Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect (original) (raw)
Abstract
The adzuki bean beetle, Callosobruchus chinensis, is triple-infected with distinct lineages of Wolbachia endosymbiont, wBruCon, wBruOri, and wBruAus, which were identified by their wsp (Wolbachia surface protein) gene sequences. Whereas wBruCon and wBruOri caused cytoplasmic incompatibility of the host insect, wBruAus did not. Although wBruCon and wBruOri were easily eliminated by antibiotic treatments, wBruAus persisted over five treated generations and could not be eliminated. The inheritance pattern of wBruAus was, surprisingly, explained by sex-linked inheritance in male-heterozygotic organisms, which agreed with the karyotype of C. chinensis (2_n_ = 20, XY). Quantitative PCR analysis demonstrated that females contain around twice as much wsp titer as males, which is concordant with an X chromosome linkage. Specific PCR and Southern blot analyses indicated that the wBruAus-bearing strain of C. chinensis contains only a fraction of the Wolbachia gene repertoire. Several genome fragments of wBruAus were isolated using an inverse PCR technique. The fragments exhibited a bacterial genome structure containing a number of ORFs typical of the α-proteobacteria, although some of the ORFs contained disruptive mutations. In the flanking region of ftsZ gene, a non-long terminal repeat (non-LTR) retrotransposon sequence, which is typical of insects but not found from bacteria, was present. These results strongly suggest that wBruAus has no microbial entity but is a genome fragment of Wolbachia endosymbiont transferred to the X chromosome of the host insect.
For a long time, horizontal gene transfers between genomes of different organisms have been believed to be rather exceptional and idiosyncratic phenomena. However, recent accumulation of microbial genome data has revealed an exciting view of evolution where horizontal gene transfers have commonly taken place between unrelated prokaryotic lineages in a dynamic manner (1–3). Now it is widely accepted that horizontal gene transfer is an important and universal pathway for bacteria to reorganize their genome and to quickly acquire novel features such as drug resistance, pathogenicity, metabolic properties, and others (3–5). On the other hand, horizontal gene transfers between prokaryote and eukaryote are still regarded as unusual, except for those derived from mitochondria and chloroplasts (6, 7), organelles of prokaryotic endosymbiont origin (8, 9).
Mechanisms of horizontal gene transfer between prokaryotes, such as transformation, transduction and conjugation, are relatively well understood (1, 3). In contrast, mechanisms underlying prokaryote–eukaryote gene transfer are unknown, although several hypothetical models have been considered. One model is the “food hypothesis,” which applies to prokaryotic genes identified in phagocytic unicellular eukaryotes like Trichomonas and Entamoeba (10–12). Unicellular eukaryotes often live close to prokaryotes and frequently use them as food, which means that they have a constant exposure to prokaryotic DNA (13, 14). However, this model appears not applicable to multicellular eukaryotes like animals, as foreign prokaryotes are not easily accessible to the germ line cells because of germ–soma separation and absence of active phagocytosis. An alternative model, the “endosymbiont hypothesis” that rests on a permanent contact between eukaryotic host cells and inhabiting microbial associates, may apply to these cases. In fact, mitochondria and chloroplasts have experienced gene transfer to the nucleus accompanied by drastic reduction in their genome size at early stages of their endosymbiotic evolution (6, 7). In plants and animals, transfer of mitochondrial genes to the nuclear genome has been shown to be a currently ongoing process (15, 16). It appears meaningful that obligate endosymbiotic bacteria tend to exhibit a remarkably reduced genome size in comparison with their free-living relatives (17–20). However, no case of horizontal gene transfer from prokaryotic endosymbiont to eukaryotic host has been described to date.
Members of the genus Wolbachia are rickettsia-like endocellular bacteria associated with insects and other invertebrates. Infection with Wolbachia often causes a wide range of effects on the reproduction and physiology of arthropod hosts such as cytoplasmic incompatibility, parthenogenesis, feminization, male killing, etc. Through host generations, Wolbachia are inherited solely in the maternal lineage by transovarial transmission (21–23). Therefore, the life cycle of Wolbachia has an intimate association with germ line cells of the host. Here we report an unprecedented case of prokaryote–eukaryote horizontal gene transfer: a genome fragment from the Wolbachia endosymbiont has been transferred to the X chromosome of a beetle.
Materials and Methods
Materials.
Three strains of Callosobruchus chinensis (Coleoptera: Bruchidae) were used in this study. Strain jC was triple-infected with wBruCon, wBruOri, and wBruAus (infection status, COA; ref. 24). Strains k10 and r13 were two isofemale lines derived from two wild populations, kkC98 and mrC98, respectively (24). These strains were double-infected with wBruCon and wBruOri (infection status, CO). These insects were maintained on adzuki seeds at 30°C and 70% relative humidity in a long-day regimen of 16 h light/8 h dark.
