Intratracheal aspirates | mip gene, 5S rRNA | 1 patient, 8 samples positive with both primers | 169 |
| Serum, urine | 5S rRNA gene | 28 patients, 18 positive | 210 |
| Serum, urine | 700-bp fragment | Serum, 12 of 41 samples positive; urine, 6 of 20 samples positive | 206 |
| Respiratory secretions (BAL, sputum, pleural fluid, lung tissue, bronchial washing) | 16S rRNA gene | 212 specimens; 31 of 31 culture-positive samples positive; 12 culture-negative samples positive by PCR, 4 confirmed by sequencing | 51 |
| BAL | mip gene, 5S rRNA | 43 specimens (LightCycler) | 131 |
| Respiratory secretions (BAL, sputum) | 16S rRNA | 77 samples, 2 culture-positive, both PCR-positive (LightCycler) |
A very limited number of laboratories test for legionellae by PCR at this time. The few studies which have been conducted indicate that the PCR detection of legionellae infections has a moderate sensitivity and a high specificity. Legionella DNA has been detected in respiratory secretions such as BAL, pharyngeal swabs, nasopharyngeal swabs, peripheral blood mononuclear cells, urine, and serum (136, 151, 156, 193, 202, 209, 210, 229, 253).
The PCR procedures described above have used either visualization of DNA amplicons by ethidium bromide staining in agarose gels or reverse dot blot hybridization to probes immobilized on nylon membranes with biotinylated primers. Other PCR assays targeting the 16S rDNA gene have been reported to be highly sensitive and specific (51). However, a number of false-positive results were obtained, revealing homology to Acinetobacter spp. or an unidentified proteobacterium. False-positive reactions have also occurred with the commercially available tests and when using primers for 16S rDNA, which reacted with Gemella spp. (R. Benson, personal communication).
Recently, several researchers have reported on the use of real-time PCR combined with the use of a hybridization probe to confirm the product identity for rapid detection of legionellae in clinical specimens (13, 131). This method reduces the risk of cross-contamination, reduces the time required to process samples, and eliminates the need for sample analysis after PCR. These assays promise increased sensitivity and specificity, although further validation is required. Limited studies and preliminary data suggest that PCR may add to the diagnostic repertoire, but this procedure may lack the sensitivity and specificity to provide anything other than supplemental data for the clinician.
Subtyping of Legionella spp.
L. pneumophila serogroup 1 comprises a fairly heterogeneous group of organisms that account for most of the cases of legionellosis in the United States. L. pneumophila serogroup 1 can be divided into a number of subtypes by various techniques. These procedures are used to match environmental isolates with patient isolates obtained from outbreak investigations of legionellosis. Because of the diversity within L. pneumophila serogroup 1, clinical and environment isolates must be matched by molecular techniques to adequately identify environmental sources of disease.
Initially, legionellae were identified to the serogroup level during investigations of legionellosis. This form of serologic subtyping uses polyvalent or monoclonal antisera and may be adequate for identifying reservoirs of some of the more uncommon legionellae causing disease. The variety of strains and distribution of L. pneumophila serogroup 1 necessitate more elaborate subtyping procedures to discriminate among these bacteria. Several research groups have developed monoclonal antibodies for this purpose (220, 272, 283). An international panel of seven monoclonal antibodies (MAb) was proposed in 1986 (155). Although much information has been gained through the use of this panel, several of the cell lines have been lost, and most of these reagents are no longer available. Use of these monoclonal antibodies has identified 12 “type” strains within L. pneumophila serogroup 1 (16). Monoclonal antibodies produced by Helbig et al. and labeled the Dresden Legionella LPS MAb, along with MAb 3 described by Joly et al., allow the subtyping of L. pneumophila serogroup 1 into 15 phenons (137, 155). Recent investigations have shown that the use of monoclonal antibodies may be inadequate for discriminating between disease-causing strains and other environmental isolates of L. pneumophila serogroup 1 (228).
Newer molecular techniques such as pulsed-field gel electrophoresis (PFGE) and arbitrarily primed PCR (AP-PCR) are able to discriminate within monoclonal subtypes of L. pneumophila serogroup 1 and identify sources of disease-causing strains (116). These techniques appear to complement the use of monoclonal antibodies and are the most recent of several techniques that separate strains based on DNA polymorphism (251). Researchers have used a variety of other techniques to discriminate between isolates of legionellae. The use of electrophoretic alloenzyme typing (multilocus enzyme typing) is able to subtype L. pneumophila into more than 40 subtypes (252). This technique was used to subdivide the Philadelphia 1 monoclonal subgroup of L. pneumophila serogroup 1 into five electrophoretic types (252).
Plasmid analysis has been used to subtype legionellae, but results have shown this technique to have limited use. In one investigation, the clinical strains were plasmidless and the environmental strains contained plasmids (39, 190, 191, 260). Restriction fragment length polymorphism (RFLP) has been used to subtype Legionella either alone or in combination with rRNA or DNA probing of the DNA digest (ribotyping) (56, 116, 249, 250, 275). The RFLP procedure has been further modified to use PCR amplification of a specific locus, such as the 16S-23S spacer region, followed by restriction enzyme digestion, with the resultant products separated by agarose gel electrophoresis (275).
Other procedures used to subtype L. pneumophila include detection of repetitive elements within DNA, amplified fragment length polymorphism, and sequence-based typing (gyrA gene) (56, 86, 113, 116, 176, 177, 197, 274, 276).
A comparison of AP-PCR and PFGE with strains from seven outbreaks of legionellosis indicated that PFGE provided slightly better discrimination (228). Both techniques offer better discrimination and are less labor intensive than other techniques such as SDS-PAGE, ribotyping, and multilocus enzyme electrophoresis. Discrimination is improved when AP-PCR and PFGE are used in conjunction with monoclonal antibody typing. Even so, caution must be used when interpreting results of subtyping for the identification of an outbreak reservoir. A recent study determined that strains isolated from non-outbreak-related sources were identical to the clinical isolates by four subtyping procedures (AP-PCR, PFGE, monoclonal antibody, and multilocus enzyme electrophoresis) (171).
Amplified fragment length polymorphism (AFLP) represents the current state-of-the-art subtyping procedure (274). The technique has two variations: one uses a single restriction enzyme and a selective primer, and the other employs two restriction enzymes and corresponding primers. The latter method has been modified and termed infrequent-restriction-site PCR (233). The DNA is digested with a restriction enzyme(s), and the restriction fragments are ligated to specially constructed adapters. Subsequent PCR with primers specific for the ligated adapters allows the amplification of a subset of the digested DNA. The primers can be labeled with a fluorescent tag to allow analysis of the fragments with an automated sequencer, or the fragments can be separated by gel electrophoresis and stained with ethidium bromide. A comparison of PFGE and AFLP on 48 unrelated and epidemiologically related strains of L. pneumophila serogroup 1 indicated that the method was rapid, versatile, and reproducible, provided useful discrimination, and was easier to perform than PFGE (233). An intercenter study by the European Working Group on Legionella Infections comprising 13 laboratories using a standardized protocol demonstrated that the method was highly reproducible and epidemiologically concordant with good discrimination. The method is to be adopted as the first standardized typing method for the investigation of travel-associated Legionnaires' disease in Europe (105).
EPIDEMIOLOGIC TRENDS
Incidence
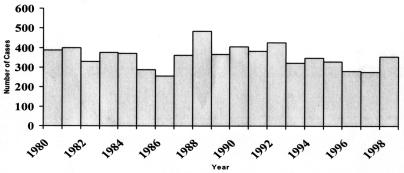
Accurate data are not available to assess whether any trends have occurred in the incidence of Legionnaires' disease. In the United States between 1980 and 1998, an average of 356 cases were reported to CDC each year, with no trend (Fig. 2) (A. L. Benin and R. E. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 873, 2001). This is a fraction of the 8,000 to 18,000 cases estimated to occur each year (200). There are many reasons why reporting does not reflect the true incidence of disease. Clinicians may fail to perform testing for Legionnaires' disease. As empirical therapy for community-acquired pneumonia has changed to cover a wider array of infectious agents, the use of diagnostic testing to determine pneumonia etiology may have declined. For Legionnaires' disease, this is a problem not only for the individual patient who is denied the benefit of targeted therapy, but for the community as a whole, since each case of Legionnaires' disease may represent a sentinel event heralding an outbreak.
FIG. 2.
Legionnaires' disease, 1980 to 1998; annual number of cases reported.
Cases of Legionnaires' disease that are diagnosed in hospitals may not get reported to state and local officials. This may be due to a lack of awareness that Legionnaires' disease is reportable in all states (except Oregon and West Virginia). It may also be due to concerns that cases of nosocomial Legionnaires' disease frequently prompt litigation. States may in turn not report to CDC. Faced with competing priorities, health departments may not have personnel to complete case report forms and submit in a timely fashion or at all.
The vast majority of cases of Legionnaires' disease reported in the United States are sporadic. During the 1980s, 11% of all cases were associated with an outbreak: 37% of possible nosocomial cases and 4% of community-acquired cases (199). In studies assessing the etiology of hospitalized cases of community-acquired pneumonia, the proportion due to Legionnaires' disease has ranged from 2 to 15% (41). Risk factors for Legionnaires' disease include increasing age, smoking, male sex, chronic lung disease, hematologic malignancies, end-stage renal disease, lung cancer, immunosuppression, and diabetes (199). CDC surveillance data indicate that persons with human immunodeficiency virus (HIV) infection are at greater risk of Legionnaires' disease than the general population (199); however, legionellae are rarely detected in studies of pulmonary disease among cohorts of HIV patients (30, 121). This may be due to prophylaxis of HIV-infected patients with trimethoprim-sulfamethoxazole, an antimicrobial agent with activity against Legionella species.
Data from a population-based study investigating the etiology of cases of pneumonia requiring hospitalization demonstrated that cases occur throughout the year, with no clear seasonal predilection (200). However, reports to CDC increase twofold in the summer (199), possibly because of increased testing (199). Outbreaks in the United States associated with cooling towers frequently occur in the summer and fall (26). Nosocomial cases occur year-round, with no seasonal pattern.
Treatment
Empirical therapy for persons hospitalized with community-acquired pneumonia should include coverage for Legionnaires' disease (19). Delay in starting appropriate therapy has been associated with increased mortality (132). Overall, mortality for Legionnaires' disease in the United States has been declining during the 1990s, possibly due to changes in empirical therapy for community-acquired pneumonia (A. L. Benin and R. E. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 873, 2001). Historically, erythromycin has been the drug of choice for Legionnaires' disease. Because of the difficulty in amassing enough cases of Legionnaires' disease in one institution and the cost and complexity of multicenter studies, it is unlikely that clinical trials will ever be conducted to determine whether other agents are better. In vitro data suggest that azithromycin and many fluoroquinolone agents have superior activity against Legionella species. Additionally, these agents have fewer side effects than erythromycin (70). Azithromycin and levofloxacin are licensed by the Food and Drug Administration for the treatment of Legionnaires' disease and are considered preferable to erythromycin. There is debate as to whether rifampin provides additional benefit to patients with Legionnaires' disease (70, 261). The use of rifampin in addition to the newer regimens is not supported by in vitro data (70) and is not recommended by the Infectious Diseases Society of America (19).
Travel-Related Legionnaires' Disease in the United States
Travel is an underappreciated factor in the acquisition of Legionnaires' disease in the community. This is surprising given that the largest outbreak in U.S. history and the one for which this disease was named was travel associated (104). The 1976 outbreak among American Legion members was not typical of more recent travel-associated clusters in that it was explosive and easily recognizable.
A more typical travel-related outbreak occurred in Georgia in 1999 (21). A 64-year-old woman was diagnosed with Legionnaires' disease in New York after returning from a wedding in Georgia. Five of her extended family members in Massachusetts developed Pontiac fever after returning from the same wedding. Extensive case finding revealed one other case of Legionnaires' disease and six cases of Pontiac fever. A cohort study implicated the hotel whirlpool spa as the source of the outbreak. The investigation allowed measures to be taken that may have prevented additional cases from occurring.
