Increased Mucosal Transmission but Not Enhanced Pathogenicity of the CCR5-Tropic, Simian AIDS-Inducing Simian/Human Immunodeficiency Virus SHIVSF162P3 Maps to Envelope gp120 (original) (raw)
Abstract
Through rapid serial transfer in vivo, the chimeric CCR5-tropic simian/human immunodeficiency virus SHIVSF162 evolved from a virus that is nonpathogenic and poorly transmissible across the vaginal mucosa to a variant that still maintains CCR5 usage but which is now pathogenic and establishes intravaginal infection efficiently. To determine whether envelope glycoprotein gp120 is responsible for increased pathogenesis and transmissibility of the variant SHIVSF162P3, we cloned and sequenced the dominant envelope gene (encoding P3 gp120) and characterized its functions in vitro. Chimeric SHIVSF162 virus expressing P3 gp120 of the pathogenic variant, designated SHIVSF162PC, was also constructed and assessed for its pathogenicity and mucosal transmissibility in vivo. We found that, compared to wild-type SHIVSF162 gp120, P3 gp120 conferred in vitro neutralization resistance and increased entry efficiency of the virus but was compromised in its fusion-inducing capacity. In vivo, SHIVSF162PC infected two of two and two of three rhesus macaques by the intravenous and intravaginal routes, respectively. Nevertheless, although peak viremia reached 106 to 107 RNA copies per ml of plasma in some infected animals and was associated with depletion of gut-associated CD4+ lymphocytes, none of the animals maintained a viral set point that would be predictive of progression to disease. Together, the data from this study suggest a lack of correlation between entry efficiency and cytopathic properties of envelope glycoproteins with viral pathogenicity. Furthermore, whereas env gp120 contains the determinant for enhanced mucosal transmissibility of SHIVSF162P3, the determinant(s) of its increased virulence may require additional sequence changes in env gp41 and/or maps to other viral genes.
Infection of rhesus macaques with chimeric simian/human immunodeficiency virus (SHIV) has provided a model system not only for evaluation of vaccines and antiviral therapies (6) but for studies of HIV infection and persistence as well, providing important insights into the pathogenic process of disease development (26, 27, 29, 33-35, 48, 61, 69). In particular, this model has been used extensively to examine the role of HIV type 1 (HIV-1) envelope (Env) glycoproteins in the development of fatal immunodeficiency disease (22, 28, 30, 36, 60, 61, 69). Env determines viral entry, tropism, neutralization susceptibility, fusogenicity, and cytopathicity in vitro (4, 45). Additionally, epidemiological and virological studies in humans have demonstrated a correlation between disease progression and emergence of cytopathic, syncytium-inducing (SI) HIV-1 variants (12, 15, 23, 65, 72). Viruses isolated early in the course of disease typically use CCR5 as a coreceptor (R5 viruses) and are non-syncytium-inducing (NSI), whereas viruses isolated late in the course of disease commonly use either CXCR4 alone (X4 viruses) or both CCR5 and CXCR4 (R5X4 viruses) and are SI (16). Consistent with this observation, enhanced pathogenicity of X4 and R5X4 SHIVs in vivo has been linked to envelope gp120-determined in vitro properties that include increased cytopathicity and fusogenicity, in addition to efficient viral entry and resistance to neutralizing antibody (8-10, 21, 37, 70).
Less well defined, however, are envelope properties associated with R5 viruses from individuals who progress to AIDS in the absence of a coreceptor switch. Overbaugh and colleagues demonstrated with the SIV system that variants isolated from SIVmne infection of macaques late in the disease stage show increased fitness for replication in the host that correlates with antigenic and phenotypic changes in the envelope glycoprotein such as neutralization resistance and cytopathicity, but not SI capacity (38, 64). The envelope glycoprotein is also an important component of SIVsmPBj pathogenesis, conferring enhanced virus replication kinetics and infectivity (25, 56). For HIV, studies in the histoculture system indicated an equal degree of replication and cytopathicity of early and late R5 viruses (40), an observation that is in agreement with findings from one (5) but not another (67) study using the SCID-hu xenograft model. The limited range of host target cells and the absence of an intact immune system in these ex vivo organ culture systems, however, restrict their utility in assessing the role of tissue tropism and antigenic changes in viral pathogenesis. Until recently, the lack of R5-tropic SHIVs that evolve from a nonpathogenic to pathogenic phenotype has prevented the use of this model system to characterize, phenotypically and antigenically, the HIV-1 envelope-determined properties that might be associated with increased virulence of R5-tropic isolates.
We previously constructed SHIVSF162 using the envelope gene of the nonpathogenic, macrophage-tropic, and CCR5-using primary HIV-1SF162 strain (49). SHIVSF162 infected rhesus macaques efficiently by the intravenous route but did not maintain a high viral set point and did not induce disease (27, 49). Surprisingly, R5-SHIVSF162 was unable to establish infection by the intravaginal route (50). Upon rapid serial intravenous passage of the virus, a variant designated SHIVSF162P3 was derived that maintained CCR5 coreceptor usage but exhibited enhanced pathogenicity (27). Two of two animals inoculated with cell-free SHIVSF162P3 by the intravenous route were infected, with one sustaining a high viral set point and progressing to simian AIDS (SAIDS) at 26 weeks postinfection (wpi) (data not shown). Significantly, cell-free intravaginal (IVAG) inoculation of rhesus macaques with SHIVSF162P3 also resulted in high-titer virus replication and induction of SAIDS in four of seven infected macaques (26) (Table 1). Increased virulence and mucosal transmissibility of the R5-tropic SHIVSF162 after rapid serial passage of the virus in macaques therefore provided an opportunity to address whether the envelope gp120 contains the determinants for these properties in an animal infection model that closely mimics that of humans, and if so, to identify the associated phenotypic and antigenic changes in envelope glycoproteins. To this end, we cloned and sequenced a major variant env gp120, termed P3 gp120, from SHIVSF162P3 and assessed its functions. Chimeric virus containing P3 env gp120 in the genomic backbone of parental SHIVSF162 was also generated and assessed for its pathogenicity and transmissibility in rhesus macaques.
TABLE 1.
Comparison of SHIVSF162 and SHIVSF162P3 infection in IVAG challenge of rhesus macaques
| Virus | No. infected/no. challenged | Clinical outcome | Reference |
|---|---|---|---|
| SHIVSF162 clone | 0/3 | None | 50 |
| SHIVSF162P3 isolate | 7/9 | Seroconversion, gradual depletion of CD4+ PBMC, SAIDS, severe mucosal CD4+-T-cell loss in 4 of 7 infected animals | 26; unpublished data |
MATERIALS AND METHODS
Cells.
