Genomic Variability of O Islands Encoding Tellurite Resistance in Enterohemorrhagic Escherichia coli O157:H7 Isolates (original) (raw)
Abstract
Strains of Escherichia coli causing enterohemorrhagic colitis belonging to the O157:H7 lineage are reported to be highly related. Fifteen strains of E. coli O157:H7 and 1 strain of E. coli O46:H− (nonflagellated) were examined for the presence of potassium tellurite resistance (Ter). Ter genes comprising terABCDEF were shown previously to be part of a pathogenicity island also containing integrase, phage, and urease genes. PCR analysis, both conventional and light cycler based, demonstrated that about one-half of the Ter E. coli O157:H7 strains (6 of 15), including the Sakai strain, which has been sequenced, carried a single copy of the Ter genes. Five of the strains, including EDL933, which has also been sequenced, contained two copies. Three other O157:H7 strains and the O46:H− strain did not contain the Ter genes. In strains containing two copies, the Ter genes were associated with the serW and serX tRNA genes. Five O157:H7 strains resembled the O157 Sakai strain whose sequence contained one copy, close to serX, whereas in one isolate the single copy was associated with serW. There was no correlation between Ter and the ability to produce Shiga toxin ST1 or ST2. The Ter MIC for most strains, containing either one or two copies, was 1,024 μg/ml, although for a few the MIC was intermediate, 64 to 128 μg/ml, which could be increased to 512 μg/ml by pregrowth of strains in subinhibitory concentrations of potassium tellurite. Reverse transcriptase PCR analysis confirmed that in most strains Ter was constitutive but that in the rest it was inducible and involved induction of terB and terC genes. Only the terB, -C, -D, and -E genes are required for Ter. The considerable degree of homology between the ter genes on IncH12 plasmid R478, which originated in Serratia marcescens, and pTE53, from an E. coli clinical isolate, suggests that the pathogenicity island was acquired from a plasmid. This work demonstrates diversity among E. coli O157:H7 isolates, at least as far as the presence of Ter genes is concerned.
Escherichia coli O157:H7 is of major interest in clinical practice, food safety, and evolutionary biology. Although only recognized as a human pathogen in 1982 (16), it has rapidly become prominent as an important cause of food-related and waterborne outbreaks of hemorrhagic colitis throughout the world (13). In a proportion of patients infection progresses to hemolytic-uremic syndrome, characterized by acute renal failure, microangiopathic hemolytic anemia, and thrombocytopenia (13). Each of these secondary sequelae carries significant mortality rates (18).
The genome sequences of two strains of enterohemorrhagic E. coli O157:H7 were recently completed (6, 14). Comparisons between laboratory E. coli strain K-12 MG1655 (2) and EDL933, an E. coli O157:H7 strain (14), demonstrated that they have a complex relationship since their divergence 4.5 million years ago (14). Homology between the two is interrupted by the presence of hundreds of islands of inserted DNA. K islands are DNA segments present in MG1655 (K-12) but absent from EDL933 (O157:H7), whereas O islands (OI) are present only in EDL933. In E. coli EDL933 two OI, designated OI 43 and OI 48, contain integrase, phage, tellurite resistance (Ter), and urease genes (14). One of these islands had been identified previously by cosmid cloning (19) in E. coli O157:H7 strains and in more distantly related serotypes. Tarr and coworkers (19) termed this region the tellurite resistance- and adherence-conferring island (TAI) and demonstrated that the associated iron-regulated homologue adhesin (encoded by iha) rendered laboratory E. coli capable of adhering to epithelial cells. Since the island was found in E. coli O157:H7 but was absent from nontoxigenic E. coli O55:H7 and toxigenic E. coli O157:H− (flagellum−), pathogenic E. coli O157:H7 is believed to have acquired the TAI only recently (19).
At least five Ter determinants have been identified by genetic analysis and DNA sequencing, all apparently unrelated to one another at the DNA or protein level (see reference 20 for review). High-level Ter is a characteristic marker encoded on all incompatibility group HI2 (IncHI2) plasmids (except for R476b) and on all IncHII plasmids (20). This high-level Ter (MIC ≈ 1,024 μg/ml) has been extensively studied in pMER610 and R478 and has been shown to be associated with several ter genes (terZABCDEF) (7, 24). This gene cluster also encodes resistance to infection by various bacteriophages, such as λ and T5, known as phage inhibition (Phi), as well as resistance to pore-forming colicins (PacB) (23, 24, 25). The mechanism of ter gene cluster Ter is not understood, and none of the precise functions of the proteins in the ter gene cluster are understood. Several bacterial species (both gram positive and gram negative) have been shown to contain one or more ter genes (20). The finding of two ter OI within the O157:H7 EDL933 genome prompted us to examine ter gene expression and the number and location of ter OI in both toxigenic and nontoxigenic E. coli O157:H7 isolates from different geographic locations. We report here that there is considerable plasticity in the presence, location, and expression of the ter OI in E. coli O157:H7, which we refer to here as Ter O islands, as we have not investigated the associated adhesin gene (iha) in this study.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli O157:H7 strains were grown at 37°C on brain heart infusion (BHI) medium or Mueller-Hinton medium (Difco, Detroit, Mich.). The O157:H7 serotype strains were also grown on Biosynth culture medium (BCM) O157:H7(+) medium (Biochemica and Synthetica, Naperville, Ill.). All solid medium contained 1.5% agar (Invitrogen, Gaithersburg, Md.). The E. coli strains used in this study are listed in Table 1. Unless otherwise indicated, restriction endonucleases and other molecular biology enzymes were supplied by Invitrogen.
TABLE 1.
