Mitochondrial Fusion in Human Cells Is Efficient, Requires the Inner Membrane Potential, and Is Mediated by Mitofusins (original) (raw)
Abstract
Mitochondrial fusion remains a largely unknown process despite its observation by live microscopy and the identification of few implicated proteins. Using green and red fluorescent proteins targeted to the mitochondrial matrix, we show that mitochondrial fusion in human cells is efficient and achieves complete mixing of matrix contents within 12 h. This process is maintained in the absence of a functional respiratory chain, despite disruption of microtubules or after significant reduction of cellular ATP levels. In contrast, mitochondrial fusion is completely inhibited by protonophores that dissipate the inner membrane potential. This inhibition, which results in rapid fragmentation of mitochondrial filaments, is reversible: small and punctate mitochondria fuse to reform elongated and interconnected ones upon withdrawal of protonophores. Expression of wild-type or dominant-negative dynamin-related protein 1 showed that fragmentation is due to dynamin-related protein 1-mediated mitochondrial division. On the other hand, expression of mitofusin 1 (Mfn1), one of the human Fzo homologues, increased mitochondrial length and interconnectivity. This process, but not Mfn1 targeting, was dependent on the inner membrane potential, indicating that overexpressed Mfn1 stimulates fusion. These results show that human mitochondria represent a single cellular compartment whose exchanges and interconnectivity are dynamically regulated by the balance between continuous fusion and fission reactions.
INTRODUCTION
The morphology and distribution of mitochondria differ significantly between the cells of different species and tissues. In addition, mitochondrial volume and morphology vary in function of cellular metabolism, are modulated during cell cycle and development, and during apoptosis (Stevens, 1981; Tzagoloff, 1982; Bereiter-Hahn and Voth, 1994; Church and Poyton, 1998; Diaz et al., 1999; Frank et al., 2001). Live microscopy has revealed that mitochondrial morphology is continuously remodeled by fission and fusion (Nunnari et al., 1997; Rizzuto et al., 1998). In yeast, selective inhibition of either process significantly modifies mitochondrial size and interconnectivity (Bleazard et al., 1999; Sesaki and Jensen, 1999). Among the best known proteins involved in mitochondrial dynamics are Fzo/mitofusin, a transmembrane GTPase involved in fusion (Hales and Fuller, 1997; Hermann et al., 1998; Santel and Fuller, 2001; Rojo et al., 2002), and Dnm1p/dynamin-related protein 1 (Drp1), a dynamin-related protein involved in fission (Bleazard et al., 1999; Pitts et al., 1999; Smirnova et al., 2001).
In yeast, mitochondrial fusion has been demonstrated by the diffusion and/or mixing of different matrix proteins during mating of haploid cells (Azpiroz and Butow, 1993; Nunnari et al., 1997). In contrast, mitochondrial fusion has not been studied with similar assays in human cells, and it is not known to what extent the apparent fusion events observed by live microscopy correspond to the formation of intermitochondrial junctions (Bakeeva et al., 1978; Amchenkova et al., 1988), to outer membrane fusion, or to complete fusion of double membranes. In addition, we ignore the extent, kinetics, and requirements of fusion and fission, which determine the degree of mitochondrial interconnectivity and the efficiency of molecular exchanges between mitochondria.
The available reports are contradictory on the nature of this compartment. Mitochondria have been reported 1) to exist as independent and heterogeneous organelles, 2) to belong to functionally different mitochondrial (sub)populations, and 3) to form interconnected mitochondrial network(s) (Bakeeva et al., 1978; Amchenkova et al., 1988; Rizzuto et al., 1998; De Giorgi et al., 2000; Park et al., 2001; Collins et al., 2002). The connection between mitochondrial filaments is highly relevant for mitochondrial function, because it allows energy transmission between different cellular regions (Amchenkova et al., 1988) and determines the size and dynamics of the mitochondrial Ca2+ pool(s) (Rizzuto et al., 1998; De Giorgi et al., 2000; Park et al., 2001; Collins et al., 2002). In addition, the efficacy of fusion-mediated exchanges governs the complementation between mitochondrial DNA (mtDNA) molecules. Numerous mtDNA mutations have been described in association with human diseases. Most of them are heteroplasmic, i.e., mutated mtDNA molecules coexist with wild-type ones within one cell. The severity of these diseases depends on the amount of wild-type mtDNA, which above a relatively low threshold, permits normal mitochondrial activity and clinical presentation (reviewed in Shoubridge, 1994; Lightowlers et al., 1997). Complementation between normal and mutant mtDNA requires molecular exchanges between mitochondria and therefore efficient mitochondrial fusion. Evidence for mtDNA complementation and recombination has been found in the budding yeast (Dujon, 1981), but complementation in mammals is the matter of debate. According to contradicting reports, complementation between different mtDNA mutants is either rare (Enriquez et al., 2000) or requires 10–14 d to become efficient (Ono et al., 2001). These reports indicate that mitochondrial fusion in mammalian cells may be rare or slow, or not accompanied by molecular exchanges.
Herein, we demonstrate that mitochondrial fusion mediates the efficient exchange of soluble matrix proteins and modulates mitochondrial size and morphology in human cell lines. Fusion proceeds in the absence of a functional respiratory chain and after significant ATP depletion, but is abolished by dissipation of the inner membrane potential. Inhibition of fusion results in rapid (≤4 h) fragmentation of mitochondria by Drp1-mediated fission. Repolarization of the inner membrane restores mitochondrial fusion and leads to rapid reformation of mitochondrial filaments. We further show that the stimulation of mitochondrial fusion by a human mitofusin leads to increased mitochondrial interconnectivity.
MATERIALS AND METHODS
Reagents
Carbonyl cyanide m-chlorophenyl hydrazone (cccp), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (fccp), oligomycin, and trichostatin A were obtained from Sigma-Aldrich (St. Louis, MO); WGA-AlexaFluor 350 was from Molecular Probes (Eugene, OR); G418 was from Invitrogen (Carlsbad, CA); polyethylene glycol (PEG) 1500 was from BDH (Poole, Dorset, United Kingdom); and staurosporine was from Alexis (Läufelfingen, Switzerland). The stock solutions of cccp (10 mM in dimethyl sulfoxide [DMSO]), deoxyglucose (2 M in DMEM without glucose), fccp (10 mM in DMSO), oligomycin (1 mg/ml in ethanol), and trichostatin A (1 mM in DMSO) were stored at −20°C. The stock solution of staurosporine (1 mM in DMSO) was stored at 4°C. All other substances were obtained and handled as described previously (Bakker et al., 2000; Rojo et al., 2000, 2002).
Cloning and Mutagenesis
Cloning was performed according to standard procedures, and all polymerase chain reaction (PCR) products were verified by sequencing. Cloning of mitofusins Mfn1 and Mfn2 and of mitochondrial green fluorescent protein (mtGFP) has been described previously (Rojo et al., 2002). A version of the red fluorescent protein (RFP) DsRed1 targeted to the mitochondrial matrix (mtRFP) was constructed as described for mtGFP (Rojo et al., 2002). The cDNA encoding human Drp1 (accession number AF000430, bases 73–2283) was amplified by reverse transcription-PCR from total RNA of human skin fibroblasts by using Superscript II RT (Invitrogen) and Expand High Fidelity polymerase (Roche Applied Science, Indianapolis, IN). The PCR primers were appended with restriction sites and the restriction-digested PCR product was cloned into pCB6 (Rojo et al., 2000). The G1-motif of Drp1 was mutated (K38A) with QuikChange (Stratagene, La Jolla, CA). Expression plasmids encoding hemagglutinin (HA)-tagged OMP25 and a GFP molecule targeted to the mitochondrial outer membrane with the transmembrane domain of OMP25 (GFPOM) (Nemoto and De Camilli, 1999) were kindly provided by Pietro De Camilli (Yale University, New Haven, CT).
