Differential N-Linked Glycosylation of Human Immunodeficiency Virus and Ebola Virus Envelope Glycoproteins Modulates Interactions with DC-SIGN and DC-SIGNR (original) (raw)
Abstract
The C-type lectins DC-SIGN and DC-SIGNR [collectively referred to as DC-SIGN(R)] bind and transmit human immunodeficiency virus (HIV) and simian immunodeficiency virus to T cells via the viral envelope glycoprotein (Env). Other viruses containing heavily glycosylated glycoproteins (GPs) fail to interact with DC-SIGN(R), suggesting some degree of specificity in this interaction. We show here that DC-SIGN(R) selectively interact with HIV Env and Ebola virus GPs containing more high-mannose than complex carbohydrate structures. Modulation of _N_-glycans on Env or GP through production of viruses in different primary cells or in the presence of the mannosidase I inhibitor deoxymannojirimycin dramatically affected DC-SIGN(R) infectivity enhancement. Further, murine leukemia virus, which typically does not interact efficiently with DC-SIGN(R), could do so when produced in the presence of deoxymannojirimycin. We predict that other viruses containing GPs with a large proportion of high-mannose _N_-glycans will efficiently interact with DC-SIGN(R), whereas those with solely complex _N_-glycans will not. Thus, the virus-producing cell type is an important factor in dictating both _N_-glycan status and virus interactions with DC-SIGN(R), which may impact virus tropism and transmissibility in vivo.
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) enter cells through a fusion reaction mediated by the viral envelope glycoprotein (Env) that is triggered when Env binds CD4 and a coreceptor, generally the chemokine receptors CCR5 or CXCR4 (reviewed in reference 49). Although the presence of these receptors is required for infection by primary HIV type 1 (HIV-1) strains, attachment of virus to the cell surface can be facilitated by interactions with other cell surface molecules and is typically rate limiting for virus infection in vitro (reviewed in references 24 and 53). Attachment can occur through interactions between Env and cell surface molecules such as heparan sulfate proteoglycans or by cellular proteins incorporated into the virus membrane that interact with their physiologic ligands on the cell surface. How virus attaches to the cell surface may alter infection kinetics, efficiency, and possibly viral tropism (53).
HIV bound to dendritic cells (DCs) can be transmitted to T cells with high efficiency, raising the possibility that DCs play an important role in virus transmission and dissemination in vivo (6, 18). The C-type lectin DC-SIGN (DC-specific ICAM-3 grabbing nonintegrin) binds HIV gp120 with high affinity (8) and is expressed at high levels on some types of DCs, accounting in part for the ability of these cells to capture and transmit HIV (18). DC-SIGN is a homotetrameric, type II membrane protein that contains at its C terminus a carbohydrate recognition domain (CRD) (8, 14, 39). DC-SIGN-positive cells are abundant in both human and rhesus macaque rectal and vaginal mucosa, and all HIV-1, HIV-2, and SIV strains studied to date bind DC-SIGN (18, 26, 43). A related molecule, termed DC-SIGNR (for DC-SIGN related) or L-SIGN (for liver/lymph node-specific ICAM-3 grabbing nonintegrin), is expressed on endothelial cells in the liver, lymph node, gastrointestinal tract, and placenta and exhibits a binding profile similar to that of DC-SIGN (4, 26, 45, 51). Although DC-SIGN does not serve as a receptor for HIV entry, it does promote efficient HIV infection of CD4+ T cells either in trans (18) or in cis (31). If mucosal DCs play an important role in HIV transmission, then it will be important to determine exactly how HIV interacts with DC-SIGN and other attachment factors.
In the present study, we found that DC-SIGN and DC-SIGNR [collectively referred to as DC-SIGN(R)] selectively bound HIV gp120 molecules that were enriched in high-mannose oligosaccharides. We confirmed earlier studies that showed that Env from HIV produced in macrophages contains N-linked carbohydrate structures that are more complex than those found on virus produced in peripheral blood mononuclear cells (PBMCs) (33, 34, 55). This differential glycosylation of HIV Env affected interactions with DC-SIGN(R) in that HIV derived from T-cell lines or PBMCs was bound and transmitted well by these lectins, whereas HIV derived from macrophages was bound and transmitted poorly. DC-SIGN(R) can also enhance the infection of Ebola virus glycoprotein (GP) pseudovirions (2, 50a). We found that differences in Ebola virus GP _N_-glycan composition affected the efficiency of DC-SIGN(R) enhancement. We also show that the hepatic asialoglycoprotein receptor (ASGP-R), which interacts with galactose moieties (38), can enhance infection by Ebola virus GP in a manner dictated by the _N_-glycan status of GP. Thus, N-linked carbohydrate content and composition determine the virus-binding specificity and transmission efficiency by DC-SIGN(R) and perhaps other C-type lectins. Interactions between viral GPs and these attachment factors in vivo will likely be dependent in part upon the cell type in which virus is produced, and we predict that other viruses containing a large proportion of high-mannose _N_-glycans may interact with DC-SIGN(R) efficiently.