Serotype, tellurite resistance, and Shiga toxin status of E. coli strains used in this study
| DT no. | Original strain no. | Serotype | Shiga toxin status | Tellurite MIC (μg/ml) | Location; yr of isolationa |
|---|---|---|---|---|---|
| DT2889 | 97-0233b | O157:H7 | stx2 | 1,024 | Calgary, AB, Canada; 1997 |
| DT2890 | 97-0254b | O157:H7 | stx1, stx2 | 1,024 | Calgary, AB, Canada; 1997 |
| DT2891 | 97-0313b | O157:H7 | stx1, stx2 | 1,024 | Regina, SK, Canada; 1997 |
| DT2892 | 97-0311b | O157:H7 | stx1, stx2 | 2-4 | Moose Jaw, SK, Canada; 1997 |
| DT2893 | 97-0379b | O157:H7 | stx1 | 1,024 | Beaumont, AB, Canada; 1997 |
| DT2894 | 97-213b | O157:H7 | stx2 | 1,024 | Japan; 1997 |
| DT2895 | 97-157b | O46:NM | TVT+h | 2-4 | Canada; 1997 |
| DT2896 | 96-1577b | O157:H7 | stx2 | 1,024 | Rome, Italy; 1996 |
| DT2897 | 96-1661b | O157:H7 | stx2 | 1-2 | Winnipeg, MB, Canada; 1996 |
| DT2898 | 93-2534b | O157:H7 | 1,024 | St. Catherine's, ON, Canada; 1996 | |
| DT2899 | 96-1095b | O157:H7 | 4 | Burnaby, BC, Canada; 1996 | |
| DT2998 | 3098-98c | O157:H7 | stx1, stx2 | 64 | Indiana; 1998 |
| DT2999 | 3099-98c | O157:H7 | stx1, stx2 | 64 | Indiana; 1998 |
| DT3000 | 3100-98c | O157:H7 | stx1, stx2 | 64 | Indiana; 1998 |
| NAg | EDL933d | O157:H7 | stx1, stx2 | 64-128 | Michigan; 1982 |
| NA | Sakaie | O157:H7 | stx1, stx2 | 128 | Sakai, Japan; 1996 |
| NA | DH5αf | K-12 | NA | <1 | NA |
Detection of Shiga toxins.
Both PCR and Vero cell assays were used to detect production of Shiga toxins in E. coli O157:H7 strains by methods described previously (15).
Tellurite susceptibility testing.
Tellurite MICs for the strains were determined in the following manner. Cultures were grown to a concentration of 108/ml in BHI broth. A volume of 100 μl from a 5-ml culture was diluted in 5 ml of 1× phosphate-buffered saline (Oxoid, Nepean, Ontario, Canada), and 10 μl of this dilution was spotted onto plates containing increasing twofold concentrations of potassium tellurite (Sigma Chemical Co., St. Louis, Mo.) ranging from 1 to 1,024 μg/ml. The plates were incubated at 37°C overnight. The MIC was determined as the lowest concentration of Te to totally inhibit bacterial growth. For MICs of tellurite-induced cultures, cultures were grown to a concentration of 108 cells/ml in BHI broth containing 1 μg of potassium tellurite/ml. The cell suspensions were diluted and spotted onto tellurite plates as described above.
Tellurite induction experiments.
Reverse transcriptase PCR (RT-PCR) was done on selected strains with and without a sublethal concentration of potassium tellurite. The strains were grown to mid-log phase in a 1-μg/ml concentration of tellurite. RNA was isolated from these samples by the techniques described below. cDNA synthesis and PCR analysis of all the ter genes were done for these RNA samples.
RNA preparation.
Total RNA was prepared from selected strains of E. coli O157:H7 with the RNeasy Midi kit in accordance with the manufacturer's directions (Qiagen, Mississauga, Ontario, Canada) or the Stratagene Mini-Prep RNA isolation kit, also in accordance with the manufacturer's directions. The amount of RNA obtained was quantitated by using an Ultraspec 4000 (Pharmacia, Uppsala, Sweden).
PCR.
PCR primers were designed for the seven ter genes found in E. coli O157:H7 from the sequence listed under accession no. AE005174. The primers and their predicted product lengths are listed in Table 2. The pfkA gene, which encodes phosphofructokinase and which, as a housekeeping gene, is present in all strains, was used as a positive control in PCRs and to determine that complete DNA transfer from membrane to Southern blots had occurred. To try to determine where the islands had integrated, primers were designed around the serW and serX genes. If the island had inserted, no PCR product would be observed. Primers were also designed for just inside the 3′ end of a TAI and were named Ter-island (forward) and Ter-island (reverse). These were used in conjunction with the serW and serX primers as an added control for the negative PCRs. The PCRs were done on a DNA thermal cycler 480 (Perkin-Elmer, Mississauga, Ontario, Canada). The reaction conditions were denaturing at 94°C for 45 s, annealing at 55°C for 1 min 30 s, and extension at 72°C for 1 min 30 s. This was repeated for 30 cycles.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′-3′) | Predicted product (bp) |
|---|---|---|
| TerA1 (forward) | TAT CGT TTC AGC GGT TAT TC | 1,228 |
| TerA2 (reverse) | TGG CGG GTC AGT TCG TCA C | |
| TerB1 (forward) | AGG CCG TGA CGA ACT GAC C | 286 |
| TerB2 (reverse) | TCG CAA CGG CAA TAC CAA CAC G | |
| TerC1 (forward) | TCC TGG CGC TGA AAG AT | 1,240 |
| TerC2 (reverse) | GAA ACA CTC ATA AAA TAA CCT CTT | |
| TerD1 (forward) | AGT AAA GCA GCT CCG TCA AT | 396 |
| TerD2 (reverse) | CCG AAC AGC ATG GCA GTC T | |
| TerE1 (forward) | TAA AAG GCG GCA ACG TAT CTC TGA | 353 |
| TerE2 (reverse) | CCG TCC CGT TGT CGT TGT TGT AA | |
| TerF1 (forward) | TTA CAA TCC GGA CAA AAC A | 244 |
| TerF2 (reverse) | CAA TGA CAA CGG TGA TCG | |
| TerW1 (forward) | TTC TCT ACC GCT TCA CTT | 214 |
| TerW2 (reverse) | TCA AAT ACA GCA AGG CAG | |
| TerZ1 (forward) | GAC GGT ATC ACT CAG CAA AGA ATC | 416 |
| TerZ2 (reverse) | TAC GGC GCA GCG AAG AAA TAA C | |
| PfkA (forward) | GTG GCG GTA CGT TCC TCG GTT CT | 762 |
| PfkA (reverse) | TTT TTC GCG CAG TCC AGC CAG TC | |
| SerW (forward) | TCG GGG AAG GTA AGG AT | 491 |
| SerW (reverse) | TTG TGA TAT GTA TGA AGT | |
| SerX (forward) | TTT TTC TAT TGT CGA TTC CTC T | 363 |
| SerX (reverse) | GAT CTA CAA AGG CCA CCA GCA | |
| Ter-island (forward) | GAC AAA CTC TCC GGG ATA ACT CA | 356 |
| Ter-island (reverse) | TGC GGG TGC TGG TGT GGG ATA A | |
| TerC-F1 | TGG TAT TGC CGT TGC GAA | 270 |
| TerC-R1 | GCA CCG TGG TGG ATG TAG A | |
| GyrB-F2 | CCG CTG GAT CAC GAG TTT ATC | 129 |
| GyrB-R2 | GCT GGC TAC CGG CTG ACG AC |
RT-PCR.