This outbreak demonstrates many of the characteristics of typical travel-related Legionnaires' disease outbreaks that make detection very difficult: low attack rates, long incubation periods, dispersal of persons away from the source of the infection, and inadequate surveillance. Discovery of the outbreak was serendipitous; cases occurred within one family from two states, with no other common exposures. If one of the clinicians caring for the two persons who developed Legionnaires' disease had not tested for the disease, the outbreak would not have been detected. At the time of diagnosis, none of the patients was still in Georgia. If the Georgia Division of Public Health had not been contacted, the outbreak would have been missed. And if the state health department had not contacted CDC, the outbreak would not have been investigated.
Although there are two national surveillance systems for Legionnaires' disease in the United States, neither is able to detect clusters of travel-associated Legionnaires' disease. The National Electronic Telecommunications System for Surveillance, an electronic system that collects data on Legionnaires' disease in addition to all reportable diseases, is timely but does not collect information on travel. The Legionnaires' Disease Reporting System, a paper-based system that collects detailed information on travel as well as other risk factors for Legionnaires' disease, is very insensitive and is used mainly for tracking major epidemiologic trends.
The true burden of travel-related legionellosis in the United States is not clear. Although 21% of cases reported to CDC through the paper-based Legionnaires' disease reporting system indicate an out-of-home stay during the incubation period (CDC, unpublished data), a case-control study of risk factors for sporadic Legionnaires' disease in Ohio found that the rate of travel was similar between patients and controls (262). Unfortunately, this study did not look in detail at travel-related activities to determine whether particular exposures when traveling or returning home were associated with Legionnaires' disease. Further studies are needed to determine the proportion of disease attributable to travel and to design appropriate prevention strategies.
Detection of travel-related outbreaks offers the potential for providing new prevention opportunities. The European Working Group on Legionella Infections conducts surveillance for travel-related Legionnaires' disease in Europe (149) and has been quite successful at identifying clusters of travel-associated Legionnaires' disease (149). In 1999, the European Working Group on Legionella Infections detected 29 clusters among 63 travelers in 16 countries (F. Lever, E. Slaymaker, C. Joseph, and C. Bartlett, 5th International Conference on Legionella, abstr. 40, 2000). Twelve of these clusters would not have been detected without this surveillance scheme in place because they involved two travelers from different countries.
In response to threats of bioterrorism and emerging infectious diseases, CDC is in the process of developing a system of a national electronic surveillance that will be rapid and efficient (152). The National Electronic Disease Surveillance System, when implemented, will be a logical platform for conducting travel-related surveillance for Legionnaires' disease.
Community Outbreaks and Prevention
During the past 3 years, the epidemiology of Legionnaires' disease has been dominated by the occurrence of several large outbreaks, two linked to cooling towers and one linked to a whirlpool spa. In April 2000, a large outbreak of Legionnaires' disease occurred among persons visiting the newly constructed aquarium in Melbourne, Australia (9). By June, 119 persons were confirmed to have Legionnaires' disease, and four (3.6%) persons died. This outbreak was due to a new cooling tower which had recently come on line. It demonstrated that even new cooling tower systems pose a risk when coming on line and that decontamination procedures should be followed. The American Society of Heating, Refrigeration, and Air-Conditioning Engineers (ASHRAE) has issued guidelines for the control of Legionella in buildings; they describe maintenance procedures that should be performed when bringing cooling towers back on line (8).
In 1999, an outbreak in the Netherlands among attendees at a flower show resulted in 133 confirmed and 55 probable cases of Legionnaires' disease (58). Eleven percent of cases died (C. Navarro, A. Garcia-Fulgueiras, J. Kool, C. Joseph, J. Lee, C. Pelaz, and O. Tello, update on the outbreak of Legionnaires' disease in Murcia, Spain, Eurosurveillance Wkly., http://www.eurosurv.org/update/). This outbreak was due to a contaminated whirlpool spa on display at a consumer product fair attached to the flower show (33). Whirlpool spas have been implicated previously in outbreaks of Legionnaires' disease (21, 22, 153) and Pontiac fever (115, 186, 195, 255). These outbreaks involved spas that were in use. In-store whirlpool spa displays have also been implicated in the past, but never with an outbreak of this magnitude (22). This outbreak demonstrates the importance of biocide treatment for all spas that will be in use, whether or not they will be occupied by bathers (8, 43).
Recently, a large community-wide outbreak of Legionnaires' disease that affected more than 750 persons was reported from Spain. Of these, 310 were confirmed to have legionellosis, making this the largest Legionnaires' disease outbreak ever reported. Remarkably, only one death has been reported (C. Navarro, A. Garcia-Fulgueiras, J. Kool, C. Joseph, J. Lee, C. Pelaz, and O. Tello, update on the outbreak of Legionnaires' disease in Murcia, Spain, Eurosurveillance Wkly., http://www.eurosurv.org/update/). A cooling tower is suspected as the source of the outbreak.
The outbreaks in Australia and Spain had very low case fatality rates compared with the outbreak in the Netherlands. It is possible that differences in strain virulence, host factors, or case ascertainment account for this difference in mortality. Another intriguing possibility is that diagnostic and treatment practices in the various countries played a role. It has been reported that the choice of antibiotics for community-acquired pneumonia differs between these countries, with agents active against Legionella being used empirically in Spain and Australia and beta-lactam antibiotics being used empirically in the Netherlands (C. Navarro, A. Garcia-Fulgueiras, J. Kool, C. Joseph, J. Lee, C. Pelaz, and O. Tello, update on the outbreak of Legionnaires' disease in Murcia, Spain, Eurosurveillance Wkly., http://www.eurosurv.org/update/). In the United States, case fatality rates for reported cases of Legionnaires' disease have declined during the 1990s, a time when empirical use of fluoroquinolones and azithromycin increased for patients hospitalized with pneumonia and when cases were diagnosed earlier because of use of the urine antigen test (A. L. Benin and R. E. Besser, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 873, 2001).
The occurrence of large community outbreaks of Legionnaires' disease can be reduced with the adoption of guidelines for the proper maintenance of cooling towers and other aerosol-generating devices (8, 43, 55). The prevention of sporadic cases is more difficult and will require increased understanding of the route of transmission. A first step, however, is the identification of cases of sporadic disease through the use of proper diagnostic tests. For patients being hospitalized with community-acquired pneumonia who have risk factors for Legionnaires' disease, this should include sputum culture and urine antigen testing. Urine antigen testing alone will miss cases due to non-L. pneumophila serogroup 1 strains; from 1980 to 1989, these cases represented 28% of reported disease (199).
Nosocomial Outbreaks and Prevention
Health care-associated Legionnaires' disease continues to present a significant public health problem. Although the magnitude of the problem is difficult to measure, reports of outbreaks continue to abound. Hospitals represent ideal locations for Legionnaires' disease transmission: at-risk individuals are present in large numbers; plumbing systems are frequently old and complex, favoring amplification of the organism; and water temperatures are often reduced to prevent scalding of patients.
To reduce the likelihood of Legionnaires' disease transmission in health care facilities, CDC recommends a strategy focusing on proper maintenance of water systems, universal testing of patients with nosocomial pneumonia with appropriate tests, and investigation of situations in which transmission has been shown to occur (44). At present, prevention efforts are inadequately implemented in the majority of health care facilities for a variety of reasons.
In 2000, ASHRAE issued guidelines for maintaining water systems to minimize the likelihood of Legionnaires' disease transmission (8). These guidelines recommend maintaining water systems at temperatures unfavorable for the amplification of legionellae. However, many states have regulations that limit the water temperature in health care facilities as a means of reducing scalding injuries (194). In Great Britain, elevated water temperatures were recommended as the primary means of preventing legionellosis in hospitals in 1991 (55). Thermostatic mixing valves are used to reduce the likelihood of scalding. Adoption of these guidelines has resulted in a dramatic decline in nosocomial Legionnaires' disease without increased reports of scalding. Adoption of the ASHRAE guidelines could dramatically reduce the likelihood that legionellae will be amplified in a water system, thereby diminishing the risk of transmission.
The second part of prevention involves clinical surveillance. Currently, a minority of institutions in the United States perform universal Legionella testing of at-risk patients with nosocomial pneumonia (95). Infection control personnel should educate medical staff to heighten suspicion for health care-associated Legionnaires' disease, especially in high-risk patients, and to implement routine testing policies. Appropriate diagnostic testing should be available on site in institutions with high-risk patients and should include Legionella culturing and urine antigen testing. Introduction of routine appropriate testing has led to the identification of previously undetected transmission (173, 179). Only when transmission is recognized can appropriate steps be taken to prevent additional cases.
When cases of Legionnaires' disease are identified, epidemiologic and environmental investigations should be undertaken to determine the means of transmission. Detailed reviews of the procedures for conducting these investigations have been published previously (44).
There has been some debate in the literature regarding the role of environmental culturing for Legionella in the absence of disease transmission (7, 42, 44; Report of the Maryland Scientific Working Group to Study Legionella in Water Systems in Healthcare Institutions, Maryland State Dept. Health Hygiene UR, http://www.dhmh.state.md.us/html/legionella.htm). While opinions differ as to the role of routine periodic environmental monitoring, there is agreement that all health care facilities should have a control strategy in place and that the strategy adopted should vary based on a risk assessment. This recommendation has recently been adopted by the Joint Council on Accreditation of Healthcare Organizations (Standard EC.1.7, www.jcaho.org/standard/stds2002_mpfrm.html). The risk assessment should take into account the types of patients treated, the age and complexity of the potable water system, previous occurrence of Legionnaires' disease, and type of biocide being used.
Potential Impact of Monochloramine
An increasing volume of epidemiologic and laboratory data have begun to accumulate which suggest that the use of monochloramine as a biocide in municipal water systems is more effective than chlorine in preventing transmission of Legionella and has the potential to greatly reduce the incidence of Legionnaires' disease in health care facilities. If these results are reproduced with prospective studies, a new era in Legionnaires' disease prevention may arrive.
In 1999, Kool et al. reported a case-control study assessing the risk of a hospital having a drinking water-associated outbreak of Legionnaires' disease, based on the type of disinfectant used by the municipality (172). Hospitals that had experienced an outbreak were nine times more likely to be in municipalities that used chlorine rather than monochloramine as the drinking water disinfectant. These results suggest that ≈90% of these outbreaks could be prevented by a change at the water treatment plant.
Another study by Kool et al. looked at risk factors for Legionella colonization in hospitals in three Texas counties (170). Once again, they found that hospitals in municipalities that used chlorine as the residual disinfectant were significantly more likely to have legionellae isolated from the water supply.
To assess whether municipal disinfection with monochloramine was associated with a lower occurrence of health care-associated Legionnaires' disease, Heffelfinger et al. conducted a survey of hospital epidemiologists (J. D. Heffelfinger, J. L. Kool, S. K. Fridkin, V. J. Fraser, J. C. Carpenter, J. Hageman, B. A. Kupronis, and W. C. Gaines, Program Abstr. 4th Decennial Conf. Hospital Infections, Atlanta, Ga., 2000). They collected information on the occurrence of outbreaks of Legionnaires' disease, hospital characteristics, including the presence of a transplant unit, and type of hospital and municipal water disinfection. Hospitals using chlorine as the residual disinfectant were significantly more likely to have cases of health care-associated Legionnaires' disease. Based on these data, more than 70% of health care-associated Legionnaires' disease cases might not have occurred if monochloramine had been used instead of free chlorine for residual disinfection.