Human osteosarcoma cells expressing CD4 and either human or rhesus CCR5 (HOS.CD4.CCR5) were gifts from N. Landau (Salk Institute, La Jolla, Calif.) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 0.5 μg of puromycin per ml, and penicillin-streptomycin. The CEMx174 5.25 M7 cell line (also a generous gift from N. Landau), expressing both CD4 and CCR5, was stably transduced with a long terminal repeat (LTR)-luciferase and LTR-green fluorescent protein cassette and was maintained in RPMI-10% FBS containing 200 μg of G418 (Life Technologies, Carlsbad, Calif.)/ml, 200 μg of hygromycin (Invitrogen, Carlsbad, Calif.)/ml, and 0.5 μg of puromycin (Sigma-Aldrich, St. Louis, Mo.)/ml. 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS and penicillin-streptomycin.
Amplification of SHIVSF162P3 gp120.
RNA from cell-free SHIVSF162P3 virus supernatants (26) was isolated with the Qiagen HIV RNA isolation kit per the manufacturer's instructions (Qiagen, Chatsworth, Calif.). RNA was reverse transcribed with SuperScript reverse transcriptase (Life Technologies) using a random hexamer primer (Amersham Pharmacia, Piscataway, N.J.) and was amplified by hot-start PCR under the following buffer conditions: 15 mM Tris-HCl (pH 8), 50 mM KCl, 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphate cocktail, and 2.5 U of Ampli_Taq_ Gold (Roche, Branchburg, N.J.). Reactions were incubated at 94°C for 10 min, followed by 5 cycles of 94°C for 60 s, 55°C for 60 s, and 72°C for 2 min and then 25 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1.5 min. Amplification primers were ED5 and ED12, corresponding to nucleotides 811 to 835 and 2011 to 2041 of the V1 and V5 domains of HIV-1SF162 env (17), respectively (GenBank accession no. M8428). PCR products were cloned with the TOPO TA cloning kit (Invitrogen) per the manufacturer's instructions, and direct automated sequencing of cloned gp120 was carried out (Rockefeller University, New York, N.Y.). Nested PCR for the detection of env V3 sequences was performed on the product of ED5-ED12 PCR, using the V3 loop-specific primers ES7 (nt 1253 to 1272) and ES8 (nt 1878 to 1898) (17). Five amplification cycles were performed as follows: 94°C for 1 min, 45°C for 45 s, 72°C for 1 min. Immediately following, 30 cycles of amplification were carried out (94°C for 30 s, 48°C for 30 s, 72°C for 1 min).
Plasmid constructs and virus production.
From DNA sequencing of 10 TOPO TA V1-V5 gp120 clones, a dominant sequence which was represented in 7 clones was selected for further analysis. A 1.1-kb _Bsg_I-Dra_III DNA fragment encompassing the V1 to V5 domains of gp120 was prepared and exchanged in place of the corresponding sequences in the envelope expression vector pCAGGS-SF162_env (11). Cotransfections of 293T cells with the NL4.3-Luc-E−R− vector, an _env_-deficient luciferase reporter plasmid (14), were carried out using the DMRIE-C reagent (Life Technologies) to generate reporter viruses capable of single-round replication as previously described (11). Reporter viruses were harvested at 72 h posttransfection, and p24 Gag antigen concentration was determined by enzyme-linked immunosorbent assay (Abbott, Abbot Park, Ill.). The V1-V5 P3 env gp120 sequence was also used to replace corresponding sequences in the 3′ SHIVSF162 genome (49). Cotransfection of 293T cells with the 5′ SIV genome (49) was followed by cocultivation with human peripheral blood mononuclear cells (PBMCs) or CEMx174 5.25 M7 cells to rescue molecular clone SHIVSF162PC virus. SHIVSF162PC for animal infection studies was propagated and titrated in CEMx174 5.25 M7 cells.
Entry and neutralization assays.
HOS.CD4.CCR5 cells were seeded in 96-well plates 16 h before infection. Cells were pretreated with Polybrene (2 μg/ml) for 30 min at 37°C, and reporter viruses were incubated with cells for 4 to 5 h at 37°C, following which fresh medium was added. Entry was quantified by luciferase activity detection 72 h after infection (Promega, Madison, Wis.). For neutralization assays, viruses were incubated in the absence or presence of various dilutions of antisera, monoclonal antibodies, or T-20 for 1 h at 37°C before infection. Neutralization was calculated by the amount of entry in the presence of relative to that in the absence of antibody, serum, or T-20, as previously described (51). T-20, a fusion peptide corresponding to the second heptad repeat of gp41 (76), was obtained from D. Ho as a generous gift from D. P. Bolognesi (Trimeris Inc., Durham, N.C.).
Fusion assay.
Fusogenicity mediated by the binding of gp120 expressed on 293T cells to CD4 on CEMx174 5.25 M7 cells was assessed as previously described (9). Briefly, 293T cells were cotransfected with pCAGGS_env_ constructs and pTatSF13, a plasmid encoding the HIV transactivator protein Tat (44). Transfected cells were incubated for 48 h with CEMx174 5.25 M7 cells at a ratio of 3:5. Fusion of transfected cells with CEMx174 5.25 M7 cells activated expression from the LTR-luciferase cassette maintained in the latter cells. Cells were collected and lysed 4 and 8 h after coculture. Lysates were analyzed for luciferase activity and normalized by cell surface gp120 expression that was determined by immunoprecipitation followed by immunoblot analysis (not shown). Controls included 293T cells transfected with pTatSF13 alone and CEMx174 5.25 M7 cells cultivated in the absence of 293T cells, from which background luciferase was calculated and subtracted from all samples.
Animal infections.