MATERIALS AND METHODS
Plasmids.
HIV-1 HXBc2, HIV-2/vcp, and SIVmac316 gp120 plasmids have been described previously (35a). Bridget A. Puffer (University of Pennsylvania) provided SIVmac239 gp120 cloned in pcDNA3 (Invitrogen). The HIV-2/sbl/isy env gene from KF-3 (23) was cloned into pSP73 (Promega) at _Hin_dIII and _Eco_RV sites. A stop codon at the gp120-gp41 cleavage site was generated by QuikChange (Stratagene). Plasmids expressing CD4, CXCR4, CCR5, and CCR5/N13D (25, 35); DC-SIGN, DC-SIGNR, rhesus DC-SIGN, pigtailed DC-SIGN, and murine DC-SIGN (3, 43-45); EboZ-GP and EboS-GP (50); and vesicular stomatitis virus G (VSV-G) and amphotropic murine leukemia virus (MLV) Env (pHit 456) have been described (50a). The ASGP-R1 subunit of the hepatic ASGP-R was PCR amplified from a human liver cDNA library (Clontech) and was subcloned into pcDNA6-B (Invitrogen) by using _Nhe_I and _Sae_II to give a C-terminal V5/His tag. The _env_-defective pNL4-3.Luc.R−E− luciferase reporter plasmid has been described (7).
A proviral clone expressing HIV-2/vcp Env (35) was constructed by first cloning the HIV-2/ROD/A env gene from pACR23 (46) into pCR3.1 (Invitrogen) with _Hin_dIII and _Bam_HI. The HIV-2/vcp env gene in pCR3.1 (11) was recloned into pGEM-4 (Promega) with _Eco_RI. The untranslated region 5′ of the HIV-2/VCP ATG start site was changed to that of ROD/A by using QuikChange, allowing exchange of the ROD/A env gene in pCR3.1 with that of HIV-2/vcp by using _Bsm_BI and _Bam_HI. VCP Env was then inserted into pACR23 with _Sac_I and _Bam_HI as described previously (46), generating pACR23.VCP.Env.
Cells and viruses.
QT6, 293T, HeLa, and DC-SIGN, and DC-SIGNR 293 T-Rex cells (43) were grown in Dulbecco modified Eagle medium-high glucose-10% fetal bovine serum (DMEM-10). SupT1, CEMx174, THP, and THP-DC-SIGN cells were grown in RPMI 1640-10% fetal bovine serum (RPMI-10). THP and THP-DC-SIGN cells (18) were provided by Dan Littman (New York University). SupT1/CCR5 cells (37) were grown in RPMI-10, with 300 ng of puromycin/ml. PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation of fresh blood obtained under an Institutional Review Board-approved protocol and were stimulated with phytohemagglutinin for 3 days, washed, and cultured in RPMI-10 with 20 U of interleukin-2/ml. Monocytes were obtained as described previously (54). Monocyte-derived macrophages (MDMs) were obtained by culturing monocytes for 7 days in RPMI-10% human serum (Sigma). Immature monocyte-derived DCs (MDDCs) were obtained by culturing monocytes for 7 days in RPMI-10 with 50 ng of granulocyte-macrophage colony-stimulating factor and 100 ng of interleukin-4/ml (54). Viral stocks were harvested from infected cells at peak infection, clarified of cell debris by centrifugation, and filtered through 0.22-μm (pore-size) filters. HIV-2/vcp stock was generated by electroporating SupT1 cells with pACR23.VCP.Env. HIV-1 Ba-L was also generated in the presence of mannosidase I inhibitor, deoxymannojirimycin (DMJ) (Sigma) by incubating SupT1/CCR5 cells overnight with Ba-L, pelleting the cells, washing the cells with RPMI-10, resuspending the cells in RPMI-10 containing 2.5 mM DMJ, and then harvesting the virus as described above.
HIV and SIV gp120 proteins.
HIV and SIV gp120 proteins were generated as described previously (35a). 293T cells were infected for 1 h at 37°C with vaccinia virus WR for HXBc2 and VCP gp120 or vTF1.1 expressing T7 RNA polymerase (1) for SBL/ISY, SIVmac239, and SIVmac316 gp120. Infected cells were then calcium phosphate transfected with the appropriate gp120 plasmid for 4 h. HIV-1 JR-FL gp120 was generated with vBD6 (9). After transfection or vBD6 infection, the medium was replaced with DMEM-10 with 100 μg of rifampin/ml (DMEM-10-R). After overnight expression at 37°C, medium containing gp120 was clarified by centrifugation. Proteins were also expressed in the presence of mannosidase inhibitors (1.0 mM DMJ or 0.1 mM swainsonine [Sigma]).
Receptor binding assays.