The RNA isolated as described above was treated with DNase I to remove residual DNA. cDNA synthesis was done with Superscript II in accordance with the manufacturer's protocols. Briefly, the reverse primer from each primer pair was used as the primer to create cDNA by using the cellular RNA as a template. The reaction was carried out at 42°C for 50 min. PCR was performed on the resulting cDNA using the primers listed in Table 2 and the same PCR conditions as above.
Preparations of agarose-embedded E. coli chromosomal DNA for pulsed-field gel electrophoresis (PFGE).
Agarose-embedded chromosomal DNA was prepared from E. coli strains (Table 1) grown overnight at 37°C on BHI agar plates using a modification of previously described protocols (4; S.-L. Liu, University of Calgary, personal communication) (Bio-Rad Laboratories [1992]). Approximately 2 loopfuls of culture were resuspended in 0.5 ml of cell suspension solution (10 mM Tris-HCl [pH 7.2], 20 mM NaCl, 100 mM EDTA) and kept at 70°C while 1.4% agarose was melted and cooled to 70°C. Agarose (0.5 ml) was added to the bacterial suspension and quickly mixed by pipetting three or four times before drawing the mixture into a 1-ml syringe with the needle adapter cutoff. The gel was allowed to solidify at room temperature for 20 min before the entire agarose rod was cut into 1-mm-thick slices. All of the following incubation steps were carried out with gentle shaking. The agarose slices were placed in 3 ml of cell lysis solution (10 mM Tris-HCl [pH 7.2], 50 mM NaCl, 100 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 0.5% _N_-laurylsarcosine, sodium salt) and incubated for 2 h at 70°C. The cell lysis solution was removed, and the slices were washed twice with wash solution (20 mM Tris-HCl [pH 8.0], 50 mM EDTA) for 15 min at room temperature. Wash solution was removed, and 3 ml of proteinase K solution (1.0 mg of proteinase K/ml, 100 mM EDTA [pH 8.0], 0.2% SDS, 1% _N_-laurylsarcosine, sodium salt) was added to the slices for incubation at 37°C for at least 72 h. Slices were washed in wash solution as described above and incubated in 3 ml of phenylmethylsulfonyl fluoride (PMSF) solution (1 mM PMSF in wash solution) for 2 h at room temperature. Two additional washes in wash solution were followed by a wash in storage solution (2 mM Tris-HCl [pH 8.0], 5 mM EDTA) for 15 min at room temperature. Agarose slices were then stored in a fresh aliquot of storage solution at 4 to 8°C.
Restriction enzyme digestion of E. coli agarose slices for PFGE.
One agarose slice of each E. coli strain was washed twice with the appropriate restriction enzyme buffer and then incubated for 2 h at 37°C with 50 U of _Spe_I (50 U/μl; New England BioLabs Inc., Mississauga, Ontario, Canada), 28 U of _Avr_II (4 U/μl; New England BioLabs Inc.), or 30 U of _Xba_I (20 U/μl; Roche Applied Sciences).
Electrophoresis.
PFGE was carried out on the digested agarose using a modification of previously described protocols (4; manual supplied with Bio-Rad CHEF DR II apparatus). Each agarose slice (one from each E. coli strain) was loaded into the wells of a 1% agarose gel prepared in 0.5× TBE buffer (45 mM Tris, 45 mM borate, 1 mM EDTA, pH 8.3). A half slice of lambda ladder PFG marker (NO340s; New England BioLabs Inc.) was loaded on both sides of the gel. The loaded gel was placed into the electrophoresis chamber of a CHEF DR II pulsed-field electrophoresis system (Bio-Rad, Hercules, Calif.) containing 0.5× TBE buffer previously cooled to 14°C. The gel was run at a voltage gradient of 6.0 V/cm for 19 h at an initial switch time of 2.2 s and a final switch time of 54.2 s.
Band visualization and image capture.
The gel was stained for 30 min with ethidium bromide (0.5 μg/ml). DNA bands were visualized by placing the gel on a UV Fisher Biotech transilluminator (312 nm) (model FBTI-614; Fisher Scientific, Nepean, Ontario, Canada) connected to a Kodak (New Haven, Conn.) Electrophoresis Documentation and Analysis System 120 interfaced with a Compaq Pentium III computer. The image was captured with Kodak Digital Science 1D LE software, version 3.02, and printed with Corel Photo-Paint, version 8.232.
Southern hybridization.