Cell Culture and Transfection
HeLa cells and osteosarcoma cells 143B and 143B-ρ0 were maintained as described previously (Bakker et al., 2000; Rojo et al., 2000). Cells were transfected with the calcium phosphate technique (Jordan et al., 1996), and stable transfectants expressing mtGFP or mtRFP were obtained by selection with 0.6 mg/ml G418. Stably transfected clones of HeLa and 143B ρ0-cell were incubated with 1 μM trichostatin A during 24–48 h to increase mtGFP expression (Shima et al., 1997; Condreay et al., 1999). Trichostatin A was removed from the medium 12–24 h before PEG-mediated cell fusion to avoid other effects of trichostatin A (aberrant morphology, cell cycle arrest, and/or apoptosis; (Hoshikawa et al., 1994; Medina et al., 1997). Cells maintained high mtGFP levels and reacquired a normal morphology within this time period. In transient transfection experiments, cells were fixed 30–48 h after addition of DNA. To ensure the coexpression of Drp1 and Drp1K38A in mtGFP-labeled cells (Figure 6), the plasmids encoding Drp1 molecules were transfected in a threefold excess (4.5 μg of pCB6-Drp1/Drp1K38A and 1.5 μg of pCB6-mtGFP per 35-mm well). In all other experiments, plasmids were cotransfected in equal amounts (2 μg of each plasmid per 35-mm well). Unless otherwise indicated, drugs and dyes were added to cells at the following concentrations: 10 μM cccp, 20 μg/ml cycloheximide, 40 mM deoxyglucose, 5 μM fccp, 5 μg/ml JC-1, 2.5 μM oligomycin, 0.2 μM rotenone, and 1 μM staurosporine. A medium containing pyruvate but devoid of glucose was used for deoxyglucose treatment. All other drugs were added to standard culture medium.
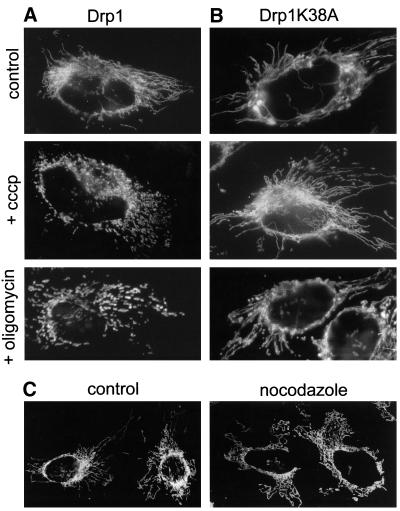
Figure 6.
Inhibition of fusion triggers Drp1-mediated mitochondrial fragmentation and is reversed upon repolarization of the inner membrane. (A and B) HeLa cells were cotransfected with plasmids expressing mtGFP and either wild-type Drp1 (A) or dominant-negative Drp1K38A (B) as described in MATERIALS AND METHODS. After 36 h, cells were fixed under control conditions or after a 4-h treatment with cccp or oligomycin. (A) Expression of wild-type Drp1 does not modify mitochondrial morphology and dynamics: mitochondria remain tubular under control conditions and fragment upon addition of cccp or oligomycin. (B) Expression of dominant-negative Drp1K38A prevents drug-induced fragmentation of mitochondrial filaments. (C) HeLa cells expressing mtRFP were treated for 4 h with cccp to fragment mitochondria. Cells were then transferred to cccp-free medium (control) or to cccp-free medium containing nocodazole. Fragmented mitochondria fuse and reassemble into filaments in the presence (control) and absence (nocodazole) of microtubules.
Cell Fusion
For cell fusion, cells carrying differently labeled mitochondria were mixed and plated on glass coverslips 16–40 h before cell fusion. Cycloheximide (20 μg/ml) was added 30 min before fusion and kept in all solutions used subsequently to inhibit protein synthesis (see below). The protocol for PEG-mediated fusion of adherent cells (Borer et al., 1989) was slightly modified. Briefly, 70–100% confluent cells in a 35-mm culture dish were washed with minimal essential medium (MEM) without serum and incubated for 45–60 s with 750 μl of a prewarmed (37°C) solution of PEG 1500 (50% [wt/vol] in MEM). Cells were then washed extensively with MEM containing 10% serum and transferred to prewarmed culture medium. Postfixation labeling of the plasma membrane with WGA-AlexaFluor350 (WGA) was used to confirm cell fusion and polykaryon formation.
Measurement of ATP Levels, Protein Synthesis, and JC-1, GFP, and RFP Fluorescence
To measure the relative ATP content and the mitochondrial inner membrane potential (ΔΨm)-dependent fluorescence of JC-1 aggregates, cells in 35-mm wells were incubated the last 15 min of the drug treatment with 5 μg/ml JC-1 (37°C, 5% CO2), washed with phosphate-buffered saline (PBS), trypsinized, and recovered in a final volume of 1 ml of PBS containing 10% fetal calf serum. A 100-μl aliquot was immediately mixed with 900 μl of lysis-solution (ATP Bioluminescence Assay kit; Roche Applied Science) and stored at −20°C. The ATP-content was then measured as described by the manufacturer. The remaining 900 μl was immediately used to measure the red fluorescence of JC-1 aggregates (excitation 490 nm, emission 590 nm) in an SFM 25 fluorometer (Kontron, Zürich, Switzerland). Cells were then recovered by centrifugation, washed with PBS, and the amount of cellular protein was quantified with bicinchoninic acid (Pierce Chemical, Rockford, IL) by using bovine serum albumin as a standard. The ATP levels and the JC-1 fluorescence were normalized to equal amounts of protein.
The ability of cycloheximide to inhibit protein synthesis was determined by measuring the incorporation of [35S]Met/Cys into proteins precipitated with methanol/chlorophorm (Wessel and Flugge, 1984). Cycloheximide (20 or 100 μg/ml) inhibited ∼90 or ∼95% of protein synthesis both in HeLa and in 143B cells. The total amount of mtGFP and mitochondrial red fluorescent protein (mtRFP) was determined after cell trypsinization and solubilization in an SFM25 fluorometer (Kontron) by measuring GFP/RFP fluorescence (excitation/emission: GFP = 473/509 nm, RFP = 558/583). Inhibition of 90% protein synthesis with 20 μg/ml cycloheximide for 4 h decreased cellular GFP or RFP fluorescence by ∼10 or ∼2%, respectively. This indicates that after a 4-h period only ∼2–10% of the total GFP/RFP fluorescence originates from neosynthesis. In a standard 4-h fusion experiment, when 90% of protein synthesis is inhibited with 20 μg/ml cycloheximide, the proportion of newly synthesized mtGFP/mtRFP represents ∼0.2–1% of total mtGFP/mtRFP content.