Antibiotic Treatment.
Antibiotic treatment was conducted by feeding the insects on artificial beans containing tetracycline or rifampicin. A mixture of milled adzuki bean powder and either tetracycline or rifampicin [0.03% (wt/wt)] was kneaded with an aliquot of water, shaped into small balls that mimicked bean grains, freeze-dried, and coated with collodion to imitate the peel of beans. The insects were allowed to oviposit and maintained on the artificial beans.
Examination of Inheritance.
Offspring of mating experiments were subjected to specific PCR detection of wBruCon, wBruOri, and wBruAus by using specific reverse PCR primers wspConR, wspOriR, and wspAusR, in combination with universal forward primer wspF, as described (24).
Quantitative PCR.
Real-time fluorescence detection quantitative PCR was performed using TaqMan PCR and an ABI Prism 7700 Sequence Detection System (PE Applied Biosystems) as described (24, 25). To estimate the titer of the respective three Wolbachia, copy number of the wsp gene was quantified. A double-fluorescence-labeled probe for detection, TQwspPRB, was targeted to a conserved region of the three types of wsp sequences. Highly specific amplifying primers were designed to variable flanking regions between the three sequences: TQwspCF and TQwspCR; TQwspOF and TQwspOR; and TQwspAF and TQwspAR (24, 25). For standardized comparison between males and females of different body size and DNA content, titer of wBruAus in each total DNA sample (wsp copies per microliter) was normalized by DNA concentration (nanograms per microliter) in the same sample.
Detection of Wolbachia Genes.
PCR detection of Wolbachia genes was conducted using the following primers as described: wspF and wspR for wsp (24); ftsF and ftsR for ftsZ (26); groEfl and groErl for groE (27); and wp16SF and wp16SR for 16S rDNA (28). Southern blot detection of Wolbachia genes was conducted using the probes for atpD, cytB, ftsZ, gltA, glyA, gyrA, Isw1, lipA, polA, rpoB, sdhA, sucD, thrA, tufA, and wsp, which were kindly provided by S. Masui (RIKEN Center for Developmental Biology, Kobe, Japan).
Cloning of Wolbachia Genome Fragments.
Total DNA of the insects was digested with _Bam_HI or _Hin_dIII, and self-ligated using T4 DNA ligase. The circularized DNA products were subjected to nested long-inverted PCR by using outward specific primers for Wolbachia genes. PCR was conducted using LA _Taq_DNA polymerase (TaKaRa, Otsu, Japan) under a temperature profile of 94°C for 2 min followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 68°C for 5 min, and a final extension step at 60°C for 5 min. For flanking sequence of wsp gene, primers wsp3FINV (5′-TACAACCAAACGCACCACCGCCAGCTATAA-3′) and TQwspAF (24) were used for the first PCR, and primers wsp3FINV and wsp5RINV (5′-ACACCTTTGGCAACTGCTGTGAGTAGTCAA-3′) were used for the second PCR. For 5′ flanking sequence of ftsZ gene, primers fts3F (5′-GAGAAATTTAAGTGGCCCTATAGTCAC-3′) and fts5R (5′-AAACAGAGTCATATCTCCGCCACCAGT-3′) were used for the first PCR, and primers fts3F and fts5R2 (5′-AAGCCCTGGCATGACCACCAAGTCAGT-3′) were for the second PCR. The sizes of these PCR products were consistent with the results of Southern blot analysis. To obtain 3′ flanking sequence of ftsZ gene, standard PCR was conducted using primers fts3F and wCZC (5′-CCAAGCCATTGTAAAGATGATTTAAGAACAT-3′). These amplified products were cloned into pT7Blue vector (Novagen), and purified plasmids containing the clones were subjected to DNA sequencing.
Source of the Genome Data of Wolbachia sp. from Drosophila melanogaster.
Preliminary sequence data of Wolbachia sp. from D. melanogaster were obtained from the Institute for Genomic Research through the web site at www.tigr.org.
Results and Discussion
Peculiar Properties of wBruAus.