Agarose gels from PFGE were transferred onto nitrocellulose membranes in accordance with previously described methods (22). Briefly, the gel was first soaked in a 1/50 dilution of concentrated HCl for 30 min. This was followed by a 30-min soak in denaturing buffer (1.5 M NaCl, 0.5 M NaOH). The gel was then placed in neutralizing buffer (2 M NaCl, 1 M Tris-HCl, pH 5.5) for 30 to 45 min. Transfer was done overnight by gravity transfer with 20× SSC solution (3 M NaCl, 0.3 M sodium citrate, pH 7.0). terC and pfkA DNA was amplified by PCR from the EDL933 template with primers shown in Table 2. The PCR was run on a 1.5% agarose gel, and the resulting band was cut out after ethidium bromide staining. DNA was purified from the gel slice with the Qiagen gel extraction kit in accordance with the manufacturer's protocols. The probe was labeled with [α-32P]dCTP (Amersham) by using a random primer labeling kit (Invitrogen) in accordance with the manufacturer's protocols. The probe and membrane were hybridized in hybridization buffer (5 ml of formamide, 2.5 ml of 20× SSC, 0.5 ml of 10% SDS, 40 μl of 0.25 M EDTA, 1 ml of 10× PM (10× PM is 20 mg of Ficoll, 20 mg of bovine serum albumin, 20 mg of polyvinylpyrrolidone, 0.5 ml of 20× SSC, and 0.5 ml of dH2O), 250 μl of a 100-μg/ml solution of herring sperm DNA, 960 μl of Milli-Q water, and the radiolabeled probe) at 42°C overnight in a Hybaid hybridization oven. After hybridization, the blot was washed (5× SSC, 0.1% SDS, 1 mM EDTA) for 1 h at 42°C and exposed to Kodak BioMax MS film.
Light cycler-based PCR amplification of terC.
DNA was extracted with a High Pure PCR template preparation kit (Roche Diagnostics, Mannheim, Germany). Amplification of terC and internal standard gyrB was performed in 18 μl of Light Cycler Fast Start DNA Master SYBR Green I (Roche Diagnostics) containing 3.0 mM MgCl2. PCR was performed in glass capillaries in the Roche Light Cycler (software version 3.5). Primers TerC-F1 and TerC-R1 (Tibmolbiol, Berlin, Germany) and GyrB-F2 and GyrB-R2 (Table 2) were used at a concentration of 0.5 M. Thirty-two samples were run in parallel by performing 45 cycles of repeated denaturation (10 s at 95°C), annealing (10 s at 55°C), and extension (20 s at 72°C). After the final cycle, melting-point analysis of all samples and controls was performed at 65°C.
Light cycler-based quantification of target DNA.
A standard curve was prepared for the determination of the concentration of DNA by using 10-fold serial dilutions of PCR amplicons of terC and gyrB prepared by amplifying E. coli DNA (Sakai strain). As an external standard, a 105 dilution corresponding to 8.67 × 106 copies/μl for terC and 1.32 × 105 copies/μl for gyrB was used for each run. The cycle numbers of the logarithmic-linear phase were plotted against the logarithm of the concentration of template DNA. The concentrations of E. coli DNA for terC and gyrB were calculated by comparing the cycle numbers of the logarithmic-linear phase of the samples with the cycle numbers of the external standards, as described previously (10).
RESULTS
Positions of Ter OI 43 and OI 48 in E. coli EDL933.
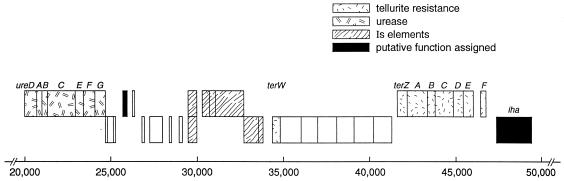
Sequence analysis of E. coli EDL933 (14) has shown that two OI of 87,547 bp comprise 92 open reading frames (ORFs), including integrase, phage, tellurite resistance, urease, and adhesin genes. Both OI are inserted 2 bp beyond a tRNA gene; OI 43 is close to serX, and OI 48 is close to serW. The two islands are about 0.5 Mb apart at positions 1.05 Mb and 1.540 Mb, respectively (14). In EDL933, the two islands are identical, with not even a single base difference between them. Figure 1 shows genes relevant to the study present in OI 43 AND OI 48.
FIG. 1.
Map of OI 43 and 48 from E. coli O157: H7 strain EDL933 (14). Genes of interest encode urease, a ribosomal protein, tellurite resistance, an insertion sequence, and adhesin (iha). A putative ribosomal protein gene (unlabeled) downstream of the urease operon is 96% identical to E. coli K-12 ORF ygkM. The overall size of the islands is 87,562 bp. The region from bp 20500 to 50000 is shown.
Ter gene in E. coli EDL933 OI 43 and OI 48.
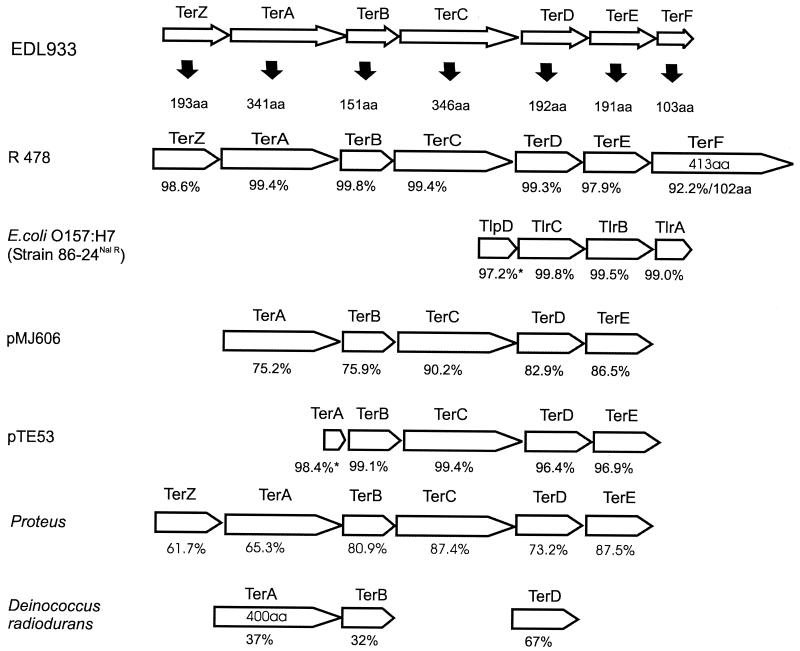
The ter genes involved in Ter in plasmid R478 and E. coli EDL933 are shown in Fig. 2. The terZABCDEF genes are present in R478, and six of the genes (terZABCDE) are also present in E. coli EDL933. However, a portion of terF is missing in E. coli EDL933, with only 103 predicted encoded amino acids compared with a total of 413 predicted amino acids encoded by the gene present in plasmid R478. This likely indicates that TerF is not essential for tellurite resistance, and terF was not present in pMJ606, a clone encoding Ter derived from IncH12 plasmid pMER610 (7).
