The Transformation Suppressor Pdcd4 Is a Novel Eukaryotic Translation Initiation Factor 4A Binding Protein That Inhibits Translation (original) (raw)
Abstract
Pdcd4 is a novel transformation suppressor that inhibits tumor promoter-induced neoplastic transformation and the activation of AP-1-dependent transcription required for transformation. A yeast two-hybrid analysis revealed that Pdcd4 associates with the eukaryotic translation initiation factors eIF4AI and eIF4AII. Immunofluorescent confocal microscopy showed that Pdcd4 colocalizes with eIF4A in the cytoplasm. eIF4A is an ATP-dependent RNA helicase needed to unwind 5′ mRNA secondary structure. Recombinant Pdcd4 specifically inhibited the helicase activity of eIF4A and eIF4F. In vivo translation assays showed that Pdcd4 inhibited cap-dependent but not internal ribosome entry site (IRES)-dependent translation. In contrast, Pdcd4D418A, a mutant inactivated for binding to eIF4A, failed to inhibit cap-dependent or IRES-dependent translation or AP-1 transactivation. Recombinant Pdcd4 prevented eIF4A from binding to the C-terminal region of eIF4G (amino acids 1040 to 1560) but not to the middle region of eIF4G(amino acids 635 to 1039). In addition, both Pdcd4 and Pdcd4D418A bound to the middle region of eIF4G. The mechanism by which Pdcd4 inhibits translation thus appears to involve inhibition of eIF4A helicase, interference with eIF4A association-dissociation from eIF4G, and inhibition of eIF4A binding to the C-terminal domain of eIF4G. Pdcd4 binding to eIF4A is linked to its transformation-suppressing activity, as Pdcd4-eIF4A binding and consequent inhibition of translation are required for Pdcd4 transrepression of AP-1.
Initiation of protein synthesis in eukaryotic cells is a multistep process leading to the assembly of ribosomes and Met-tRNAi at the initiation codon of an mRNA (12, 14). The rate-limiting step of this process is the binding of the 40S ribosomal subunit to mRNA. Several eukaryotic translation initiation factors (eIFs), including the eIF4F complex, participate in this process. Translation initiation factor eIF4F is a multiple-subunit complex comprising eIF4A, eIF4E, and eIF4G.
eIF4A is an ATP-dependent RNA helicase belonging to the DEAD box protein family (25) that has nine highly conserved motifs shared with other DEAD box proteins. The RNA helicase activity of eIF4A is further increased by eIF4B, eIF4H, or as a subunit of eIF4F (1, 41, 43, 45). Mutations in the nine motifs greatly reduce the RNA binding ability, ATPase activity, or helicase activity of eIF4A and produce inhibition of translation (37). eIF4A is thought to catalyze the unwinding of mRNA secondary structure at the 5′ untranslated region, allowing the 40S ribosomal subunit to bind the mRNA and scan in a 5′-to-3′ direction, searching for the initiation codon (14). In mammals, three eIF4A isoforms have been identified. eIF4AI and eIF4AII encoded by two different genes are highly (91%) identical in amino acid sequence (33). eIF4AI and eIF4AII are functionally indistinguishable and exchangeable, performing similar kinetics of incorporation into eIF4F (50). The third factor, eIF4AIII, is less identical to eIF4AI (≈65%) and functions as a translation inhibitor (23).
eIF4G functions as a scaffold containing several translation initiation factor binding sites, including the sites for cap-binding protein eIF4E (27) and for eIF4A (16). eIF4E is required for cap-dependent translation and binds to the N-terminal one-third of eIF4G (amino acids 1 to 634). Cleavage of this domain from eIF4G results in inhibition of cap-dependent translation (13). Two eIF4A binding sites in eIF4G are located within the middle one-third (amino acids 635 to1039) and the C-terminal one-third (amino acids 1040 to 1560) (32). The middle one-third of eIF4G is sufficient for cap-independent 5′-end-dependent translation (8) and internal ribosome entry site (IRES)-mediated translation (26). The C-terminal one-third of eIF4G has been reported to serve as a regulatory domain for translation (32).
Pdcd4 was found in a differential display analysis of mouse epidermal JB6 variants to be highly expressed in transformation-resistant (P−) but not in transformation-susceptible (P+) cells (5). Expression of the pdcd4 gene is upregulated during apoptosis in response to several inducers (46) and downregulated by topoisomerase inhibitor treatment (34). No causal relationship to apoptosis or to topoisomerase inhibitor-induced cytotoxicity has been reported. The reduction of Pdcd4 in P− cells by overexpression of antisense pdcd4 is accompanied by acquisition of a transformation-susceptible phenotype (5). Conversely, overexpression of sense pdcd4 in stably transfected P+ cells renders them resistant to tumor promoter-induced transformation, indicating that elevated expression of Pdcd4 protein is sufficient to inhibit transformation (49).
In order to elucidate the molecular target(s) of Pdcd4, we performed a yeast two-hybrid analysis with pdcd4 cDNA as bait. Translation initiation factors eIF4AI and eIF4AII were identified as binding partners of Pdcd4. Pdcd4 inhibits eIF4A helicase activity and inhibits translation in vivo. Interestingly, Pdcd4 prevents eIF4A from binding to the C-terminal region of eIF4G, and Pdcd4 itself binds to the middle region of eIF4G, suggesting that Pdcd4 functions as a novel regulatory factor for translation initiation.
MATERIALS AND METHODS
Construction of plasmids.
pdcd4 cDNA was excised from pcDNA-Pdcd4 (49) and ligated into the _Eco_RI and _Sma_I sites of the pGBKT7 DNA binding domain vector (Clontech), and named pGBKT7-Pdcd4. This plasmid was the bait construct used in the yeast two-hybrid assay. Plasmids pAcHLT-A-Pdcd4 and pAcGHLT-A-Pdcd4 were used for generating recombinant His-Pdcd4 and glutathione _S_-transferase (GST)-Pdcd4, respectively, in Sf-9 cells. pdcd4 cDNA was inserted into the _Eco_RI and _Pst_I sites of the pAcHLT-A or pAcGHLT-A vector (Pharmingen). The plasmid Xpress-eIF4A was used to generate an Xpress-tagged eIF4A. eIF4A cDNA was inserted into the _Bam_HI and _Xho_I sites of pcDNA4/Max (Invitrogen). Plasmids pCMV-BD-Pdcd4 and pCMVAD-eIF4A were used for mammalian two-hybrid analysis with pdcd4 cDNA and eIF4A cDNA inserted into the _Eco_RI and _Xba_I sites of the pCMV-DB and the _Bam_HI and _Xho_I sites of the pCMV-AD vectors (Stratagene), respectively. All constructs were subsequently sequenced to confirm in-frame fusion of pdcd4.
Yeast two-hybrid screen and assay.
pGBKT7-Pdcd4 was transformed into yeast strain PJ69-2A and mated with the pretransformed mouse brain Matchmaker cDNA library (Clontech) according to the manufacturer's protocol. The yeast cells were selected by their growth on Leu-, Try-, His-, and Ade-free plates for 7 days. The colonies from these plates were subsequently assayed for β-galactosidase expression three times with a β-galactosidase liftover assay according to the manufacturer's protocol (Clontech). The library plasmids from Leu+/Trp+/His+/Ade+ β-galactosidase-positive clones were isolated and sequenced.
Recombinant protein expression and purification.
His-Pdcd4 or GST-Pdcd4 was expressed in insect Sf-9 cells. Sf-9 cells were infected with recombinant virus at a multiplicity of infection of 5 and cultured for 72 h at 27°C. Cells were then lysed in lysis buffer [25 mM sodium phosphate (pH 8), 300 mM NaCl, 1% Triton X-100], and expressed proteins were purified by glutathione-Sepharose resins (Pharmingen) or Ni-agarose affinity resins (Qiagen). The protein concentrations were determined by comparison of band intensity with bovine serum albumin as the standard on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) following staining with Coomassie blue.
His-eIF4A, GST-eIF4G(672-1065), and GST-eIF4G(1201-1445) were expressed in Escherichia coli BL21(DE3) and purified on Ni-agarose resin (Qiagen) as described previously (32, 38).
Immunoprecipitation and pull-down assays.
JB6 P+ cells were transfected with plasmid pcDNA-Pdcd4 (49) or Xpress-eIF4A plasmid with Lipofectamine (Invitrogen). After 48 h, cells were harvested and lysed in lysis buffer [20 mM HEPES-KOH (pH 7.6), 100 mM KCl, 0.5 mM EDTA, 20% glycerol, 0.5% Triton X-100, 50 μg of RNase A per ml, and 1× protease inhibitor cocktail (Boehringer Mannheim)]. For immunoprecipitation, eIF4A antibody (0.1 volume of supernatant) was added to cell lysates from cells transfected with plasmid pcDNA-Pdcd4 and incubated for 1 h at 4°C. Ten microliters of protein A-Sepharose beads (CL-4B; Phamacia) were washed with 250 μl of lysis buffer twice, added to the cell lysates, and rotated for 2 h at 4°C.
For the GST pull-down assay, 50 μg of recombinant GST-Pdcd4 was added to lysates of JB6 P+ cells that had been transiently transfected with Xpress-eIF4A plasmid and incubated for 1 h at 4°C. Ten microliters of glutathione-Sepharose beads (Pharmingen) were washed with 250 μl of lysis buffer twice, added to the cell lysates, and rotated for 2 h at 4°C. The beads were washed three times with 250 μl of lysis buffer and boiled in SDS sample buffer. The bound proteins were resolved by SDS-PAGE and detected by Pdcd4 or Xpress antibodies.