All infections were carried out in adult rhesus macaques (Macaca mulatta) individually housed at the Tulane National Primate Research Center in compliance with its Guide for the Care and Use of Laboratory Animals. Animals were confirmed to be serologically negative for simian type D retrovirus, SIV, and simian T-cell lymphotrophic virus prior to infection. Intravenous (i.v.) infections were carried out with 4 × 103 50% tissue culture infectious doses (TCID50) of SHIVSF162PC in two rhesus macaques (animals CB79 and T027). IVAG inoculations were performed by placing virus inocula atraumatically on the cervicovaginal mucosa of three animals (AV45, BD19, and BE72). This dose is comparable to that used previously for SHIVSF162P3 IVAG inoculations wherein four out of four animals became infected (26). Whole blood was collected at designated time intervals and plasma virus was quantified by branched DNA analysis (Bayer Diagnostics, Emeryville, Calif.). T-cell subsets (CD3+, CD4+, and CD8+) were determined by TruCount (Becton Dickinson, San Jose, Calif.). Intestinal resection surgery was carried out as previously described (27), and CD3+, CD4+, and CD8+ lymphocytes were assayed by flow cytometry (FACScalibur).
Detection of antiviral humoral responses.
SHIV-specific antibody responses were analyzed by strip immunoblot assay according to the manufacturer's instructions (Chiron, Emeryville, Calif.). This assay employs recombinant viral antigens derived from HIV-1SF2 p24 Gag, gp120 and gp41 Envs, p31 from the endonuclease portion of Pol, and p27 (Gag) from HIV-2UC1. Cross-reactivity of anti-SIV and anti-HIV antibodies in plasma with viral antigens allowed detection of the antiviral humoral response. Each independent assay was carried out with internal positive and negative controls. Reactivity was quantified relative to controls as described in Table 2.
TABLE 2.
Humoral immune response in SHIVSF162PC i.v. and IVAG infected animals
| Animal no. | Protein | Reactivity with strip in immunoblot assaya | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i.v. infected animals | IVAG infected animals | ||||||||||||||
| w2 | w4 | w7 | w9 | w12 | w19 | w24 | w33 | w3 | w4 | w5 | w6 | w8 | w12 | w16 | w20 |
| CB79 | gp120 | − | − | + | ++ | +++ | +++ | ++++ | ++++ | ||||||
| gp41 | ± | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |||||||
| p31 | − | + | ++ | ++ | ++ | ++++ | ++++ | ++++ | |||||||
| p27 | − | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |||||||
| T027 | gp120 | − | − | − | − | ± | +++ | +++ | +++ | ||||||
| gp41 | − | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |||||||
| p31 | − | − | − | ± | ± | ± | ± | ± | |||||||
| p27 | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |||||||
| AV45 | gp120 | − | − | − | − | − | − | − | − | ||||||
| gp41 | − | − | − | − | − | − | − | − | |||||||
| p31 | − | − | − | − | − | − | − | − | |||||||
| p27 | − | − | − | ± | ± | ± | ± | ± | |||||||
| BD19 | gp120 | − | − | − | − | − | − | − | − | ||||||
| gp41 | − | ± | +++ | +++ | +++ | ++++ | ++++ | ++++ | |||||||
| p31 | − | − | − | − | − | − | − | − | |||||||
| p27 | − | − | − | ± | ± | ++++ | +++ | ++++ | |||||||
| BE72 | gp120 | − | − | − | − | − | − | − | − | ||||||
| gp41 | − | + | +++ | +++ | +++ | +++ | ++++ | ++++ | |||||||
| p31 | − | − | − | − | − | − | − | − | |||||||
| p27 | − | ++ | +++ | ++ | ++ | +++ | +++ | +++ |
RESULTS
Cloning and analysis of SHIVSF162P3 env gp120 sequences.
The env gp120 sequences of SHIVSF162P3 were amplified by reverse transcription-PCR of viral RNA in culture supernatants of infected cells, which was followed by sequence analysis. Compared to parental SHIVSF162 envelope glycoprotein, the predicted sequence of the major variant of SHIVSF162P3 (P3 Env) gp120 exhibited 14 amino acid changes, 10 of which are in the variable regions (Fig. 1). Of interest are amino acid substitutions K140T and K158N, which generate two potential new glycosylation sites in the V1-V2 domain, as well as amino acid changes in the V3 region (T306P, R311K) that are likely to alter the antigenic structure of envelope gp120. env gp120 encodes many of the determinants of increased pathogenesis of HIV as well as X4 and R5 SHIVs, such as fusogenicity, entry and replication, neutralization sensitivity, and depletion of CD4+ lymphocytes (8-10, 21, 22, 36, 37, 60, 61, 70). We therefore examined the susceptibility to antibody neutralization, entry, and fusion properties mediated by P3 Env gp120 in vitro and compared them to that of wild-type SF162 Env.
FIG. 1.
Amino acid sequence alignment of V1-V5 gp120 of SF162 and serially passaged clone P3. Dashes indicate amino acid identity. Amino acid changes generating new potential sites of N-linked glycosylation are boxed.
SHIVSF162P3 envelope glycoprotein gp120 confers increased neutralization resistance.
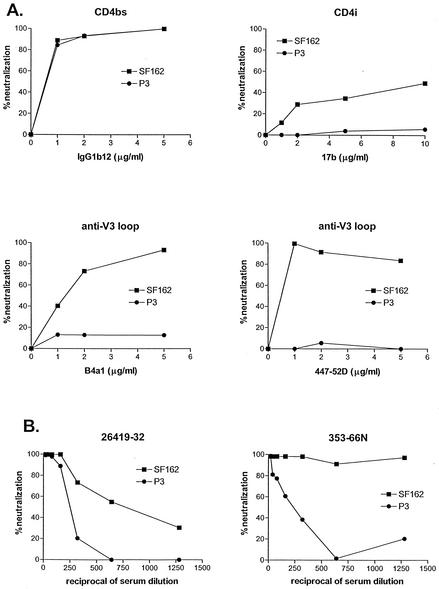
To assess whether Env gp120 amino acid changes alter susceptibility to antibody neutralization, luciferase reporter viruses pseudotyped with SHIVSF162 (SF162) or SHIVSF162P3 (P3) Env gp120 were generated and subjected to neutralization with monoclonal antibodies (MAbs) directed against conserved epitopes overlapping with the CD4 (immunoglobulin G1b12 [IgG1b12]) or coreceptor (17b) binding sites and the V3 loop (B4a1 and 447-52D). Neutralization by sera from SHIVSF162 (49)- and SHIVSF162P3 (26)-infected animals was tested as well (animals 26419 and 353-66N, respectively) (Fig. 2). No difference in susceptibility to neutralization of SF162 or P3 Env pseudotyped viruses by IgG1b12 was observed (Fig. 2A), suggestive of conformational similarity in the CD4 binding domain of wild-type and P3 envelope glycoprotein gp120s. However, neutralization of the P3 viruses by CD4i MAb 17b and anti-V3 loop MAbs B4a1 and 447-52D was significantly reduced compared to wild-type viruses. These last findings are indicative of antigenic changes in the V3 loop of SHIVSF162P3, likely as a result of substitutions in this domain (Fig. 1). Since the V3 loop constitutes part of the coreceptor-binding site on gp120 (32, 62, 71, 73, 77), this change in V3 loop conformation, in turn, could affect the structure of the 17b epitope (41, 62). P3 viruses also showed increased resistance to neutralization with sera from SHIVSF162 (serum 26419)- and SHIVSF162P3-infected animal T353 obtained at necropsy (66 weeks postinfection) (Fig. 2B). T353 is the animal from which SHIVSF162P3 was isolated (26), suggesting that some of the P3 env changes arose as a result of immune escape.