FIG. 2.
Comparison of Ter determinants from various sources. Deduced ORFs are shown, with the sizes in amino acids of the products of ORFs from E. coli O157:H7 EDL933 (14) below the filled arrows. ORFs from Serratia marcescens plasmid R478 (24) are most closely related. The percent identity of each ORF to its corresponding ORF in EDL933 is shown below. Those labeled TlpD and TlrC, -B, and -A from E. coli O157: H7 strain 86-24NalR were given this nomenclature by Tarr et al. (19). Plasmid pMJ606 contains the cloned Ter determinants from pMER610 originally isolated from an Alcaligenes sp. (7). Plasmid pTE53 originated in a clinical isolate of E. coli (3). The last two sequences are from Proteus sp. and D. radiodurans genome sequence databases.
Lack of relationship between Shiga toxin production and tellurite resistance in E. coli O157:H7 isolates.
The strains examined, along with their serotypes and ability to produce Shiga toxin, and the tellurite MICs for them are shown in Table 1. Fifteen E. coli O157:H7 strains were tested. Also included was a nonflagellated strain of E. coli O46:H− (DT2895), producing neither stx1 nor stx2, which acted as a negative control along with E. coli K-12. All strains except two were Shiga toxin positive. Nine strains produced Stx1 and contained the stx1 gene, whereas 12 produced Stx2 and contained the stx2 gene. Eight of the strains contained both stx1 and stx2. There was no relationship between Shiga toxin production and tellurite resistance. For example, of the two Shiga toxin-negative strains, the MIC for one was 1,024 μg/ml whereas the MIC for the other was 4 μg/ml. There was also no relationship between the presence of either stx1 or stx2 and level of tellurite resistance in the E. coli isolates.
Tellurite-resistant strains of E. coli O157:H7 contain ter genes.
The presence of each of the ter genes, including terF (Fig. 2), was tested by PCR analysis using the primers listed in Table 2. All the Ter strains tested were positive for terZ, terA, terB, terC, terD, terE, and terF genes (Table 3). E. coli EDL933 was used as a positive control, and E. coli DH5α was used as a negative control. The tellurite-susceptible strains (DT2892, DT2895, DT2897, and DT2899) were negative for all ter genes. The tellurite MICs for the latter strains ranged from 1 to 4 μg/ml and were slightly higher than that for the negative control, E. coli DH5α, for which the tellurite MIC was <1 μg/ml.
TABLE 3.
Number and location of tellurite resistance OI in E. coli O157:H7 strains
| Strain no. | Tellurite MIC (μg/ml) | Presence of ter genesa | Positive PCR withb: | No. of copiesc | Locationd | |
|---|---|---|---|---|---|---|
| serW primers | serX primers | |||||
| DT2889 | 1,024 | + | − | − | 2 | serW, serX |
| DT2890 | 1,024 | + | − | − | 2 | serW, serX |
| DT2891 | 1,024 | + | − | − | 2 | serW, serX |
| DT2892 | 2-4 | − | + | + | 0 | NAe |
| DT2893 | 1,024 | + | − | − | 2 | serW, serX |
| DT2894 | 1,024 | + | − | + | 1 | serW |
| DT2895 | 2-4 | − | + | + | 0 | NA |
| DT2896 | 1,024 | + | + | − | 1 | serX |
| DT2897 | 1-2 | − | + | + | 0 | NA |
| DT2898 | 1,024 | + | − | − | 2 | serW, serX |
| DT2899 | 4 | − | + | + | 0 | NA |
| DT2998 | 64 | + | + | − | 1 | serX |
| DT2999 | 64 | + | + | − | 1 | serX |
| DT3000 | 64 | + | + | − | 1 | serX |
| EDL933 | 64-128 | + | − | − | 2 | serW, serX |
| Sakai | 128 | + | + | − | 1 | serX |
| DH5α | <1 | − | + | + | 0 | NA |
To assess the presence or absence of ter genes in E. coli O157 strains, pulsed-field gel analysis of genome DNA from representative strains was undertaken by using _Xba_I, _Spe_I, and _Avr_II digestion. The nitrocellulose blots obtained from the digestion were hybridized to a terC gene probe produced by PCR. All four strains which were negative in the PCR experiments were also negative in the Southern blots, confirming our previous results (data not shown). In contrast, all strains positive in the PCR experiments with the various ter genes were positive in the Southern blots. The blots were also hybridized with a pfkA probe as a positive control for DNA transfer.
Number and location of Ter OI in E. coli O157:H7 strains.
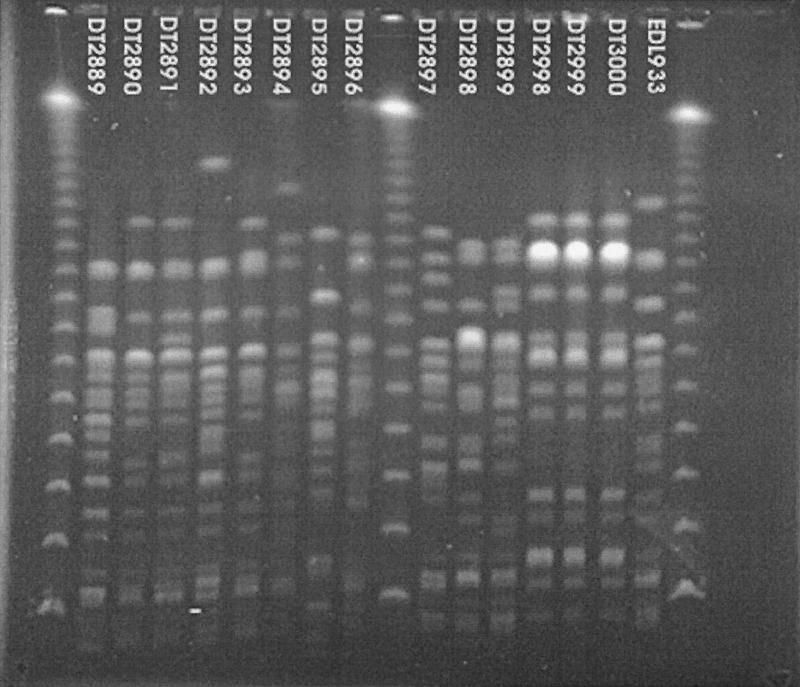
_Xba_I digests of E. coli O157:H7 DNA after PFGE are shown in Fig. 3. Fingerprints for the three Centers for Disease Control and Prevention (CDC; Atlanta, Ga.) strains (DT2998, DT2999, and DT3000) are fairly similar; these strains are from the same outbreak and have identical patterns. PFGE after digestion of EDL933 DNA resulted in two separate _Spe_I fragments of 90 and 330 kb, which hybridized with the terC probe. All other Ter O157:H7 strains contained a band at the lower position. Neither _Xba_I nor _Spe_I nor _Avr_II digestion could be used to differentiate between one or two copies of the TAI because, unlike EDL933, all other strains contained two bands which ran at the same position on pulsed-field gels and which hybridized with the terW probe. In addition Southern blotting was not sensitive enough to differentiate between one and two copies (data not shown).