Antibodies, Immunohistological Staining, and Fluorescence Microscopy
Antibodies against tubulin (1A2) were a gift of Thomas Kreis (University of Geneva, Switzerland) and antibodies against the myc (9E10) and HA (HA.11) epitopes were obtained from Eurogentec (Seraing, Belgium). Antibodies against Mfn2 have been described previously (Rojo et al., 2002). For improved covisualization of myc-tagged mitofusins and HA-OMP25 with mtGFP or mtRFP, cells were shortly fixed with paraformaldehyde (3% [wt/vol], 5 min) and permeabilized with cold methanol (−20°C, 5 min) as described previously (Rojo et al., 2002). All other immunohistochemical stainings were performed on cells fixed with paraformaldehyde (3% wt/vol, 20 min) and permeabilized with 0.1% Triton X-100 (5 min) as described previously (Rojo et al., 1997). Fixed cells were observed with an Axiophot microscope (Zeiss, Jena, Germany) and living cells with an inverted IX70 microscope equipped with a computerized shutter (Olympus, Tokyo, Japan). Images were acquired with a charge-coupled device camera and micrographs were processed with MetaView and Adobe Photoshop.
Online Supplemental Material
During in vivo microscopy, cells plated on glass bottom dishes (Willco Wells BV, Amsterdam, The Netherlands)) were maintained in MEM containing 20 mM HEPES, pH 7.4, instead of carbonate. Light from a 50-W source was attenuated with neutral density filters to 2.5–10% of intensity, and the dish was kept at 37°C with a heating plate. Total observation time was 45 min, acquisition interval was 30 s, exposure time was 0.1–0.3 s, and movie speed was 6 frames/s (3 min/s).
RESULTS
Mitochondria Fuse and Exchange Soluble Matrix Proteins
To investigate mitochondrial fusion with an assay based on content mixing, we generated stably transfected human HeLa cells expressing green or red fluorescent proteins targeted to the mitochondrial matrix (mtGFP and mtRFP). Cells with differently labeled mitochondria were fused with PEG in the presence of cycloheximide to inhibit protein synthesis (see MATERIALS AND METHODS). Cytoplasmic fusion became apparent by phase contrast microscopy after 30–45 min, and polykaryons were readily observed after 3–4 h. Preliminary experiments had shown that mitochondrial morphology and distribution were not affected by PEG-mediated cell fusion or by treatment with cycloheximide. The ΔΨm was also unaffected by these treatments, as indicated by the red fluorescence of the potential-sensitive dye JC-1 (our unpublished data; Smiley et al., 1991). Two hours after cell fusion, double-labeled mitochondria (Figure 1A) coexisted with single-labeled ones (Figure 1A, arrowheads) in the central area of the polykaryon. Four hours after cell fusion, the fraction of double-labeled mitochondria had increased, with most mitochondria still containing more of one or the other fluorescent protein (Figure 1, B and C). The different distributions of mtGFP and mtRFP within the mitochondrial compartment and the presence of single-labeled mitochondria at peripheral regions of the polykaryon revealed the original label and position of the fused cells (Figure 1, B and C). Because the mitochondrial presequence of matrix proteins is cleaved during import, fluorescent proteins cannot be exchanged via the cytosol through successive export and import reactions. Therefore, these experiments demonstrate that the outer and the inner membranes do fuse with each other and allow the exchange of fluorescent matrix proteins between mitochondria. Mitochondrial fusion being an obligatory requisite for complementation, it has been speculated that the differences between “rarely complementing” 143B cells (Enriquez et al., 2000) and “complementing” HeLa cells (Ono et al., 2001) could arise from differences in the nuclear background and the machineries available for intermitochondrial fusion and/or complementation. To investigate this hypothesis, we generated 143B cells stably expressing mtGFP or mtRFP. We observed that mitochondria of 143B cells also fuse and exchange fluorescent matrix proteins (our unpublished data), demonstrating that mitochondrial fusion is not specific to a particular human cell line.
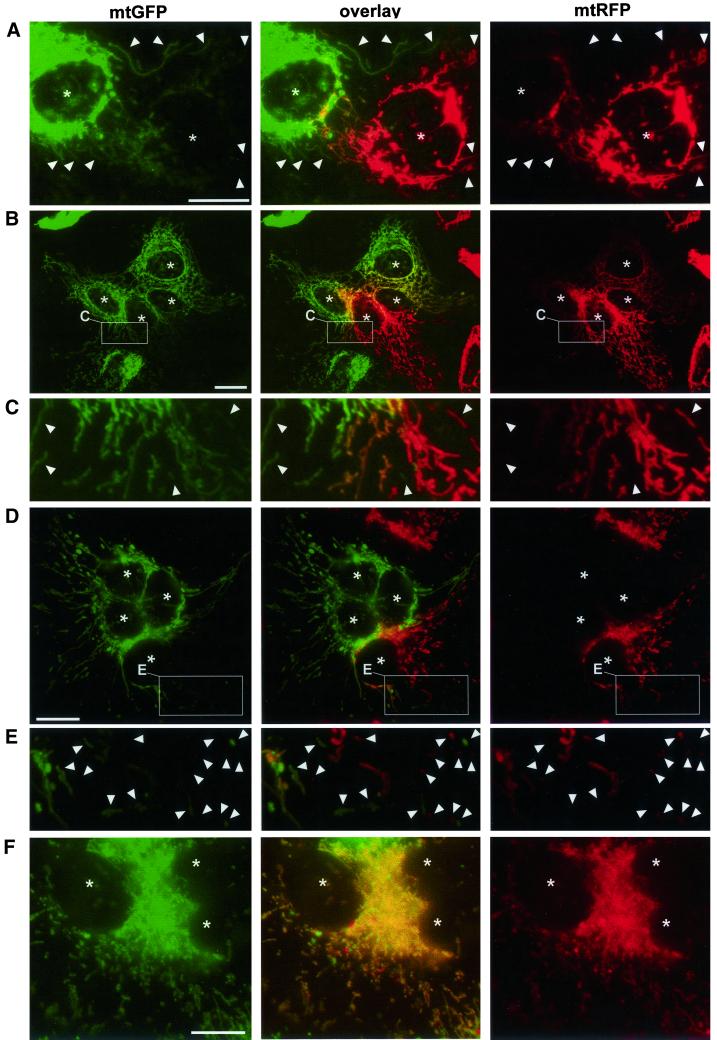
Figure 1.
Mitochondria exchange soluble matrix proteins by fusion. Differently labeled HeLa cells (A–C) or 143B-ρ0 cells (D–F) were coplated and fused with PEG in the presence of cycloheximide and fixed after further 2 h (A), 4 h (B–E), or 16 h (F). C and E are enlargements of the areas boxed in B and D, respectively. Asterisks indicate the position of polykaryon nuclei and arrowheads indicate the position of single-labeled mitochondria. The presence of double-labeled mitochondria (yellow) demonstrates the exchange of fluorescent matrix proteins by fusion. (A–E) Presence of mitochondria with a single label, even in proximity to differently labeled mitochondria, indicates that mitochondrial fusion events are not completed within 2 h (A) or 4 h (B–E). The different distribution of mtGFP and mtRFP within the double-labeled mitochondrial compartment (B) indicates that their diffusion is still ongoing. (F) Homogeneous double labeling of mitochondria indicates that fusion and diffusion have reached equilibrium 16 h after cytoplasmic fusion. Bars, 30 μm.