What is the mechanism by which monochloramine might achieve this effect? Monochloramine is more stable than chlorine and may provide a better residual disinfectant over long distribution systems (163). It also appears to penetrate the biofilms where Legionella reside more effectively than chlorine. This property has been demonstrated in studies by Donlan et al. comparing the effects of monochloramine and chlorine on _H. vermiformis_-associated L. pneumophila in a laboratory-grown potable-water biofilm (66). While both chlorine and monochloramine were effective at eradicating planktonic Legionella, monochloramine was significantly more effective than chlorine at killing Legionella in the biofilm.
Many municipalities are switching to monochloramine as the biocide of choice for their public water distribution systems. Currently, nearly 25% of municipalities use monochloramine, motivated not by Legionnaires' disease prevention but rather by a desire to minimize some of the disinfection by-products produced by chlorine. The U.S. Environmental Protection Agency issued regulations that require reductions in the levels of certain disinfection by-products that have been associated with cancer in laboratory animals (217). Monochloramine is attractive for municipal water systems because it results in reduced production of these regulated disinfection by-products.
If, in fact, many sporadic cases of Legionnaires' disease are transmitted through potable-water systems, it is plausible that the use of monochloramine could have an impact on community-acquired cases as well. Given that many municipalities are already planning to convert from chlorine to monochloramine disinfection in response to the Environmental Protection Agency regulations, there should be many opportunities to study the impact of this intervention prospectively.
FUTURE DIRECTIONS AND CONCLUSIONS
Over the past 25 years, we have learned a great deal about legionellae and legionellosis. Researchers are on the brink of fully characterizing the novel mechanism by which these bacteria infect eukaryotic cells. Understanding of this means of entry into a host cell may have unforeseen benefits. Currently, we know that legionellae can infect very diverse hosts, ranging from slime molds to protozoa to mammalian cells. There may be additional hosts yet to be discovered for these diverse intracellular organisms.
The convergence of studies on microbial biofilms and the cell-cell signaling that occurs within these communities appear particularly relevant to legionellae. We have only begun to characterize the interaction of legionellae with the microbial community. While it seems clear that legionellae are common inhabitants of biofilms, the significance of biofilms to the multiplication and distribution of legionellae remains undefined. Reducing legionellae in the biofilms that form in building water systems is a primary goal in strategies to prevent Legionnaires' disease. Application of advances in cell signaling to the study of legionellae in biofilms will lead to a better understanding of the ecology of these bacteria and to more efficient mechanisms to control this disease.
We continue to make progress in understanding the epidemiology of Legionnaires' disease. Many opportunities now exist to improve the prevention of legionellosis, especially in the United States. Computer-based reporting systems may one day provide a means of conducting timely surveillance for previously undetected outbreaks of travel-related Legionnaires' disease. New rapid diagnostic tests can improve case detection, but at a public health cost if these new methodologies replace bacterial cultures. Municipal disinfection with monochloramine may provide a means of reducing the incidence of both health care-associated and community-acquired Legionnaires' disease.
Acknowledgments
We acknowledge Claressa E. Lucas for assistance in preparation of the manuscript.
REFERENCES
- 1.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62**:**2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., B. S. Fields, and N. C. Engleberg. 1994. Protein expression by the protozoan Hartmannella vermiformis upon contact with its bacterial parasite Legionella pneumophila. Infect. Immun. 62**:**1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., L. Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64**:**3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adeleke, A., J. Pruckler, R. Benson, T. Rowbotham, M. Halablab, and B. Fields. 1996. _Legionella_-like amebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2**:**225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adeleke, A. A., B. S. Fields, R. F. Benson, M. I. Daneshvar, J. M. Purckler, R. M. Ratcliff, T. G. Harrison, R. S. Weyant, R. J. Birtles, D. Raoult, and M. A. Halablab. 2001. Legionella drozanskii sp. nov., Legionella rowbothamii sp. nov. and Legionella fallonii sp. nov.: three unusual new Legionella species. Int. J. Syst. E vol. Microbiol. 51**:**1151-1160. [DOI] [PubMed] [Google Scholar]
- 6.Aguero-Rosenfeld, M. E., and P. H. Edelstein. 1988. Retrospective evaluation of the Du Pont radioimmunoassay kit for detection of Legionella pneumophila serogroup 1 antigenuria in humans. J. Clin. Microbiol. 26**:**1775-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allegheny County Health Department. 1997. Approaches to prevention and control of Legionella infection in Allegheny County health care facilities, 2nd ed. Allegheny County Health Department, Pittsburgh, Pa.
- 8.American Society of Heating, Refrigeration, and Air-Conditioning Engineers. 2000. ASHRAE Guideline 12-2000 — minimizing the risk of legionellosis associated with building water systems. American Society for Heating, Refrigeration, and Air-Conditioning Engineers, Inc., Atlanta, Ga.
- 9.Anonymous. 2000. Outbreak of Legionnaires' disease associated with an aquarium in Australia. CDR Wkly. 10**:**161. [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Aragon, V., S. Kurtz, and N. P. Cianciotto. 2001. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69**:**177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and _pilD_-deficient Legionella pneumophila. Infect. Immun. 68**:**1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballard, A. L., N. K. Fry, L. Chan, S. B. Surman, J. V. Lee, T. G. Harrison, and K. J. Towner. 2000. Detection of Legionella pneumophila using a real-time PCR hybridization assay. J. Clin. Microbiol. 38**:**4215-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bangsborg, J. M., G. Shand, E. Pearlman, and N. Hoiby. 1991. Cross-reactive Legionella antigens and the antibody response during infection. APMIS 99**:**854-865. [PubMed] [Google Scholar]
- 15.Bangsborg, J. M., G. H. Shand, K. Hansen, and J. B. Wright. 1994. Performance of four different indirect enzyme-linked immunosorbent assays (ELISAs) to detect specific IgG, IgA, and IgM in Legionnaires' disease. APMIS 102**:**501-508. [DOI] [PubMed] [Google Scholar]
- 16.Barbaree, J. M. 1993. Selecting a subtyping technique for use in investigations of Legionellosis epidemics, p. 169-172. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 17.Barbaree, J. M., B. S. Fields, J. C. Feeley, G. W. Gorman, and W. T. Martin. 1986. Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl. Environ. Microbiol. 51**:**422-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barka, N., J. P. Tomasi, and S. Stadtsbaeder. 1986. ELISA using whole Legionella pneumophila cell as antigen. Comparison between monovalent and polyvalent antigens for the serodiagnosis of human legionellosis. J. Immunol. Methods 93**:**77-81. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett, J. G., S. F. Dowell, L. A. Mandell, T. M. File, Jr., D. M. Musher, and A. M. Fine. 2000. Practice guidelines for the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 31**:**347-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellinger-Kawahara, C., and M. A. Horwitz. 1990. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J. Exp. Med. 172**:**1201-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benin, A., R. Benson, K. Arnold, A. Fiore, P. Cook, K. Williams, B. Fields, and R. Besser. 2002. An outbreak of travel-associated Legionnaires' disease and Pontiac fever and the need for enhanced surveillance for travel-associated legionellosis in the United States. J. Infect. Dis. 185**:**237-243. [DOI] [PubMed] [Google Scholar]
- 22.Benkel, D. H., E. M. McClure, D. Woolard, J. V. Rullan, G. B. Miller, S. R. Jenkins, J. H. Hershey, R. F. Benson, J. M. Pruckler, E. W. Brown, M. S. Kolczak, R. L. Hackler, B. S. Rouse, and R. F. Breiman. 2000. Outbreak of Legionnaires' disease associated with a display whirlpool spa. Int. J. Epidemiol. 29**:**1092-1098. [DOI] [PubMed] [Google Scholar]
- 23.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13**:**90-99. [PubMed] [Google Scholar]
- 24.Benson, R. F., P. W. Tang, and B. S. Fields. 2000. Evaluation of the Binax and Biotest urinary antigen kits for detection of Legionnaires' disease due to multiple serogroups and species of Legionella. J. Clin. Microbiol. 38**:**2763-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson, R. F., W. L. Thacker, B. B. Plikaytis, and H. W. Wilkinson. 1987. Cross-reactions in Legionella antisera with Bordetella pertussis strains. J. Clin. Microbiol. 25**:**594-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bentham, R. H., and C. R. Broadbent. 1993. A model for autumn outbreaks of Legionnaires' disease associated with cooling towers, linked to system operation and size. Epidemiol. Infect. 111**:**287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berdal, B. P., C. E. Farshy, and J. C. Feeley. 1979. Detection of Legionella pneumophila antigen in urine by enzyme-linked immunospecific assay. J. Clin. Microbiol. 9**:**575-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7**:**7-19. [DOI] [PubMed] [Google Scholar]
- 29.Bernander, S., B. Gastrin, S. Lofgren, and A. M. Olinder-Nielsen. 1994. Legionella urinary antigen in early disease. Scand. J. Infect. Dis. 26**:**777-778. [DOI] [PubMed] [Google Scholar]
- 30.Blatt, S. P., M. J. Dolan, C. W. Hendrix, and G. P. Melcher. 1994. Legionnaires' disease in human immunodeficiency virus-infected patients: eight cases and review. Clin. Infect. Dis. 18**:**227-232. [DOI] [PubMed] [Google Scholar]
- 31.Bornstein, N., N. Janin, G. Bourguignon, M. Surgot, and J. Fleurette. 1986. Prevalence of anti-Legionella antibodies in a healthy population and in patients with tuberculosis or pneumonia. Isr. J. Med. Sci. 22**:**740-744. [PubMed] [Google Scholar]
- 32.Bornstein, N., D. Marmet, M. H. Dumaine, M. Surgot, and J. Fleurette. 1991. Detection of flagella in 278 Legionella strains by latex reagent sensitized with antiflagellum immunoglobulins. J. Clin. Microbiol. 29**:**953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boshuizen, H. C., S. E. Neppelenbroek, H. van Vliet, J. F. Schellekens, J. W. Boer, M. F. Peeters, and M. A. Spaendonck. 2001. Subclinical Legionella infection in workers near the source of a large outbreak of legionnaires disease. J. Infect. Dis. 184**:**515-518. [DOI] [PubMed] [Google Scholar]
- 34.Bosshardt, S. C., R. F. Benson, and B. S. Fields. 1997. Flagella are a positive predictor for virulence in Legionella. Microb. Pathog. 23**:**107-112. [DOI] [PubMed] [Google Scholar]
- 35.Boswell, T. C. 1996. Serological cross reaction between Legionella and Campylobacter in the rapid microagglutination test. J. Clin. Pathol. 49**:**584-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boswell, T. C., and G. Kudesia. 1992. Serological cross-reaction between Legionella pneumophila and Campylobacter in the indirect fluorescent antibody test. Epidemiol. Infect. 109**:**291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64**:**668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner, D. J., A. G. Steigerwalt, and J. E. McDade. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann. Intern. Med. 90**:**656-658. [DOI] [PubMed] [Google Scholar]
- 39.Brown, A., M. Lema, C. A. Ciesielski, and M. J. Blaser. 1985. Combined plasmid and peptide analysis of clinical and environmental Legionella pneumophila strains associated with a small cluster of Legionnaires' disease cases. Infection 13**:**163-166. [DOI] [PubMed] [Google Scholar]
- 40.Buesching, W. J., R. A. Brust, and L. W. Ayers. 1983. Enhanced primary isolation of Legionella pneumophila from clinical specimens by low-pH treatment. J. Clin. Microbiol. 17**:**1153-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler, J., and R. Breiman. 1998. Legionellosis, p. 355-376. In A. Evans and P. Brachman (ed.), Bacterial infections in humans: epidemiology and control, 3rd ed. Plenum Medical Book Company, New York, N.Y.
- 42.Centers for Disease Control and Prevention. 2000. CDC/IDSA/ASBMT guidelines for the prevention of opportunistic infections in hematopoietic stem cell transplant recipients. Morb. Mortal. Wkly. Rep. 49:RR-1**:**1-128. [PubMed] [Google Scholar]
- 43.Centers for Disease Control and Prevention. 1995. Final recommendations to minimize transmission of Legionnaires' disease from whirlpool spas on cruise ships. Centers for Disease Control and Prevention, Atlanta, Ga.