A cell surface binding assay was used as described previously (35a) and adapted to analyze Env from viruses. QT6 quail receptor cells were generated by infection with vTF1.1, transfection with T7 promoted receptor plasmids, and expression overnight at 37°C in DMEM-10-R. Cells were then detached from flasks, pelleted, and resuspended in 200 μl of gp120 containing supernatant, with or without 100 nM soluble CD4 (sCD4) or 500 μl of virus supernatant supplemented with 100 μg of rifampin/ml and then incubated for 1 h at 37°C. For studies with MDDCs, gp120 containing supernatant was optionally supplemented with anti-CD4 monoclonal antibody (MAb) 19 (11) and/or mannan (Sigma). Cells were washed with 4°C phosphate-buffered saline containing Ca2+ and Mg2+ and lysed with 4°C lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris [pH 8.0], and Complete protease inhibitor cocktail [Roche Molecular Biochemicals]). Lysates were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotted for bound gp120 with rabbit serum 1169 (25) for HIV-1 vaccinia gp120, J1 (42) for HIV-1 virus gp120, or MAb DA6 (10) for HIV-2 and SIV Env. The appropriate anti-rabbit or anti-mouse peroxidase conjugate was then used (Jackson Immunoresearch Laboratories), and Western blots were visualized by using enhanced chemiluminescence and autoradiography film.
Glycosylation analysis.
Cell lysates from HIV gp120 binding assays or Env from viruses were diluted in MES buffer (20 mM morpholineethanesulfonic acid, 130 mM NaCl, and 10 mM CaCl2) precipitated overnight with _Galanthus nivalis_-agarose beads (Sigma), washed with MES buffer, treated with neuraminidase, endo-β-_N_-acetylglucosaminidase H (Endo H), or peptide N-glycosidase F (PNGase F) according to the recommendations of the manufacturer (New England Biolabs), and then analyzed by SDS-PAGE and Western blot and visualized as described above.
HIV transmission assays.
Twofold serial dilutions of HIV-1 Ba-L grown in PBMCs MDMs, SupT1/CCR5, or SupT1/CCR5 cells with 2.5 mM DMJ were incubated in RPMI-10 or 5 × 104 MDDCs, THP cells, or THP-DC-SIGN cells for 2 h at 37°C. Then, 5 × 104 SupT1/CCR5 target cells were added and cocultured for 5 days. Cocultures were then passaged onto 105 SupT1/CCR5 cells. After another 3 days of infection, cultures were again passaged to establish a productively infected culture. After a total of 11 days, infection was assessed by p24 enzyme-linked immunosorbent assay, and the last dilution yielding a productive infection for each condition and virus source was noted. The fold difference in dilution factor over the control condition (THP versus THP-DC-SIGN or media versus MDDCs) was used as a measure of infectivity enhancement.
Pseudotype infection assays.
Ebola virus or MLV Env pseudovirions were generated by cotransfecting 293T cells with Env expression plasmids and pNL4-3.Luc.R−E− as described previously (7) in the presence or absence of 2.5 mM DMJ. Additionally, MDMs were used to generate EboZ-GP pseudovirions by transducing 5-day-old MDMs with pNL4.3.Luc.R−E− pseudotyped with VSV-G at a multiplicity of infection (MOI) of 100. After 24 to 48 h., the cells were transduced for 6 h with an adenovirus expressing EboZ-GP at an MOI of 50 (50). Cells were extensively washed and incubated in fresh medium for a further 24 h before pseudovirions were harvested. For target cells, either receptor-transfected 293T cells expressed for 48 h, DC-SIGN or DC-SIGNR 293 T-Rex cells induced with 0.01 μg of doxycycline/ml overnight, or control 293 T-Rex cells were used. Target cells were infected with either 500 50% tissue culture infective dose(s) (TCID50; determined on 293T cells) or normalized p24 values. After 2 days, luciferase expression was quantified by using a commercial kit (Promega).
Plant lectin analysis of Ebola virus GP.
Ebola GP pseudovirions were filtered through a 0.45-μm (pore-size) filter and pelleted through a 20% sucrose cushion at 40,000 rpm for 1 h in an SW41 rotor. Pelleted virions were lysed in cold radioimmunoprecipitation assay buffer (1% NP-40, 0.5% deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris [pH 8.0]), precipitated with lectin-biotin conjugates (Vector Labs) and streptavidin-agarose (Sigma), and analyzed by SDS-PAGE and Western blot for GP with a polyclonal rabbit serum raised against the soluble form of GP. The lectins used and their specificities were as follows: Vivia villosa lectin (terminal _N_-acetylgalactosamine), Ricinus communiz agglutinin (galactose), concanavalin A (α-linked mannose, glucose), Datura stramonium lectin (_N_-acetylglucosamine-_N_-acetyllactosamine), Erythrina cristagalli lectin [galactosyl(β1-4)_N_-acetylglucosamine], wheat germ agglutinin (_N_-acetylglucosamine), Galanthus nivalis lectin (nonreducing end of terminal α-linked mannose), peanut agglutinin [galactosyl(β1-3)_N_-acetylgalactosamine], Jacalin [galactosyl(β1-3)_N_-acetylgalactosamine], and Ulex europaeus agglutinin (fucose).
RESULTS
Binding of HIV and SIV gp120 to DC-SIGN(R) results in molecules differing in N-linked carbohydrates.