In recent studies (24, 25), it has been demonstrated that the adzuki bean beetle, C. chinensis, is triple-infected with distinct lineages of Wolbachia, called wBruCon, wBruOri, and wBruAus. Among them, wBruAus showed peculiar properties. First, the titer of wBruAus (≈107 wsp copies equivalent per adult female) was smaller by an order of magnitude than that of wBruCon and wBruOri (≈108 wsp copies equivalent per adult female). Secondly, infection with wBruAus showed no detectable reproductive symptoms of the host insect, although infections with wBruCon and wBruOri caused significant levels of cytoplasmic incompatibility. Thirdly, although the titer of wBruAus in oocytes and unfertilized eggs was extremely low (<102 _wsp_ copies equivalent per egg), the _Wolbachia_ was stably inherited through host generations in the laboratory, and the infection rate in natural populations was >97% on average. In this study, further unusual properties of wBruAus atypical of endosymbiotic bacteria emerged.
Antibiotic Resistance of wBruAus.
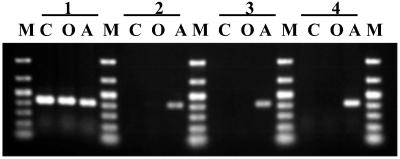
In an attempt to obtain _Wolbachia_-free C. chinensis, a triple-infected strain, jC, was reared with artificial beans containing tetracycline or rifampicin. Fig. 1 shows the results of specific PCR detection of wBruCon, wBruOri, and wBruAus after tetracycline treatments. Tetracycline treatment of only one generation was sufficient to eliminate wBruCon and wBruOri. On the other hand, wBruAus persisted throughout five generations of tetracycline treatment and could not be eliminated. After the treatment, the insects were transferred to and maintained on normal beans. The insects, named a strain jCAus, contained wBruAus but were completely free of wBruCon and wBruOri. The same patterns of tetracycline sensitivity, resistant wBruAus and sensitive wBruCon and wBruOri, were found in other strains of C. chinensis (data not shown). The same results were obtained from rifampicin treatments (data not shown). Thus, it was shown that wBruAus is exceptionally resistant to tetracycline and rifampicin, although Wolbachia and other endosymbiotic bacteria are, in general, susceptible to these antibiotics.
Fig 1.
Specific PCR detection of Wolbachia before and after tetracycline treatment. wBruCon, wBruOri, and wBruAus were detected by PCR using specific primers. 1, untreated; 2, after treatment for 1 generation; 3, after treatment for 5 generations; 4, 10 generations after treatment for 1 generation. C, wBruCon; O, wBruOri; A, wBruAus; M, DNA size markers (1,000, 700, 500, 400, 300, 200, and 100 bp from top to bottom). Each sample contains whole DNA from an adult female. Although data from only one insect for each treatment were shown, reproducibility of the results was confirmed for more than 10 individuals.
X-Linked Inheritance of wBruAus.
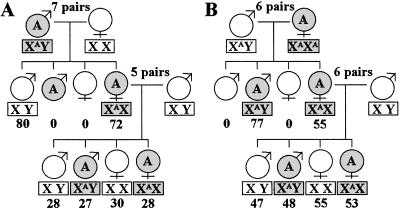
In mating experiments between C. chinensis strains of different infection status, we observed the inheritance of the three Wolbachia. As expected, wBruCon and wBruOri were maternally inherited to the offspring. Unexpectedly, however, we found that wBruAus was passed to the offspring not only maternally but also paternally. Fig. 2 shows the inheritance of wBruAus in reciprocal crosses between COA insects and CO insects. The crosses between COA fathers and CO mothers produced all CO males and, surprisingly, all COA females. When these COA females were mated with CO males, wBruAus exhibited 1:1 segregation in both males and females (Fig. 2A). The crosses between CO fathers and COA mothers produced all COA offspring, superficially showing typical maternal inheritance. However, when the COA female offspring were mated with CO males, wBruAus did not exhibit maternal inheritance but 1:1 segregation in both males and females (Fig. 2B). Notably, all these patterns were perfectly explained by sex-linked inheritance in male-heterozygotic organisms (Fig. 2), which agreed with the karyotype of C. chinensis (2_n_ = 20, XY; ref. 29; T.F., unpublished data).
Fig 2.
Inheritance of wBruAus. (A) Cross between wBruAus-bearing (COA) males and wBruAus-free (CO) females. (B) Cross between wBruAus-free (CO) males and wBruAus-bearing (COA) females. Shade indicates presence of wBruAus. The inheritance patterns are in agreement with X-linked inheritance. Sex chromosome types deduced are shown in rectangles. XA means the X chromosome carrying wBruAus. Numbers beneath the rectangles are the number of offspring obtained.
Sex-Linked Difference in Titer of wBruAus.