- 44.Centers for Disease Control and Prevention. 1997. Guidelines for prevention of nosocomial pneumonia. Morb. Mortal. Wkly. Rep. 46:RR-1**:**1-79. [PubMed] [Google Scholar]
- 45.Centers for Disease Control and Prevention. 2000. Legionnaires' disease associated with potting soil—California, Oregon, and Washington, May-June 2000. Morb. Mortal. Wkly. Rep. 49**:**777-778. [PubMed] [Google Scholar]
- 46.Chang, F. Y., J. E. Stout, and V. L. Yu. 1996. Assessment of enzyme immunoassay versus radioimmunoassay for detection of Legionella pneumophila serogroup 1 antigen in frozen urine specimens. J. Clin. Microbiol. 34**:**2628-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cianciotto, N. P., J. M. Bangsborg, B. I. Eisenstein, and N. C. Engleberg. 1990. Identification of _mip_-like genes in the genus Legionella. Infect. Immun. 58**:**2912-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, G. B. Toews, and N. C. Engleberg. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 57**:**1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89**:**5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67**:**4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cloud, J. L., K. C. Carroll, P. Pixton, M. Erali, and D. R. Hillyard. 2000. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol. 38**:**1709-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins, M. T., S. N. Cho, N. Høiby, F. Espersen, L. Baek, and J. S. Reif. 1983. Crossed immunoelectrophoretic analysis of Legionella pneumophila serogroup 1 antigens. Infect. Immun. 39**:**1428-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins, M. T., F. Espersen, N. Hoiby, S. N. Cho, A. Friss-Moller, and J. S. Reif. 1983. Cross-reactions between Legionella pneumophila (serogroup 1) and twenty-eight other bacterial species, including other members of the family Legionellaceae. Infect. Immun. 39**:**1441-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins, M. T., J. McDonald, N. Høiby, and O. Aalund. 1984. Agglutinating antibody titers to members of the family Legionellaceae in cystic fibrosis patients as a result of cross-reacting antibodies to Pseudomonas aeruginosa. J. Clin. Microbiol. 19**:**757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Commission, H. S. 1991. The prevention or control of legionellosis (including legionnaires' disease). Health and Safety Executive, London, England.
- 56.De, Z., and T. G. Harrison. 1999. Molecular typing of Legionella pneumophila serogroup 1 by pulsed-field gel electrophoresis with SfiI and comparison of this method with restriction fragment-length polymorphism analysis. J. Med. Microbiol. 48**:**269-278. [DOI] [PubMed] [Google Scholar]
- 57.DeForges, L., P. Legrand, J. Tankovic, C. Brun-Buisson, P. Lang, and C. J. Soussy. 1999. Case of false-positive results of the urinary antigen test for Legionella pneumophila. Clin. Infect. Dis. 29**:**953-954. [DOI] [PubMed] [Google Scholar]
- 58.Den Boer, J. W., E. P. F. Yzerman, J. Schellekens, K. D. Lettinga, H. C. Boshuizen, J. E. Van Steenbergen, A. Bosman, S. Van den Hof, H. A. Van Vliet, M. F. Peeters, R. J. Van Ketel, P. Speelman, J. L. Kool, and M. A. E. Conyn-Van Spaendonck. 2002. A large outbreak of Legionnaires' disease at a flower show, the Netherlands, 1999. Emerg. Infect. Dis. 8**:**37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Ory, F., J. M. Echevarria, C. Pelaz, A. Tellez, M. A. Mateo, and J. Lopez. 2000. Detection of specific IgM antibody in the investigation of an outbreak of pneumonia due to Legionella pneumophila serogroup 1. Clin. Microbiol. Infect. 6**:**64-68. [DOI] [PubMed] [Google Scholar]
- 60.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58**:**2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doebbeling, B. N., M. J. Bale, F. P. Koontz, C. M. Helms, R. P. Wenzel, and M. A. Pfaller. 1988. Prospective evaluation of the Gen-Probe assay for detection of legionellae in respiratory specimens. Eur. J. Clin. Microbiol. Infect. Dis. 7**:**748-752. [DOI] [PubMed] [Google Scholar]
- 62.Domínguez, J. A., N. Galí, L. Matas, P. Pedroso, A. Hernandez, E. Padilla, and V. Ausina. 1999. Evaluation of a rapid immunochromatographic assay for the detection of Legionella antigen in urine samples. Eur. J. Clin. Microbiol. Infect. Dis. 18**:**896-898. [DOI] [PubMed] [Google Scholar]
- 63.Domínguez, J. A., N. Galí, P. Pedroso, A. Fargas, E. Padilla, J. M. Manterola, and L. Matas. 1998. Comparison of the Binax Legionella urinary antigen enzyme immunoassay (EIA) with the Biotest-Legionella-urine antigen EIA for detection of Legionella antigen in both concentrated and nonconcentrated urine samples. J. Clin. Microbiol. 36**:**2718-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Domínguez, J. A., J. M. Manterola, R. Blavia, N. Sopena, F. J. Belda, E. Padilla, M. Gimenez, M. Sabria, J. Morera, and V. Ausina. 1996. Detection of Legionella pneumophila serogroup-1 antigen in nonconcentrated urine and urine concentrated by selective ultrafiltration. J. Clin. Microbiol. 34**:**2334-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Domínguez, J. A., L. Matas, J. M. Manterola, R. Blavia, N. Sopena, F. J. Belda, E. Padilla, M. Gimenez, M. Sabria, J. Morera, and V. Ausina. 1997. Comparison of radioimmunoassay and enzyme immunoassay kits for detection of Legionella pneumophila serogroup 1 antigen in both concentrated and nonconcentrated urine samples. J. Clin. Microbiol. 35**:**1627-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donlan, R., R. Murga, J. Carpenter, E. Brown, R. Besser, and B. Fields. 2002. Monochloramine disinfection of biofilm-associated Legionella pneumophila in a potable water model system, p. 406-410. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. American Society for Microbiology, Washington, D.C.
- 67.Dournon, E. 1988. Isolation of legionellae from clinical specimens, p. 13-30. In T. G. Harrison and A. G. Taylor (ed.), A laboratory manual for Legionella. John Wiley and Sons, New York, N.Y.
- 68.Drozanski, W. 1956. Fatal bacterial infection in soil amoebae. Acta Microbiol. Pol. 5**:**315-317. [PubMed] [Google Scholar]
- 69.Dwyer, D. E., V. L. Gibbons, L. M. Brady, and A. L. Cunningham. 1988. Serological reaction to Legionella pneumophila group 4 in a patient with Q fever. J. Infect. Dis. 158**:**499-500. [PubMed] [Google Scholar]
- 70.Edelstein, P. H. 1998. Antimicrobial chemotherapy for Legionnaires' disease: time for a change. Ann. Intern. Med. 129**:**328-330. [DOI] [PubMed] [Google Scholar]
- 71.Edelstein, P. H. 1982. Comparative study of selective media for isolation of Legionella pneumophila from potable water. J. Clin. Microbiol. 16**:**697-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edelstein, P. H. 1997. Detection of antibodies to Legionella spp., p. 502-509. In N. R. Rose, E. C. de Macario, J. D. Folds, H. C. Lane, and R. M. Nakamura (ed.), Manual of clinical laboratory immunology, vol. 5. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 73.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14**:**298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edelstein, P. H. 1987. The laboratory diagnosis of Legionnaires' disease. Semin. Respir. Infect. 2**:**235-241. [PubMed] [Google Scholar]
- 75.Edelstein, P. H. 1993. Laboratory diagnosis of Legionnaires' disease: an update from 1984, p. 7-11. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 76.Edelstein, P. H. 1993. Legionnaires' disease. Clin. Infect. Dis. 16**:**741-749. [DOI] [PubMed] [Google Scholar]
- 77.Edelstein, P. H., K. B. Beer, J. C. Sturge, A. J. Watson, and L. C. Goldstein. 1985. Clinical utility of a monoclonal direct fluorescent reagent specific for Legionella pneumophila: comparative study with other reagents. J. Clin. Microbiol. 22**:**419-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edelstein, P. H., R. N. Bryan, R. K. Enns, D. E. Kohne, and D. L. Kacian. 1987. Retrospective study of Gen-Probe rapid diagnostic system for detection of legionellae in frozen clinical respiratory tract samples. J. Clin. Microbiol. 25**:**1022-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edelstein, P. H., and M. A. C. Edelstein. 1989. Evaluation of the Merifluor-Legionella immunofluorescent reagent for identifying and detecting 21 Legionella species. J. Clin. Microbiol. 27**:**2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edelstein, P. H., R. M. McKinney, R. D. Meyer, M. A. C. Edelstein, C. J. Krause, and S. M. Finegold. 1980. Immunologic diagnosis of Legionnaires' disease: cross-reactions with anaerobic and microaerophilic organisms and infections caused by them. J. Infect. Dis. 141**:**652-655. [DOI] [PubMed] [Google Scholar]
- 81.Edelstein, P. H., J. B. Snitzer, and J. A. Bridge. 1982. Enhancement of recovery of Legionella pneumophila from contaminated respiratory tract specimens by heat. J. Clin. Microbiol. 16**:**1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Engleberg, N. C., C. Carter, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57**:**1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fallon, R. J., and W. H. Abraham. 1982. Polyvalent heat-killed antigen for the diagnosis of infection with Legionella pneumophila. J. Clin. Pathol. 35**:**434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farshy, C. E., D. D. Cruce, G. C. Klein, H. W. Wilkinson, and J. C. Feeley. 1979. Immunoglobulin specificity of the microagglutination test for the Legionnaires' disease bacterium. Ann. Intern. Med. 90**:**690. [DOI] [PubMed] [Google Scholar]
- 85.Farshy, C. E., G. C. Klein, and J. C. Feeley. 1978. Detection of antibodies to legionnaires disease organism by microagglutination and micro-enzyme-linked immunosorbent assay tests. J. Clin. Microbiol. 7**:**327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feddersen, A., H. G. Meyer, P. Matthes, S. Bhakdi, and M. Husmann. 2000. GyrA sequence-based typing of Legionella. Med. Microbiol. Immunol. 189**:**7-11. [DOI] [PubMed] [Google Scholar]
- 87.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10**:**437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feeley, J. C., G. W. Gorman, R. E. Weaver, D. C. Mackel, and H. W. Smith. 1978. Primary isolation media for Legionnaires' disease bacterium. J. Clin. Microbiol. 8**:**320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferguson, D. A., Jr., and W. R. Mayberry. 1987. Differentiation of Legionella species by soluble protein patterns in polyacrylamide slab gels. Microbios 52**:**105-114. [PubMed] [Google Scholar]
- 90.Fields, B. S. 1997. Legionellae and Legionnaires' disease, p. 666-675. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 91.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4**:**286-290. [DOI] [PubMed] [Google Scholar]
- 92.Fields, B. S. 1992. Procedures for the recovery of Legionella from the environment, vol. 2. U.S. Department of Health and Human Services, Atlanta, Ga.