Mammalian two-hybrid assay of protein-protein binding.
In the mammalian two-hybrid assay, a luciferase reporter becomes activated when a DNA binding domain (BD) fusion protein binds to an activation domain (AD) fusion protein. pdcd4 and eIF4A cDNAs were inserted into pCMV-BD (Stratagene) and pCMV-AD (Stratagene), respectively. Then 104 JB6 P+ cells were seeded in 24-well plates in Eagle's minimal essential medium (EMEM) with 4% fetal bovine serum. Cells were transfected with pCMV-BD-Pdcd4 (5 to 50 ng), pCMV-AD-eIF4A (5 to 50 ng), Gal4-luciferase reporter gene (25 ng), and thymidine kinase (TK)-Renilla luciferase gene (10 ng) with 2 μl of Lipofectamine (Invitrogen) for 4 h, and cells were then incubated with fresh EMEM with 4% fetal bovine serum. After 48 h, cells were lysed in 1× passive lysis buffer (Promega), and luciferase activity was measured as previously described (49).
Immunohistochemistry analysis of Pdcd4 and eIF4A localization.
JB6 P− cells were grown on no. 1 glass coverslips in EMEM containing 4% fetal bovine serum, washed twice in phosphate-buffered saline, fixed in 4% paraformaldehyde, and permeabilized in 0.5% Triton X-100. Cells were incubated with a 1:200 dilution of a rabbit polyclonal anti-eIF4A antibody. After being washed three times with phosphate-buffered saline, the slides were incubated with goat anti-rabbit immunoglobulin G coupled to fluorescein isothiocyanate (Molecular Probes). After being washed three more times, the cells were incubated with anti-Pdcd4 antibody previously labeled with Alexa Fluor 568 (Molecular Probes) at a dilution of 1:100. The cells were washed twice with phosphate-buffered saline, and nuclei were stained with 4′,6′-diamidino-2-phenylindole (1:10,000). The cells were washed twice with phosphate-buffered saline and then mounted with antifading mounting fluid (Prolong; Molecular Probes). Fluorescence images were obtained with the 63× objective of a Zeiss LSM 410 confocal microscope at the National Cancer Institute Image Analysis Lab.
For quantification, we used image analysis to detect and quantify the colocalization. The analysis consisted of first calculating the probability that real colocalization of the two proteins existed and, if so, then calculating the amount of colocalization of each protein relative to the other. The first step in the analysis was to crop the image to the region where both proteins were allowed to exist. In this case the region was the cytoplasm of a cell. In the following text, “image” refers only to this region.
The probability that real colocalization existed, in addition to the apparent random colocalization seen when the signals from two proteins overlapped in the color image, was calculated as follows. First, the Pearson correlation coefficient (r) of the two images, which measures the degree of similarity of the patterns of the proteins in the two images, was calculated (30). The r value ranges from 0 to 1; 1 indicates a high similarity of the patterns, suggesting real colocalization of the two proteins, whereas 0 indicates only random colocalization. Next, the spatial arrangement of the protein pattern in one of the images was randomized in order to destroy any real (nonrandom) colocalization between the two proteins, followed by calculating r again between the randomized region and the region in the image of the other protein (_r_ran). This process was repeated hundreds of times in order to obtain a distribution of _r_ran values. In the final step, the initial r from the pair of images (that were not randomized) was compared to the distribution of _r_ran values. If r was significantly higher than the _r_ran values, it can be concluded that there is a significant probability that real colocalization was present. The probability for each cell that the colocalization was the result of a random overlap was estimated to be less than 0.1%.
Because it was concluded that real colocalization was present, the amount and locations of real colocalization were estimated based on the assumption that positions where the proteins were colocalized most likely corresponded to locations in the images where the same point in both images had high intensity. (A point in an image is called a pixel, and in fluorescence images the intensity at a given pixel is approximately proportional to the concentration of protein at the equivalent position in the specimen.) The estimation was performed by successively removing pixel pairs from the images, starting with the pair with the highest intensity and stopping when r calculated for the remaining pixels was 0 (implying that only random colocalization remained). The pixels removed were marked as those corresponding to real protein colocalization. Summing the intensities of the colocalized pixels in its image and dividing the sum by the intensities of all the pixels in its image calculated the degree of colocalization for each protein.
Helicase activity assay.
The unwinding of duplex RNA was performed as described previously (44). In brief, eIF4A was incubated with or without Pdcd4 in a 20-μl reaction which contained 2 nM RNA duplex, 1 mM ATP, 1 mM MgCl2, 20 mM HEPES-KOH (pH 7.5), 70 mM KCl, 2 mM dithiothreitol, and 1 mg of bovine serum albumin per ml. The sequence of the long RNA strand was5′-GGGAGAAAAACAAAACAAAACAAAACUAGCACCGUAAAGCACGC-3′, and the sequence of the short, 32P-radiolabeled strand was 5′-GCUUUACGGUGC-3′ or 5′-GCUUUACGGUGCU-3′. The duplex contained 12 bp or 13 bp.
Reaction mixes were incubated for 15 min at 35°C, and the reactions were stopped with 5 μl of a solution containing 50% glycerol, 2% SDS, 20 mM EDTA, and 0.05% each bromophenol blue and xylene cyanol dyes. Duplex and single strands were resolved by gel electrophoresis on 15% native polyacrylamide gels at 4°C for about 2 h at 200 V in 1× TBE (Tris-borate-EDTA) buffer. Radioactivity was determined with an Ambis radioanalytic scanner, and the resulting data were quantified as previously described (44). eIF4A, eIF4B, and eIF4F were purified from rabbit reticulocyte lysate. Yeast Ded1p was provided by Eckhard Jankowsky (Department of Biochemistry, Case Western Reserve University). Analysis of Pdcd4 alone indicated that there was no nuclease or phosphatase contamination (data not shown).
In vitro translation.
Ten microliters of nuclease-treated rabbit reticulocyte lysate (Promega) were mixed with 1 μl of SUPERase In (40 U, Ambion), 1 μl of an amino acid mixture lacking methionine (1 mM each of the other amino acids), 0.2 μg of bicistronic CAT/EMCV/LUC (47) mRNA, 15 μCi of [35S]methionine, and His-Pdcd4 protein (0 to 4.8 μg). The bicistronic CAT/EMCV/LUC mRNA contains chloramphenicol acetyltransferase (CAT) and luciferase reporter genes. Translation of the CAT open reading frame is cap dependent, whereas the luciferase open reading frame is encephalomyocarditis virus (EMCV) IRES dependent.
The reaction mixture was added to buffer A [20 mM HEPES-KOH (pH 7.6), 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol] to a final volume of 20 μl and incubated at 30°C for 1 h. The products of translation were resolved by SDS-PAGE, fixed with 40% methanol-7% acetic acid, and treated with Amplify (Amersham). The intensity of bands was determined by a Storm 850 Phosphorimager (Molecular Dynamics).
Transient-transfection assays of in vivo translation and of AP-1-dependent transcription.
An in vivo translation assay of the bicistronic reporter system was based on that described previously (28). Briefly, 5 × 104 JB6 RT101 cells were seeded in a six-well plate in EMEM with 4% fetal bovine serum. After transfection with 0 to 2 μg of pcDNA-Pdcd4 (or pcDNA-Pdcd4D418A), 0.2 μg of the bicistronic reporter system, pcDNA-CAT/EMCV/LUC, and 10 ng of the TK-Renilla luciferase gene, cells were allowed to recover for 12 to 18 h in EMEM with 4% fetal bovine serum. Cells were then serum starved with 0.2% fetal bovine serum in EMEM for 24 h and incubated in EMEM with 4% fetal bovine serum for an additional 24 h. Cells were harvested for CAT assay and luciferase assay. CAT activity was measured with the Quan-T-CAT kit (Amersham). The radioactivity was determined with a Beckman LS 3801. Luciferase activity was measured as described above. Transfection efficiency was normalized to Renilla luciferase activity.
Transient transfection of the 4× AP-1-luciferase reporter and assay of tumor promoter-induced activation of AP-1-dependent transcription were done as previously described (49). Transfection efficiency was normalized to Renilla luciferase activity. In this assay, nonresponsiveness of other promoter-luciferase constructs establishes that transcription, not luciferase translation, is being measured.
In vitro binding assay.
Fifty micrograms of GST-eIF4G(672-1065) or GST-eIF4G(1201-1445) recombinant protein immobilized on a 10-μl bed volume of glutathione-Sepharose resin was incubated with 5 μg of His-eIF4A in the presence or absence of 5 μg of His-Pdcd4 in 10 μl of binding buffer [20 mM Tris-HCl (pH 7.5), 100 mM KCl, 2.5 mM Mg Cl2, 0.1 mM EDTA, 10% glycerol, 0.4% Triton X-100] on ice for 20 min. The beads were washed three times with 250 μl of binding buffer and boiled in SDS sample buffer. The bound proteins were resolved by SDS-PAGE and detected by immunoblot with penta-His antibody (Qiagen), GST antibody (Santa Cruz Biotechnology), or Pdcd4 antibody, as indicated. The Pdcd4 peptide antibody recognizes a single band of 64 kDa. This Pdcd4 antibody has high specificity, since this 64-kDa band was erased by the peptide that was used to generate this antibody. (49).