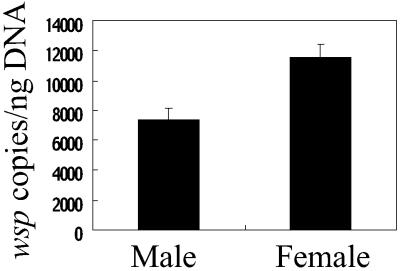
From the antibiotic insensitivity and the X-linked inheritance, a possibility was suggested that wBruAus has no microbial entity but is a bacterial genome fragment associated with X chromosome of the host insect. If so, female insects should contain twice as much titer of wBruAus as male insects. Fig. 3 shows the titer of wBruAus in adult males and females of C. chinensis in terms of wsp gene copies per nanogram of total insect DNA. As expected, females contained about twice as much wBruAus titer as males, which confirmed the idea of the X chromosome linkage.
Fig 3.
Sex-linked difference in titer of wBruAus. Titers of wBruAus in adult males and females were quantified in terms of wsp gene copies per nanogram of total insect DNA. The difference was statistically significant (Mann–Whitney U test, P < 0.001).
Specific PCR Detection of Wolbachia Genes from the Strain jCAus.
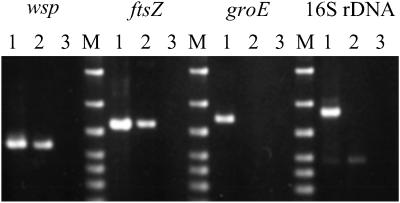
In addition to wsp, we attempted PCR detection of several Wolbachia genes, ftsZ, groE, and 16S rDNA, in the strain jCAus, which contains only wBruAus (Fig. 4). Whereas wsp and ftsZ were amplified by PCR, groE and 16S rDNA were not detected.
Fig 4.
Specific PCR detection of Wolbachia genes from the original triple-infected strain jC and the tetracycline-treated, wBruAus-bearing strain jCAus. Lanes 1, jC; lanes 2, jCAus; lanes 3, no template control; lanes M, DNA size markers (1,500, 1,000, 700, 500, 400, 300, and 200 bp from top to bottom). A faint band in lane 2 of the 16S rDNA panel is due to a nonspecific PCR product.
Southern Blot Detection of Wolbachia Genes from the Strain jCAus.
Furthermore, we attempted to detect Wolbachia genes in the strain jCAus by using Southern blotting. The following 15 genes were targeted: atpD, cytB, ftsZ, gltA, glyA, gyrA, Isw1, lipA, polA, rpoB, sdhA, sucD, thrA, tufA, and wsp. Whereas all of the genes were detected from the strain jC, only three genes, ftsZ, gyrA, and wsp, were detected from the strain jCAus (data not shown). These results indicated that the strain jCAus contains only a fraction of the Wolbachia gene repertoire, supporting the idea that the wBruAus strain in fact is a genome fragment of Wolbachia located on the host chromosome.
Structure of the Wolbachia Genome Fragment in the Strain jCAus.
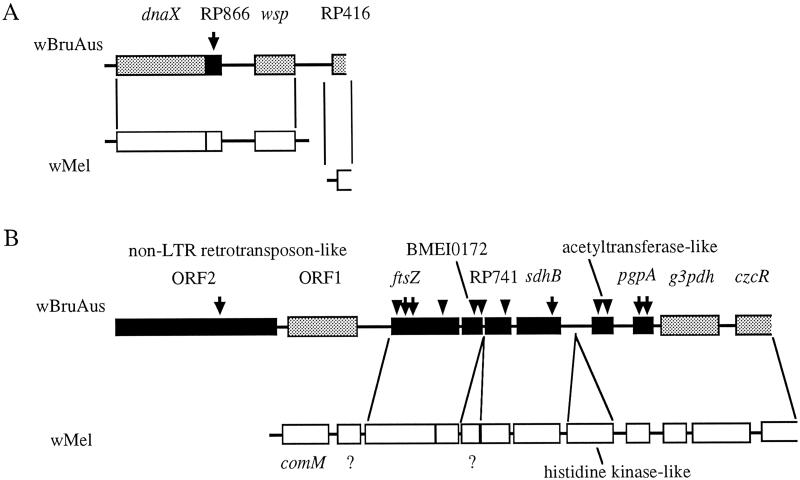
We cloned and sequenced the amplified fragments of wsp, ftsZ, and gyrA genes from the total DNA of the strain jCAus. In addition, we successfully isolated large DNA fragments flanking wsp and ftsZ genes by using an inverse PCR technique. A 4,141-bp fragment containing wsp (Fig. 5A), an 11,400-bp fragment containing ftsZ (Fig. 5B), and a 373-bp fragment of gyrA were obtained. These DNA fragments encoded bacterial genes that showed high sequence similarity to those of the α-proteobacteria, such as Wolbachia, Rickettsia, and Brucella. In the 4.1-kb fragment, four bacterial ORFs were identified: dnaX, wsp, and two ORFs homologous to RP866 and RP416 in the genome of Rickettsia prowazekii (17). In the 11.4-kb fragment, seven bacterial ORFs were detected: ftsZ, sdhB, pgpA, g3pdh, czcR, an ORF homologous to RP741 in the genome of R. prowazekii, and an ORF homologous to BMEI0172 in the genome of Brucella melitensis (30). One ORF showed a partial weak homology to acetyltransferase genes from various bacteria, archea, and plants.