FIG. 3.
PFGE of E. coli O157:H7 DNA digested with _Xba_I. The strain numbers are shown at the top of the gel. Lambda phage size markers were run at each end of the gel and in the central well.
Therefore, to determine the number of copies of the Ter OI, light cycler-based quantitation of terC genes using the procedure described by Loeffler et al. (10) with the gyrB gene (always present in one copy per chromosome equivalent) as a control was employed. The results are shown in Table 3. To identify the location of the Ter OI in each strain, PCR analysis was undertaken with primers specific for tRNA genes serX and serW. EDL933 has been shown to contain Ter OI located two bases beyond the tRNA genes (14), whereas the Sakai strain contains one Ter OI (6). It was not possible to amplify a complete copy of either tRNA gene by using DNA from EDL933 and five other Ter OI strains (Table 3), demonstrating that a Ter OI is associated with both serX and serW genes. Only the serW primer was amplified in four other strains (Table 3), which is similar to results for the Sakai strain and which places the Ter OI close to serX. In DT2894 only the serX primer was amplified, indicating that the Ter OI is located close to serW.
Since the location of the OI by PCR depends on a negative result, i.e., the inability to amplify serW and serX genes, additional PCRs were undertaken using primers inside the Ter OI as a positive control. These primers (Table 2) hybridize specifically to the Ter OI. E. coli EDL933 was used as a positive control, with DH5α as a negative control. PCR with the Ter-island primers (Table 2) produced a band of the predicted length for each of the strains that contain the Ter OI (data not shown). These results were used in combination with the results from the serW and serX primers to determine the locations of the Ter OI in the strains.
There appeared to be no relationship between the Te MIC and the presence of one or two copies of the Ter genes. For example, the Te MIC for E. coli DT2889 and DT2893 is 1,024 μg/ml and these strains contain two copies of the Ter genes, whereas the MIC for DT2896 is identical but the strain contains only one Ter OI. Similarly, the MICs for both the Sakai strain and DT2998 were intermediate, 64 to 128 μg/ml, and the strains had one copy of the Ter genes, whereas the MIC for EDL933 was similar but the strain contained two copies.
Expression of ter genes.
RT-PCR was used to examine the expression of ter genes by the production mRNA transcripts. All strains were tested first in the absence of tellurite. Those strains which were negative for expression of some ter genes were then retested in the presence and absence of tellurite to determine if tellurite induction plays a role in the expression of Ter and could explain the difference in MICs. Table 4 contains results of RT-PCR experiments for representative strains. Strains of E. coli O157:H7 for which the tellurite MIC was 1,024 μg/ml (e.g., DT2889) expressed terBCDEFZ genes. Those without ter genes as shown by PCR and PFGE (for example, DT2895 and DT2899) were negative.
TABLE 4.
Tellurite resistance gene expression as determined by RT-PCR
| Strain | Expression ofd: | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| terA | terB | terC | terD | terE | terF | terZ | ||||||||
| +Te | −Te | +Te | −Te | +Te | −Te | +Te | −Te | +Te | −Te | +Te | −Te | +Te | −Te | |
| DT2889a | − | − | + | + | + | + | + | + | + | + | + | + | + | + |
| DT2895b | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DT2899b | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DT2998c | − | − | + | − | + | − | + | + | + | + | + | − | + | + |
| DT2999c | − | − | + | − | + | − | + | + | + | + | + | − | + | + |
| DT3000c | − | − | + | − | + | − | + | + | + | + | + | − | + | + |
| EDL933c | − | − | + | − | + | − | + | + | + | + | + | − | + | + |
| DH5αb | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
In contrast, E. coli strains for which the tellurite MICs were intermediate, DT2998, DT2999, and DT3000 (MIC = 64 μg/ml) and EDL933 (MIC = 64 to 128 μg/ml) showed different patterns of transcription among the ter genes. For all four strains terD, terE, and terZ were transcribed both in the presence and absence of tellurite. The terB, terC, and Δ_terF_ genes were transcribed only in the presence of a low concentration (1 μg/ml) of tellurite. This result suggests that these genes are regulated by tellurite and may account for the lower MICs for these strains. When the strains were retested to determine tellurite MICs by growing cells overnight in subinhibitory concentrations of potassium tellurite (1 μg/ml), tellurite MICs which had been intermediate increased to 512 μg/ml. This group of strains comprised DT2998, DT2999, DT3000, and EDL933. MICs for the other strains did not change. None of the strains was found to transcribe the terA gene, either with or without tellurite induction. This gene is probably not required for Ter expression.
To determine if inducible versus constitutive behavior of the ter genes was a result of DNA sequence changes upstream of the Ter genes, we sequenced ∼200 bp upstream of the ATG start site of the terA, terB, terC, terD, terE, and terF genes for DT2889 (constitutive) and DT2998 (inducible). We found no differences in either sequence compared with the published sequence of EDL933 (inducible). Inducible versus constitutive Ter may therefore reflect changes in an unrelated part of the chromosome, such as the loss of a repressor.