A Functional Respiratory Chain Is Dispensable for Mitochondrial Fusion
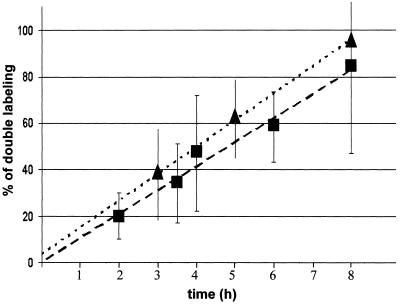
Studies on intermitochondrial complementation have been performed with respiration-deficient cell lines devoid of mtDNA (ρ0-cells) or with ρ0-derived cells that had been repopulated with mutant mtDNA from patients (Enriquez et al., 2000; Ono et al., 2001). To investigate whether a functional respiratory chain is necessary for mitochondrial fusion, we generated 143B-ρ0 cells stably expressing mtGFP or mtRFP. The 143B-ρ0 cells often contained a higher proportion of short and/or punctate mitochondria than HeLa and 143B cells (Figure 1, D–F). Four hours after cytoplasmic fusion, fused (double-labeled) and nonfused (single-labeled) mitochondria coexisted within polykaryons, the different distributions of mtGFP and mtRFP indicating the original label and position of the fused cells (Figure 1, D and E). In contrast, 16 h after cytoplasmic fusion, mitochondria were homogeneously double labeled (Figure 1F). To estimate the efficiency and kinetics of mitochondrial fusion, the proportion of double-labeled mitochondria was determined on polykaryons fixed at different times after PEG treatment. Both in HeLa cells and in 143B-ρ0 cells, mitochondrial fusion seems to be an efficient process that progresses linearly with time, reaches 70% content mixing after 8 h, and is predicted to mediate full content mixing after 10–12 h (Figure 2). These results demonstrate that mitochondrial fusion proceeds equally well in respiratory-competent HeLa cells and in respiratory-deficient 143B-ρ0 cells, and thus that a functional respiratory chain is dispensable for mitochondrial fusion.
Figure 2.
Mitochondrial fusion achieves complete mixing of matrix contents. Heterokaryons obtained as described in Figure 1 were fixed at different times after PEG-mediated cytoplasmic fusion. For each time point, the percentage of area with double-labeled mitochondria was estimated in 20–30 polykaryons. The mean values and the SEs are plotted as a function of time. The percentage of double labeling increases with time and is predicted to reach 100% 10–12 h after cytoplasmic fusion. The kinetics of fusion is similar for respiration-competent HeLa cells (squares, r = 0.95) and for respiration-deficient 143B-ρ0 cells (triangles, r = 0.98).
Inner Membrane Potential Is Required for Mitochondrial Fusion
The observation that respiration-deficient and respiration-competent mitochondria fuse with similar kinetics was relatively unexpected, because mitochondrial fusion most probably represents an energy-dependent process. Because the available clones of ρ0-cells (HeLa-ρ0 and 143B-ρ0) maintain a ΔΨm that allows the import of nuclear-encoded proteins and the maintenance of several mitochondrial functions (Buchet and Godinot, 1998), we used specific inhibitors of energy metabolism to investigate the energetic requirements of mitochondrial fusion. We used 1) oligomycin to inhibit the mitochondrial ATP-synthetase; 2) the protonophore cccp to dissipate the ΔΨm and inhibit mitochondrial ATP-synthesis; and 3) deoxyglucose to sequester cytosolic ATP, block the glycolytic pathway, and inhibit cytosolic ATP synthesis.
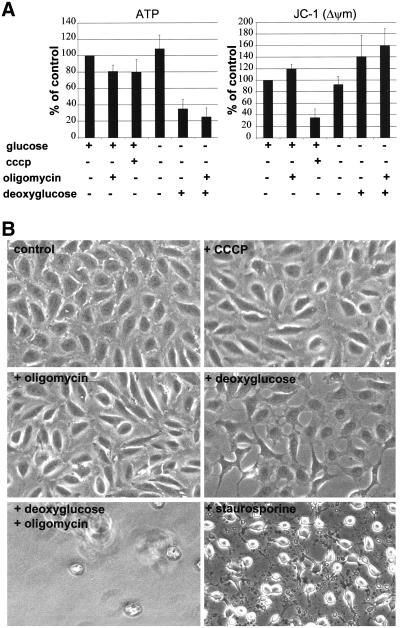
After a 45-min treatment, oligomycin and cccp had lowered cellular ATP levels by 20%, deoxyglucose by 60%, and a combined treatment with oligomycin and deoxyglucose by 80% (Figure 3A). In contrast, withdrawal of glucose alone did not lower the cellular ATP content. Measurement of ΔΨm with the JC-1 dye (Smiley et al., 1991) showed that oligomycin led to a slight increase in ΔΨm and that cccp induced significant depolarization of the inner membrane (Figure 3A); the other treatments were without effect or even led to an increase of JC-1 fluorescence (Figure 3A). To further investigate the toxicity of these drugs, cells were visualized by phase contrast microscopy. Overall, cell morphology was unaffected after 45 min, but differences were clearly visible after 3 h. In contrast to control cells and to cells treated with cccp or oligomycin, deoxyglucose-treated cells seemed to retract (Figure 3B). The combined treatment with deoxyglucose and oligomycin provoked massive cell detachment after 1–2 h, leading to the absence of adherent cells after 3 h (Figure 3B). None of these treatments provoked the apoptotic morphology observed upon treatment with 1 μM staurosporine (Figure 3B). The relatively low toxicity of mitochondria-specific drugs was confirmed by the observation that cccp-treated cells remained viable for up to 24 h and that their mitochondria retained cytochrome c (our unpublished data), as described previously (Lim et al., 2001).
Figure 3.
Inhibitor-mediated modulation of cellular ATP and of the ΔΨm. (A) HeLa cells treated with the indicated drugs for 45 min were trypsinized and subjected to measurement of ATP content and of ΔΨm-dependent fluorescence of JC-1 aggregates. Values were normalized to equal amounts of cellular protein and are expressed as percentage of control. Mean values and SEs are derived from three independent experiments. All treatments lead to a variable reduction of cellular ATP, but only cccp induces a significant reduction of the ΔΨm-dependent fluorescence of JC-1 aggregates. (B) HeLa cells treated with the indicated drugs for 3 h were fixed and visualized by phase contrast microscopy. Cellular morphology seems normal upon treatment with oligomycin or cccp, but cells retract upon addition of deoxyglucose. Cells have detached from the substrate after combined treatment with oligomycin and deoxyglucose, and none of these treatments provokes the apoptotic morphology induced by staurosporine.