RESULTS
Identification of eIF4A as a Pdcd4-interacting protein in a yeast two-hybrid screen.
Overexpression of Pdcd4 in JB6 P+ cells suppresses tetradecanoyl phorbol acetate-induced transformation and inhibits the AP-1 activation required for transformation (49). However, ectopic introduction of AP-1 proteins (Fra-1, JunD, or c-Jun) does not relieve inhibition of AP-1-dependent transcription by Pdcd4. In addition, recombinant GST-Pdcd4 did not pull down Fra-1 or c-Fos from tetradecanoyl phorbol acetate-treated P+ cell lysates (data not shown), indicating that Pdcd4 does not physically interact with AP-1 proteins.
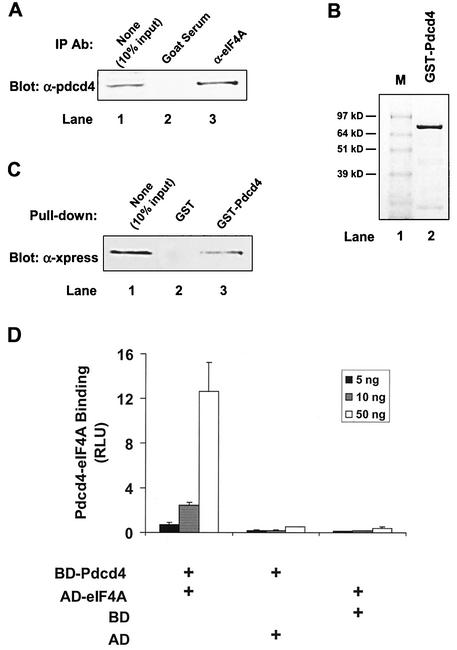
In order to identify the binding partner(s) of Pdcd4, we performed a yeast two-hybrid screen of a mouse brain MATCHMAKER cDNA library with full-length Pdcd4 as the bait. After several iterations of β-galactosidase liftover assays, the DNA sequences of clones 1 and 19 of 31 positive clones were identical to translation initiation factor eIF4AI and eIF4AII, respectively. Since eIF4AII is preferentially expressed in the brain (33), the higher number of eIF4AII-positive clones observed was expected. To further confirm this interaction, we performed a coimmunoprecipitation with eIF4A antibody. As shown in Fig. 1A, eIF4A antibody but not preimmune serum coprecipitated Pdcd4 from lysates of (low-Pdcd4) JB6 P+ cells transfected with the Pdcd4 expression plasmid. We generated recombinant GST-Pdcd4 from baculovirus-infected Sf-9 cells and purified it with glutathione-Sepharose (Fig. 1B). GST-Pdcd4 was able to pull down eIF4A from lysates of JB6 P+ cells transfected with the Xpress-tagged eIF4A expression plasmid (Fig. 1C).
FIG. 1.
Identification of Pdcd4 binding to eIF4A. (A) Coimmunoprecipitation of Pdcd4 with eIF4A. JB6 P+ cell lysates isolated following transient transfection with Pdcd4 expression plasmid were immunoprecipitated (IP) with goat serum (lane 2) or eIF4A antibody (Ab) (lane 3). The immunoprecipitates were resolved by SDS-10% PAGE followed by immunoblotting with Pdcd4 antibody. Lane 1 shows one-tenth of the cell lysates. (B) Coomassie blue staining of GST-Pdcd4. One microgram of GST-Pdcd4 expressed in SF-9 cells and purified from a glutathione column as described in Materials and Methods was loaded onto SDS-PAGE (10%) and stained with SimpleBlue (Invitrogen). Lane M, protein molecular size markers. (C) GST pull-down of eIF4A with Pdcd4. JB6 P+ cell lysates isolated following transient transfection with Xpress-tagged eIF4A expression plasmid were pulled down with GST (lane 2) or GST-Pdcd4 (lane 3). The bound proteins were resolved by SDS-10% PAGE followed by immunoblotting with Xpress antibody. Lane 1 shows one-tenth of the cell lysates. (D) Mammalian two-hybrid assay of Pdcd4 binding to eIF4A. Various amounts (5, 10, and 50 ng) of plasmid pCMV-BD-Pdcd4 (or its empty vector, pCMV-DB) and pCMV-AD-eIF4A (or its empty vector, pCMV-AD) along with the Gal4-luciferase reporter gene were cotransfected into JB6 P+ cells. After 48 h, cells were lysed, and the luciferase activity was measured. The luciferase activity from the cells with 5 ng of pCMV-BD-Pdcd4 and 5 ng of pCMV-AD-eIF4A was designated as 1. These experiments were repeated three times, each with five independent transfections, and representative data are shown. Results are expressed as the mean ± standard deviation. RLU, relative luciferase units.
To demonstrate that the association of Pdcd4 and eIF4A occurred in vivo, we used a mammalian two-hybrid system. The pdcd4 cDNA and eIF4A cDNA were fused with the Gal4 DNA-binding domain (pCMV-BD) and NF-κB activation domain (pCMV-AD), respectively. Since both plasmids pCMV-BD and pCMV-AD contain nuclear localization signals, Gal4-Pdcd4 and NF-κB-eIF4A fusion proteins are able to translocate into nuclei. As shown in Fig. 1D, the level of luciferase expression was greatly increased when pCMV-BD-Pdcd4 (bait) and pCMV-AD-eIF4A (prey) were cotransfected. Transfection of pCMV-BD-Pdcd4 along with pCMV-AD (empty vector) or of pCMV-BD (empty vector) with pCMV-AD-eIF4A showed only the background level of luciferase activity. These results indicate that Pdcd4 and eIF4A physically interact in vivo and in vitro.
Pdcd4 colocalizes with eIF4A in the cytoplasm.
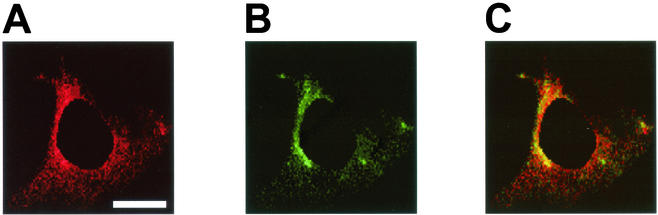
Human H731 is a homolog of Pdcd4 reported to be expressed in the cytoplasm, the nucleus, or both (51, 52). However, based on the data shown above, we would expect Pdcd4 to associate with eIF4A in the cytoplasm. In order to address this question and to further confirm the Pdcd4-eIF4A interaction in vivo, we used multicolor confocal immunofluorescence microscopy to ascertain the subcellular localizations of Pdcd4 and eIF4A in JB6 P− cells. Both red-fluorescent Pdcd4 (Fig. 2A) and green-fluorescent eIF4A (Fig. 2B) displayed diffuse cytoplasmic expression with concentrated perinuclear distribution. Although all cells displayed colocalization in the cytoplasm, the perinuclear region appeared to have the most concentrated colocalization (Fig. 2C). The antibody binding to Pdcd4 and eIF4A was successfully erased by recombinant His-Pdcd4 protein and native eIF4A protein purified from rabbit reticulocyte lysate, respectively (data not shown).
FIG. 2.
Immunofluorescent detection of colocalization of Pdcd4 and eIF4a in JB6 P− cells. P− cells were immunostained for Pdcd4 (A) and eIF4A (B) and viewed by confocal microscopy. (C) The merged images of panels A and B display a yellow color indicative of colocalization of the two proteins to the perinuclear region of the cytoplasm. The cell shown is a representative example of multiple P− cells. Of seven randomly selected cells, 63% ± 5% of total Pdcd4 was colocalized with the eIF4A, and 65% ± 6% of total eIF4A was colocalized with Pdcd4 in the cytoplasm. The nucleus, which did not show either Pdcd4 or eIF4A staining, was masked for the purpose of limiting the area and therefore the stringency by which the spatial statistical algorithm could test whether the colocalization was the result of random overlap. The estimated probability that the colocalization was due to random overlap was 0.1%. Bar, 15 μm.
Based on the analysis of seven randomly selected cells, the average Pearson correlation in the cytoplasm between the two proteins was 55% ± 11% (30). The Pearson correlation was used as a cutoff value to select which pixels in the cytoplasm were colocalized. Within the cytoplasm, 63% ± 6% of all Pdcd4 proteins and 65% ± 6% of all eIF4A proteins were determined to be compartmentalized within the perinuclear region. Upon selection of the colocalized pixels in the perinuclear region, the approximate percent colocalization for each protein was determined by dividing the sum of all colocalized pixel intensities by the total intensity of each component within the region. By doing so, 54 ± 20% of all Pdcd4 proteins within the perinuclear region were determined to be colocalized with eIF4A. A similar analysis of all eIF4A within the perinuclear region revealed that 58% ± 21% of all eIF4A proteins were colocalized with Pdcd4. Thus, the ratio of Pdcd4-bound eIF4A to free eIF4A was about 1:1. Small shifts away from this Pdcd4 bound to unbound ratio could be functionally significant.
Pdcd4 inhibits the helicase activity of eIF4A.