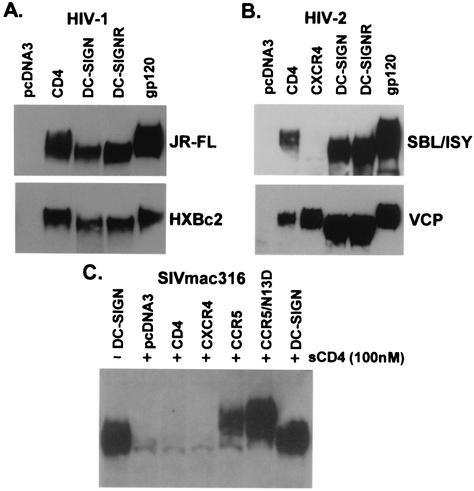
To analyze gp120-receptor interactions, we produced HIV and SIV gp120 and bound it to cells expressing CD4, chemokine receptors, DC-SIGN, or DC-SIGNR. Soluble CD4 was used as necessary for chemokine receptor binding. After removal of the unbound protein, we analyzed receptor-bound gp120 by SDS-PAGE and Western blot. We found that gp120 bound to DC-SIGN(R) or rhesus macaque, pigtailed macaque, or murine DC-SIGN migrated at a lower molecular weight than when bound to CD4 or chemokine receptors (Fig. 1 and data not shown). This observation was made for all HIV-1, HIV-2, and SIV gp120 proteins tested, although the change in molecular weight was less pronounced for HIV-1 gp120. To determine whether the more rapid migration of gp120 bound to DC-SIGN(R) relative to that bound to CD4/chemokine receptor was due to differences in carbohydrates or protein backbone, receptor-bound gp120 was recovered by precipitation and treated with PNGase F, an enzyme that removes all N-linked carbohydrate structures. After PNGase F treatment, all gp120 proteins migrated equivalently and at the expected size of ca. 60K for a fully deglycosylated protein (Fig. 2A and C). Thus, gp120 bound to DC-SIGN(R) migrated more quickly than gp120 bound to CD4/chemokine receptors due to differences in N-linked carbohydrates.
FIG. 1.
HIV-1, HIV-2, and SIV gp120 binding to CD4, chemokine receptors, or DC-SIGN(R). gp120 produced in 293T cells was bound to receptor expressing cells with or without sCD4 as indicated, and the bound protein was analyzed by SDS-PAGE and Western blot. Unbound gp120 is also shown in panels A and B (lanes labeled gp120). (A) HIV-1 gp120 proteins JR-FL and HXBc2 were bound to CD4, DC-SIGN, DC-SIGNR, or no receptor (pcDNA3). (B) HIV-2 gp120 proteins SBL/ISY and VCP were additionally evaluated on CXCR4. VCP, a CD4 independent Env, can bind directly to CXCR4, whereas SBL/ISY gp120 cannot. (C) SIVmac316 gp120 was evaluated on CCR5 or CCR5 with an aspartic acid at position 13 (CCR5/N13D), which confers more efficient binding of SIV gp120 (36).
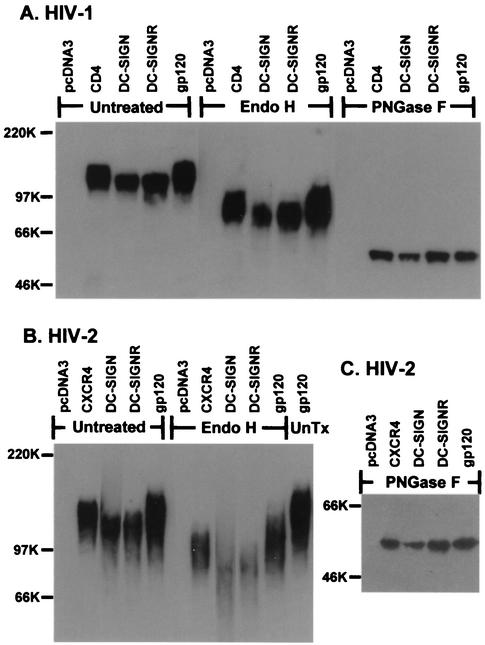
FIG. 2.
Glycosidase analysis of receptor bound HIV-1 or HIV-2 gp120. (A) HIV-1 HXBc2 or (B) HIV-2 VCP gp120 was bound to cells expressing the indicated receptor. Cells were then washed and lysed. The gp120 in lysates was precipitated, digested with Endo H, PNGase F, or left untreated (UnTx), and then analyzed by SDS-PAGE and Western blot. Input gp120 (lanes labeled gp120) was also digested to serve as a control.
HIV and SIV gp120 bound to DC-SIGN(R) is enriched in high-mannose carbohydrates.