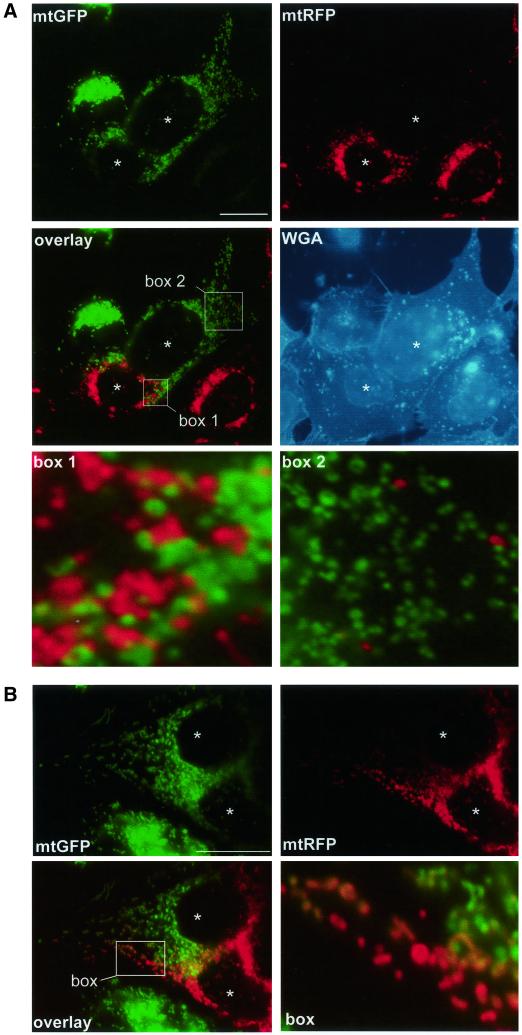
We then performed the assay of mitochondrial fusion in the presence of cccp, deoxyglucose, or oligomycin. Cells expressing mtGFP or mtRFP were preincubated with the respective drugs for 30 min, fused with PEG, and fixed after further 4 h in drug-containing medium. Mitochondrial tubules were shorter and numerous mitochondria appeared as punctate structures, especially upon incubation with cccp (Figure 4). In the presence of cccp, differently labeled mitochondria were mixed in the central region of the polykaryon (Figure 4A, box 1), and a few red mitochondria had moved to the region of the polykaryon containing green mitochondria (Figure 4A, box 2). However, and despite their proximity, mitochondria did not mix their fluorescent proteins (Figure 4A), indicating that mitochondrial fusion was inhibited. Live microscopy of cccp-treated cells revealed that mitochondria remained mobile, reached and maintained close vicinity, but did not seem to fuse (online supplemental material, Movie 1). In contrast, ATP depletion with oligomycin (Figure 4B) or deoxyglucose (our unpublished data) did not abolish mitochondrial fusion. Nevertheless, double-labeled mitochondria were somewhat less abundant after ATP depletion with deoxyglucose (our unpublished data) or oligomycin (Figure 4B) than in control cells (Figure 1). Together, these data demonstrate that mitochondrial fusion can proceed after significant reduction of cellular ATP levels (with oligomycin or deoxyglucose) but that dissipation of ΔΨm (with cccp) leads to complete inhibition of mitochondrial fusion.
Figure 4.
ΔΨm is necessary for mitochondrial fusion. Differently labeled 143B cells (A) or HeLa cells (B) were coplated and fused in the presence of 10 μM cccp (A) or 2.5 μM oligomycin (B) and cells were fixed 4 h after cell fusion. Drugs were added to the culture medium 30 min before PEG-mediated cell fusion and kept throughout the experiment. Enlargements of the boxed areas are shown separately. (A) In the presence of cccp, mitochondria loose their tubular morphology and appear as punctate structures. Despite thorough intermixing (overlay, box1) and high mobility (overlay, box 2), mitochondria remain single labeled. Postfixation labeling of the plasma membrane with WGA confirms polykaryon formation and demonstrates the absence of intermitochondrial fusion. (B) Mitochondrial filaments are shorter or fragmented in the presence of oligomycin. The appearance of double-labeled mitochondria (overlay, box) demonstrates mitochondrial fusion. Bars, 30 μm.
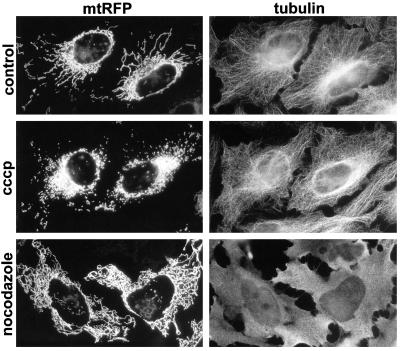
It has been reported that protonophores uncoupling oxidation from phosphorylation disrupt microtubules in cultured human cells (Maro and Bornens, 1982). We therefore investigated whether cccp-mediated mitochondrial fragmentation (Figure 4A) was a direct consequence of inner membrane depolarization (and fusion inhibition) or a secondary effect of microtubule disruption. HeLa cells expressing mtRFP were treated with cccp or with the microtubule-disrupting agent nocodazole, and tubulin was subsequently visualized with specific antibodies. Incubation with cccp led to mitochondrial fragmentation, but not to disruption of microtubules (Figure 5, cccp). In contrast, treatment with 10 μM nocodazole led to depolymerization of microtubules and to the disorganization of the mitochondrial network, but not to fragmentation of mitochondrial filaments (Figure 5, nocodazole). These results show that in HeLa cells, mitochondrial fragmentation is a direct consequence of fusion-inhibition. The fragmentation of mitochondria by oligomycin (Figures 4B and 6A) and deoxyglucose (our unpublished data) suggest that the rate of fusion is lowered upon ATP depletion. It is noteworthy that the inhibition of fusion (by inactivation of temperature-sensitive Fzo1p) also leads to mitochondrial fragmentation in yeast (Hermann et al., 1998; Rapaport et al., 1998).
Figure 5.
Inhibition of mitochondrial fusion by dissipation of ΔΨm triggers fragmentation of mitochondrial filaments. Cells expressing mtRFP were treated for 4 h with the indicated drugs, fixed, and incubated with antibodies against tubulin. Treatment with cccp leads to mitochondrial fragmentation but not to disruption of microtubules. Microtubule depolymerization with nocodazole provokes disorganization of the mitochondrial network, but not to fragmentation of tubular mitochondria.
Inhibition of Fusion Triggers Drp1-mediated Fragmentation and Is Reversed upon Repolarization of Inner Membrane
We then investigated whether mitochondrial fragmentation is due to ongoing mitochondrial division, a process mediated by Drp1 in mammals. To this end, cells were transfected with either wild-type Drp1 or a dominant-negative mutant of Drp1 (Drp1K38A) that inhibits Drp1 function (Smirnova et al., 2001). Mitochondria displayed their normal tubular morphology in cells transfected with wild-type Drp1 (Figure 6A, control). In contrast, mitochondrial length and connectivity were slightly higher in cells transfected with Drp1K38A (Figure 6B, control). The two types of transfected cells were treated during 4 h with cccp, oligomycin, or deoxyglucose. The mitochondrial fragmentation normally induced by these treatments also occurred in cells expressing the wild-type Drp1 (Figure 6A). In contrast, mitochondrial fragmentation was inhibited in cells expressing dominant-negative Drp1K38A (Figure 6B). This demonstrates that, upon inhibition of fusion, mitochondrial filaments are fragmented by Drp1-mediated fission.
Because the inner mitochondrial membrane reacquires a normal ΔΨm after cccp-removal, we investigated whether fusion inhibition by cccp was also reversible. Live microscopy revealed that, upon removal of cccp, punctate mitochondria fuse and form tubules (online supplemental material, Movie 2). Indeed, mitochondria reacquired their normal filamentous morphology, which is indistinguishable from those of control cells, 4 h after cccp washout (Figure 6C, control). The capacity of punctate mitochondria to fuse and form filaments upon cccp removal allowed us to investigate the requirements of mitochondrial fusion more precisely. Cells treated for 4 h with cccp were transferred to a medium devoid of cccp but containing cycloheximide or nocodazole. Cells were fixed 4 h after cccp removal and analyzed by fluorescence microscopy. Mitochondrial filaments were formed by mitochondrial fusion in the absence of protein synthesis (our unpublished data) or of a functional microtubule cytoskeleton (Figure 6C, nocodazole). These results validate the fusion experiments performed in the presence of cycloheximide (Figures 1, 2, and 4) and show that microtubules are not required for mitochondrial fusion. Together, these experiments demonstrate that fusion and fission are continuous processes and that their balance determines the length and interconnectivity of mitochondrial filaments in human cells.