- 93.Fields, B. S., S. R. Fields, J. N. Loy, E. H. White, W. L. Steffens, and E. B. Shotts. 1993. Attachment and entry of Legionella pneumophila in Hartmannella vermiformis. J. Infect. Dis. 167**:**1146-1150. [DOI] [PubMed] [Google Scholar]
- 94.Finkelstein, R., P. Brown, W. A. Palutke, B. B. Wentworth, J. G. Geiger, G. D. Bostic, and J. D. Sobel. 1993. Diagnostic efficacy of a DNA probe in pneumonia caused by Legionella species. J. Med. Microbiol. 38**:**183-186. [DOI] [PubMed] [Google Scholar]
- 95.Fiore, A. E., J. C. Butler, T. G. Emori, and R. P. Gaynes. 1999. A survey of methods used to detect nosocomial legionellosis among participants in the National Nosocomial Infections Surveillance System. Infect. Control Hosp. Epidemiol. 20**:**412-416. [DOI] [PubMed] [Google Scholar]
- 96.Fiore, A. E., J. P. Nuorti, O. S. Levine, A. Marx, A. C. Weltman, S. Yeager, R. F. Benson, J. Pruckler, P. H. Edelstein, P. Greer, S. R. Zaki, B. S. Fields, and J. C. Butler. 1998. Epidemic Legionnaires' disease two decades later: old sources, new diagnostic methods. Clin. Infect. Dis. 26**:**426-433. [DOI] [PubMed] [Google Scholar]
- 97.Fischer, G., H. Bang, B. Ludwig, K. Mann, and J. Hacker. 1992. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPlase) activity. Mol. Microbiol. 6**:**1375-1383. [DOI] [PubMed] [Google Scholar]
- 98.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41**:**9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flournoy, D. J., K. A. Belobraydic, S. L. Silberg, C. H. Lawrence, and P. J. Guthrie. 1988. False positive Legionella pneumophila direct immunofluorescent monoclonal antibody test caused by Bacillus cereus spores. Diagn. Microbiol. Infect. Dis. 9**:**123-125. [DOI] [PubMed] [Google Scholar]
- 100.Formica, N., G. Tallis, B. Zwolak, J. Camie, M. Beers, G. Hogg, N. Ryan, and M. Yates. 2000. Legionnaires' disease outbreak: Victoria's largest identified outbreak. Commun. Dis. Intel. 24**:**199-202. [PubMed] [Google Scholar]
- 101.Fox, K. F., and A. Brown. 1989. Application of numerical systematics to the phenotypic differentiation of legionellae. J. Clin. Microbiol. 27**:**1952-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fox, K. F., and A. Brown. 1993. Properties of the genus Tatlockia. Differentiation of Tatlockia (Legionella) maceachernii and micdadei from each other and from other legionellae. Can. J. Microbiol. 39**:**486-491. [DOI] [PubMed] [Google Scholar]
- 103.Fraser, D. W. 1984. Sources of legionellosis, p. 277-280. In A. Balows, C. Thornsberry, J. C. Feeley, and W. Jakubowski (ed.), Proceedings of the Second International Symposium on Legionella. American Society for Microbiology, Washington, D.C.
- 104.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297**:**1189-1197. [DOI] [PubMed] [Google Scholar]
- 105.Fry, N. K., J. M. Bangsborg, S. Bernander, J. Etienne, B. Forsblom, V. Gaia, P. Hasenberger, D. Lindsay, A. Papoutsi, C. Pelaz, M. Struelens, S. A. Uldum, P. Visca, and T. G. Harrison. 2000. Assessment of intercentre reproducibility and epidemiological concordance of Legionella pneumophila serogroup 1 genotyping by amplified fragment length polymorphism analysis. Eur. J. Clin. Microbiol. Infect. Dis. 19**:**773-780. [DOI] [PubMed] [Google Scholar]
- 106.Fry, N. K., S. Warwick, N. A. Saunders, and T. M. Embley. 1991. The use of 16S ribosomal RNA analyses to investigate the phylogeny of the family Legionellaceae. J. Gen. Microbiol. 137**:**1215-1222. [DOI] [PubMed] [Google Scholar]
- 107.Gao, L. Y., and Y. Abu Kwaik. 1999. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect. Immun. 67**:**862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao, L. Y., and Y. Abu Kwaik. 2000. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ. Microbiol. 2**:**79-90. [DOI] [PubMed] [Google Scholar]
- 109.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65**:**4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao, L. Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. 1998. Different fates of Legionella pneumophila pmi and mil mutants within macrophages and alveolar epithelial cells. Microb. Pathog. 25**:**291-306. [DOI] [PubMed] [Google Scholar]
- 111.Garcia, M., I. Campbell, P. Tang, and C. Krishnan. 1993. Incidence of Legionnaires' disease and report of Legionella sainthelensi serogroup 1 infection in Toronto, Canada, p. 37-39. In J. M. Barbaree, R. F. Breiman, and A. P. DuFour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 112.Garrity, G. M., A. Brown, and R. M. Vickers. 1980. Tatlockia and Fluoribacter: two new genera of organisms resembling Legionella pneumophila. Int. J. Syst. Bacteriol. 30**:**609-614. [Google Scholar]
- 113.Georghiou, P. R., A. M. Doggett, M. A. Kielhofner, J. E. Stout, D. A. Watson, J. R. Lupski, and R. J. Hamill. 1994. Molecular fingerprinting of Legionella species by repetitive element PCR. J. Clin. Microbiol. 32**:**2989-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Glick, T. H., M. B. Gregg, B. Berman, G. Mallison, W. W. Rhodes, Jr., and I. Kassanoff. 1978. Pontiac fever. An epidemic of unknown etiology in a health department. I. Clinical and epidemiologic aspects. Am. J. Epidemiol. 107**:**149-160. [DOI] [PubMed] [Google Scholar]
- 115.Goldberg, D. J., J. A. Emslie, R. J. Fallon, S. T. Green, and J. G. Wrench. 1992. Pontiac fever in children. Pediatr. Infect. Dis. J. 11**:**240-241. [DOI] [PubMed] [Google Scholar]
- 116.Gomez-Lus, P., B. S. Fields, R. F. Benson, W. T. Martin, S. P. O'Connor, and C. M. Black. 1993. Comparison of arbitrarily primed polymerase chain reaction, ribotyping, and monoclonal antibody analysis for subtyping Legionella pneumophila serogroup 1. J. Clin. Microbiol. 31**:**1940-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gosting, L. H., K. Cabrian, J. C. Sturge, and L. C. Goldstein. 1984. Identification of a species-specific antigen in Legionella pneumophila by a monoclonal antibody. J. Clin. Microbiol. 20**:**1031-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grady, G. F., and R. F. Gilfillan. 1979. Relation of Mycoplasma pneumoniae seroreactivity, immunosuppression, and chronic disease to Legionnaires' disease. A twelve-month prospective study of sporadic cases in Massachusetts. Ann. Intern. Med. 90**:**607-610. [DOI] [PubMed] [Google Scholar]
- 119.Gray, J. J., K. N. Ward, R. E. Warren, and M. Farrington. 1991. Serological cross-reaction between Legionella pneumophila and Citrobacter freundii in indirect immunofluorescence and rapid microagglutination tests. J. Clin. Microbiol. 29**:**200-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Green, P. N., and R. S. Pirrie. 1993. A laboratory apparatus for the generation and biocide efficacy testing of Legionella biofilms. J. Appl. Bacteriol. 74**:**388-393. [DOI] [PubMed] [Google Scholar]
- 121.Gutierrez Rodero, F., V. Ortiz de la Tabla, C. Martinez, M. M. Masia, E. Chiner, and J. L. Calpe. 1995. Legionnaires' disease in patients infected with human immunodeficiency virus. Clin. Infect. Dis. 21**:**712-713. [DOI] [PubMed] [Google Scholar]
- 122.Hackman, B. A., J. F. Plouffe, R. F. Benson, B. S. Fields, and R. F. Breiman. 1996. Comparison of Binax Legionella urinary antigen EIA kit with Binax Ria urinary antigen kit for detection of Legionella pneumophila serogroup 1 antigen. J. Clin. Microbiol. 34**:**1579-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hagele, S., R. Kohler, H. Merkert, M. Schleicher, J. Hacker, and M. Steinert. 2000. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell. Microbiol. 2**:**165-171. [DOI] [PubMed] [Google Scholar]
- 124.Hagele, S., J. Hacker, and B. C. Brand. 1998. Legionella pneumophila kills human phagocytes but not protozoan host cells by inducing apoptotic cell death. FEMS Microbiol. Lett. 169**:**51-58. [DOI] [PubMed] [Google Scholar]
- 125.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33**:**721-731. [DOI] [PubMed] [Google Scholar]
- 126.Harrison, T. G., E. Dournon, and A. G. Taylor. 1987. Evaluation of sensitivity of two serological tests for diagnosing pneumonia caused by Legionella pneumophila serogroup 1. J. Clin. Pathol. 40**:**77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Harrison, T. G., and A. G. Taylor. 1982. Diagnosis of Legionella pneumophila infections by means of formolised yolk sac antigens. J. Clin. Pathol. 35**:**211-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Harrison, T. G., and A. G. Taylor. 1988. The diagnosis of Legionnaires' disease by estimation of antibody levels, p. 113-135. In T. G. Harrison and A. G. Taylor (ed.), A laboratory manual for Legionella, vol. 1. John Wiley and Sons, Ltd., Chichester, England. [Google Scholar]
- 129.Harrison, T. G., and A. G. Taylor. 1988. A laboratory manual for Legionella. Wiley, New York, N.Y.
- 130.Harrison, T. G., and A. G. Taylor. 1982. A rapid microagglutination test for the diagnosis of Legionella pneumophila (serogroup 1) infection. J. Clin. Pathol. 35**:**1028-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hayden, R. T., J. R. Uhl, X. Qian, M. K. Hopkins, M. C. Aubry, A. H. Limper, R. V. Lloyd, and F. R. Cockerill. 2001. Direct detection of Legionella species from bronchoalveolar lavage and open lung biopsy specimens: comparison of LightCycler PCR, in situ hybridization, direct fluorescence antigen detection and culture. J. Clin. Microbiol. 39**:**2618-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heath, C. H., D. I. Grove, and D. F. Looke. 1996. Delay in appropriate therapy of _Legionella_-pneumonia associated with increased mortality. Eur. J. Clin. Microbiol. Infect. Dis. 15**:**286-290. [DOI] [PubMed] [Google Scholar]
- 133.Hebert, G. A. 1981. Hippurate hydrolysis by Legionella pneumophila. J. Clin. Microbiol. 13**:**240-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reference deleted.