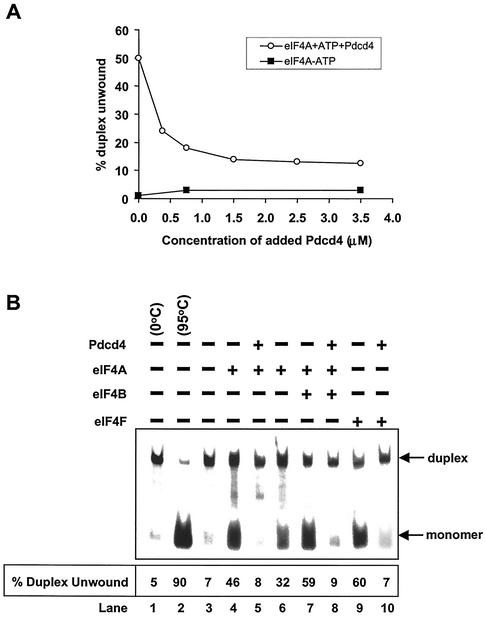
To address whether Pdcd4 alters the eIF4A helicase activity, we performed a helicase activity assay with recombinant His-Pdcd4, native eIF4A, and a synthetic duplex RNA. The helicase activity of eIF4A was determined by measuring the unwinding of a 12- or 13-bp RNA duplex (42, 44). As shown in Fig. 3A, recombinant His-Pdcd4 inhibited eIF4A helicase activity in a concentration-dependent manner. Helicase activity of eIF4A was enhanced by eIF4B or as a subunit of eIF4F (1, 41, 43). However, in the presence of recombinant His-Pdcd4, eIF4B was not able to stimulate the eIF4A helicase activity (Fig. 3B and 3C). Pdcd4 inhibited not only the free eIF4A helicase activity, but also the helicase activity when eIF4A was a subunit of eIF4F (Fig. 3B and 3C). Recombinant His-Pdcd4 inhibited eIF4F helicase activity in a concentration-dependent manner (Fig. 3C). These results indicate that Pdcd4 functions as a dominant eIF4A inhibitor.
FIG. 3.
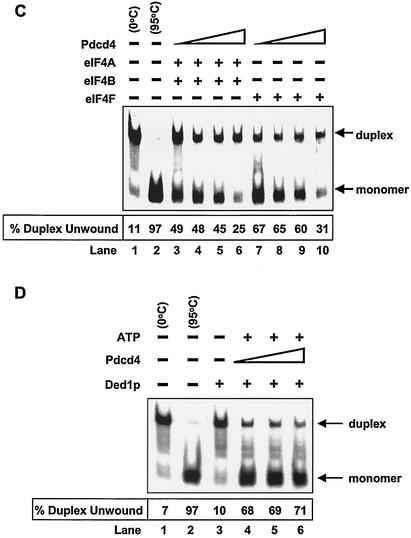
Inhibition of eIF4A RNA helicase activity by Pdcd4. (A) Pdcd4 inhibits eIF4A helicase activity. Unwinding of a 2 nM RNA duplex (12 bp; Δ_G_ = −21.4 kcal/mol) by 1.5 μM eIF4A (open circles) was performed as described in Materials and Methods. As a control (solid squares), eIF4A was incubated with the duplex in the absence of ATP. Separate controls indicated that there was no unwinding by Pdcd4 in the presence of 1 mM ATP (not shown). After the 15-min incubation at 35°C, unwinding was quantitated by gel electrophoresis and subsequent analysis with an Ambis radioanalytic scanner. (B) Pdcd4 inhibits eIF4F helicase activity and eIF4B does not stimulate eIF4A helicase activity in the presence of Pdcd4. A 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) was incubated with the following proteins: 13.5 pmol of eIF4A (lane 4), 13.5 pmol of eIF4A plus 27 pmol of Pdcd4 (lane 5), 6.75 pmol of eIF4A (lane 6), 6.75 pmol of eIF4A plus 6.75 pmol of eIF4B (lane 7), 6.75 pmol of eIF4A plus 6.75 pmol of eIF4B plus 13.5 pmol of Pdcd4 (lane 8), 6.75 pmol of eIF4F (lane 9), or 6.75 pmol of eIF4F plus 13.5 pmol of Pdcd4 (lane 10) for 15 min at 35°C. Samples were separated and quantitated as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C; lane 3, duplex RNA incubated under the same conditions without ATP and proteins for 15 min at 35°C. (C) Pdcd4 inhibits helicase activities of eIF4A plus eIF4B and eIF4F in a concentration-dependent manner. Unwinding of a 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) by eIF4A (6.75 pmol) plus eIF4B (6.75 pmol) without (lane 3) or with increasing concentrations of Pdcd4 (3.4 to 13.5 pmol, lanes 4 to 6) or by eIF4F (6.75 pmol) without (lane 7) or with increasing concentrations of Pdcd4 (3.4 pmol to 13.5 pmol, lanes 8 to 10) was performed as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C. (D) Pdcd4 does not inhibit Ded1p helicase activity. Unwinding of a 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) by Ded1p helicase (0.5 pmol) in the absence (lane 4) or presence (lanes 5 and 6, 1 and 2 pmol, respectively) of Pdcd4 was performed as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C.
In the absence of eIF4A, recombinant His-Pdcd4 was not able to unwind the RNA duplex (in either the presence or absence of ATP), indicating that Pdcd4 does not have helicase activity (data not shown). Recombinant His-Pdcd4 did not inhibit the helicase activity of RNA helicase Ded1p up to a 4:1 Pdcd4-Ded1p molar ratio (Fig. 3D). Ded1p is a DEAD box family helicase that is essential for translation initiation in Saccharomyces cerevisiae (17). In contrast, Pdcd4 appeared to inhibit eIF4A at about a 1:1 molar ratio of eIF4A to His-Pdcd4. At a 4:1 His-Pdcd4 to eIF4A molar ratio, Pdcd4 inhibited the helicase activities of eIF4A and eIF4F by approximately 70% (Fig. 3C, lanes 6 and 10). The inhibition noted in Fig. 3A is similar to that observed with more stable duplexed RNA (i.e., 18 to 20 bp), which was also inhibited at a less than a 1:1 ratio (42). Thus, Pdcd4 specifically suppresses eIF4A helicase activity.
Pdcd4 inhibits translation.
The helicase activity of eIF4A is critical for translation. Mutational inactivation of the helicase activity of eIF4A inhibits protein translation in vitro (36). Since Pdcd4 inhibits the helicase activity of eIF4A (Fig. 3), Pdcd4 would be predicted to inhibit protein translation. A test of this hypothesis measured cap- and IRES-dependent translation in vitro with a capped bicistronic CAT/EMCV/LUC mRNA in a rabbit reticulocyte lysate assay to which recombinant His-Pdcd4 was added. Translation of the CAT open reading frame is cap dependent, whereas translation of the luciferase open reading frame is EMCV IRES-dependent. Since nickel resin-bound proteins purified from uninfected Sf-9 cells did not affect in vitro translation, the proteins copurified with His-Pdcd4 from nickel resin were assumed not to inhibit translation (data not shown).
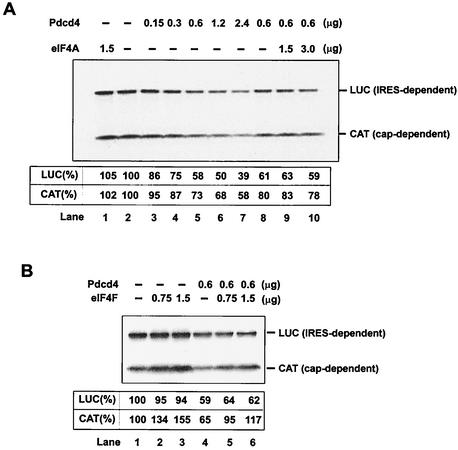
Addition of native eIF4A to the reticulocyte lysate did not affect either cap- or IRES-dependent translation (Fig. 4A, compare lanes 1 and 2), in agreement with previous observations (36) that addition of eIF4A did not alter the rate of translation. Addition of recombinant His-Pdcd4 (0.15 to 2.4 μg) to the reticulocyte lysate inhibited both cap-dependent and IRES-dependent translation in a concentration-dependent manner (Fig. 4A, lanes 2 to 7). Addition of eIF4F (0.75 and 1.5 μg) purified from rabbit reticulocyte lysates produced a partial recovery of cap-dependent translation but not of IRES-dependent translation (Fig. 4B). However, addition of native eIF4A (1.5 μg and 3.0 μg) purified from rabbit reticulocyte lysates did not relieve the inhibition of either CAT or luciferase translation by recombinant His-Pdcd4 (0.6 μg) (Fig. 4A, lanes 8 to 10), suggesting that the mechanism of inhibition of translation by Pdcd4 is not simply to quench or sequester eIF4A activity.
FIG. 4.
Inhibition of in vitro translation by Pdcd4. (A) Rabbit reticulocyte lysate was preincubated with eIF4A alone (lane 1), with increasing amounts of Pdcd4 (0.15 to 4.8 μg, lanes 2 to 7), or with Pdcd4 and increasing amounts of eIF4A (lanes 8 to 10) for 5 min at 30°C prior to the addition of the bicistronic CAT/EMCV/LUC mRNA (0.2 μg). Translation was performed in a total volume of 20 μl as described in Materials and Methods. The band intensity was determined by Phosphorimager. The value obtained for both cap- and IRES-dependent translation in the absence of added Pdcd4 and eIF4A was designated as 100%. (B) Rabbit reticulocyte lysate was incubated with eIF4F (lanes 1 to 3) or eIF4F and Pdcd4 (lanes 4 to 6) for 5 min at 30°C prior to the addition of the bicistronic CAT/EMCV/LUC mRNA (0.2 μg). Translation was performed in a total volume of 20 μl as described in Materials and Methods. The band intensity was determined by Phosphorimager. The value obtained for both cap- and IRES-dependent translation in the absence of added Pdcd4 and eIF4F was designated as 100%.