Fig 5.
Structure of the genome fragments of wBruAus obtained by inverse PCR, aligned with genome sequences of wMel, a strain of Wolbachia from D. melanogaster. (A) A 4.1-kb fragment containing wsp gene. (B) An 11.4-kb fragment containing ftsZ gene. Arrows indicate the position of intermittent stop codons. Arrowheads show the position of frame-shift substitutions. Filled ORFs contain either stop codons or frame-shift substitutions, whereas shaded ORFs are structurally intact. A non-LTR retrotransposon-like sequence was located upstream of the ftsZ gene. All ORFs on wMel genome fragments are structurally intact. Question marks indicate unidentified ORFs.
Non-LTR Retrotransposon-Like Sequence in the Wolbachia Genome Fragment.
Notably, a non-LTR retrotransposon-like sequence, which showed a significant similarity to I and You elements from Drosophila species (31) and MosquI elements from Aedes aegypti (32), was identified in the upstream region of ftsZ. The sequence contained two ORFs typical of non-LTR retrotransposons: ORF1, encoding a protein of unknown function, and ORF2, encoding a protein with endonuclease and reverse transcriptase activities (31). Deduced amino acid sequence similarities between the retrotransposon-like sequence and a You element (accession no. AJ302712) were 41% in ORF1 and 47% in ORF2, respectively. I element is known to be involved in the I_–_R system of hybrid dysgenesis in D. melanogaster (33). A number of I and You elements, which constitute a distinct clade in the non-LTR retrotransposon phylogeny (31), have been identified in the genomes of D. melanogaster and other species. Because non-LTR retrotransposons of this family have been found in insect genomes but not in bacterial genomes, this finding may favor the idea that the Wolbachia genome fragment is located on a chromosome of the host insect.
Comparison with the Wolbachia Genome from D. melanogaster.
The genome fragments of wBruAus were compared with the preliminary genome sequences of wMel, a strain of Wolbachia from D. melanogaster, determined by the Institute for Genomic Research (Fig. 5). Gene arrangements on the genome fragments were, in general, conserved between wBruAus and wMel, although several differences were identified. In wBruAus wsp was next to RP416, whereas in wMel these genes were not on the same genome fragment. Only in wMel was a histidine kinase-like sequence found between sdhB and acetyltransferase-like sequence, and was an unidentified ORF located between BMEI0172 and RP741. In the upstream region of ftsZ, where non-LTR retrotransposon-like sequence was identified in wBruAus, comM and an unidentified ORF were found in wMel. These results indicated that the genome fragments of wBruAus certainly retain the Wolbachia genome structure.
Horizontal Gene Transfer from Wolbachia Endosymbiont to X Chromosome of Host Insect.
From all these results taken together, it is concluded that wBruAus, a strain of Wolbachia identified from C. chinensis, has no bacterial entity but is a genome fragment of Wolbachia horizontally transferred to the genome of the host insect. The Wolbachia genome fragment is at least 11 kb (excluding the non-LTR retrotransposon region), probably much larger, in size, containing a number of bacterial ORFs, and located on the X chromosome of the host. Because the genome size of Wolbachia is established to be 0.95–1.66 Mb (34), the transferred gene fragment may account for >1% of the whole genome of the original bacterium. Thus far, there have been several reports of putative prokaryote–eukaryote horizontal gene transfers in which a gene highly homologous to a prokaryotic one was detected in the nuclear genome of eukaryotes (10–12). However, evolutionary processes underlying these putative gene transfers were difficult to infer because the events are likely to have occurred quite anciently and the original structures of the transferred genes are no longer retained. Therefore, our finding provides unprecedented evidence of horizontal gene transfer from an endosymbiotic prokaryote to a host multicellular eukaryote in that (i) the transfer must be a recent event, (ii) the donor bacterium is unequivocally identified to be Wolbachia, (iii) a large fragment of bacterial genome is transferred, (iv) the structure of the transferred bacterial genome is highly preserved, and (v) the location of the transferred genome fragment is identified to be X chromosome.
Are Wolbachia Genes on Host Chromosome Functional?