HIV Env contains a large number of _N_-glycans that are differentially processed in the Golgi, resulting in a mixture of complex, hybrid, and high-mannose carbohydrate structures (21, 40, 41). To determine how N-linked carbohydrates on gp120 bound to DC-SIGN(R) differed from those on gp120 bound to CD4/chemokine receptors, we treated bound gp120 proteins with Endo H, an enzyme that only removes high-mannose/hybrid _N_-glycans and not complex carbohydrate structures. After Endo H treatment, the migration differences between gp120 bound to DC-SIGN(R) and gp120 bound to CD4/chemokine receptors remained (Fig. 2A and B). Likewise, neuraminidase treatment of bound gp120 molecules to remove sialic acid residues, which are located on terminal branches of complex _N_-glycans, did not eliminate the molecular weight shift (data not shown). Thus, we concluded that DC-SIGN(R)-bound gp120 molecules contained a higher number of high-mannose and/or fewer complex carbohydrate structures, resulting in more rapid migration in SDS-PAGE.
DC-SIGN(R) selectively bind HIV and SIV gp120 molecules enriched in high-mannose carbohydrates.
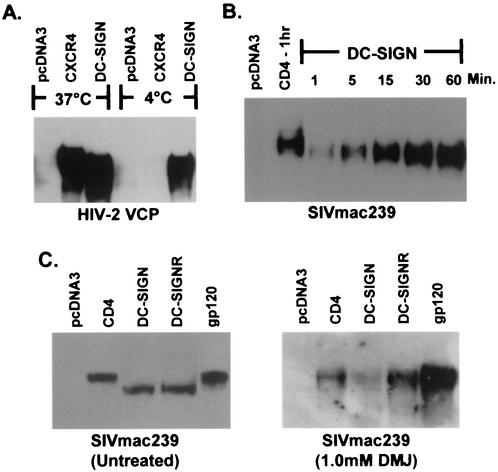
Since gp120 bound to DC-SIGN can be rapidly internalized (8, 29) and may enter into cellular compartments involved in protein processing for subsequent presentation by major histocompatibility complex (MHC) molecules (12), the _N_-glycans of gp120 bound to DC-SIGN could potentially be modified or removed by glycosidases or DC-SIGN(R) itself. Alternatively, DC-SIGN(R) could selectively bind gp120 molecules enriched in high-mannose _N_-glycans, which would migrate more quickly in SDS-PAGE than gp120 molecules containing a greater proportion of complex carbohydrate structures. To investigate these possibilities, we bound gp120 to DC-SIGN at 4°C with sodium azide and found that gp120 bound DC-SIGN well and that the molecular weight shift was retained (Fig. 3A), indicating that internalization was not required for this effect. We also found that gp120 could bind DC-SIGN(R) within 1 min and that bound protein always migrated at a lower molecular weight (Fig. 3B and data not shown). Although other structural changes may take place upon gp120 binding to DC-SIGN(R), these results make it unlikely that active modification of _N_-glycans on gp120 occurs upon binding to DC-SIGN(R) and are more consistent with selective binding of gp120 proteins enriched in high-mannose _N_-glycans from the heterogeneous, differentially glycosylated gp120 population.
FIG. 3.
Temperature, kinetic, and differential glycosylation analysis of receptor bound HIV-2 or SIV gp120. gp120 produced in 293T cells was bound to receptor expressing cells and analyzed as described in Fig. 1. (A) VCP gp120 was bound to cells expressing CXCR4, DC-SIGN, or no receptor (pcDNA3) for 1 h either at 37 or at 4°C in the presence of 0.1% sodium azide. (B) SIVmac239 gp120 was bound to cells expressing CD4 or DC-SIGN at 37°C for 1 to 60 min as indicated. (C) SIVmac239 gp120 generated in the absence (left panel) or presence (right panel) of 1.0 mM DMJ was bound to cells expressing CD4, DC-SIGN, DC-SIGNR, or no receptor (pcDNA3) and then analyzed by SDS-PAGE and Western blot. Unbound gp120 (lanes labeled gp120) is also shown. Results with HIV-2 or SIV gp120s are shown because of the more pronounced molecular weight change with these proteins; however, results obtained with HIV-1 gp120s were similar (data not shown).
DC-SIGN(R) binding characteristics of differentially glycosylated HIV and SIV gp120 proteins.
To test the hypothesis that DC-SIGN(R) selectively bind HIV and SIV gp120 proteins that are enriched in high-mannose _N_-glycans, we manipulated the carbohydrate composition of gp120 by using a mannosidase I inhibitor (DMJ) or a mannosidase II inhibitor (swainsonine), generating proteins containing only high-mannose or high-mannose/hybrid oligosaccharides, respectively (28). All gp120 proteins tested made in the presence of DMJ bound CD4/chemokine receptor and DC-SIGN(R) and migrated identically in SDS-PAGE (Fig. 3C and data not shown). Thus, there was no selective binding of gp120 when only high-mannose _N_-glycans were present. In contrast, gp120 made in the presence of swainsonine migrated more rapidly when bound to DC-SIGN(R) than to CD4/chemokine receptor, indicating that hybrid _N_-glycans can affect DC-SIGN(R) binding in a manner similar to complex carbohydrate structures (data not shown). The mannosidase inhibitors were effective since gp120 produced in DMJ was resistant to neuraminidase but fully sensitive to Endo H, whereas gp120 produced in swainsonine was sensitive to both neuraminidase and Endo H (data not shown and Fig. 4E).