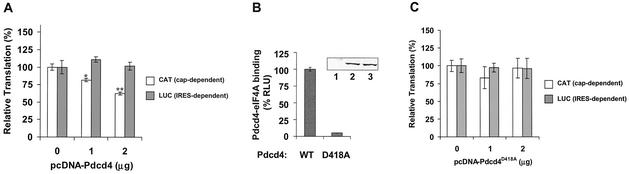
To investigate whether Pdcd4 inhibits translation in vivo, the pcDNA-Pdcd4 expression plasmid and the cytomegalovirus-driven bicistronic CAT/IRES/LUC reporter system, pcDNA-CAT/EMCV/LUC, were transiently cotransfected into (low-Pdcd4) JB6 RT101 cells. The effect of Pdcd4 on translation was measured by CAT and luciferase activity. Transfection with increasing concentrations of pcDNA-Pdcd4 DNA produced a concentration-dependent decrease in translation of the CAT reporter (Fig. 5A). Transfection of 2 μg of pcDNA-Pdcd4 DNA inhibited CAT expression (cap-dependent translation) by 40 to 45%. It is noteworthy that inhibiting protein translation by 50% is sufficient to alter a cell's physiological function (48). For example, tumstatin inhibits cap-dependent translation in endothelial cells by 25 to 45%, with the result that activation of phosphatidylinositol 3-kinase and Akt kinase was inhibited and apoptosis was stimulated (28, 29). In contrast, Pdcd4 did not inhibit luciferase expression (IRES-dependent translation) at a concentration of up to 2 μg of pcDNA-Pdcd4 DNA, suggesting that Pdcd4 preferentially inhibits cap-dependent translation in vivo. Greater selectivity was seen in vivo than in vitro, a not uncommon observation. Since the CAT and luciferase reporter genes are located on the same bicistronic mRNA, and since luciferase expression was unaffected, it follows that CAT transcription was also unaffected by Pdcd4. Thus, the inhibition of cap-dependent CAT expression by Pdcd4 is occurring at the level of translation. A Pdcd4 mutant, Pdcd4D418A, in which glutamine 418 was changed to alanine showed a dramatic decrease in binding to eIF4A when expressed in JB6 RT101 cells (Fig. 5B). Transfection of pcDNA-Pdcd4D418A DNA with the bicistronic reporter system into RT101 cells produced no inhibition of CAT or luciferase expression (cap-dependent and IRES-dependent translation, respectively) (Fig. 5C). These results indicate that binding of Pdcd4 to eIF4A is required for Pdcd4 to inhibit translation in vivo.
FIG. 5.
Pdcd4 inhibits translation in vivo. (A and C) The plasmid pcDNA-CAT/EMCV/LUC reporter system (0.2 μg) was transiently transfected with (A) pcDNA-Pdcd4 (0 to 2 μg) or (C) pcDNA-Pdcd4D418A (0 to 2 μg) into JB6 RT101 cells. Total DNA was maintained at 2.2 μg by adding pcDNA3.1+ vector DNA. After transfection, cells were serum starved (0.2% fetal bovine serum) for 24 h and then incubated with normal medium (4% fetal bovine serum) for an additional 24 h. The CAT and luciferase activities from the cells with 0 μg of pcDNA-Pdcd4 (A) or pcDNA-Pdcd4D418A (C) transfection was designated as 100%. These experiments were repeated three times in triplicate, and representative data are shown. Results are expressed as mean ± standard deviation. * and ** indicate significant differences compared with the control as determined by Student's t test (*, <0.005; **, <0.0001). (B) Pdcd4D418A mutant does not bind to eIF4A. Plasmid pCMV-BD-Pdcd4 (50 ng) (wild type [WT]) or pCMV-BD-Pdcd4D418A (50 ng) (D418A) was transiently transfected with pCMV-AD-eIF4A (50 ng) and Gal4-luciferase reporter DNA (25 ng) into JB6 RT101 cells. After 48 h, cells were lysed and luciferase activity was measured. The luciferase activity of wild-type Pdcd4 was designated as 100%. These experiments were repeated three times with five independent transfections each, and representative data are shown. Results are expressed as mean ± standard deviation. The inset shows an immunoblot of RT101 cells transiently transfected with pcDNA3.1+ (lane 1), pcDNA-Pdcd4 (lane 2), or pcDNA-Pdcd4D418A (lane 3) and detected with Pdcd4 antibody.
Pdcd4D418 does not suppress AP-1-dependent transcription in JB6 P+ cells.
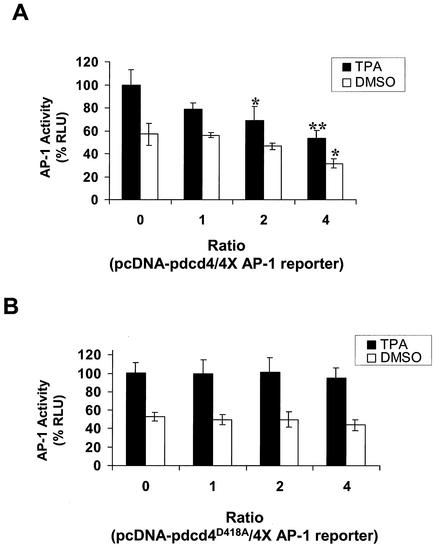
To determine whether Pdcd4 binding to eIF4A is physiologically significant, we asked whether the Pdcd4D418A mutant is defective in inhibiting AP-1-dependent transcription. Wild-type Pdcd4 inhibits AP-1-dependent transcriptional activation but not NF-κB or ornithine decarboxylase activation in JB6 P+ cells (49). AP-1 transactivation is one of the few molecular events known to be required for tumor promoter-induced neoplastic transformation in JB6 P+ cells (15) and in mouse skin tumor promotion in vivo (53), and AP-1 activation is the only transformation-relevant molecular event known to be targeted by Pdcd4 (49). Inhibition of AP-1-dependent transcription by dominant negative c-jun suppresses tumor promoter-induced transformation in JB6 cells (9) and tumorigenesis in mouse skin (53).
As shown in Fig. 6A, transient cotransfection of wild-type Pdcd4 expression plasmid with the 4× AP-1-LUC reporter into JB6 P+ cells inhibited both basal and tetradecanoyl phorbol acetate-induced AP-1-dependent luciferase expression in a concentration-dependent manner, in agreement with previous observations (49). In contrast, cotransfection of Pdcd4D418A expression plasmid with the 4× AP-1 reporter did not inhibit basal or tetradecanoyl phorbol acetate-induced AP-1-dependent luciferase expression (Fig. 6B). Wild-type Pdcd4 inhibits the luciferase expression driven by the 4× AP-1 promoter but not that driven by the NF-κB or serum response element promoter (49). Since these three luciferase reporter plasmids have identical sequences in their 5′untranslated regions, there is no basis for differential Pdcd4 effects at the level of luciferase translation_._ Thus, Pdcd4 is acting, albeit indirectly, to inhibit AP-1-dependent transcription. Because Pdcd4D418A does not bind to eIF4A (Fig. 5B), the results shown in Fig. 6 suggest that Pdcd4 binding to eIF4A is required for suppression of AP-1-dependent transcription.
FIG. 6.
Pdcd4D418A does not inhibit AP-1-dependent transcription. JB6 P+ cells were transfected with 0.2 μg of the 4× AP-1 luciferase reporter gene and increasing amounts (0 to 0.8 μg) of pcDNA-Pdcd4 (A) or pcDNA-Pdcd4D418A (B). Total DNA was maintained at 1.0 μg by adding pcDNA3.1+ vector DNA. The luciferase activity of cells treated with tetradecanoyl phorbol acetate (TPA) and without pcDNA-Pdcd4 or pcDNA-Pdcd4D418A was designated as 100%. These experiments were repeated three times in triplicate, and representative data are shown. Results are expressed as mean ± standard deviation. * and ** indicate significant differences compared with the control (tetradecanoyl phorbol acetate or dimethyl sulfoxide [DMSO] treatment following transfection with 0 μg of pcDNA-Pdcd4) as determined by Student's t test (*, <0.005; **, <0.0001).
Pdcd4 prevents eIF4A association with the C-terminal one-third but not the middle one-third of eIF4G.