DISCUSSION
Tellurite has been used for more than 80 years in selective media for the isolation of pathogens including Corynebacterium diphtheriae, Staphylococcus aureus, and Shigella spp. (20). Potassium tellurite is linked to the presence of plasmids in enteric bacteria (17) and has been shown to be encoded almost exclusively by IncHI2 or IncHII plasmids (20, 21). Various studies have successfully used Ter as a marker for preliminary identification of IncHI2 plasmids (1, 21). Biosynth (BCM) agar which contains tellurite is frequently used to screen for E. coli O157:H7 isolates (26).
The complete genome sequence of E. coli O157:H7 strain EDL933 contains two OI encoding Ter (14), whereas another strain of E. coli O157:H7 (Sakai), also completely sequenced, contains only a single Ter determinant (6). In this study, we have confirmed by PCR studies and PFGE analysis that most strains of E. coli O157:H7 (12 out of 15) contain at least one copy of the Ter OI in which are located the ter genes as shown in Fig. 2. Three E. coli O157:H7 strains (DT2892, DT2897, and DT2899) did not contain any ter genes. Tarr et al. (19) identified in E. coli O157:H7 strain 86-24 an island encoding Ter and containing iha, designated TAI (Fig. 2). These authors suggested that TAI was acquired relatively recently after it acquired the O157 rfb cluster and the _stx2_-carrying bacteriophage.
Most of the strains examined in this study (DT2889 to DT2899, excluding non-O157 strain DT2895 in Table 1) are unable to ferment sorbitol and fail to produce β-d-glucuronidase. These features distinguish them from the majority of fecal E. coli strains belonging to other serotypes (8, 13). In contrast DT2998, DT2999, and DT3000, obtained from CDC, were isolated from the same E. coli outbreak in Indiana in 1998. All three strains are β-d-glucuronidase positive, are slow sorbitol fermenters (day 3 or 4), and are also urease positive by day 2 or 3 (N. Strockbine, CDC, personal communication). These three strains are also indistinguishable based on PFGE patterns by _Bln_I and _Spe_I digestion (N. Strockbine, personal communication) and by _Xba_I (Fig. 3) and _Avr_II digestion (this study; data not shown). All other isolates are urease negative. The three Indiana isolates are related, and all contain one Ter OI located close to serX. In contrast all other isolates examined in this study are unrelated to one another.
Recently elegant models to describe the evolution of E. coli O157:H7 strains have been published (5, 8); these models are based on octamer-based genome scanning, marker sorting, and DNA sequence analysis. Markers used were ability to ferment sorbitol, loss of β-d-glucuronidase activity, and acquisition of TAI (19). Evolution in E. coli O157 was considered to depend on “gene acquisition events, movement of insertion sequences, and movement or recombination within prophage, implying that the conserved polymorphisms comprise a mosaic of mutation, recombination, and acquisition events fixed by one or more selective bottlenecks” (8). Although little sequencing was undertaken, acquisition of one or two copies of the Ter OI is clear from our study and points out the considerable diversity of the 15 E. coli O157:H7 strains examined in the context of the location and number of Ter OI.
E. coli O157:H7 may have acquired the Ter OI such as R478, which has also been shown to confer adherence on E. coli J62 (12, 19). Examination of Fig. 2 demonstrates that R478 shows the greatest predicted amino acid identity with EDL933 when the predicted TerZ, -A, -B, -C, -D, -E, and -F proteins are compared. R478 was originally isolated from Serratia marcescens. Burian et al. (3) reported a large plasmid, pTE53, which encoded high-level Ter and which may be an IncHI2 plasmid, in a clinical isolate of E. coli. The Ter determinate was cloned and sequenced (3) and is highly homologous to Ter determinants of other IncHI2 plasmids (M. Rooker and D. E. Taylor, unpublished data). Nevertheless, a number of other species contain Ter genes, including Yersinia pestis (20) and Deinococcus radiodurans (11), which appear less closely related. Although the MIC of tellurite for D. radiodurans is 1,024 μg/ml (M. Rooker and D. E. Taylor, unpublished data), this bacterium does not contain terZ, terC, or terE. Since there is significant homology among the terC, terD, and terE genes (25), some functions may be encoded by redundant genes. Alternatively, products of other genes within the D. radiodurans genome may substitute for the missing ORFs.
Cloning and transposon mutagenesis of derivatives of plasmid pTE53 have suggested that only terB, terC, terD, and terE are required for Ter (9). That previous work and the present study indicate that neither terA nor terF genes are required for expression of Ter. Kormitakova et al. (9) pointed out that their insertion mutagenesis does not differentiate between the requirement for terB, -C, -D, and -E genes and transcription of the genes from a common promoter upstream of terB. The RT-PCR data in the present study (Table 4) are consistent with a common transcriptional regulatory region for strains for which the MIC is high, such as DT2889. The plasmids examined, R478, PMER610, and pTE53, are associated with high MICs, and several ter genes that they carry may also be coordinately regulated (9).
Tellurite MICs for E. coli O157:H7 strains that are intermediate (64 to 128 μg/ml) increase to 512 μg/ml when the strains are pregrown in low concentrations of tellurite. Treatment with tellurite induces the production of mRNA from terB, terC, and terF. These RT-PCR results for E. coli O157:H7 strains resistant to intermediate levels of tellurite are not consistent with a common transcriptional unit and imply that separate promoters occur before terC, terD, terE, and terZ as well as before terB, all of which appear to be regulated by tellurite. Since terF and terZ are not required for Ter, there is a possibility that other genes are induced in response to tellurite. It is possible that tellurite induces a general stress response or that the ter genes have some additional functions not yet recognized.
As has been pointed out previously (20), none of the functions of TerZ, -A, -B, -C, -D, -E, and -F are known, although several of these proteins show homology with the _Dictyostelium_-type cyclic AMP-binding protein family (23). We know, however, that potassium tellurite (K2TeO3) is metabolized to form intracellular crystals of black metallic tellurium, which are often deposited just inside the inner membrane. Resistant cells grow as black colonies on relatively high concentrations of K2TeO3.