Among 12 ORFs identified on the Wolbachia genome fragment (excluding the non-LTR retrotransposon region), more than half, 7 ORFs, contained stop codons or frame-shift substitutions, although these ORFs in the wMel genome were structurally intact (Fig. 5). Preliminary RT-PCR analyses showed that wsp and ftsZ are not transcribed in the strain jCAus (data not shown). These results indicate that most of the Wolbachia genes became pseudogenes on or after the horizontal transfer and were no longer functional. However, it is unknown whether all of the transferred Wolbachia genes are inactivated, or whether some of them survive on the host chromosome. It will be necessary to clone the whole Wolbachia genome fragment, to determine its structure, and to examine the expression of all ORFs on the fragment.
Process of Symbiont–Host Horizontal Gene Transfer?
At present, neither mechanism nor process involved in the _Wolbachia_–insect horizontal gene transfer is understood. Through generations of host insects, Wolbachia are maternally inherited via infection of developing oocytes. In eggs, Wolbachia cells localize at the posterior pole where germ cells develop (35), and are closely associated with astral microtubules of the host cells during mitosis (36). Therefore, although speculative, it is conceivable that these intimate associations of Wolbachia with proliferating germ cells have provided a favorable condition for the observed horizontal gene transfer.
Evolutionary Origin?
Absence of Wolbachia in other Callosobruchus species (37) suggests that wBruAus was acquired by the ancestor of C. chinensis through horizontal transmission from an unrelated host. In the DNA databases, a wsp sequence from the tephritid fruit fly Dacus destillatoria (accession no. AF295344) shows a very high similarity (98.9%) to the wsp gene of wBruAus, although a biological connection between the fruit fly and the bruchid beetle is obscure. Probably, the acquisition of bacterial wBruAus preceded the transfer of its genome fragment to the host chromosome. If so, a worldwide survey of C. chinensis populations might lead to the discovery of relict bacterial wBruAus, which would provide further insights into the evolutionary origin and process of the horizontal gene transfer.
Mechanism of Maintenance in Host Populations?
In all local populations of C. chinensis examined in Japan, the frequency of wBruAus, which can be considered to be “chromosomal Wolbachia,” was consistently >90% (24). At present, the mechanism whereby the Wolbachia genome fragment on the host chromosome prevails in populations is a mystery. Fixation by chance through drift cannot be ruled out but appears unlikely because of the consistent prevalence in many local populations. The chromosomal Wolbachia might be able to increase its frequency by hitchhiking with coexisting bacterial Wolbachia that cause cytoplasmic incompatibility. The chromosomal Wolbachia might be tightly linked to genes that confer a positive fitness effect to the insect, or genes that enhance its own transmission in a selfish manner like meiotic drive genes (38). Alternatively, the chromosomal Wolbachia itself might behave as a selfish genetic element like Medea, a maternal-effect chromosomal factor known from flour beetles (39). The last possibility is intriguing in that the Medea phenotype and _Wolbachia_-induced cytoplasmic incompatibility can be explained by the same “poison–antidote” or “modification–rescue” mechanism (40, 41).
Other Symbiont–Host Horizontal Gene Transfers to Be Found?
Previous studies have reported that genetic recombination can occur between co-infecting Wolbachia strains (42, 43). In this study, we first demonstrated that genetic materials can be exchanged between Wolbachia and host insect. It is unknown whether the _Wolbachia_–host gene transfer is an orphan exception, or whether other cases are to be found. It should be noted that, in previous extensive survey of infection and diversity of Wolbachia (44–46), only PCR detection was conducted without examining the inheritance pattern of the genes. Considering that insects are the most diverse eukaryotic group in the terrestrial ecosystem (47) and that infection frequency of Wolbachia in natural insect populations reaches ≈20–70% worldwide (45, 46), it would not be surprising if future careful studies revealed other cases of _Wolbachia_–host gene transfer. Other endosymbiotic systems such as Buchnera in aphids, in which the obligate symbiont genome exhibits remarkable degeneration and reduction (18, 20), might conceal similar horizontal gene transfer events. Genome sequencing of Arabidopsis thaliana revealed a continuous stretch of nearly 75% of mitochondrial genome located on chromosome 2 of the plant (48). Similarly, other cases of symbiont–host gene transfer might come from genome sequencing projects of various eukaryotic organisms now in progress.
Evolutionary Implications.
It has been pointed out that _Wolbachia_-induced cytoplasmic incompatibility can promote reproductive isolation of host insects (49, 50). If genes of Wolbachia responsible for cytoplasmic incompatibility are transferred to the host genome in a functional form, this would also reinforce reproductive isolation and ultimately lead to speciation. Endosymbiotic associations with microorganisms have been suggested to act as a source of evolutionary innovations for their hosts (51). Implications of the ongoing genome transfer between the symbiont and host can be far-reaching in this context.