FIG. 2.
Neutralization of luciferase reporter viruses. (A) Neutralization by MAbs against the CD4 binding site (CD4bs), CD4 induced site (CD4i), and the V3 loop (anti-V3 loop). (B) Neutralization by sera isolated from a macaque infected with SHIVSF162 (serum 26419-32) and SHIVSF162P3 (serum 353-66N). Percent neutralization was determined by luciferase expression by viruses infected in the presence relative to the absence of antibodies. Three independent assays were carried out and representative neutralization curves are shown.
P3 Env gp120 confers increased viral entry and susceptibility to T-20 but is defective in its fusion capacity.
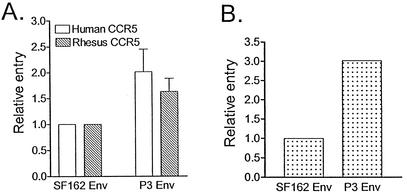
The ability of wild-type and P3 Env gp120 to mediate viral entry was assessed in single-round infection of HOS.CD4.CCR5 cells expressing human or rhesus macaque CCR5 and in the CEMx174 5.25 M7 cell line. We found that P3 Env gp120 conferred between 1.6- and 3-fold increased entry relative to SF162 Env in all of the three cell types tested (Fig. 3). Neither envelope was able to mediate entry into HOS.CD4.CXCR4 cells, indicating that passage-associated mutations in env did not confer expansion or a switch to CXCR4 usage (not shown).
FIG. 3.
Relative entry of luciferase reporter viruses expressing SF162 gp120 or P3 gp120. (A) Entry of HOS.CD4.CCR5 cells. (B) Entry of CEMx174 5.25 M7 cells. Data for relative entry of HOS.CD4.CCR5 cells are the means and standard errors of six independent experiments, while those for CEMx174 5.25 M7 cells are the averages of two independent experiments.
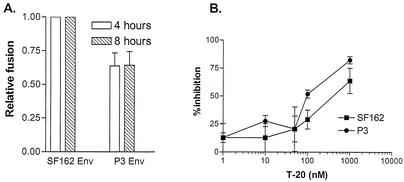
Fusogenic capacity mediated by the two envelopes was also examined. For these studies, 293T cells expressing envelope glycoproteins and Tat were cocultivated with the CEMx174 5.25 M7 indicator cell line transduced with an LTR-Luc reporter cassette, and fusion mediated by the envelopes was quantified by luciferase activity (Fig. 4A). We found that the extent of fusion increased over time in all cultures, reaching saturation at 16 h postcocultivation (data not shown). At both time points examined (4 and 8 h after coculture), however, the fusogenic capacity of P3 Env gp120 was found to be reduced compared to that of wild-type SF162 Env gp120 (Fig. 4A). Relative to SF162 Env gp120, P3 Env gp120 displayed 60% fusogenic activity. Western blotting confirmed that similar amounts of plasma membrane-associated gp120s were expressed on transfected 293T cells, indicating that decreased fusion mediated by P3 Env gp120 was not due to reduced envelope surface expression.
FIG. 4.
Fusogenic capacity and susceptibility to T-20. (A) Fusion of SF162 and P3 gp120-expressing 293T cells with CEMx174 5.25 M7 cells. Results shown are the means and standard errors from three independent experiments. (B) Inhibition of luciferase reporter virus fusion with HOS.CD4.CCR5 cells by T-20. Viruses and T-20 were incubated for 1 h at 37°C before addition to cells, and inhibition relative to viruses in the absence of drug was calculated. The means and standard errors from three independent experiments are shown.
The difference in fusogenic properties of the wild-type and P3 envelopes was further evaluated by assessing the ability of T-20, a fusion peptide inhibitor, to block infection mediated by these envelopes (76). High resistance of HIV to T-20 has been linked to greater CCR5 binding and, by inference, greater 17b epitope exposure that results in a more efficient fusion process (20, 43). Thus, sensitivity to neutralization with T-20 is also used as a surrogate marker for coreceptor binding affinity and 17b epitope exposure (18-20). We found that T-20 inhibited single-round infection mediated by P3 Env gp120 to a greater extent than that by wild-type SF162 gp120 (Fig. 4B). A 50% inhibition of entry mediated by SF162 gp120 requires 400 nM T-20, compared to the 100 nM concentration required for P3 gp120. These findings are consistent with the relative fusogenic capacity of the two envelopes (Fig. 4A) and are in agreement with the greater resistance of P3 Env pseudotype viruses to neutralization with the 17b MAb (Fig. 3). Furthermore, since the gp41 domains of the wild-type and P3 pseudotype viruses used in these studies are identical, the difference in susceptibility to neutralization by an inhibitor targeted to the gp41 domain supports a functional interaction between gp120 and gp41 that modulates the kinetics of the fusion process (18, 19). Whether the difference in V3 loop conformation of SF162 and P3 Env gp120 alters coreceptor affinity and hence T-20 sensitivity requires further investigation.
i.v. infection with the SHIVSF162PC molecular clone.