Stimulation of Mitochondrial Fusion by Mitofusin 1 Requires Inner Membrane Potential
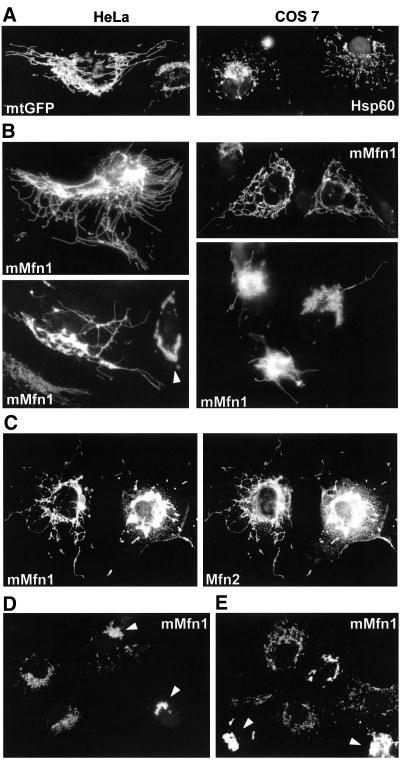
The proteins known to directly participate in mitochondrial fusion (Fzo1p/Mfn and Ugo1p) localize to the mitochondrial outer membrane (Rapaport et al., 1998; Sesaki and Jensen, 2001; Rojo et al., 2002). To investigate whether and how the activity of such outer membrane proteins is modulated by the inner membrane potential, we studied Mfn1, a mammalian Fzo homolog that has not been characterized yet (Santel and Fuller, 2001; Rojo et al., 2002). On expression, myc-tagged Mfn1 was targeted to mitochondria, where it colocalized with mtGFP (our unpublished data) and severely modified mitochondrial morphology and distribution. In HeLa cells, expression of Mfn1 led to the appearance of mitochondria that were frequently branched and interconnected, even at the cell periphery (Figure 7B, HeLa). Mfn1 expression also increased the length and interconnectivity of mitochondria in COS7 cells (Figure 7B, COS7). In numerous cells, mitochondrial filaments tended to accumulate in the perinuclear region and long mitochondrial tubules grew out of such mitochondrial bundles (Figure 7B). These mitochondrial profiles differ significantly from those observed upon fission inhibition with Drp1K38A (Figure 6B). At very high expression levels, Mfn1 led to mitochondrial clustering (Figure 7B, arrowhead), as described previously for Mfn2 (Santel and Fuller, 2001; Rojo et al., 2002). Close analysis of Mfn2 transfectants also revealed cells with elongated and branched mitochondria, but their proportion was very low (our unpublished data). To investigate the specificity of the profiles induced by excess Mfn1, we expressed two other outer membrane proteins: OMP25, a protein that can cluster mitochondria via its PDZ domain; and GFPOM, a GFP molecule targeted to mitochondria by the transmembrane domain of OMP25 (Nemoto and De Camilli, 1999). Neither protein induced the appearance of elongated and interconnected mitochondria (our unpublished data), demonstrating that the capacity to increase the length and interconnectivity of mitochondria is specific for Mfn1. The coexpression of Mfn1 and Mfn2 did not change the morphologies and frequencies of Mfn-specific profiles (Figure 7C). Immunofluorescence with antibodies against the myc-tag and with antibodies against the N-terminal domain of Mfn2 (Rojo et al., 2002) revealed that mitofusins colocalize on elongated and interconnected filaments as well as on clustered mitochondria (Figure 7C). This indicates that it is not the absence of equivalent amounts of Mfn1 and Mfn2 that limits the appearance of elongated and branched mitochondria and/or provokes mitochondrial clustering.
Figure 7.
Inner membrane potential is required for the stimulation of mitochondrial fusion by excess mitofusin 1. (A) Control mitochondrial morphology in mtGFP-transfected HeLa cells and in untransfected COS7 cells (Hsp60). (B) Transfection of myc-tagged Mfn1 (mMfn1) induces the appearance of highly elongated and frequently branched mitochondria. Long mitochondrial filaments grow out of perinuclear bundles of mitochondria and mitochondria become clustered at high expression levels (arrowhead). (C) On cotransfection, myc-tagged Mfn1 and untagged Mfn2 colocalize on elongated and branched mitochondria as well as on perinuclear mitochondrial clusters. (D) Myc-tagged Mfn1 is targeted to mitochondria in the presence of cccp, clusters mitochondria at high expression levels (arrowheads), but does not increase mitochondrial length and interconnectivity. The treatment with cccp started before expression of exogenous Mfn1 (9 h after transfection) and lasted for 31 h. (E) Elongated and branched mitochondria are fragmented in Mfn1-transfected cells treated with cccp. The treatment with cccp started after overexpression of exogenous Mfn1 (36 h after transfection) and lasted for 4 h.
The disappearance of punctate mitochondria and the increase in mitochondrial length and interconnectivity suggest that excess Mfn1 stimulates fusion. To investigate this hypothesis, we analyzed the influence of ΔΨm on this process. Cells were transfected with Mfn1 and mtGFP and cccp was added either 9 h after transfection (before expression of exogenous proteins; Figure 7D) or 36 h after transfection (after Mfn1 overexpression; Figure 7E). In both cases, cells were fixed and analyzed by immunofluorescence 40 h after transfection. In the absence of ΔΨm, mtGFP behaved like a matrix protein (Neupert, 1997) and remained cytosolic (our unpublished data), whereas Mfn1 behaved as an outer membrane protein (Shore et al., 1995) and was targeted to mitochondria (Figure 7D). Mfn1 retained the capacity to cluster mitochondria at high expression levels, but did not mediate the apparition of elongated and interconnected mitochondrial filaments (Figure 7D). The inhibition of fusion (for 4 h) in cells that had been expressing Mfn1 (for 36 h) led to the disappearance of elongated and interconnected mitochondria, but not to the dissociation of mitochondrial clusters (Figure 7E). These results show that Drp1-mediated mitochondrial division remains active in Mfn1-transfected cells and thus that Mfn1 increases mitochondrial length and interconnectivity by the stimulation of ΔΨm-dependent fusion.
DISCUSSION
The capacity of mitochondria to exchange soluble matrix proteins demonstrates that mitochondria fuse with each other by the merging of inner and outer mitochondrial membranes. The complete exchange of matrix proteins by fusion shows that mitochondrial fusion is efficient and that the mitochondrial matrix represents a single cellular compartment. The mitochondria of the HeLa cells used in this work appeared as long and interconnected filaments, in agreement with the work of others (Rizzuto et al., 1998; De Giorgi et al., 2000). The smaller size of HeLa mitochondria in other reports (Collins et al., 2002) may be related to the fact that HeLa cells have diverged into sublines since the establishment of the founder “uncloned” HeLa cells (Herrnstadt et al., 2002). We found that mitochondrial fusion also occurred in 143B and 143B-ρ0 cells and was therefore not restricted to a particular cell line. Furthermore, the kinetics of intermitochondrial fusion was similar in HeLa cells and in 143B-ρ0 cells, which are devoid of a functional respiratory chain and posses significantly smaller mitochondria. Nevertheless, the fact that complete intermixing of matrix contents required 10–12 h indicates that mitochondrial fusion is a relatively slow process. Separate mitochondria will thus remain heterogeneous and behave as independent entities during shorter time periods, as reported previously (Park et al., 2001; Collins et al., 2002).