Acknowledgments
We thank A. Sugimura, S. Kumagai, and K. Sato for technical and secretarial assistance; S. Masui for Wolbachia gene probes; and S. L. O'Neill for reading the manuscript. Preliminary sequence data of Wolbachia sp. from D. melanogaster, which was accomplished with support from the National Institutes of Health, were obtained from the Institute for Genomic Research through the web site www.tigr.org. This research was supported by the Program for Promotion of Basic Research Activities for Innovation Biosciences (ProBRAIN) of the Bio-Oriented Technology Research Advancement Institution. N.K. was supported by the Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Abbreviations
- non-LTR retrotransposon, non-long terminal repeat retrotransposon
- wsp, Wolbachia surface protein
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. AB080664, AB080665, and AB081842).
References
- 1.Ochman H., Lawrence, J. G. & Groisman, E. A. (2000) Nature 405**,** 299-304. [DOI] [PubMed] [Google Scholar]
- 2.Koonin E. V., Makarova, K. S. & Aravind, L. (2001) Annu. Rev. Microbiol. 55**,** 709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman F., (2002) Lateral DNA Transfer: Mechanisms and Consequences (Cold Spring Harbor Lab. Press, Plainview, NY).
- 4.Mazel D. & Davies, J. (1999) Cell. Mol. Life Sci. 56**,** 742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacker J. & Kaper, J. B. (2000) Annu. Rev. Microbiol. 54**,** 641-679. [DOI] [PubMed] [Google Scholar]
- 6.Martin W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M. & Kowallik, K. V. (1998) Nature 393**,** 162-165. [DOI] [PubMed] [Google Scholar]
- 7.Gray M. W., Burger, G. & Lang, B. F. (1999) Science 283**,** 1476-1481. [DOI] [PubMed] [Google Scholar]
- 8.Margulis L., (1970) Origin of Eukaryotic Cells (Yale Univ. Press, New Haven, CT).
- 9.Margulis L., (1981) Symbiosis in Cell Evolution (Freeman, San Francisco).
- 10.de Koning A. P., Brinkman, F. S. L., Jones, S. J. M. & Keeling, P. J. (2000) Mol. Biol. Evol. 17**,** 1769-1773. [DOI] [PubMed] [Google Scholar]
- 11.Field J., Rosenthal, B. & Samuelson, J. (2000) Mol. Microbiol. 38**,** 446-455. [DOI] [PubMed] [Google Scholar]
- 12.Andersson J. O., Doolittle, W. F. & Nesbo, C. L. (2001) Science 292**,** 1848-1850. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle W. F. (1998) Trends Genet. 14**,** 307-311. [DOI] [PubMed] [Google Scholar]
- 14.Berg O. G. & Kurland, C. G. (2000) Mol. Biol. Evol. 17**,** 951-961. [DOI] [PubMed] [Google Scholar]
- 15.Palmer J. D., Adams, K. L., Cho, Y., Parkinson, C. L., Qiu, Y. L. & Song, K. (2000) Proc. Natl. Acad. Sci. USA 97**,** 6960-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensasson D., Zhang, D.-X., Hartl, D. L. & Hewitt, G. M. (2001) Trends Ecol. Evol. 16**,** 314-321. [DOI] [PubMed] [Google Scholar]
- 17.Andersson S. G. E., Zomorodipour, A., Andersson, J. O., Sicheritz-Pontén, T., Alsmark, U. C. M., Podowski, R. M., Näslund, A. K., Eriksson, A.-S., Winkler, H. H. & Kurland, C. G. (1998) Nature 396**,** 133-140. [DOI] [PubMed] [Google Scholar]
- 18.Shigenobu S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. (2000) Nature 407**,** 81-86. [DOI] [PubMed] [Google Scholar]
- 19.Ochman H. & Moran, N. A. (2001) Science 292**,** 1096-1098. [DOI] [PubMed] [Google Scholar]
- 20.Andersson S. G. E. & Kurland, C. G. (1998) Trends Microbiol. 6**,** 263-268. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill S. L., Hoffmann, A. A. & Werren, J. H., (1997) Influential Passengers: Inherited Microorganisms and Arthropod Reproduction (Oxford Univ. Press, Oxford).