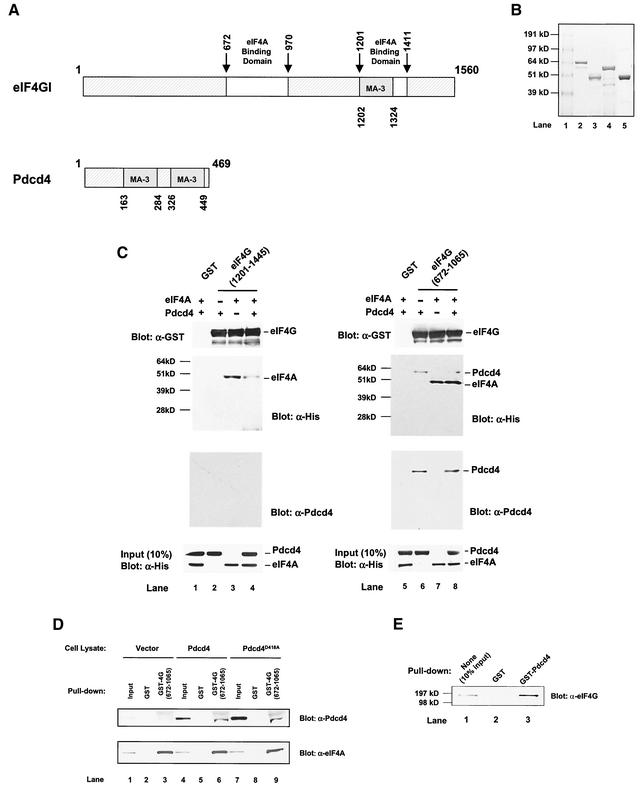
Analysis of the Pdcd4 protein sequence reveals a recognizably conserved feature, two α-helical MA-3 domains (also designated MI domains) (2, 39). The function of the MA-3 domain is not well understood. This domain is also found in the C-terminal one-third of eIF4G and located within the second eIF4A binding region (32) (Fig. 7A). This location suggests that the MA-3 domain may play an important role in the binding of eIF4A. If this hypothesis is correct, Pdcd4 should be able to compete with the C-terminal one-third of eIF4G for binding to eIF4A.
FIG. 7.
Prevention of eIF4A binding to the C-terminal but not to the middle one-third of eIF4G by Pdcd4: Pdcd4 binds to the middle one-third of eIF4G. (A) Structures of eIF4G1 and Pdcd4. The numbers refer to the size (in amino acids) of eIF4G1 and Pdcd4 and to the locations of the eIF4A binding domain and MA-3 domain (2, 32, 40). eIF4A binding domains (open box and arrows) in eIF4G1 are indicated schematically. The MA-3 domains (grey box) in eIF4G1 and Pdcd4 are indicated schematically. (B) Coomassie blue staining of recombinant GST-eIF4G(672-1065), GST-eIF4G(1201-1445), His-eIF4A, and His-Pdcd4. Three micrograms of each recombinant GST-eIF4G(672-1065) (lane 2), GST-eIF4G(1201-1445) (lane 3), His-Pdcd4 (lane 4), and His-eIF4A (lane 5) was resolved by SDS-PAGE and stained with SimpleBlue (Invitrogen). Lane 1, proteinmolecular size markers. (C) In vitro binding assay. Bovine liver GST (lanes 1 and 5), recombinant GST-eIF4G(1201-1445) (lanes 2 to 4), or GST-eIF4G(672-1065) (lanes 6 to 8) was immobilized on glutathione-Sepharose beads and incubated with 5 μg of His-Pdcd4 only (lanes 2 and 6), 5 μg of His-eIF4A only (lanes 3 and 7), or 5 μg of both His-Pdcd4 and His-eIF4A (lanes 1, 4, 5, and 8) on ice for 10 min. After being washed with binding buffer, the bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting with GST antibody (first panel), penta-His antibody (second panel), or Pdcd4 antibody (third panel). Ten percent of input His-Pdcd4 and His-eIF4A proteins were subjected to SDS-PAGE followed by immunoblotting with penta-His antibody (fourth panel). GST-eIF4G(672-1065) and GST-eIF4G(1201-1445) immobilized on glutathione-Sepharose beads were shown as similar amounts. (D) Pulldown of Pdcd4 and Pdcd4D418A with GST-eIF4G(627-1065). JB6 P+ cell lysates isolated following transient transfection with pcDNA3.1+ (lanes 1 to 3), pcDNA-Pdcd4 (lanes 4 to 6), or pcDNA-Pdcd4D418A (lanes 7 to 9) were pulled down with GST (lanes 2, 5, and 8) or GST-eIF4G(627-1065) (lanes 3, 6, and 9). The bound proteins were resolved by SDS-10% PAGE followed by immunoblotting with Pdcd4 or eIF4A antibodies. (E) GST pulldown of endogenous eIF4G with Pdcd4. JB6 P+ cell lysate was pulled down with GST (lane 2) or GST-Pdcd4 (lane 3). The bound proteins were resolved by SDS-10% PAGE followed by immunoblotting with eIF4G antibody. Lane 1 shows one-tenth of the cell lysate.
To address this question, we performed an in vitro binding assay that used recombinant His-Pdcd4, recombinant His-eIF4A (32), GST-eIF4G(672 to 1065) (middle region) (32), and GST-eIF4G(1201 to 1445) (C-terminal region) (32) (Fig. 7B). GST-eIF4G(672-1065) and GST-eIF4G(1201-1445) were immobilized on glutathione-Sepharose beads and incubated with His-eIF4A in the presence and absence of His-Pdcd4. The bound protein was resolved by SDS-PAGE and analyzed by immunoblotting with GST, penta-His, and Pdcd4 antibodies (Fig. 7C). His-eIF4A bound to GST-eIF4G(1201-1445) (Fig. 7C, lane 3), but His-Pdcd4 did not (Fig. 7C, lane 2). Addition of His-Pdcd4 along with eIF4A to the binding reaction mixture that included eIF4G(1201-1445)-bound beads (ratio of His-eIF4A to His-Pdcd4 of 1:1) dramatically decreased (five- to sixfold, as determined by densitometry) the association of His-eIF4A with GST-eIF4G(1201-1445) (Fig. 7C, compare lanes 3 and 4). However, addition of His-Pdcd4 with eIF4A to the reaction mixture that included GST-eIF4G(672-1065)-immobilized beads showed an unchanged level of association of His-eIF4A with GST-eIF4G(672-1065) (Fig. 7C, compare lanes 7 and 8). These results indicate that Pdcd4 prevents eIF4A from binding to the C-terminal one-third of eIF4G but does not prevent eIF4A from binding to the middle one-third of eIF4G.
Pdcd4 Binds to the middle one-third of eIF4G.
Figure 7C shows that His-Pdcd4 also associates with eIF4G(672-1065) in the absence or presence of eIF4A (lanes 6 and 8, anti-His- and anti-Pdcd4 immunoblots). The independence of Pdcd4 and eIF4A binding to the middle domain of eIF4G suggests that the Pdcd4 binding site on eIF4G(672-1065) is different from the site on eIF4G(672-1065) that binds to eIF4A.
To seek independent verification that Pdcd4 is able to bind to the middle domain of eIF4G, we performed a GST pulldown. As shown in Fig. 6D, GST-eIF4G(672-1065) was able to pull down eIF4A from lysates of JB6 P+ cells transiently transfected with pcDNA3.1 (vector), pcDNA-Pdcd4 (Pdcd4) or pcDNA-Pdcd4D418A (Pdcd4D418A) (Fig. 6D, lanes 3, 6, and 9; anti-eIF4A immunoblot). GST-eIF4G(672-1065) also pulled down Pdcd4 and Pdcd4D418 from lysates of cells transfected with pcDNA-Pdcd4 and pcDNA-Pdcd4D418A, respectively (Fig. 6D, lanes 6 and 9, anti-Pdcd4 immunoblot). Since Pdcd4D418A does not bind to eIF4A (Fig. 5B), the results shown in Fig. 7D indicate that Pdcd4 binding to the eIF4G middle domain occurs independently of eIF4A-Pdcd4 binding. We also used GST-Pdcd4 to test whether Pdcd4 binds to full-length endogenous eIF4G. As expected, GST-Pdcd4 pulled down endogenous eIF4G from JB6 P+ cell lysates (Fig. 7E, lane 3) but GST did not (Fig. 7E, lane 2). Binding of the human Pdcd4 homolog H731 to eIF4G has also been reported (18).
The GST-eIF4G(672-1065) and GST-eIF4G(1201-1445) immobilized on glutathione-Sepharose beads used in the above experiments were analyzed by immunoblotting with GST antibody to indicate that similar amounts were bound (Fig. 7C). In summary, Pdcd4 interferes with eIF4A binding to the C-terminal but not to the middle-domain binding site on eIF4G, and Pdcd4 itself binds to the middle domain of eIF4G. The domains on Pdcd4 that bind to eIF4A and to eIF4G appear to be distinct.
DISCUSSION
This study demonstrates that the transformation suppressor Pdcd4 physically associates with the translation initiation factor eIF4A, resulting in inhibition of helicase activity (Fig. 3) and translation (Fig. 4 and 5). The inhibition of translation requires Pdcd4 binding to eIF4A, as Pdcd4D418A, a mutant inactivated for binding to eIF4A, has no effect on translation (Fig. 5C). These findings are in agreement with previous observations that mutational inhibition of the helicase and/or ATPase activity of eIF4A inhibits translation (35-37, 47). Therefore, Pdcd4 is not only a novel eIF4A binding protein, it is the first example of a protein that inhibits translation through inactivation of eIF4A activity.