FIG. 4.
Receptor binding and glycosylation characteristics of Env from viruses grown in T-cell lines, PBMCs, pr MDMs or in the presence of DMJ. Supernatant from HIV- or SIV-infected cells was used in receptor binding assays. Bound gp120 was analyzed by SDS-PAGE and Western blot. HIV-1, HIV-2, and SIV grown in T-cell lines (A), PBMCs (B), or MDMs (C) were bound to cells expressing CD4, DC-SIGN, DC-SIGNR, or no receptor. (D and E) HIV-1 Ba-L virus from PBMCs or MDMs (D) or SupT1/CCR5 cells (E) grown in the presence or absence of 2.5 mM DMJ (absence labeled as UnTx and presence labeled as DMJ) was digested with Endo H (H) or PNGase F (F) or left untreated (UnTx) and then analyzed by SDS-PAGE and Western blot for gp120.
DC-SIGN(R) binding characteristics of HIV and SIV grown in T-cell lines, PBMCs and macrophages.
HIV grown in T-cell lines, PBMCs and MDMs differs in N-linked glycosylation of Env (33, 34, 55) with gp120 from virus grown in MDMs containing _N_-acetyllactosamine repeats on complex chains, resulting in a more heterogeneous pattern of glycosylation compared to gp120 from virus grown in PBMCs (33, 34, 55). We confirmed that gp120 from HIV and SIV grown from T-cell lines and PBMCs was more homogeneous, migrated more quickly in SDS-PAGE, and was more sensitive to Endo H digestion than when produced in MDMs, findings consistent with a greater number of high-mannose carbohydrates (Fig. 4D and data not shown). When PNGase F was used, the proteins migrated at the same position, showing that differences in migration were indeed due to differences in N-linked carbohydrates (Fig. 4D and data not shown). When tested in binding assays, gp120 obtained from virus infected T-cell lines and PBMCs bound efficiently to CD4 and DC-SIGN(R) (Fig. 4A and B). In contrast, gp120 from virus-infected MDMs bound CD4, but the level of binding to DC-SIGN(R) was below the level of detection in this assay (Fig. 4C). We conclude that the more complex structures present on Env from MDM-derived virus prevent efficient interactions with DC-SIGN(R).
HIV from PBMCs is transmitted by DC-SIGN more efficiently than MDM-derived HIV.
To test whether differential glycosylation of HIV Env from virus derived from PBMCs or MDMs had any effect on the ability of DC-SIGN to enhance infectivity, we performed transmission assays in a limiting-dilution format and found that THP-DC-SIGN cells could only enhance infectivity of virus grown from PBMCs and not from MDMs (Table 1), supporting the notion that the complex carbohydrate structures on Env from MDM-derived virus reduce efficient interactions with DC-SIGN. Remarkably, when HIV was produced in the presence of DMJ, allowing formation of only high-mannose carbohydrate structures, transmission by THP-DC-SIGN cells was enhanced >2,000-fold compared to 20-fold enhancement for virus grown in the absence of DMJ (Table 1). Endo H digestion confirmed that such treatment was efficient (Fig. 4E). In contrast to THP-DC-SIGN cells, we found that MDDCs could enhance the infectivity of virus grown from both PBMCs and MDMs (Table 1), suggesting that something other than DC-SIGN on DCs is able to enhance virus transmission.
TABLE 1.
Transmission of HIV-1 Ba-L to SupT1/CCR5 T cells in trans
| Virus sourcea | Comparison | Infectivity enhancementb |
|---|---|---|
| PBMCs | THP vs THP-DC-SIGN | 6 (22, 23) |
| MDMs | THP vs THP-DC-SIGN | 0 (0, 0) |
| SupT1/CCR5 cells | THP vs THP-DC-SIGN | 20 (23, 25) |
| SupT1/CCR5-2.5 mM DMJ cells | THP vs THP-DC-SIGN | 2,560 (210, 212) |
| PBMCs | Media vs MDDC | 34 (22, 26) |
| MDMs | Media vs MDDC | 20 (23, 25) |
Characteristics of HIV and SIV gp120 binding to MDDCs.
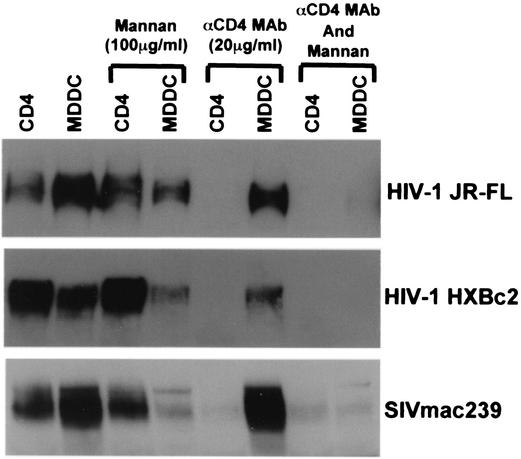
Because MDDCs were able to transmit virus derived from MDMs, which interacts poorly with DC-SIGN, we analyzed the gp120 binding properties of MDDCs. Since MDDCs also contain CD4, we assessed gp120 binding in the presence of an anti-CD4 MAb, as well as mannan, which blocks gp120 binding to DC-SIGN (8, 18, 43). We found that MDDCs did not selectively bind gp120 molecules containing larger numbers of high-mannose carbohydrate chains (Fig. 5). Neither the anti-CD4 MAb nor mannan could completely inhibit gp120 binding to MDDC alone but only in combination (Fig. 5). These results further support the notion that MDDCs have other modalities in addition to DC-SIGN to bind HIV.