- 22.Werren J. H. (1997) Annu. Rev. Entomol. 42**,** 587-609. [DOI] [PubMed] [Google Scholar]
- 23.Stouthamer R., Breeuwer, J. A. J. & Hurst, G. D. D. (1999) Annu. Rev. Microbiol. 53**,** 71-102. [DOI] [PubMed] [Google Scholar]
- 24.Kondo N., Ijichi, N., Shimada, M. & Fukatsu, T. (2002) Mol. Ecol. 11**,** 167-180. [DOI] [PubMed] [Google Scholar]
- 25.Ijichi N., Kondo, N., Matsumoto, R., Shimada, M., Ishikawa, H. & Fukatsu, T. (2002) Appl. Environ. Microbiol. 68**,** 4074-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holden P. R., Brookfield, J. F. Y. & Jones, P. (1993) Mol. Gen. Genet. 240**,** 213-220. [DOI] [PubMed] [Google Scholar]
- 27.Masui S., Sasaki, T. & Ishikawa, H. (1997) Zool. Sci. 14**,** 701-706. [DOI] [PubMed] [Google Scholar]
- 28.O'Neill S. L., Giordano, R., Colbert, A. M. E., Karr, T. L. & Robertson, H. M. (1992) Proc. Natl. Acad. Sci. USA 89**,** 2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takenouchi Y. (1955) Jpn. J. Genet. 30**,** 7-9. [Google Scholar]
- 30.DelVecchio V. G., Kapatral, V., Redkar, R. J., Patra, G., Mujer, C., Los, T., Ivanova, N., Anderson, I., Bhattacharyya, A., Lykidis, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99**,** 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berezikov E., Bucheton, A. & Busseau, I. (2000) Genome Biol. 1**,** 0012.1-0012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu Z. & Hill, J. J. (1999) Mol. Biol. Evol. 16**,** 1675-1686. [DOI] [PubMed] [Google Scholar]
- 33.Bucheton A., Paro, R., Sang, H. M., Pelisson, A. & Finnegan, D. J. (1984) Cell 38**,** 153-163. [DOI] [PubMed] [Google Scholar]
- 34.Sun L. V., Foster, J. M., Tzertzinis, G., Ono, M., Bandi, C., Slatko, B. E. & O'Neill, S. L. (2001) J. Bacteriol. 183**,** 2219-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadfield S. J. & Axton, J. M. (1999) Nature 402**,** 482. [DOI] [PubMed] [Google Scholar]
- 36.Kose H. & Karr, T. L. (1995) Mech. Dev. 51**,** 275-288. [DOI] [PubMed] [Google Scholar]
- 37.Kondo N., Shimada, M. & Fukatsu, T. (1999) Zool. Sci. 16**,** 955-962. [Google Scholar]
- 38.Lyttle T. W. (1993) Trends Genet. 9**,** 205-210. [DOI] [PubMed] [Google Scholar]
- 39.Beeman R. W., Friesen, K. S. & Denell, R. E. (1992) Science 256**,** 89-92. [DOI] [PubMed] [Google Scholar]
- 40.Bull J. J., Molineux, I. J. & Werren, J. H. (1992) Science 256**,** 65. [DOI] [PubMed] [Google Scholar]
- 41.Hurst L. D. & McVean, G. T. (1996) Proc. R. Soc. London Ser. B 263**,** 97-104. [Google Scholar]
- 42.Werren J. H. & Bartos, J. D. (2001) Curr. Biol. 11**,** 431-435. [DOI] [PubMed] [Google Scholar]
- 43.Jiggins F. M., Schulenburg, J. H. G., Hurst, G. D. D. & Majerus, M. E. N. (2001) Proc. R. Soc. London Ser. B 268**,** 1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werren J. H., Windsor, D. & Guo, L. (1995) Proc. R. Soc. London Ser. B 262**,** 197-204. [Google Scholar]
- 45.Werren J. H. & Windsor, D. M. (2000) Proc. R. Soc. London Ser. B 267**,** 1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeyaprakash A. & Hoy, M. A. (2000) Insect Mol. Biol. 9**,** 393-405. [DOI] [PubMed] [Google Scholar]
- 47.Wilson E. O., (1989) Biodiversity (Natl. Acad. Press, Washington, DC).
- 48.Lin X. Y., Kaul, S. S., Rounsley, S., Shea, T. P., Benito, M. I., Town, C. D., Fujii, C. Y., Mason, T., Bowman, C. L., Barnstead, M., et al. (1999) Nature 402**,** 761-768. [DOI] [PubMed] [Google Scholar]
- 49.Hurst G. D. D. & Schilthuizen, M. (1998) Heredity 80**,** 2-8. [Google Scholar]
- 50.Bordenstein S. R., O'Hara, F. P. & Werren, J. H. (2001) Nature 409**,** 707-710. [DOI] [PubMed] [Google Scholar]
- 51.Margulis L. & Fester, R., (1991) Symbiosis as a Source of Evolutionary Innovation (MIT Press, Cambridge, MA). [PubMed]