- 135.Helbig, J., S. A. Uldum, P. C. Luck, and T. G. Harrison. 2001. Detection of Legionella pneumophila antigen in urine samples by the Binax NOW. J. Med. Microbiol. 50**:**509-516. [DOI] [PubMed] [Google Scholar]
- 136.Helbig, J. H., T. Engelstadter, M. Maiwald, S. A. Uldum, W. Witzleb, and P. C. Luck. 1999. Diagnostic relevance of the detection of Legionella DNA in urine samples by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 18**:**716-722. [DOI] [PubMed] [Google Scholar]
- 137.Helbig, J. H., J. B. Kurtz, M. C. Pastoris, C. Pelaz, and P. C. Luck. 1997. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J. Clin. Microbiol. 35**:**2841-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Helbig, J. H., B. Ludwig, P. C. Luck, A. Groh, W. Witzleb, and J. Hacker. 1995. Monoclonal antibodies to Legionella Mip proteins recognize genus- and species-specific epitopes. Clin. Diagn. Lab. Immunol. 2**:**160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Helsel, L. O., W. F. Bibb, C. A. Butler, P. S. Hoffman, and R. M. McKinney. 1988. Recognition of a genus-wide antigen of Legionella by a monoclonal antibody. Curr. Microbiol. 16**:**201-208. [Google Scholar]
- 140.Holliday, M. 1983. Counterimmunoelectrophoresis in the serodiagnosis of Legionnaires' disease. J. Clin. Pathol. 36**:**466-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Holliday, M. G. 1980. The diagnosis of Legionnaires' disease by counterimmunoelectrophoresis. J. Clin. Pathol. 33**:**1174-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Holliday, M. G. 1990. Use of latex agglutination technique for detecting Legionella pneumophila (serogroup 1) antibodies. J. Clin. Pathol. 43**:**860-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hookey, J. V., N. A. Saunders, N. K. Fry, R. J. Birtles, and T. G. Harrison. 1996. Phylogeny of legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for _Legionella_-like amoebal pathogens. Int. J. Syst. Bacteriol. 46**:**526-531. [Google Scholar]
- 144.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158**:**1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158**:**2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36**:**27-33. [DOI] [PubMed] [Google Scholar]
- 147.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99**:**1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66**:**441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hutchinson, E. J., C. A. Joseph, and C. L. R. Bartlett. 1996. EWGLI: a European surveillance scheme for travel associated legionnaires' disease. Eurosurvey 1**:**37-39. [DOI] [PubMed] [Google Scholar]
- 150.Ingram, J. G., and J. F. Plouffe. 1994. Danger of sputum purulence screens in culture of Legionella species. J. Clin. Microbiol. 32**:**209-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jaulhac, B., M. Nowicki, N. Bornstein, O. Meunier, G. Prevost, Y. Piemont, J. Fleurette, and H. Monteil. 1992. Detection of Legionella spp. in bronchoalveolar lavage fluids by DNA amplification. J. Clin. Microbiol. 30**:**920-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jernigan, D. B. 2001. Electronic laboratory-based reporting: opportunities and challenges for surveillance. Emerg. Infect. Dis. 7(Suppl. 3)**:**538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jernigan, D. B., J. Hofmann, M. S. Cetron, C. A. Genese, J. P. Nuorti, B. S. Fields, R. F. Benson, R. J. Carter, P. H. Edelstein, I. C. Guerrero, S. M. Paul, H. B. Lipman, and R. Breiman. 1996. Outbreak of Legionnaires' disease among cruise ship passengers exposed to a contaminated whirlpool spa. Lancet 347**:**494-499. [DOI] [PubMed] [Google Scholar]
- 154.Jimenez-Lucho, V., M. Shulman, and J. Johnson. 1994. Bordetella bronchiseptica in an AIDS patient cross-reacts with Legionella antisera. J. Clin. Microbiol. 32**:**3095-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Joly, J. R., R. M. McKinney, J. O. Tobin, W. F. Bibb, I. D. Watkins, and D. Ramsay. 1986. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 23**:**768-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jonas, D., A. Rosenbaum, S. Weyrich, and S. Bhakdi. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J. Clin. Microbiol. 33**:**1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kallings, I., and K. Nordstrom. 1983. The pattern of immunoglobulins with special reference to IgM in Legionnaires' disease patients during a 2 year follow-up period. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 255**:**27-32. [PubMed] [Google Scholar]
- 158.Kashuba, A. D., and C. H. Ballow. 1996. Legionella urinary antigen testing: potential impact on diagnosis and antibiotic therapy. Diagn. Microbiol. Infect. Dis. 24**:**129-139. [DOI] [PubMed] [Google Scholar]
- 159.Katz, S. M., and J. M. Hammel. 1987. The effect of drying, heat, and pH on the survival of Legionella pneumophila. Ann. Clin. Lab. Sci. 17**:**150-156. [PubMed] [Google Scholar]
- 160.Kazandjian, D., R. Chiew, and G. L. Gilbert. 1997. Rapid diagnosis of Legionella pneumophila serogroup 1 infection with the Binax enzyme immunoassay urinary antigen test. J. Clin. Microbiol. 35**:**954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kessler, H. H., F. F. Reinthaler, A. Pschaid, K. Pierer, B. Kleinhappl, E. Eber, and E. Marth. 1993. Rapid detection of Legionella species in bronchoalveolar lavage fluids with the EnviroAmp Legionella PCR amplification and detection kit. J. Clin. Microbiol. 31**:**3325-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.King, C. H., B. S. Fields, E. B. Shotts, Jr., and E. H. White. 1991. Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect. Immun. 59**:**758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kirmeyer, G., G. Foust, G. Pierson, J. Simmler, and M. MeChevalier. 1993. Optimizing chloramine treatment. American Water Works Research Foundation, Denver, Colo.
- 164.Klein, G. C. 1980. Cross-reaction to Legionella pneumophila antigen in sera with elevated titers to Pseudomonas pseudomallei. J. Clin. Microbiol. 11**:**27-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Klein, G. C., W. L. Jones, and J. C. Feeley. 1979. Upper limit of normal titer for detection of antibodies to Legionella pneumophila by the microagglutination test. J. Clin. Microbiol. 10**:**754-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kohler, R. B., L. J. Wheat, M. L. French, P. L. Meenhorst, W. C. Winn, Jr., and P. H. Edelstein. 1985. Cross-reactive urinary antigens among patients infected with Legionella pneumophila serogroups 1 and 4 and the Leiden 1 strain. J. Infect. Dis. 152**:**1007-1012. [DOI] [PubMed] [Google Scholar]
- 167.Kohler, R. B., W. C. Winn, Jr., and L. J. Wheat. 1984. Onset and duration of urinary antigen excretion in Legionnaires' disease. J. Clin. Microbiol. 20**:**605-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kohler, R. B., S. E. Zimmerman, E. Wilson, S. D. Allen, P. H. Edelstein, L. J. Wheat, and A. White. 1981. Rapid radioimmunoassay diagnosis of Legionnaires' disease: detection and partial characterization of urinary antigen. Ann. Intern. Med. 94**:**601-605. [DOI] [PubMed] [Google Scholar]
- 169.Koide, M., and A. Saito. 1995. Diagnosis of Legionella pneumophila infection by polymerase chain reaction. Clin. Infect. Dis. 21**:**199-201. [DOI] [PubMed] [Google Scholar]
- 170.Kool, J. L., D. Bergmire-Sweat, J. C. Butler, E. W. Brown, D. J. Peabody, D. S. Massi, J. C. Carpenter, J. M. Pruckler, R. F. Benson, and B. S. Fields. 1999. Hospital characteristics associated with colonization of water systems by Legionella and risk of nosocomial legionnaires' disease: a cohort study of 15 hospitals. Infect. Control Hosp. Epidemiol. 20**:**798-805. [DOI] [PubMed] [Google Scholar]
- 171.Kool, J. L., U. Buchholz, C. Peterson, E. W. Brown, R. F. Benson, J. M. Pruckler, B. S. Fields, J. Sturgeon, E. Lehnkering, R. Cordova, L. M. Mascola, and J. C. Butler. 2000. Strengths and limitations of molecular subtyping in a community outbreak of Legionnaires' disease. Epidemiol. Infect. 125**:**599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kool, J. L., J. C. Carpenter, and B. S. Fields. 1999. Effect of monochloramine disinfection of municipal drinking water on risk of nosocomial Legionnaires' disease. Lancet 353**:**272-277. [DOI] [PubMed] [Google Scholar]
- 173.Kool, J. L., A. E. Fiore, C. M. Kioski, E. W. Brown, R. F. Benson, J. M. Pruckler, C. Glasby, J. C. Butler, G. D. Cage, J. C. Carpenter, R. M. Mandel, B. England, and R. F. Breiman. 1998. More than 10 years of unrecognized nosocomial transmission of legionnaires' disease among transplant patients. Infect. Control Hosp. Epidemiol. 19**:**898-904. [DOI] [PubMed] [Google Scholar]
- 174.Kusnetsov, J. M., P. J. Keskitalo, H. E. Ahonen, A. I. Tulkki, I. T. Miettinen, and P. J. Martikanen. 1994. Growth of Legionella and other heterotrophic bacteria in a circulating cooling water system exposed to ultraviolet irradiation. J. Appl. Bacteriol. 77**:**461-466. [DOI] [PubMed] [Google Scholar]
- 175.Laussucq, S., D. Schuster, W. J. Alexander, W. L. Thacker, H. W. Wilkinson, and J. S. Spika. 1988. False-positive DNA probe test for Legionella species associated with a cluster of respiratory illnesses. J. Clin. Microbiol. 26**:**1442-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lawrence, C., E. Ronco, S. Dubrou, R. Leclercq, C. Nauciel, and P. Matsiota-Bernard. 1999. Molecular typing of Legionella pneumophila serogroup 1 isolates from patients and the nosocomial environment by arbitrarily primed PCR and pulsed-field gel electrophoresis. J. Med. Microbiol. 48**:**327-333. [DOI] [PubMed] [Google Scholar]
- 177.Ledesma, E., M. L. Camaro, E. Carbonell, T. Sacristan, A. Marti, S. Pellicer, J. Llorca, P. Herrero, and M. A. Dasi. 1995. Subtyping of Legionella pneumophila isolates by arbitrarily primed polymerase chain reaction. Can. J. Microbiol. 41**:**846-848. [DOI] [PubMed] [Google Scholar]
- 178.Lema, M., and A. Brown. 1983. Electrophoretic characterization of soluble protein extracts of Legionella pneumophila and other members of the family Legionellaceae. J. Clin. Microbiol. 17**:**1132-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lepine, L. A., D. B. Jernigan, J. C. Butler, J. M. Pruckler, R. F. Benson, G. Kim, J. L. Hadler, M. L. Cartter, and B. S. Fields. 1998. A recurrent outbreak of nosocomial legionnaires' disease detected by urinary antigen testing: evidence for long-term colonization of a hospital plumbing system. Infect. Control Hosp. Epidemiol. 19**:**905-910. [PubMed] [Google Scholar]
- 180.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31**:**959-970. [DOI] [PubMed] [Google Scholar]
- 181.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66**:**1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lindsay, D. S., W. H. Abraham, and R. J. Fallon. 1994. Detection of mip gene by PCR for diagnosis of Legionnaires' disease. J. Clin. Microbiol. 32**:**3068-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lisby, G., and R. Dessau. 1994. Construction of a DNA amplification assay for detection of Legionella species in clinical samples. Eur. J. Clin. Microbiol. Infect. Dis. 13**:**225-231. [DOI] [PubMed] [Google Scholar]
- 184.Lo Presti, F., S. Riffard, H. Meugnier, M. Reyrolle, Y. Lasne, P. A. D. Grimont, F. Grimont, R. F. Benson, D. J. Brenner, A. G. Steigerwalt, J. Etienne, and J. Freney. 2001. Legionella gresilensis sp. nov. and Legionella beliardensis sp. nov., isolated from water in France. Int. J. Syst. E vol. Microbiol. 51**:**1949-1957. [DOI] [PubMed] [Google Scholar]
- 185.Lo Presti, F., S. Riffard, H. Meugnier, M. Reyrolle, Y. Lasne, P. A. Grimont, F. Grimont, F. Vandenesch, J. Etienne, J. Fleurette, and J. Freney. 1999. Legionella taurinensis sp. nov., a new species antigenically similar to Legionella spiritensis. Int. J. Syst. Bacteriol. 49**:**397-403. [DOI] [PubMed] [Google Scholar]
- 186.Luttichau, H. R., C. Vinther, S. A. Uldum, J. Moller, M. Faber, and J. S. Jensen. 1998. An outbreak of Pontiac fever among children following use of a whirlpool. Clin. Infect. Dis. 26**:**1374-1378. [DOI] [PubMed] [Google Scholar]