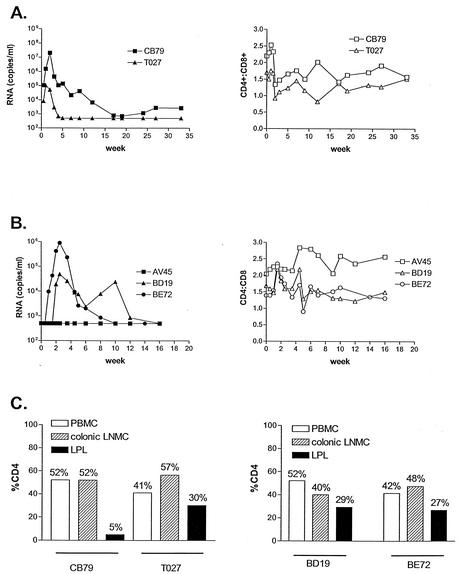
To determine whether SHIVSF162P3 gp120 contains the genetic determinant for increased pathogenicity, chimeric virus expressing the P3 env gp120, termed SHIVSF162PC, was constructed in the genomic backbone of SHIVSF162 for animal infection studies. Two rhesus macaques were inoculated by the i.v. route with the molecular clone SHIVSF162PC. Peak viremia reached 107.3 RNA copies/ml of plasma for animal CB79 and 105 RNA copies/ml of plasma for animal T027. Viremia for both animals declined thereafter, becoming undetectable (<500 copies/ml of plasma) in T027 by 6 wpi and remaining undetectable at 33 wpi (Fig. 5A). For animal CB79, the viral load established a set point of 102 to 103 copies/ml by 33 wpi (Fig. 5A). In both animals, peripheral CD4+ counts dropped during acute infection but rebounded to an equilibrium CD4+/CD8+ ratio that ranged from 1.25 to 2. Gut resection surgery and fluorescence-activated cell sorter analysis of T-cell subsets during acute infection (day 13) revealed significant depletion of jejunal CD4+ lymphocytes only in animal CB79, which displayed high peak viremia (5%) (Fig. 5C). The percentage of CD4+ lymphocytes in both the periphery and colonic lymph nodes of CB79, however, remained in the normal range (52%), indicating selective depletion in the gut-associated mucosa (Fig. 5C).
FIG. 5.
i.v. and IVAG infection of macaques. (A) i.v. infection of rhesus macaques with SHIVSF162PC. (B) IVAG infection of rhesus macaques with SHIVSF162PC. Filled symbols indicate viral loads expressed as RNA copies per milliliter, while empty symbols depict CD4/CD8 cell ratios. (C) Percentages of CD4+ lymphocytes from PBMCs, colonic lymph node mononuclear cells (LNMC), and lamina propria (LPL) during acute infection. The percentage of CD4+ lymphocytes in the intestinal tract of uninfected macaques has been shown to be in the range of 34 to 57% (75).
IVAG infection with the SHIVSF162PC molecular clone.
To evaluate the mucosal transmissibility of SHIVSF162PC, three animals were inoculated with the virus by the IVAG route. Two animals, BD19 and BE72, showed peak viremia of 104.7 and 106 RNA copies/ml, respectively, but plasma viral load was undetectable in animal AV45 (Fig. 5B). Viral DNA was not detected by nested PCR from any samples of PBMCs or lymph node mononuclear cells from AV45 during the first 12 weeks postinoculation, indicating that this animal remained uninfected (data not shown). Both BD19 and BE72 cleared their virus by 12 wpi. No significant peripheral CD4+-T-cell loss was detected in either animal, with CD4+/CD8+ ratios falling in the normal range of baseline values during the course of infection. Gut resection surgery and quantitation of CD4+ lymphocytes performed on animals BD19 and BE72 at day 35 also revealed minimal depletion of jejunal CD4+ cells (29 and 27% compared to baseline values of 26 and 35%, respectively). Consistent with findings in the i.v. infected animals, no diminution of CD4+ lymphocytes in either the periphery or colonic lymph node compartments was observed in the two IVAG infected animals.
Humoral immune responses against SHIVSF162PC.
Strip immunoblotting was performed to assess the sero-status of the inoculated animals (Table 2). Both i.v. infected animals seroconverted at 4 wpi, mounting a strong antibody response to gp41 and p27 (Table 2). The breadth of antibody responses increased over time, and by 12 wpi, reactivity against all four HIV/SIV Env, Pol, and Gag proteins tested was detected in animals CB79 and T027 (Table 2). Weaker antibody responses were observed in the IVAG infected animals, with anti-gp41 and anti-p27 antibodies predominating in animals BD19 and BE72 by 12 wpi (Table 2). Animal AV45, in which plasma viral RNA and cell-associated proviral DNA have been consistently undetectable, mounted a weak anti-p27 antibody response at 6 wpi that persisted up to the last time point tested (20 wpi).
DISCUSSION
We sought to determine whether the genetic determinant(s) for enhanced pathogenicity and increased mucosal transmissibility of R5-SHIVSF162P3 maps to the surface envelope glycoprotein gp120 and to identify the associated antigenic and phenotypic envelope properties. We found that the envelope glycoprotein gp120 of R5-SHIVSF162P3 conferred resistance to antibody neutralization (Fig. 2) and mediated more efficient entry than its wild-type counterpart SHIVSF162 (Fig. 3) but was compromised in its fusogenic capacity (Fig. 4). In vivo studies demonstrated that whereas chimeric SHIVSF162 virus expressing SHIVSF162P3 Env gp120 infected macaques with comparable efficiency by the i.v. and IVAG routes, it failed to establish a high viral set point (Fig. 5 and Table 2). Since the concentration of virus in the plasma after seroconversion predicts the rate of disease progression for both HIV and SIV infections (47, 53), effective resolution of acute viremia observed in the SHIVSF162PC-infected animals suggests that the infections are unlikely to be pathogenic. Indeed, with the exception that the molecular clone SHIVSF162PC can now traverse the vaginal mucosa, the virological and immunological parameters of SHIVSF162PC infection in rhesus macaques parallel those reported for infection with the nonpathogenic parental clone SHIVSF162 (27, 49) (Table 1). Although the limited number of animals inoculated with SHIVSF162PC does not permit statistical analysis of infection rate, our data do indicate that the envelope gp120 of SHIVSF162P3 contains the determinants for enhanced transmission of the virus across the mucosal barrier but not for its increased virulence compared to the parental clone SHIVSF162.