FIG. 5.
HIV and SIV gp120 binding to MDDCs. gp120 produced in 293T cells was bound to CD4 expressing QT6 cells or MDDCs with or without 100 μg of mannan or 20 μg of anti-CD4 MAb 19 (αCD4 MAb)/ml and then analyzed by SDS-PAGE and Western blot.
_N_-Glycan status of Ebola virus GP determines efficiency of DC-SIGN(R) and ASGP-R enhancement of Ebola GP pseudovirions.
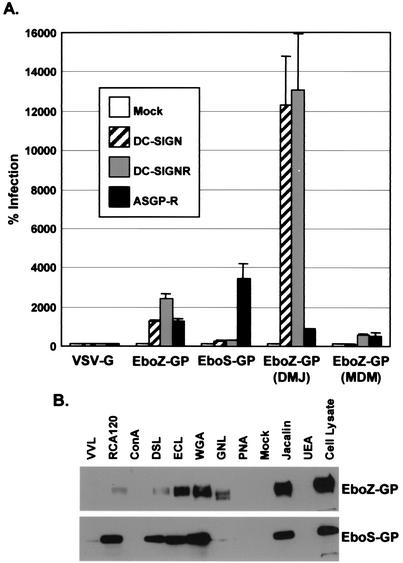
It was recently shown that DC-SIGN(R) bind Ebola virus GP and enhance infection of macrophages and endothelial cells (2, 50a). Since Ebola virus GP can be differentially glycosylated (15), we investigated whether differences in _N_-glycans also affect Ebola virus GP-DC-SIGN(R) interactions. We found that Ebola virus Zaire GP (EboZ-GP) pseudovirions could be enhanced by DC-SIGN(R) to a greater degree than Ebola virus Sudan GP (EboS-GP) pseudovirions (Fig. 6A). By using a panel of lectins with known carbohydrate specificities, we found that this effect correlated with EboZ-GP containing more high-mannose _N_-glycans than EboS-GP (Fig. 6B, GNL lane), a finding which is consistent with an earlier study (15). As for HIV, production of pseudovirions in the presence of DMJ resulted in dramatically increased infectivity enhancement by DC-SIGN(R) (Fig. 6A).
FIG. 6.
Infection by Ebola virus GP pseudovirions and analysis of GP glycosylation. (A) 293T cells mock transfected or transfected with DC-SIGN, DC-SIGNR, or ASGP-R were infected with 500 TCID50 of HIV-luciferase reporter viruses pseudotyped with VSV-G, EboZ-GP, EboS-GP, or EboZ-GP generated in the presence of 2.5 mM DMJ or MDM-derived EboZ-GP. Values are represented as the percent infection, calculated by using luciferase activity normalized to mock-transfected cells. Mean values plus the standard error of the mean are represented. (B) EboZ-GP and EboS-GP obtained from pseudovirions were incubated with the indicated lectin-biotin conjugates and then precipitated with streptavidin-agarose and analyzed by SDS-PAGE and Western blot for GP. Lectins are identified as follows: Vivia villosa lectin (VVL), Ricinus communiz agglutinin (RCA120), concanavalin A (ConA), Datura stramonium lectin (DSL), Erythrina cristagalli lectin (ECL), wheat germ agglutinin (WGA), Galanthus nivalis lectin (GNL), peanut agglutinin (PNA), Jacalin, and Ulex europaeus agglutinin (UEA). Unbound cell lysate is also shown.
Another C-type lectin, ASGP-R, can serve as a receptor for Marburg virus on hepatocytes (5). We found that ASGP-R can also enhance the infectivity of Ebola virus GP pseudovirions (Fig. 6A). This effect was inhibitable by asialofetuin but not fetuin (data not shown), a finding which was expected since ASGP-R binds galactose on asialylated carbohydrate structures (38). Interestingly, EboS-GP pseudovirions were enhanced to a greater degree than EboZ-GP pseudovirions, which correlated with their degree of GP galactosylation (Fig. 6B, RCA120 lane). Modulation of EboZ-GP _N_-glycans with DMJ decreased ASGP-R enhancement since high-mannose _N_-glycans do not have galactose moieties (Fig. 6A), although the enhancement was not entirely negated likely due to the presence of asialylated O-linked carbohydrates, which would have galactose available for ASGP-R binding. Evidence for this mechanism is provided by the fact that the O-glycan rich mucin-like domain of Ebola virus GP can be removed without negatively affecting infectivity (50) and that Ebo-GP pseudovirions lacking this domain were significantly less enhanced by ASGP-R (data not shown).