- 187.MacDonell, M. T., and R. R. Colwell. 1987. The nucleotide sequence of the 5S rRNA from Legionella pneumophila. Nucleic Acids Res. 15**:**1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Macfarlane, J. T., A. C. Miller, W. H. Roderick Smith, A. H. Morris, and D. H. Rose. 1984. Comparative radiographic features of community acquired Legionnaires' disease, pneumococcal pneumonia, mycoplasma pneumonia, and psittacosis. Thorax 39**:**28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mahbubani, M. H., A. K. Bej, R. Miller, L. Haff, J. DiCesare, and R. M. Atlas. 1990. Detection of Legionella with polymerase chain reaction and gene probe methods. Mol. Cell. Probes 4**:**175-187. [DOI] [PubMed] [Google Scholar]
- 190.Maher, W. E., M. F. Para, and J. F. Plouffe. 1987. Subtyping of Legionella pneumophila serogroup 1 isolates by monoclonal antibody and plasmid techniques. J. Clin. Microbiol. 25**:**2281-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maher, W. E., J. F. Plouffe, and M. F. Para. 1983. Plasmid profiles of clinical and environmental isolates of Legionella pneumophila serogroup 1. J. Clin. Microbiol. 18**:**1422-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Maiwald, M., J. H. Helbig, and P. C. Luck. 1998. Laboratory methods for the diagnosis of Legionella infections. J. Microbiol. Methods 33**:**59-79. [Google Scholar]
- 193.Maiwald, M., M. Schill, C. Stockinger, J. H. Helbig, P. C. Luck, W. Witzleb, and H. G. Sonntag. 1995. Detection of Legionella DNA in human and guinea pig urine samples by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 14**:**25-33. [DOI] [PubMed] [Google Scholar]
- 194.Mandel, A., M. Sprauer, D. Sniadack, and S. Ostroff. 1993. State regulation of hospital water temperature. Infect. Control Hosp. Epidemiol. 14**:**642-645. [DOI] [PubMed] [Google Scholar]
- 195.Mangione, E. J., R. S. Remis, K. A. Tait, H. B. McGee, G. W. Gorman, B. B. Wentworth, P. A. Baron, A. W. Hightower, J. M. Barbaree, and C. V. Broome. 1985. An outbreak of Pontiac fever related to whirlpool use, Michigan 1982. JAMA 253**:**535-539. [PubMed] [Google Scholar]
- 196.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 89**:**9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Marrie, T. J., S. Tyler, G. Bezanson, C. Dendy, and W. Johnson. 1999. Analysis of Legionella pneumophila serogroup 1 isolates by pulsed-field gel electrophoresis. J. Clin. Microbiol. 37**:**251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Marshall, L. E., T. C. Boswell, and G. Kudesia. 1994. False positive Legionella serology in Campylobacter infection: Campylobacter serotypes, duration of antibody response and elimination of cross-reactions in the indirect fluorescent antibody test. Epidemiol. Infect. 112**:**347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Marston, B. J., H. B. Lipman, and R. F. Breiman. 1994. Surveillance for legionnaires' disease. Risk factors for morbidity and mortality. Arch. Intern. Med. 154**:**2417-2422. [PubMed] [Google Scholar]
- 200.Marston, B. J., J. F. Plouffe, T. M. File, B. A. Hackman, S. J. Salstrom, H. B. Lipman, M. S. Kolczak, and R. F. Breiman. 1997. Incidence of community-acquired pneumonia requiring hospitalization—results of a population-based active surveillance study in Ohio. Arch. Intern. Med. 157**:**1709-1718. [PubMed] [Google Scholar]
- 201.Matsiota-Bernard, P., E. Pitsouni, N. Legakis, and C. Nauciel. 1994. Evaluation of commercial amplification kit for detection of Legionella pneumophila in clinical specimens. J. Clin. Microbiol. 32**:**1503-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Matsiota-Bernard, P., S. Waser, and G. Vrioni. 2000. Detection of Legionella pneumophila DNA in urine and serum samples from patients with pneumonia. Clin. Microbiol. Infect. 6**:**223-225. [DOI] [PubMed] [Google Scholar]
- 203.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297**:**1197-1203. [DOI] [PubMed] [Google Scholar]
- 204.McIntyre, M., J. B. Kurtz, and J. B. Selkon. 1990. Prevalence of antibodies to 15 antigens of Legionellaceae in patients with community-acquired pneumonia. Epidemiol. Infect. 104**:**39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mermel, L. A., S. L. Josephson, C. H. Giorgio, J. Dempsey, and S. Parenteau. 1995. Association of Legionnaires' disease with construction: contamination of potable water? Infect. Control Hosp. Epidemiol. 16**:**76-81. [DOI] [PubMed] [Google Scholar]
- 206.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63**:**2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Morrill, W. E., J. M. Barbaree, B. S. Fields, G. N. Sanden, and W. T. Martin. 1990. Increased recovery of Legionella micdadei and Legionella bozemanii on buffered charcoal yeast extract agar supplemented with albumin. J. Clin. Microbiol. 28**:**616-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Moss, C. W., and S. B. Dees. 1979. Further studies of the cellular fatty acid composition of Legionnaires' disease bacteria. J. Clin. Microbiol. 9**:**648-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Murdoch, D. R., and S. T. Chambers. 2000. Detection of Legionella DNA in peripheral leukocytes, serum, and urine from a patient with pneumonia caused by Legionella dumoffii. Clin. Infect. Dis. 30**:**382-383. [DOI] [PubMed] [Google Scholar]
- 210.Murdoch, D. R., E. J. Walford, L. C. Jennings, G. J. Light, M. I. Schousboe, A. Y. Chereshsky, S. T. Chambers, and G. I. Town. 1996. Use of the polymerase chain-reaction to detect Legionella DNA in urine and serum samples from patients with pneumonia. Clin. Infect. Dis. 23**:**475-480. [DOI] [PubMed] [Google Scholar]
- 211.Murga, R., T. S. Forster, E. Brown, J. M. Pruckler, B. S. Fields, and R. M. Donlan. 2001. The role of biofilms in the survival of Legionella pneumophila in a model potable water system. Microbiology 147**:**3121-3126. [DOI] [PubMed] [Google Scholar]
- 212.Musso, D., and D. Raoult. 1997. Serological cross-reactions between Coxiella burnetii and Legionella micdadei. Clin. Diagn. Lab. Immunol. 4**:**208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nagai, H., and C. Roy. 2001. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20**:**5962-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nagington, J., T. G. Wreghitt, J. O. Tobin, and A. D. Macrae. 1979. The antibody response in Legionnaires' disease. J. Hyg. 83**:**377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Reference deleted.
- 216.Neumeister, B., M. Susa, B. Nowak, E. Straube, G. Ruckdeschel, J. Hacker, and R. Marre. 1995. Enzyme immunoassay for detection of antibodies against isolated flagella of Legionella pneumophila. Eur. J. Clin. Microbiol. Infect. Dis. 14**:**764-767. [DOI] [PubMed] [Google Scholar]
- 217.Office of Water, U.S. Environmental Protection Agency. 1999. 40 CFR parts 9, 141, and 142. National primary drinking water regulations: disinfectants and disinfection byproducts. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 218.Ormsbee, R. A., M. G. Peacock, G. L. Lattimer, L. A. Page, and P. Fiset. 1978. Legionnaires' disease: antigenic peculiarities, strain differences, and antibiotic sensitivities of the agent. J. Infect. Dis. 138**:**260-264. [DOI] [PubMed] [Google Scholar]
- 219.Orrison, L. H., W. F. Bibb, W. B. Cherry, and L. Thacker. 1983. Determination of antigenic relationships among legionellae and non-legionellae by direct fluorescent-antibody and immunodiffusion tests. J. Clin. Microbiol. 17**:**332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Para, M. F., and J. F. Plouffe. 1983. Production of monoclonal antibodies to Legionella pneumophila serogroups 1 and 6. J. Clin. Microbiol. 18**:**895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Pasculle, A. W., J. C. Feeley, R. J. Gibson, L. G. Cordes, R. L. Myerowitz, C. M. Patton, G. W. Gorman, C. L. Carmack, J. W. Ezzell, and J. N. Dowling. 1980. Pittsburgh pneumonia agent: direct isolation from human lung tissue. J. Infect. Dis. 141**:**727-732. [DOI] [PubMed] [Google Scholar]
- 222.Pasculle, A. W., G. E. Veto, S. Krystofiak, K. McKelvey, and K. Vrsalovic. 1989. Laboratory and clinical evaluation of a commercial DNA probe for the detection of Legionella spp. J. Clin. Microbiol. 27**:**2350-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Payne, N. R., and M. A. Horwitz. 1987. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J. Exp. Med. 166**:**1377-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pine, L., J. R. George, M. W. Reeves, and W. K. Harrell. 1979. Development of a chemically defined liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 9**:**615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Plikaytis, B. B., G. M. Carlone, C. P. Pau, and H. W. Wilkinson. 1987. Purified 60-kilodalton Legionella protein antigen with _Legionella_-specific and nonspecific epitopes. J. Clin. Microbiol. 25**:**2080-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Plouffe, J. F., T. M. File, Jr., R. F. Breiman, B. A. Hackman, S. J. Salstrom, B. J. Marston, and B. S. Fields. 1995. Reevaluation of the definition of Legionnaires' disease: use of the urinary antigen assay. Community Based Pneumonia Incidence Study Group. Clin. Infect. Dis. 20**:**1286-1291. [DOI] [PubMed] [Google Scholar]
- 227.Pruckler, J. M., R. F. Benson, M. Moyenuddin, W. T. Martin, and B. S. Fields. 1995. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect. Immun. 63**:**4928-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pruckler, J. M., L. A. Mermel, R. F. Benson, C. Giorgio, P. K. Cassiday, R. F. Breiman, C. G. Whitney, and B. S. Fields. 1995. Comparison of Legionella pneumophila isolates by arbitrarily primed PCR and pulsed-field gel electrophoresis: analysis from seven epidemic investigations. J. Clin. Microbiol. 33**:**2872-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ramirez, J. A., S. Ahkee, A. Tolentino, R. D. Miller, and J. T. Summersgill. 1996. Diagnosis of Legionella pneumophila, Mycoplasma pneumoniae, or Chlamydia pneumoniae lower respiratory-infection using the polymerase chain-reaction on a single throat swab specimen. Diagn. Microbiol. Infect. Dis. 24**:**7-14. [DOI] [PubMed] [Google Scholar]
- 230.Ramirez, J. A., and J. T. Summersgill. 1994. Rapid tests for the diagnosis of Legionella infections. J. Ky. Med. Assoc. 92**:**62-65. [PubMed] [Google Scholar]
- 231.Ratcliff, R. M., S. C. Donnellan, J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1997. Interspecies sequence differences in the Mip protein from the genus Legionella: implications for function and evolutionary relatedness. Mol. Microbiol. 25**:**1149-1158. [DOI] [PubMed] [Google Scholar]
- 232.Ratcliff, R. M., J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36**:**1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Riffard, S., F. LoPresti, F. Vandenesch, F. Forey, M. Reyrolle, and J. Etienne. 1998. Comparative analysis of infrequent-restriction-site PCR and pulsed-field gel electrophoresis for epidemiological typing of Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 36**:**161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rigby, E. W., J. F. Plouffe, B. A. Hackman, D. S. Hill, R. F. Benson, and R. F. Breiman. 1997. Stability of Legionella urinary antigens over time. Diagn. Microbiol. Infect. Dis. 28**:**1-3. [DOI] [PubMed] [Google Scholar]
- 235.Rogers, J., A. B. Dowsett, P. J. Dennis, J. V. Lee, and C. W. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60**:**1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Rogers, J., A. B. Dowsett, and C. W. Keevil. 1995. A paint incorporating silver to control mixed biofilms containing Legionella pneumophila. J. Ind. Microbiol. 15**:**377-383. [DOI] [PubMed] [Google Scholar]
- 237.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabeling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58**:**2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Roig, J., C. Domingo, and J. Morera. 1994. Legionnaires' disease. Chest 105**:**1817-1825. [DOI] [PubMed] [Google Scholar]
- 239.Rossier, O., and N. Cianciotto. 2001. Type II secretion is a subset of the _pilD_-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69**:**2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22**:**678-689. [PubMed] [Google Scholar]