The difference in clinical outcome of infection with clone SHIVSF162PC and isolate SHIVSF162P3 could be due to greater genetic complexity of the latter than of the former. However, for pathogenic X4 and R5X4 SHIVs (SHIVKU1, SHIV89.6P, and SHIV33A), the genetic determinants for enhanced pathogenicity in vivo have been mapped to specific changes in the external glycoprotein gp120 (8, 21, 22, 28, 37). Sequence changes in this region of the viruses are associated with a set of envelope-determined properties in vitro that include efficient entry, increased membrane fusion capability, and resistance to antibody neutralization (8-10, 22, 42). Furthermore, the ability of the HIV-1 envelope glycoproteins to fuse membranes has been shown to contribute to the capacity of the pathogenic dual-tropic SHIVKB9 to deplete CD4+ T lymphocytes in vivo (21). Whether increased envelope fusogenicity also contributes to the development of disease was not directly addressed in this study, but since it is generally believed that progressive loss of CD4+ T lymphocytes underlies the development of AIDS in HIV-1-infected individuals, a link between fusogenicity and disease development was implicated. In this regard, we found that compared to envelope gp120 of wild-type SHIVSF162, the gp120 of SHIVSF162PC mediates more-efficient entry and confers neutralization resistance (Fig. 2 and 3) but is deficient in its fusogenic capacity (Fig. 4A). Thus, it is conceivable that the underlying basis for the nonpathogenic nature of infection associated with R5-SHIVSF162PC is its reduced fusogenicity, suggestive of an association between envelope fusogenicity and disease pathogenicity and implicating a common mechanism of disease for X4, dual-tropic, and R5 SHIVs. Nevertheless, several observations raise the possibility that another mechanism(s) might also be involved.
Although a hallmark of X4 and R5X4 viruses is their SI capacity, R5 viruses, even those isolated from individuals during late-stage disease, are generally not SI in vitro (39, 66). Furthermore, whereas the P3 envelope gp120 is compromised in its fusion capacity, it is cytopathic in vivo (Fig. 5). In the animal in which replication of virus expressing this glycoprotein was robust (i.e., CB79 [Fig. 5A]), a transient drop in peripheral CD4+-T-cell count and dramatic depletion of gut-associated CD4+ T lymphocytes were observed (Fig. 5C), suggesting that in vitro fusogenicity was not predictive of in vivo cytopathicity. A similar discordance between syncytium-forming ability in vitro and cytopathicity ex vivo has also been reported for HIV infections (5, 40, 67, 74) and for SIVmne infection of macaques (38). In the latter experimental animal infection system, increased replication fitness and cytopathicity of this R5 virus, and not its SI capacity in vitro, correlate with increased virulence in the host. Thus, although fusogenicity may very well contribute to pathogenicity of R5 viruses, deficiencies in other properties of SHIVSF162P3 Env gp120 that are not readily apparent or are subtle in vitro could also play a role in the nonpathogenic nature of SHIVSF162PC infection in vivo.
What could these other deficiencies be that are associated with P3 env gp120? It is unlikely to be replication fitness since the P3 Env gp120 mediated more-efficient entry than parental SF162 Env. A possibility to consider is the ability to escape immune recognition. Although compared to wild-type SHIVSF162, SHIVSF162PC is resistant to neutralization by sera from SHIVSF162 (serum 26419)- and SHIVSF162P3 (serum 353-66N)-infected animals, the degree of resistance is less pronounced than what has been reported for autologous neutralization of the pathogenic X4 SHIVSF33A2 molecular clone (9, 10) and is not to the level seen for the pathogenic isolate SHIVSF162P3 (unpublished data). There are mounting data in support of a robust and effective antibody response that arises early in HIV, SIV, and SHIV infections and contributes to the control of virus replication (1, 2, 7, 10, 13, 38, 46, 70; D. D. Richman, T. Wrin, S. Little, and C. Petropoulous, Abstr. 9th Conf. Retrovir. Opportun. Infect., abstr. LB5, 2002). Furthermore, passive immunization as well as vaccine studies in animal models demonstrated that antibodies conferred partial protection (3, 31, 52, 57, 68). Lastly, a relationship between neutralization sensitivity of the viral Env protein and reduced pathogenicity has been reported for the SIV system (59). Thus, the relative susceptibility of the molecular clone SHIVSF162PC compared to the isolate SHIVSF162P3 to neutralization antibody responses in vivo may render it easier to control. Further experiments are required to address this possibility and to evaluate its impact on viral pathogenesis.
Regardless of the basis for its nonpathogenicity, the envelope gp120 of SHIVSF162P3 does confer increased transmission across the vaginal mucosa compared to that of SHIVSF162. The basis for the enhanced mucosal transmissibility of SHIVSF162PC compared to that of SHIVSF162 is not clear. Miller et al. (55) suggested that virus replication is a predictor of the efficiency of vaginal transmission. In this regard, increased entry mediated by P3 Env gp120 is likely to translate into increased replicative capacity, and perhaps, efficient vaginal transmission. Alternatively, we recently reported increased binding of P3 Env gp120 compared to SF162 Env gp120 to DC-SIGN, a type C lectin present on dendritic cells that interacts with gp120 (50). It has been suggested that DC-SIGN plays an important role in HIV transmission and dissemination at the mucosal surfaces by capturing and transporting the virus to peripheral lymph nodes (24, 58, 63). Increased binding to DC-SIGN, therefore, could translate into more-efficient transmission and dissemination across mucosal barriers. Lastly, it is also possible that increased attachment of SHIVSF162PC to cells lining the mucosa may facilitate transmission across this membrane. The recent observation that CCR5-tropic but not CXCR4-tropic HIV bound to intestinal epithelial cells via galactosylceramide warrants investigation (54). It may be of interest to determine whether enhanced binding of this receptor is associated with the P3 Env gp120.
In conclusion, we show that gp120 of the pathogenic isolate SHIVSF162P3 contains the determinant of its transmissibility across the vaginal mucosa but not of its enhanced pathogenicity. Whereas envelope-mediated entry efficiency and/or increased binding to DC-SIGN could be linked to efficient vaginal transmission, increased membrane fusion capacity may be required for viral pathogenesis. Further studies will clarify whether recapitulation of the full pathogenic potential of the CCR5-tropic, SAIDS-inducing SHIVSF162P3 requires amino acid changes in envelope glycoprotein gp41 in addition to gp120 or maps to other viral genes.
Acknowledgments
We are grateful to Allen Mayer for critical reading of the manuscript. We thank Dennis Burton, Lisa Cavacini, James Robinson, and Susan Zolla-Pazner for monoclonal antibodies.
This work was supported by NIH grants CA72822, AI46980, and AI41945 and by TRPRC base grant RR00164. M.H. is a recipient of a Partnership Fellowship of the Canadian Foundation for AIDS Research and the Medical Research Council of Canada.