Wild-type Pdcd4 but not the Pdcd4D418A mutant inhibited AP-1-dependent transcription (Fig. 6). Inhibition of AP-1 is sufficient to inhibit tumor promotion, both in the JB6 cell model and in mouse skin carcinogenesis in vivo (9, 53). The lack of eIF4A binding by the mutant Pdcd4 appears to disable the inhibition of translation and consequently to disable the transrepression of AP-1 that contributes to Pdcd4's suppression of transformation. Pdcd4D418A is not inactivated for all its activities, as it retains the ability to bind the middle domain of eIF4G (Fig. 7D). Taken together, these results suggest that the loss of binding to eIF4A results in the loss of a transformation-relevant function.
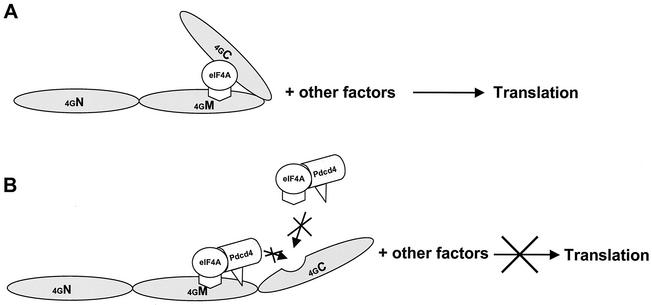
How does Pdcd4 inhibit translation? A model to explain the mechanism underlying the translational inhibition by Pdcd4 must take into account the findings that (i) Pdcd4 inhibits the helicase activity of eIF4A, (ii) Pdcd4 blocks eIF4A binding to the C-terminal one-third of eIF4G, (iii) Pdcd4 binds to the middle one-third of eIF4G independently of eIF4A binding, and (iv) the Pdcd4 domains for binding to eIF4A and the middle domain of eIF4G are distinct (Fig. 8). The C-terminal domain of eIF4G is a regulatory domain for translation, and its binding to eIF4A has been shown to greatly enhance translation (32). Therefore, prevention of eIF4A binding to the C-terminal domain by Pdcd4 (Fig. 7C) contributes to inhibition of translation.
FIG. 8.
Model of how Pdcd4 inhibits translation. (A) A sandwich model of eIF4A binding to eIF4G is shown (32). The eIF4A molecule binds to the middle and C-terminal one-third of eIF4G. (B) Model for Pdcd4 inhibition of translation. Pdcd4 inhibits the helicase activity of eIF4A. This inactivated eIF4A molecule is further trapped by Pdcd4 on the middle one-third of eIF4G. This process will block eIF4A association-dissociation cycling and keep the eIF4F complex inactivated. In addition, Pdcd4 also prevents from eIF4A binding to the C-terminal one-third of eIF4G. This will prevent eIF4A from stimulating the activity of eIF4F.
eIF4A exchanges between free eIF4A and eIF4A bound in the eIF4F complex (36). Biochemical and kinetic studies have shown that eIF4A may proceed through two to three association-dissociation cycles at the beginning of translation initiation to unwind double-stranded RNA (42, 43). Blocking this process inhibits translation. Pdcd4 binding to the middle one-third of eIF4G independent of eIF4A (Fig. 7C and 7D) suggests that eIF4A may be trapped by Pdcd4 on the middle domain of eIF4G, thus blocking the association-dissociation cycle of eIF4A through eIF4F. The observation (Fig. 4A) that addition of eIF4A to the rabbit reticulocyte lysate does not relieve inhibition of translation by Pdcd4 further supports this model. eIF4A, when trapped in a Pdcd4-inactivated form bound to eIF4G, would not be displaced by added free eIF4A. Further testing of this model will be important.
Our data do not indicate the stoichiometry for the ratio of eIF4G and eIF4A. One molecule of eIF4A may associate with one molecule of eIF4G to form a “sandwich,” as proposed by Morino et al. (32) (Fig. 8A), or two molecules of eIF4A may associate with one molecule of eIF4G. Recently, two groups have proposed a stoichiometry for eIF4G to eIF4A. Korneeva et al. (19), using surface plasmon resonance techniques and recombinant eIF4G proteins, showed a 1:2 ratio for eIF4G to eIF4A. On the other hand, Li et al. (24), using immunoprecipitation of endogenous and tagged eIF4A, concluded that 1:1 was the ratio for eIF4G to eIF4A.
Two human Pdcd4 homologs, H731-L and H731, are 96% and 93% identical, respectively, in amino acid sequence to mouse Pdcd4. H731-L and H731 are alternative transcripts of the same gene. H731 lacks 11 amino acids in the N-terminal region that are present in H731-L and Pdcd4. H731 was identified and isolated with the Pr-28 antibody, which recognizes a nuclear antigen in proliferating cells (31). Recent studies by Yoshinaga et al. (52) used the human H731 antibody to determine the expression and localization of H731 in several cell lines and tissues. H731 was abundantly expressed and localized in the cytoplasm of cancer cells. In normal cells, however, H731 was localized in the nuclei. These observations are in disagreement with our observations of Pdcd4 localization and differential expression. First, the level of Pdcd4 expression in the (less progressed) JB6 P− cells was about 8- to 10-fold higher than that in JB6 P+ cells (49), and transformed JB6 cells (unpublished data). Second, the results of immunofluorescent confocal microscopy analysis indicate that Pdcd4 is colocalized with eIF4A in the cytoplasm (Fig. 2). In addition, the immunoprecipitation and GST pulldowns showing that Pdcd4 physically interacts with eIF4A (Fig. 1) and eIF4G (Fig. 7) provide further support for cytoplasmic localization of Pdcd4.
It is unknown whether the two highly identical proteins, Pdcd4 and H731L, localize differently. The nuclear localization of H731 might be attributed to differential tissue specificity or to the use of different antibodies. It is noteworthy that the Pdcd4 antibody used in the present studies shows high specificity (49), whereas the H731 antibody recognizes several proteins ranging in molecular mass from 51 to 64 kDa (5).
Comparison of the Pdcd4 protein sequence with proteins in the GenBank database reveals two α-helical MA-3 domains that are located from amino acids 163 to 284 and amino acids 326 to 449 (Fig. 7A). The MA-3 domain extends over approximately 120 amino acids with 80 to 85% consensus secondary structure; although there are many conserved amino acids in the MA-3 domain, no specific consensus sequence has been reported (2, 39). In human and mouse eIF4G, the MA-3 domain is located within the second eIF4A binding domain (amino acids 1201 to 1441) (32) (Fig. 7A), implying that the MA-3 domain may play an essential role in binding eIF4A. The finding that Pdcd4 prevents eIF4A from binding to the C-terminal one-third of eIF4G (Fig. 7C) supports this hypothesis. Indeed, deletion or mutation of either MA-3 domain in the Pdcd4 protein dramatically inactivated the binding to eIF4A in a mammalian two-hybrid assay (Fig. 5B and unpublished data), indicating that the MA-3 domain is required for binding eIF4A.
Several tumors and tumor cell lines show elevated levels of translation initiation factors such as eIF4E (7), eIF4A (10), and eIF4G (3). Overexpression of eIF4E (22) or eIF4G (11) resulted in transformation of NIH 3T3 cells, suggesting that translation factors may function as oncogenes. Therefore, downregulation or inactivation of translation factors may suppress transformation. How does Pdcd4 suppress tetradecanoyl phorbol acetate-induced neoplastic transformation in JB6 cells? A small number of molecular events are known to be required for tumor promoter-induced transformation of JB6 P+ cells and tumorigenesis in vivo. Among these required molecular events are activation of transcription factors AP-1, NF-κB, and serum response element as well as ornithine decarboxylase activation (15). Of these events, wild-type Pdcd4 inhibits only AP-1 activation (49).
The mRNAs that are translational targets of Pdcd4 are unknown. One possibility is that Pdcd4 inhibits the translation of AP-1 proteins or of enzymes or coactivators required for their activation. Mitogen treatment of cells greatly stimulates the translation of a group of so-called “translationally repressed” mRNAs (4, 40). This group of mRNAs are often involved in cell proliferation. Included are mRNAs for growth factors, growth promotion genes, and proto-oncogenes (7) that contain long GC-rich 5′ untranslated regions having the potential to form stable secondary structure(s) at the 5′ end. Translation of this group of mRNAs may be inefficient and highly dependent on the eIF4A helicase activity (20, 21). Inhibiting or decreasing eIF4A helicase activity would be expected to limit translation of the translationally repressed mRNAs resulting in the suppression of cell growth or transformation. For instance, mutation of eIF4A in Schizosaccharomyces pombe inhibited translation of cdc25 but not of cdc2 and arrested cells in the G2 phase. Deletion of the 5′ untranslated region of cdc25 restored cdc25 translation (6).
In a related study, we found an inverse relationship between Pdcd4 expression and proliferation within a number of tissues, but especially the cervical epithelia of mice during estrus, which includes a cyclical period of actively proliferating cervical epithelium (A. Jansen, unpublished data). Recent studies by Svitkin et al. (47), with mRNAs varying in stability of secondary structure and eIF4A mutants, showed that the more stable the secondary structure within the 5′ untranslated region of mRNA, the lower the efficiency of translation. These results further support the idea that the requirement for eIF4A in translation is proportional to the stability of the secondary structure within the 5′ untranslated region.
In summary, suppression of eIF4A helicase activity and/or interference with eIF4A binding to eIF4G by Pdcd4 may suppress the translation of a set of mRNAs that limits the activation of AP-1 or other molecular events required for transformation in JB6 cells. Identification of the genes that are most sensitive to translational inhibition by Pdcd4 will be important.