- 241.Rowbotham, T. J. 1998. Isolation of Legionella pneumophila serogroup 1 from human feces with use of amebic cocultures. Clin. Infect. Dis. 26**:**502-503. [DOI] [PubMed] [Google Scholar]
- 242.Rowbotham, T. J. 1993. _Legionella_-like amoebal pathogens, p. 137-140. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 243.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33**:**1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ruehlemann, S. A., and G. R. Crawford. 1996. Panic in the potting shed: the association between Legionella longbeachae serogroup 1 and potting soils in Australia. Med. J. Aust. 164**:**36-38. [DOI] [PubMed] [Google Scholar]
- 245.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61**:**5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sampson, J. S., B. B. Plikaytis, C. H. Aloisio, G. M. Carlone, C. P. Pau, and A. R. Stinson. 1991. Immunologic characterization and specificity of three monoclonal antibodies against the 58-kilodalton protein of Legionella pneumophila. J. Clin. Microbiol. 29**:**836-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sampson, J. S., B. B. Plikaytis, and H. W. Wilkinson. 1986. Immunologic response of patients with legionellosis against major protein-containing antigens of Legionella pneumophila serogroup 1 as shown by immunoblot analysis. J. Clin. Microbiol. 23**:**92-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sampson, J. S., H. W. Wilkinson, V. C. Tsang, and B. J. Brake. 1983. Kinetic-dependent enzyme-linked immunosorbent assay for detection of antibodies to Legionella pneumophila. J. Clin. Microbiol. 18**:**1340-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Saunders, N. A., T. G. Harrison, A. Haththotuwa, N. Kachwalla, and A. G. Taylor. 1990. A method for typing strains of Legionella pneumophila serogroup 1 by analysis of restriction fragment length polymorphisms. J. Med. Microbiol. 31**:**45-55. [DOI] [PubMed] [Google Scholar]
- 250.Saunders, N. A., T. G. Harrison, A. Haththotuwa, and A. G. Taylor. 1991. A comparison of probes for restriction fragment length polymorphism (RFLP) typing of Legionella pneumophila serogroup 1 strains. J. Med. Microbiol. 35**:**152-158. [DOI] [PubMed] [Google Scholar]
- 251.Schoonmaker, D., T. Heimberger, and G. Birkhead. 1992. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J. Clin. Microbiol. 30**:**1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Selander, R. K., R. M. McKinney, T. S. Whittam, W. F. Bibb, D. J. Brenner, F. S. Nolte, and P. E. Pattison. 1985. Genetic structure of populations of Legionella pneumophila. J. Bacteriol. 163**:**1021-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Socan, M., D. Kese, and N. Marinic-Fisher. 2000. Polymerase chain reaction for detection of Legionellae DNA in urine samples from patients with community-acquired pneumonia. Folia Microbiol. 45**:**469-472. [DOI] [PubMed] [Google Scholar]
- 254.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68**:**2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Spitalny, K. C., R. L. Vogt, L. A. Orciari, L. E. Witherell, P. Etkind, and L. F. Novick. 1984. Pontiac fever associated with a whirlpool spa. Am. J. Epidemiol. 120**:**809-817. [DOI] [PubMed] [Google Scholar]
- 256.Starnbach, M. N., S. Falkow, and L. S. Tompkins. 1989. Species-specific detection of Legionella pneumophila in water by DNA amplification and hybridization. J. Clin. Microbiol. 27**:**1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Steele, T. W. 1989. Legionnaires' disease in South Australia, 1979-1988. Med. J. Aust. 151**:**322, 325-326, 328. [DOI] [PubMed] [Google Scholar]
- 258.Steinmetz, I., C. Rheinheimer, I. Hübner, and D. Bitter-Suermann. 1991. Genus-specific epitope on the 60-kilodalton Legionella heat shock protein recognized by a monoclonal antibody. J. Clin. Microbiol. 29**:**346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66**:**1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Stout, J. E., J. Joly, M. Para, J. Plouffe, C. Ciesielski, M. J. Blaser, and V. L. Yu. 1988. Comparison of molecular methods for subtyping patients and epidemiologically linked environmental isolates of Legionella pneumophila. J. Infect. Dis. 157**:**486-495. [DOI] [PubMed] [Google Scholar]
- 261.Stout, J. E., and V. L. Yu. 1997. Legionellosis. N. Engl. J. Med. 337**:**682-687. [DOI] [PubMed] [Google Scholar]
- 262.Strauss, W. L., J. F. Plouffe, T. M. File, Jr., H. B. Lipman, B. H. Hackman, S. J. Salstrom, R. F. Benson, and R. F. Breiman. 1996. Risk factors for domestic acquisition of Legionnaires disease. Ohio Legionnaires Disease Group. Arch. Intern. Med. 156**:**1685-1692. [PubMed] [Google Scholar]
- 263.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54**:**567-613. [DOI] [PubMed] [Google Scholar]
- 264.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63**:**3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tang, P., and C. Krishnan. 1993. Legionella antigenuria: six-year study of broad-spectrum enzyme-linked immunosorbent assay as a routine diagnostic test, p. 12-13. In J. M. Barbaree, R. F. Breiman, and A. P. DuFour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 266.Tang, P. W., and S. Toma. 1986. Broad-spectrum enzyme-linked immunosorbent assay for detection of Legionella soluble antigens. J. Clin. Microbiol. 24**:**556-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tang, P. W., S. Toma, and W. D. Rajkumar. 1989. Detection of urinary antigens of Legionella pneumophila serogroup 12 by broad-spectrum enzyme-linked immunosorbent assay. J. Clin. Microbiol. 27**:**783-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Tatlock, H. 1944. A Rickettsia-like organism recovered from guinea pigs. Proc. Soc. Exp. Biol. Med. 57**:**95-99. [Google Scholar]
- 269.Taylor, A. G., and T. G. Harrison. 1983. Serological tests for Legionella pneumophila serogroup 1 infections. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 255**:**20-26. [PubMed] [Google Scholar]
- 270.Thacker, W. L., B. B. Plikaytis, and H. W. Wilkinson. 1985. Identification of 22 Legionella species and 33 serogroups with the slide agglutination test. J. Clin. Microbiol. 21**:**779-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Tilton, R. C. 1979. Legionnaires' disease antigen detected by enzyme-linked immunosorbent assay. Ann. Intern. Med. 90**:**697-698. [DOI] [PubMed] [Google Scholar]
- 272.Tobin, J. O., I. D. Watkins, S. Woodhead, and R. G. Mitchell. 1986. Epidemiological studies using monoclonal antibodies to Legionella pneumophila serogroup 1. Isr. J. Med. Sci. 22**:**711-714. [PubMed] [Google Scholar]
- 273.Tsai, T. F., D. R. Finn, B. D. Plikaytis, W. McCauley, S. M. Martin, and D. W. Fraser. 1979. Legionnaires' disease: clinical features of the epidemic in Philadelphia. Ann. Intern. Med. 90**:**509-517. [DOI] [PubMed] [Google Scholar]
- 274.Valsangiacomo, C., F. Baggi, V. Gaia, T. Balmelli, R. Peduzzi, and J. C. Piffaretti. 1995. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J. Clin. Microbiol. 33**:**1716-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.van Belkum, A., H. Maas, H. Verbrugh, and N. Van Leeuwen. 1996. Serotyping, ribotyping, PCR-mediated ribosomal 16S-23S spacer analysis and arbitrarily primed PCR for epidemiological studies on Legionella pneumophila. Res. Microbiol. 147**:**405-413. [DOI] [PubMed] [Google Scholar]
- 276.van Belkum, A., M. Struelens, and W. Quint. 1993. Typing of Legionella pneumophila strains by polymerase chain reaction-mediated DNA fingerprinting. J. Clin. Microbiol. 31**:**2198-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Venkataraman, C., B. J. Haack, S. Bondada, and Y. Abu Kwaik. 1997. Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformis as a potential receptor for attachment and invasion by the Legionnaires' disease bacterium. J. Exp. Med. 186**:**537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Vesey, G., P. J. Dennis, J. V. Lee, and A. A. West. 1988. Further development of simple tests to differentiate the legionellas. J. Appl. Bacteriol. 65**:**339-345. [DOI] [PubMed] [Google Scholar]
- 279.Vickers, R. M., A. Brown, and G. M. Garrity. 1981. Dye-containing buffered charcoal-yeast extract medium for differentiation of members of the family Legionellaceae. J. Clin. Microbiol. 13**:**380-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Vickers, R. M., J. E. Stout, V. L. Yu, and J. D. Rihs. 1987. Manual of culture methodology for Legionella. Semin. Respir. Infect. 2**:**274-279. [PubMed] [Google Scholar]
- 281.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279**:**873-876. [DOI] [PubMed] [Google Scholar]
- 282.Walker, J. T., A. Sonesson, C. W. Keevil, and D. C. White. 1993. Detection of Legionella pneumophila in biofilms containing a complex microbial consortium by gas chromatography-mass spectrometry analysis of genus-specific hydroxy fatty acids. FEMS Microbiol. Lett. 113**:**139-144. [DOI] [PubMed] [Google Scholar]
- 283.Watkins, I. D., J. O. Tobin, P. J. Dennis, W. Brown, R. Newnham, and J. B. Kurtz. 1985. Legionella pneumophila serogroup 1 subgrouping by monoclonal antibodies-an epidemiological tool. J. Hyg. 95**:**211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Weir, S. C., S. H. Fischer, F. Stock, and V. J. Gill. 1998. Detection of Legionella by PCR in respiratory specimens using a commercially available kit. Am. J. Clin. Pathol. 110**:**295-300. [DOI] [PubMed] [Google Scholar]
- 285.Wilkinson, H. W. 1987. Hospital-laboratory diagnosis of Legionella infections. U.S. Dept. of Health and Human Services, Public Health Service, Centers for Disease Control, Atlanta, Ga.
- 286.Wilkinson, H. W., and B. J. Brake. 1982. Formalin-killed versus heat-killed Legionella pneumophila serogroup 1 antigen in the indirect immunofluorescence assay for legionellosis. J. Clin. Microbiol. 16**:**979-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wilkinson, H. W., D. D. Cruce, and C. V. Broome. 1981. Validation of Legionella pneumophila indirect immunofluorescence assay with epidemic sera. J. Clin. Microbiol. 13**:**139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Wilkinson, H. W., C. E. Farshy, B. J. Fikes, D. D. Cruce, and L. P. Yealy. 1979. Measure of immunoglobulin G-, M-, and A-specific titers against Legionella pneumophila and inhibition of titers against nonspecific, gram-negative bacterial antigens in the indirect immunofluorescence test for legionellosis. J. Clin. Microbiol. 10**:**685-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Wilkinson, H. W., B. J. Fikes, and D. D. Cruce. 1979. Indirect immunofluorescence test for serodiagnosis of Legionnaires disease: evidence for serogroup diversity of Legionnaires' disease bacterial antigens and for multiple specificity of human antibodies. J. Clin. Microbiol. 9**:**379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wilkinson, H. W., A. L. Reingold, B. J. Brake, D. L. McGiboney, G. W. Gorman, and C. V. Broome. 1983. Reactivity of serum from patients with suspected legionellosis against 29 antigens of legionellaceae and _Legionella_-like organisms by indirect immunofluorescence assay. J. Infect. Dis. 147**:**23-31. [DOI] [PubMed] [Google Scholar]
- 291.Wilkinson, H. W., J. S. Sampson, and B. B. Plikaytis. 1986. Evaluation of a commercial gene probe for identification of Legionella cultures. J. Clin. Microbiol. 23**:**217-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Williams, A., and M. S. Lever. 1995. Characterisation of Legionella pneumophila antigen in urine of guinea pigs and humans with Legionnaires' disease. J. Infect. 30**:**13-16. [DOI] [PubMed] [Google Scholar]
- 293.Yonke, C. A., H. E. Stiefel, D. L. Wilson, and B. B. Wentworth. 1981. Evaluation of an indirect hemagglutination test for Legionella pneumophila serogroups 1 to 4. J. Clin. Microbiol. 13**:**1040-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zimmerman, S. E., M. L. French, S. D. Allen, E. Wilson, and R. B. Kohler. 1982. Immunoglobulin M antibody titers in the diagnosis of Legionnaires' disease. J. Clin. Microbiol. 16**:**1007-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]