REFERENCES
- 1.Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nystrom, and E. M. Fenyo. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4**:**107-112. [DOI] [PubMed] [Google Scholar]
- 2.Arendrup, M., C. Nielsen, J. E. Hansen, C. Pedersen, L. Mathiesen, and J. O. Nielsen. 1992. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 5**:**303-307. [PubMed] [Google Scholar]
- 3.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6**:**200-206. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11**:**S3-S16. [PubMed] [Google Scholar]
- 5.Berkowitz, R. D., A. B. van't Wout, N. A. Kootstra, M. E. Moreno, V. D. Linquist-Stepps, C. Bare, C. A. Stoddart, H. Schuitemaker, and J. M. McCune. 1999. R5 strains of human immunodeficiency virus type 1 from rapid progressors lacking X4 strains do not possess X4-type pathogenicity in human thymus. J. Virol. 73**:**7817-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogers, W. M., C. Cheng-Mayer, and R. C. Montelaro. 2000. Developments in preclinical AIDS vaccine efficacy models. AIDS 14(Suppl. 3)**:**S141-S151. [PubMed] [Google Scholar]
- 7.Bradney, A. P., S. Scheer, J. M. Crawford, S. P. Buchbinder, and D. C. Montefiori. 1999. Neutralization escape in human immunodeficiency virus type 1-infected long-term nonprogressors. J. Infect. Dis. 179**:**1264-1267. [DOI] [PubMed] [Google Scholar]
- 8.Cayabyab, M., G. B. Karlsson, B. Etemad-Moghadam, W. Hofman, T. Steenbeke, M. Halloran, J. W. Fanton, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-hxbc2). J. Virol. 73**:**976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabarti, L. A., T. Ivanovic, and C. Cheng-Mayer. 2002. Properties of the surface envelope glycoprotein associated with virulence of simian-human immunodeficiency virus SHIVSF33A molecular clones. J. Virol. 76**:**1588-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer, C., A. Brown, J. M. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73**:**5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer, C., R. Liu, N. R. Landau, and L. Stamatatos. 1997. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J. Virol. 71**:**1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng-Mayer, C., D. Seto, M. Tateno, and J. A. Levy. 1988. Biologic features of HIV-1 that correlate with virulence in the host. Science 240**:**80-82. [DOI] [PubMed] [Google Scholar]
- 13.Ciurea, A., P. Klenerman, L. Hunziker, E. Horvath, B. M. Senn, A. F. Ochsenbein, H. Hengartner, and R. M. Zinkernagel. 2000. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc. Natl. Acad. Sci. USA 97**:**2749-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206**:**935-944. [DOI] [PubMed] [Google Scholar]
- 15.Connor, R. I., and D. D. Ho. 1994. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J. Virol. 68**:**4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185**:**621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delwart, E. L., and C. J. Gordon. 1997. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 12**:**348-354. [DOI] [PubMed] [Google Scholar]
- 18.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74**:**8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75**:**8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276**:**229-237. [DOI] [PubMed] [Google Scholar]
- 21.Etemad-Moghadam, B., D. Rhone, T. Steenbeke, Y. Sun, J. Manola, R. Gelman, J. W. Fanton, P. Racz, K. Tenner-Racz, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD4+-T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J. Virol. 75**:**5646-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, M. Fernandes, K. Liou, R. Gomila, J. Lee, and J. Sodroski. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 74**:**4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenyo, E. M., J. Albert, and B. Asjo. 1989. Replicative capacity, cytopathic effect and cell tropism of HIV. AIDS 3S**:**S5-S12. [DOI] [PubMed] [Google Scholar]
- 24.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100**:**587-597. [DOI] [PubMed] [Google Scholar]
- 25.Haddrick, M., C. R. Brown, R. Plishka, A. Buckler-White, V. M. Hirsch, and H. Ginsberg. 2001. Biologic studies of chimeras of highly and moderately virulent molecular clones of simian immunodeficiency virus SIVsmPBj suggest a critical role for envelope in acute AIDS virus pathogenesis. J. Virol. 75**:**6645-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harouse, J. M., A. Gettie, T. Eshetu, R. C. H. Tan, R. Bohm, J. Blanchard, G. Baskin, and C. Cheng-Mayer. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific SHIVSF162P3. J. Virol. 75**:**1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harouse, J. M., A. Gettie, R. C. H. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284**:**816-819. [DOI] [PubMed] [Google Scholar]
- 28.Harouse, J. M., A. Gettie, R. C. H. Tan, T. Eshetu, M. Ratteree, J. Blanchard, and C. Cheng-Mayer. 2001. Pathogenic determinants of the mucosally transmissible CXCR4-specific SHIVSF33A2 map to env region. J. Acquir. Immune Defic. Syndr. 27**:**222-228. [DOI] [PubMed] [Google Scholar]
- 29.Harouse, J. M., R. C. H. Tan, A. Gettie, P. Dailey, P. A. Marx, P. A. Luciw, and C. Cheng-Mayer. 1998. Mucosal transmission of pathogenic CXCR4-utilizing SHIVSF33A variants in rhesus macaques. Virology 248**:**95-107. [DOI] [PubMed] [Google Scholar]
- 30.Himathongkham, S., N. S. Halpin, J. Li, M. W. Stout, C. J. Miller, and P. A. Luciw. 2000. Simian-human immunodeficiency virus containing a human immunodeficiency virus type 1 subtype-E envelope gene: persistent infection, CD4+ T-cell depletion, and mucosal membrane transmission in macaques. J. Virol. 74**:**7851-7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76**:**7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung, C.-S., N. Vander Heyden, and L. Ratner. 1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol. 73**:**8216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igarashi, R., C. R. Brown, R. A. Byrum, Y. Nishimura, Y. Endo, R. J. Plishka, C. Buckler, A. Buckler-White, G. Miller, V. M. Hirsch, and M. A. Martin. 2002. Rapid and irreversible CD4+-T-cell depletion induced by the highly pathogenic simian/human immunodeficiency virus SHIVDH12R is systemic and synchronous. J. Virol. 76**:**379-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joag, S. V., I. Adany, Z. Li, L. Foresman, D. M. Pinson, C. Wang, E. B. Stephens, R. Raghavan, and O. Narayan. 1997. Animal model of mucosally transmitted human immunodeficiency virus type 1 disease: intravaginal and oral deposition of simian/human immunodeficiency virus in macaques results in systemic infection, elimination of CD4+ T cells, and AIDS. J. Virol. 71**:**4016-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joag, S. V., Z. Li, L. Foresman, E. B. Stephens, L. J. Zhao, I. Adany, D. M. Pinson, H. M. McClure, and O. Narayan. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70**:**3189-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson, G. B., M. Halloran, J. Li, I. W. Park, R. Gomila, K. A. Reimann, M. K. Axthelm, S. A. Iliff, N. L. Letvin, and J. Sodroski. 1997. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 71**:**4218-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson, G. B., M. Halloran, D. Schenten, J. Lee, P. Racz, K. Tenner-Racz, J. Manola, R. Gelman, B. Etemad-Moghadam, E. Desjardins, R. Wyatt, N. P. Gerard, L. Marcon, D. Margolin, J. Fanton, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 1998. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J. Exp. Med. 188**:**1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5**:**535-541. [DOI] [PubMed] [Google Scholar]