Clinical microbiologists use Biosynth agar which contains tellurite to select for Shiga toxin-producing E. coli O157:H7 (26). Since this agar contains only 0.1 μg of tellurite/ml, strains with or without urease and/or a Ter island and containing one or two copies all grow as black colonies on red agar. In contrast, E. coli K-12 forms white colonies on a yellow background. Therefore, the response of E. coli O157:H7 on Biosynth agar appears unrelated to the presence of Ter genes on the OI. Exactly which genes are responsible for a positive reaction of E. coli O157:H7 on Biosynth agar is unclear.
Many other questions relating to the tellurite determinant studied here remain. These include the functions of the individual gene products, the inducible nature of some genes, and the precise mechanism of resistance to Te and possible other unrecognized functions associated with the determinant. The questions regarding the mechanism of tellurite resistance encoded by plasmids and chromosomal genes require additional study. They should not divert attention from the main points of this work: that the Ter OI display considerable plasticity in E. coli O157:H7 in number and chromosomal location but not sequence and that their origin may well have been a plasmid present in E. coli at the time of acquisition.
Acknowledgments
We thank Cliff Clark, Frank Rodgers, and S. L. Liu for their help during the course of this study. The Sakai strain was kindly supplied by M. Yoh.
This work was supported by a grant from the Canadian Institutes of Health research (grant no. MT6200) to D.E.T. M.K. was supported by a Fellowship and D.E.T. was supported by a Medical Scientist award from the Alberta Heritage Foundation for Medical Research. Sequence analysis of E. coli O157:H7 was supported as part of the Bacterial Pathogen Genome Initiative at the University of Wisconsin, funded by NIH grant A144387.
REFERENCES
- 1.Alonso, G., C. Gomez, C. Gonzalez, and V. R. Lemoine. 2000. On the mechanism of resistance to channel-forming colicins (PacB) and tellurite, encoded by plasmid Mip233 (IncHI3). FEMS Microbiol. Lett. 192**:**257-261. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. Goeden, D. Rose, B. Mau, and Y. Shao. 1997. The complete sequence of Escherichia coli K-12. Science 277**:**1453-1462. [DOI] [PubMed] [Google Scholar]
- 3.Burian, J., N. Tu, L. Klucar, L. Guller, G. Lloyd-Jones, S. Stuchlik, P. Fejdi, P. Siekel, and J. Turna. 1998. In vivo and in vitro cloning and phenotype characterization of tellurite resistance determinant conferred by plasmid pTE53 of a clinical isolate of Escherichia coli. Folia Microbiol. 43**:**589-599. [DOI] [PubMed] [Google Scholar]
- 4.Chang, N., Q. Jiang, and D. E. Taylor. 1997. Construction of a genomic map of H. pylori by pulsed-field gel electrophoresis (PFGE), p. 165-176. In C. L. Clayton and H. L. T. Mobley (ed.), Methods in molecular medicine, Helicobacter pylori protocols. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 5.Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177**:**1750-1753. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C.-G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shingawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8(Suppl.)**:**47-52. [DOI] [PubMed] [Google Scholar]
- 7.Jobling, M. G., and D. A. Ritchie. 1988. The nucleotide sequence of a plasmid determinant for resistance to tellurium anions. Gene 662**:**245-258. [DOI] [PubMed] [Google Scholar]
- 8.Kim, J., J. Nietfeldt, J. Ju, J. Wise, N. Fegan, P. Desmarchelier, and A. K. Benson. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, β-glucuronidase-negative enterhemorrhagic Escherichia coli O157. J. Bacteriol. 183**:**6885-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kormutakova, R., L. Klucar, and J. Turna. 2000. DNA sequence analysis of the tellurite-resistance determinant from clinical strain of Escherichia coli and identification of essential genes. BioMetals 13**:**135-139. [DOI] [PubMed] [Google Scholar]
- 10.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycle system. J. Clin. Microbiol. 38**:**586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65**:**44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mignatti, P., L. Pagani, M. Perduca, and E. Romero. 1985. R factor-mediated adhesiveness to mammalian cells in E. coli K-12. Microbiologica 8**:**101-111. [PubMed] [Google Scholar]
- 13.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11**:**450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409**:**529-533. [DOI] [PubMed] [Google Scholar]
- 15.Rahn, K., J. B. Wilson, K. A. McFadden, S. C. Read, A. G. Ellis, S. A. Renwick, R. C. Clarke, and R. P. Johnson. 1996. Comparison of Vero cell assay and PCR as indicators of the presence of verocytotoxigenic Escherichia coli in bovine and human fecal samples. Appl. Environ. Microbiol. 62**:**4314-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, P. A. Hargrett, P. A. Blake, and M. L. Cohen. 1982. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308**:**681-685. [DOI] [PubMed] [Google Scholar]
- 17.Summers, A., and G. A. Jacoby. 1977. Plasmid-determined resistance to tellurium compounds. J. Bacteriol. 129**:**276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20**:**1-10. [DOI] [PubMed] [Google Scholar]
- 19.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68**:**1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7**:**111-115. [DOI] [PubMed] [Google Scholar]
- 21.Taylor, D. E., and A. O. Summers. 1979. Association of tellurium resistance and bacteriophage inhibition conferred by R plasmids. J. Bacteriol. 137**:**1430-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor, D. E., E. C. Brose, S. Kwan, and W. Yan. 1985. Mapping of transfer regions within incompatibility group HI plasmid R27. J. Bacteriol. 162**:**1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter, E. G., and D. E. Taylor. 1992. Plasmid-mediated resistance to tellurite: expressed and cryptic. Plasmid 27**:**52-64. [DOI] [PubMed] [Google Scholar]
- 24.Whelan, K. F., E. Colleran, and D. E. Taylor. 1995. Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 177**:**5016-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whelan, K. F., R. K. Sherburne, and D. E. Taylor. 1997. Characterization of a region of the IncHI2 plasmid R478 which protects Escherichia coli from toxic effects specified by components of the tellurite, phage, and colicin resistance cluster. J. Bacteriol. 179**:**63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zadik, P. M., P. A. Chapman, and C. A. Siddons. 1993. Use of tellurite for the selection of verocytotoxigenic Escherichai coli O157. J. Med. Microbiol. 39**:**155-158. [DOI] [PubMed] [Google Scholar]