We have demonstrated that dissipation of the inner membrane potential inhibits mitochondrial fusion. Protonophores did not affect the microtubule network, probably because they were used at lower concentrations (10 μM cccp) than in other studies (30 μM fccp; Maro and Bornens, 1982). The mechanisms by which inner membrane depolarization inhibit fusion remain to be determined. The inhibition of mRNA translation with cycloheximide, which blocks synthesis of nuclear-encoded mitochondrial proteins, did not impede mitochondrial fusion. Therefore, it is probably not the inhibition of protein import to the matrix and inner membrane that hampers mitochondrial fusion. It is possible that the ΔΨm is required by yet unknown proteins to catalyze the fusion of the inner membrane, maybe via ΔΨm-dependent conformational changes. Alternatively, dissipation of ΔΨm may induce modifications of mitochondrial structure that inhibit the function or the coordination of the fusion machinery. Indeed, changes in the energetic state of mitochondria induce severe modifications in the matrix compartment (Hackenbrock, 1968) and affect the size and frequency of contact sites between the inner and outer mitochondrial membranes (Knoll and Bridczka, 1983; Biermans et al., 1990). Interestingly, yeast Fzo1p is enriched in contact sites and mutant Fzo1p molecules that fail to enrich in contact sites loose the capacity to mediate fusion (Fritz et al., 2001).
Mitochondrial fusion was abolished upon dissipation of ΔΨm and partially inhibited by treatments that lowered ATP levels. It is thus possible that kinetics and efficiency of mitochondrial fusion are modulated in vivo by ΔΨm and/or ATP level. We found that the proportion of mitochondrial energy supply is relatively low in cultured HeLa cells (around 20%), in contrast to aerobic tissues in vivo, where mitochondrial respiration is the major source of ATP (Tzagoloff, 1982). The dependence of mitochondrial fusion on ΔΨm and ATP level indicates that its efficiency may be lowered in tissues of patients with mitochondrial diseases (Leonard and Schapira, 2000a,b). This may explain why complementation by wild-type mtDNA is hampered at high doses of mutant mtDNA in vivo (Boulet et al., 1992; Nakada et al., 2001). In contrast, fusion capacity should not be affected in clones of ρ0-cells repopulated with mutant mtDNA (Bakker et al., 2000; Enriquez et al., 2000; Ono et al., 2001), because they are expected to maintain a ΔΨm similar to that of parental ρ0-cells (Buchet and Godinot, 1998). Therefore, mitochondrial fusion should not be the limiting factor for functional complementation between mitochondria carrying different mutants of mtDNA. Further studies are necessary to identify and understand the factors that render complementation rare (Enriquez et al., 2000) or relatively slow (Ono et al., 2001). They may be the diffusion and exchange of complementing molecules (mtDNA, RNAs, and membrane proteins) and/or the synthesis and assembly of functional respiratory complexes.
The ability to specifically inhibit fusion (by dissipation of ΔΨm) or fission (by expression of dominant-negative Drp1K38A) and the changes induced by these treatments demonstrated that, also in mammals, mitochondrial morphology is determined by the balance of both reactions. The variability of mitochondrial length and interconnectivity in different cells and tissues probably reflects differences in the relative rates of fusion and fission. In yeast, this balance is maintained by the constitutive activity of Fzo1p and Dnm1p (Bleazard et al., 1999; Sesaki and Jensen, 1999). The function of the human orthologues (Mfn1, Mfn2, and Drp1) is less well known. The role of human Drp1 in fission has been inferred from 1) the increased mitochondrial connectivity in cells transfected with Drp1K38A, and 2) the enrichment of Drp1 at sites of mitochondrial division (Smirnova et al., 2001). The fragmentation of mitochondrial filaments upon inhibition of fusion and the capacity of Drp1K38A to inhibit fragmentation demonstrate that mammalian Drp1 is directly involved in mitochondrial division. In this work, expression of Mfn1 increased the length and interconnectivity of mitochondria in a specific and significant manner. The requirement of the inner membrane potential for the appearance and the maintenance of elongated and interconnected mitochondria demonstrate that overexpressed Mfn1 stimulates fusion. Together, these results show that Drp1 and mitofusins are the functional orthologues of yeast Fzo1p and Dnm1p and that the balance of their antagonizing activities determines mitochondrial morphology in human cells.
The efficacy of mitochondrial fusion in cultured cells, the ubiquitous expression of mitofusins (Rojo et al., 2002), and the filamentous morphology of mitochondria in several tissues (Tzagoloff, 1982) suggest that mitochondrial fusion is a constitutive and ubiquitous process in mammals. Nevertheless, it may well be that the inhibition of fusion or the stimulation of fission provokes mitochondrial fragmentation during certain stages of development and of the cell cycle. This would have important consequences for the transmission, segregation, and/or fixation of heteroplasmic mtDNA, the mechanisms of which are largely unknown (Lightowlers et al., 1997). The disintegration of mitochondrial filaments by Drp1 is also observed during apoptosis (Frank et al., 2001). Our findings also predict that mitochondrial fusion is inhibited during apoptosis, when the opening of the permeability transition pore dissipates ΔΨm (Bernardi et al., 2001). Further work will be necessary to investigate the relative contributions of stimulated fission and/or inhibited fusion to the fragmentation of mitochondria during apoptosis.
In contrast to Mfn2, which mediates massive clustering of mitochondria (Santel and Fuller, 2001; Rojo et al., 2002), Mfn1 was able to stimulate mitochondrial fusion. At present, we ignore what confers different properties to these highly conserved molecules (60% identity and 77% similarity). Stimulation of fusion was mainly observed in cells expressing intermediate levels of Mfn1. Thus, mitochondrial clustering by Mfn may become dominant at high levels of expression and mask the profiles that suggest increased mitochondrial fusion. Mfn2 may have a higher capacity to cluster mitochondria (and mask stimulated fusion) or a lower ability to stimulate fusion. Alternatively, overexpression of mitofusin(s) alone may not be sufficient to increase the activity the fusion machinery, which probably involves numerous proteins. Further work is necessary to understand mitochondrial fusion and whether and how it resembles membrane fusion in other systems (Jahn and Südhof, 1999; Heiman and Walter, 2000; Peters et al., 2001).