- 39.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118**:**681-688. [DOI] [PubMed] [Google Scholar]
- 40.Kreisberg, J. F., D. Kwa, B. Schramm, V. Trautner, R. Connor, H. Schuitemaker, J. I. Mullins, A. B. van't Wout, and M. A. Goldsmith. 2001. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J. Virol. 75**:**8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393**:**648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaBonte, J. A., T. Patel, W. Hofmann, and J. Sodroski. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J. Virol. 74**:**10690-10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaBranche, C. C., G. Galasso, J. P. Moore, D. P. Bolognesi, M. S. Hirsch, and S. M. Hammer. 2001. HIV fusion and its inhibition. Antivir. Res. 50**:**95-115. [DOI] [PubMed] [Google Scholar]
- 44.LeGuern, M., T. Shioda, J. A. Levy, and C. Cheng-Mayer. 1993. Single amino acid change in Tat determines the different rates of replication of two sequential HIV-1 isolates. Virology 195**:**441-447. [DOI] [PubMed] [Google Scholar]
- 45.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57**:**183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis, J., P. Balfe, C. Arnold, S. Kaye, R. S. Tedder, and J. A. McKeating. 1998. Development of a neutralizing antibody response during acute primary human immunodeficiency virus type 1 infection and the emergence of antigenic variants. J. Virol. 72**:**8943-8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. R. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71**:**9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luciw, P. A., C. P. Mandell, S. Himathongkham, J. Li, T. A. Low, K. A. Schmidt, K. E. S. Shaw, and C. Cheng-Mayer. 1999. Fatal immunopathogenesis by SIV/HIV-1 (SHIV) containing a variant form of the HIV-1SF33 env gene in juvenile and newborn rhesus macaques. Virology 263**:**112-127. [DOI] [PubMed] [Google Scholar]
- 49.Luciw, P. A., E. Pratt-Lowe, K. E. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. USA 92**:**7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lue, J., M. Hsu, D. Yang, P. Marx, Z. Chen, and C. Cheng-Mayer. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76**:**10299-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74**:**11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6**:**207-210. [DOI] [PubMed] [Google Scholar]
- 53.Mellors, J. W., C. R. J. Rinaldo, P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272**:**1167-1170. [DOI] [PubMed] [Google Scholar]
- 54.Meng, G., X. Wei, X. Wu, M. T. Sellers, J. M. Decker, Z. Moldoveanu, J. M. Orenstein, M. F. Graham, J. C. Kappes, J. Mestecky, G. M. Shaw, and P. D. Smith. 2002. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat. Med. 8**:**150-156. [DOI] [PubMed] [Google Scholar]
- 55.Miller, C. J., M. Marthas, J. Greenier, D. Lu, P. Dailey, and Y. Lu. 1998. In vivo replication capacity rather than in vitro macrophage tropism predicts efficiency of vaginal transmission of simian immunodeficiency virus or simian/human immunodeficiency virus in rhesus macaques. J. Virol. 72**:**3248-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novembre, F. J., P. R. Johnson, M. G. Lewis, D. C. Anderson, S. Klumpp, H. M. McClure, and V. M. Hirsch. 1993. Multiple viral determinants contribute to pathogenicity of the acutely lethal simian immunodeficiency virus SIVsmmPBj variant. J. Virol. 67**:**2466-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75**:**8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75**:**4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puffer, B. A., S. Pohlmann, A. L. Edinger, D. Carlin, M. D. Sanchez, J. Reitter, D. D. Watry, H. S. Fox, R. C. Desrosiers, and R. W. Doms. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76**:**2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I. W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus macaques. J. Virol. 70**:**6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reimann, K. A., J. T. Li, G. Voss, C. Lekutis, K. Tenner-Racz, P. Racz, W. Lin, D. C. Montefiori, D. E. Lee-Parritz, Y. Lu, R. G. Collman, J. Sodroski, and N. L. Letvin. 1996. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J. Virol. 70**:**3198-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280**:**1949-1953. [DOI] [PubMed] [Google Scholar]
- 63.Rowland-Jones, S. L. 1999. HIV: the deadly passenger in dendritic cells. Curr. Biol. 9**:**R248-R250. [DOI] [PubMed]
- 64.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72**:**209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66**:**1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schuitemaker, H., N. A. Kootstra, R. E. de Goede, F. de Wolf, F. Miedema, and M. Tersmette. 1991. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65**:**356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scoggins, R. M., J. R. J. Taylor, J. Patrie, A. B. van't Wout, H. Schuitemaker, and D. Camerini. 2000. Pathogenesis of primary R5 human immunodeficiency virus type 1 clones in SCID-hu mice. J. Virol. 74**:**3205-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5**:**204-210. [DOI] [PubMed] [Google Scholar]
- 69.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M. A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176**:**362-373. [DOI] [PubMed] [Google Scholar]
- 70.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75**:**4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71**:**7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tersmette, M., R. E. de Goede, B. J. Al, I. N. Winkel, R. A. Gruters, H. T. Cuypers, H. G. Huisman, and F. Miedema. 1988. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 62**:**2026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR5. Nature 384**:**184-187. [DOI] [PubMed] [Google Scholar]
- 74.van't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72**:**5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veazey, R. S., M. Rosenzweig, D. E. Shvetz, D. R. Pauley, M. DeMaria, L. V. Chalifoux, R. P. Johnson, and A. A. Lackner. 1997. Characterization of gut-associated lymphoid tissue (GALT) of normal rhesus macaques. Clin. Immunol. Immunopathol. 82**:**230-242. [DOI] [PubMed] [Google Scholar]
- 76.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 9**:**1051-1053. [DOI] [PubMed] [Google Scholar]
- 77.Wu, L., N. P. Gerard, R. Wyatt, S. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardins, N. W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384**:**179-183. [DOI] [PubMed] [Google Scholar]