Acknowledgments
We thank Ed Cho for technical assistance for microscopy, Eckhard Jankowsky and Wen Wang (Department of Biochemistry, Case Western Reserve University) for providing purified yeast Ded1p as a His6-tagged protein expressed from a plasmid provided by Patrick Linder, and Terry Copeland for advice and synthesis of Pdcd4 peptides.
REFERENCES
- 1.Abramson, R. D., T. E. Dever, T. G. Lawson, B. K. Ray, R. E. Thach, and W. C. Merrick. 1987. The ATP−dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 262**:**3826-3832. [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2000. Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 10**:**1172-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, C., I. Diesinger, N. Brass, H. Steinhart, H. Iro, and E. U. Meese. 2001. Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer 92**:**822-829. [DOI] [PubMed] [Google Scholar]
- 4.Brown, E. J., and S. L. Schreiber. 1996. A signaling pathway to translational control. Cell 86**:**517-520. [DOI] [PubMed] [Google Scholar]
- 5.Cmarik, J. L., H. Min, G. Hegamyer, S. Zhan, M. Kulesz-Martin, H. Yoshinaga, S. Matsuhashi, and N. H. Colburn. 1999. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. USA 96**:**14037-14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daga, R. R., and J. Jimenez. 1999. Translational control of the cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J. Cell Sci. 112**:**3137-3146. [DOI] [PubMed] [Google Scholar]
- 7.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumors: its possible role in progression of malignancies. Int. J. Biochem. Cell. Biol. 31**:**59-72. [DOI] [PubMed] [Google Scholar]
- 8.De Gregorio, E., T. Preiss, and M. W. Hentze. 1998. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA 4**:**828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Z., M. J. Birrer, R. G. Watts, L. M. Matrisian, and N. H. Colburn. 1994. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc. Natl. Acad. Sci. USA 91**:**609-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberle, J., K. Krasagakis, and C. E. Orfanos. 1997. Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro. Int. J. Cancer 71**:**396-401. [DOI] [PubMed] [Google Scholar]
- 11.Fukuchi-Shimogori, T., I. Ishii, K. Kashiwagi, H. Mashiba, H. Ekimoto, and K. Igarashi. 1997. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 57**:**5041-5044. [PubMed] [Google Scholar]
- 12.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68**:**913-963. [DOI] [PubMed] [Google Scholar]
- 13.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70**:**8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-38. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 15.Hsu, T. C., M. R. Young, J. Cmarik, and N. H. Colburn. 2000. Activator protein 1 (AP-1)- and nuclear factor κB (NF-κB)-dependent transcriptional events in carcinogenesis. Free Radic. Biol. Med. 28**:**1338-1348. [DOI] [PubMed] [Google Scholar]
- 16.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17**:**6940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iost, I., M. Dreyfus, and P. Linder. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274**:**17677-17683. [DOI] [PubMed] [Google Scholar]
- 18.Kang, M. J., H. S. Ahn, J. Y. Lee, S. Matsuhashi, and W. Y. Park. 2002. Up-regulation of PDCD4 in senescent human diploid fibroblasts. Biochem. Biophys. Res. Commun. 293**:**617-621. [DOI] [PubMed] [Google Scholar]
- 19.Korneeva, N. L., B. J. Lamphear, F. L. Hennigan, W. C. Merrick, and R. E. Rhoads. 2001. Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 276**:**2872-2879. [DOI] [PubMed] [Google Scholar]
- 20.Koromilas, A. E., A. Lazaris-Karatzas, and N. Sonenberg. 1992. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 11**:**4153-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson, T. G., B. K. Ray, J. T. Dodds, J. A. Grifo, R. D. Abramson, W. C. Merrick, D. F. Betsch, H. L. Weith, and R. E. Thach. 1986. Influence of 5′ proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J. Biol. Chem. 261**:**13979-13989. [PubMed] [Google Scholar]
- 22.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345**:**544-547. [DOI] [PubMed] [Google Scholar]
- 23.Li, Q., H. Imataka, S. Morino, G. W. Rogers, Jr., N. J. Richter-Cook, W. C. Merrick, and N. Sonenberg. 1999. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 19**:**7336-7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, W., G. J. Belsham, and C. G. Proud. 2001. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J. Biol. Chem. 276**:**29111-29115. [DOI] [PubMed] [Google Scholar]
- 25.Linder, P., P. F. Lasko, M. Ashburner, P. Leroy, P. J. Nielsen, K. Nishi, J. Schnier, and P. P. Slonimski. 1989. Birth of the D-E-A-D box. Nature 337**:**121-122. [DOI] [PubMed] [Google Scholar]
- 26.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20**:**6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15**:**4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeshima, Y., A. Sudhakar, J. C. Lively, K. Ueki, S. Kharbanda, C. R. Kahn, N. Sonenberg, R. O. Hynes, and R. Kalluri. 2002. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science 295**:**140-143. [DOI] [PubMed] [Google Scholar]
- 29.Maeshima, Y., U. L. Yerramalla, M. Dhanabal, K. A. Holthaus, S. Barbashov, S. Kharbanda, C. Reimer, M. Manfredi, W. M. Dickerson, and R. Kalluri. 2001. Extracellular matrix-derived peptide binds to αvβ3 integrin and inhibits angiogenesis. J. Biol. Chem. 276**:**31959-31968. [DOI] [PubMed] [Google Scholar]
- 30.Manders, E. M. M., F. J. Verbeek, and J. A. Aten. 1993. Measurement of colocalization of objects in dual-color confocal images. J. Microsc. 169**:**375-382. [DOI] [PubMed] [Google Scholar]
- 31.Matsuhashi, S., H. Yoshinaga, H. Yatsuki, A. Tsugita, and K. Hori. 1997. Isolation of a novel gene from a human cell line with Pr-28 MAb which recognizes a nuclear antigen involved in the cell cycle. Res. Commun. Biochem. Cell. Mol. Biol. 1**:**109-120. [Google Scholar]
- 32.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20**:**468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, P. J., and H. Trachsel. 1988. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 7**:**2097-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onishi, Y., and H. Kizaki. 1996. Molecular cloning of the genes suppressed in RVC lymphoma cells by topoisomerase inhibitors. Biochem. Biophys. Res. Commun. 228**:**7-13. [DOI] [PubMed] [Google Scholar]
- 35.Pause, A., N. Methot, and N. Sonenberg. 1993. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 13**:**6789-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pause, A., N. Methot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13**:**1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11**:**2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16**:**6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponting, C. P. 2000. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25**:**423-426. [DOI] [PubMed] [Google Scholar]
- 40.Proud, C. G. 1994. Translation: turned on by insulin. Nature 371**:**747-748. [DOI] [PubMed] [Google Scholar]
- 41.Richter-Cook, N. J., T. E. Dever, J. O. Hensold, and W. C. Merrick. 1998. Purification and characterization of a new eukaryotic protein translation factor, eukaryotic initiation factor 4H. J. Biol. Chem. 273**:**7579-7587. [DOI] [PubMed] [Google Scholar]
- 42.Rogers, G. W., Jr., W. F. Lima, and W. C. Merrick. 2001. Further characterization of the helicase activity of eIF4A: substrate specificity. J. Biol. Chem. 276**:**12598-12608. [DOI] [PubMed] [Google Scholar]
- 43.Rogers, G. W., Jr., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276**:**30914-30922. [DOI] [PubMed] [Google Scholar]
- 44.Rogers, G. W., Jr., N. J. Richter, and W. C. Merrick. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274**:**12236-12244. [DOI] [PubMed] [Google Scholar]
- 45.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eukaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10**:**1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibahara, K., M. Asano, Y. Ishida, T. Aoki, T. Koike, and T. Honjo. 1995. Isolation of a novel mouse gene, MA-3, that is induced upon programmed cell death. Gene 166**:**297-301. [DOI] [PubMed] [Google Scholar]
- 47.Svitkin, Y. V., A. Pause, A. Haghighat, S. Pyronnet, G. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7**:**382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watkins, S. J., and C. J. Norbury. 2002. Translation initiation and its deregulation during tumorigenesis. Br. J. Cancer 86**:**1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, H. S., A. P. Jansen, R. Nair, K. Shibahara, A. K. Verma, J. L. Cmarik, and N. H. Colburn. 2001. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-κB or ornithine decarboxylase transactivation. Oncogene 20**:**669-676. [DOI] [PubMed] [Google Scholar]
- 50.Yoder-Hill, J., A. Pause, N. Sonenberg, and W. C. Merrick. 1993. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J. Biol. Chem. 268**:**5566-5573. [PubMed] [Google Scholar]
- 51.Yoshinaga, H., S. Matsuhashi, J. Ahaneku, Z. Masaki, and K. Hori. 1997. Expression and identification of H731 gene product in HeLa cells. Res. Commun. Biochem. Cell Mol. Biol. 1**:**121-131. [Google Scholar]
- 52.Yoshinaga, H., S. Matsuhashi, C. Fujiyama, and Z. Masaki. 1999. Novel human PDCD4 (H731) gene expressed in proliferative cells is expressed in the small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol. Int. 49**:**1067-1077. [DOI] [PubMed] [Google Scholar]
- 53.Young, M. R., J. J. Li, M. Rincon, R. A. Flavell, B. K. Sathyanarayana, R. Hunziker, and N. Colburn. 1999. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl. Acad. Sci. USA 96**:**9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]