Supplementary Material
MBC Videos
ACKNOWLEDGMENTS
We are grateful to Frank Perez for valuable advice in live microscopy; Pietro De Camilli for OMP25-plasmids; Valérie Allamand, Bruno Miroux, and Franck Perez for critical reading of the manuscript; and Ketty Schwartz for support and constructive discussions. Research in the laboratory of A.L. is supported by Institut National de la Santé et de la Recharche Médicale and by grants from Association Française contre les Myopathies. M.R. is supported by Centre National de la Recherche Scientifique and previously by postdoctoral fellowships from Institut National de la Santé et de la Recharche Médicale and from Association Française contre les Myopathies
Abbreviations used:
cccp
carbonyl cyanide m-chlorophenyl hydrazone
ΔΨm
mitochondrial inner membrane potential
fccp
carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone
GFP
green fluorescent protein
GFPOM
green fluorescent protein targeted to the mitochondrial outer membrane
mtDNA
mitochondrial DNA
mtGFP/mtRFP
green fluorescent protein/red fluorescent protein targeted to the mitochondrial matrix
PEG
polyethylene glycol
RFP
red fluorescent protein DsRed1
Footnotes
V⃞
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
REFERENCES
- Amchenkova AA, Bakeeva LE, Chentsov YS, Skulachev VP, Zorov DB. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J Cell Biol. 1988;107:481–495. doi: 10.1083/jcb.107.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Butow RA. Patterns of mitochondrial sorting in yeast zygotes. Mol Biol Cell. 1993;4:21–36. doi: 10.1091/mbc.4.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakeeva LE, Chentsov YS, Skulachev VP. Mitochondrial framework (reticulum mitochondriale) in rat diaphragm muscle. Biochim Biophys Acta. 1978;501:349–369. doi: 10.1016/0005-2728(78)90104-4. [DOI] [PubMed] [Google Scholar]
- Bakker A, Barthelemy C, Frachon P, Chateau D, Sternberg D, Mazat JP, Lombes A. Functional mitochondrial heterogeneity in heteroplasmic cells carrying the mitochondrial DNA mutation associated with the MELAS syndrome (mitochondrial encephalopathy, lactic acidosis, and strokelike episodes) Pediatr Res. 2000;48:143–150. doi: 10.1203/00006450-200008000-00005. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Bernardi P, Petronilli V, Di Lisa F, Forte M. A mitochondrial perspective on cell death. Trends Biochem Sci. 2001;26:112–117. doi: 10.1016/s0968-0004(00)01745-x. [DOI] [PubMed] [Google Scholar]
- Biermans W, Bakker A, Jacob W. Contact site between inner and outer mitochondrial membrane: a dynamic microcompartment for creatine kinase activity. Biochim Biophys Acta. 1990;1018:225–228. doi: 10.1016/0005-2728(90)90254-2. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Boulet L, Karpati G, Shoubridge EA. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF) Am J Hum Genet. 1992;51:1187–1200. [PMC free article] [PubMed] [Google Scholar]
- Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J Biol Chem. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- Church C, Poyton RO. Neither respiration nor cytochrome c oxidase affects mitochondrial morphology in saccharomyces cerevisiae. J Exp Biol. 1998;201:1729–1737. doi: 10.1242/jeb.201.11.1729. [DOI] [PubMed] [Google Scholar]
- Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21:1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condreay JP, Witherspoon SM, Clay WC, Kost TA. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc Natl Acad Sci USA. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgi F, Lartigue L, Ichas F. Electrical coupling and plasticity of the mitochondrial network. Cell Calcium. 2000;28:365–370. doi: 10.1054/ceca.2000.0177. [DOI] [PubMed] [Google Scholar]
- Diaz G, Setzu MD, Zucca A, Isola R, Diana A, Murru R, Sogos V, Gremo F. Subcellular heterogeneity of mitochondrial membrane potential: relationship with organelle distribution and intercellular contacts in normal, hypoxic and apoptotic cells. J Cell Sci. 1999;112:1077–1084. doi: 10.1242/jcs.112.7.1077. [DOI] [PubMed] [Google Scholar]
- Dujon B. Mitochondrial genetics and functions. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces, Vol. 1: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 505–635. [Google Scholar]
- Enriquez JA, Cabezas-Herrera J, Bayona-Bafaluy MP, Attardi G. Very rare complementation between mitochondria carrying different mitochondrial DNA mutations points to intrinsic genetic autonomy of the organelles in cultured human cells. J Biol Chem. 2000;275:11207–11215. doi: 10.1074/jbc.275.15.11207. [DOI] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Fritz S, Rapaport D, Klanner E, Neupert W, Westermann B. Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J Cell Biol. 2001;152:683–692. doi: 10.1083/jcb.152.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenbrock CR. Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc Natl Acad Sci USA. 1968;61:598–605. doi: 10.1073/pnas.61.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Preston G, Andrews R, Chinnery P, Lightowlers RN, Turnbull DM, Kubacka I, Howell N. A high frequency of mtDNA polymorphisms in HeLa cell sublines. Mutat Res. 2002;501:19–28. doi: 10.1016/s0027-5107(01)00304-9. [DOI] [PubMed] [Google Scholar]
- Hoshikawa Y, Kwon HJ, Yoshida M, Horinouchi S, Beppu T. Trichostatin A induces morphological changes and gelsolin expression by inhibiting histone deacetylase in human carcinoma cell lines. Exp Cell Res. 1994;214:189–197. doi: 10.1006/excr.1994.1248. [DOI] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll G, Bridczka D. Changes in freeze-fractured mitochondrial membranes correlated to their energetic state. Dynamic interactions of the boundary membranes. Biochim Biophys Acta. 1983;733:102–110. doi: 10.1016/0005-2736(83)90095-0. [DOI] [PubMed] [Google Scholar]
- Leonard JV, Schapira AH. Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet. 2000a;355:299–304. doi: 10.1016/s0140-6736(99)05225-3. [DOI] [PubMed] [Google Scholar]
- Leonard JV, Schapira AH. Mitochondrial respiratory chain disorders II: neurodegenerative disorders and nuclear gene defects. Lancet. 2000b;355:389–394. doi: 10.1016/s0140-6736(99)05226-5. [DOI] [PubMed] [Google Scholar]
- Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- Lim ML, Minamikawa T, Nagley P. The protonophore CCCP induces mitochondrial permeability transition without cytochrome c release in human osteosarcoma cells. FEBS Lett. 2001;503:69–74. doi: 10.1016/s0014-5793(01)02693-x. [DOI] [PubMed] [Google Scholar]
- Maro B, Bornens M. Reorganization of HeLa cell cytoskeleton induced by an uncoupler of oxidative phosphorylation. Nature. 1982;295:334–336. doi: 10.1038/295334a0. [DOI] [PubMed] [Google Scholar]
- Medina V, Edmonds B, Young GP, James R, Appleton S, Zalewski PD. Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Res. 1997;57:3697–3707. [PubMed] [Google Scholar]
- Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, Hayashi JI. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Park MK, Ashby MC, Erdemli G, Petersen OH, Tepikin AV. Perinuclear, perigranular and sub-plasmalemmal mitochondria have distinct functions in the regulation of cellular calcium transport. EMBO J. 2001;20:1863–1874. doi: 10.1093/emboj/20.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Pitts KR, Yoon Y, Krueger EW, McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Rojo M, Emery G, Marjomaki V, McDowall AW, Parton RG, Gruenberg J. The transmembrane protein p23 contributes to the organization of the Golgi apparatus. J Cell Sci. 2000;113:1043–1057. doi: 10.1242/jcs.113.6.1043. [DOI] [PubMed] [Google Scholar]
- Rojo M, Legros F, Chateau D, Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton RG, Gruenberg J. Involvement of the transmembrane protein p23 in biosynthetic protein transport. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus Fusion: Dnm1p and Fzo1p Antagonistically Regulate Mitochondrial Shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore GC, McBride HM, Millar DG, Steenaart NA, Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. Eur J Biochem. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Shoubridge EA. Mitochondrial DNA diseases: histological and cellular studies. J Bioenerg Biomembr. 1994;26:301–310. doi: 10.1007/BF00763101. [DOI] [PubMed] [Google Scholar]
- Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW, Steele GD, Bo Chen L. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci USA. 1991;88:3671–3675. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van Der Bliek AM. Dynamin-related protein drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B. Mitochondrial structure. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces, Vol. 1: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 471–504. [Google Scholar]
- Tzagoloff A. Mitochondria. New York: Plenum Press; 1982. [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MBC Videos