Finally, as for HIV, Ebola virus GP pseudovirions produced in macrophages interacted with DC-SIGN poorly (Fig. 6A). However, there was still some enhancement by DC-SIGNR and ASGP-R (Fig. 6A), suggesting that there may be subtle differences in how DC-SIGN and DC-SIGNR bind oligosaccharides. Thus, as for HIV, the virus-producing cell type can affect how Ebola virus GP interacts with C-type lectins.
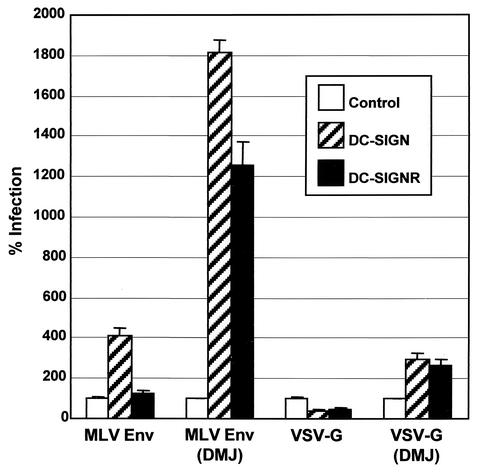
DC-SIGN(R) enhance infectivity of MLV and VSV-G pseudovirions containing high-mannose N-glycans.
If the major determinant in whether a virus will interact with DC-SIGN(R) is the _N_-glycan status of Env, then it is possible that viruses previously reported as being unable to interact with DC-SIGN(R) can be made to do so by altering _N_-glycans to the high-mannose variety. Thus, we generated MLV Env, as well as VSV-G pseudovirions, which have been shown not to interact with DC-SIGN or to do so poorly (29, 50a), in the presence of DMJ. Indeed, we found that infectivity of such pseudovirions could now be efficiently enhanced by DC-SIGN(R) (Fig. 7). MLV Env generated in the absence of DMJ did contain some high-mannose _N_-glycans, as determined by the ability to bind Galanthus nivalis lectin, as well as partial sensitivity to Endo H (data not shown), which is consistent with previous findings (22), and likely accounts for the slight infectivity enhancement of this virus by DC-SIGN (Fig. 7).
FIG. 7.
Infection by MLV Env or VSV-G pseudovirions. Doxycycline-induced DC-SIGN or DC-SIGNR 293 T-Rex cells or control 293 T-Rex cells were infected with p24 normalized HIV-luciferase reporter viruses pseudotyped with MLV Env or VSV-G generated in the presence or absence of 2.5 mM DMJ. Values are represented as the percent infection, calculated by using luciferase activity normalized to control 293 T-Rex cells. Mean values plus the standard error of the mean are represented.
DISCUSSION
The known ligands for DC-SIGN are ICAM-2, ICAM-3, and the viral Env proteins from HIV-1, HIV-2, SIV, and Ebola virus (2, 17-19, 43, 50a). These ligands all contain a relatively large number of N-linked carbohydrates. However, other viruses fail to interact with DC-SIGN even though they express heavily glycosylated GPs on their surface (29, 50a), suggesting some degree of specificity in DC-SIGN binding. Our results indicate that the ability of DC-SIGN to bind virus is dependent on the type of carbohydrate structures present on viral Envs and not merely the presence of _N_-glycans.
For HIV, Env proteins typically have 20 to 25 N-linked sites, and the resulting _N_-glycans are differentially processed in the Golgi resulting in a mixture of complex, hybrid, and high-mannose carbohydrate structures (21, 32, 40, 41). Ebola virus GP is also differentially glycosylated (15). We found that DC-SIGN(R) selectively bind HIV-1, HIV-2, and SIV Env proteins that are enriched in high-mannose _N_-glycans and that transmission of these viruses from one cell to another was much more efficient when Env contained more high-mannose _N_-glycans. Similar results were obtained with Ebola virus GP, MLV Env, and VSV-G pseudovirions. A greater proportion of high-mannose _N_-glycans versus hybrid or complex _N_-glycans on Env results in more efficient DC-SIGN(R) binding and infectivity enhancement, which fits a model in which several high-mannose _N_-glycans bind a DC-SIGN(R) tetramer, resulting in a high-avidity interaction (39). Thus, we propose that a combination of quantitative, qualitative, and conformational factors govern interactions of GPs with DC-SIGN(R): high-mannose rather than complex carbohydrate structures are needed for efficient binding, and several such high-mannose structures could lead to multiple interactions with DC-SIGN tetramers provided that these structures have the correct spatial orientation on the GP surface.
Our results with viral GPs are consistent with recently solved crystal structures of DC-SIGN(R) CRDs complexed with a pentasaccharide, revealing an interaction of the CRD with the Manα1-3[Manα1-6]Man trisaccharide (where Man is mannose) (14). In the same study, DC-SIGN(R) bound only proteins containing high-mannose _N_-glycans and not hybrid or complex _N_-glycans (14). Based on the crystal structure and modeling of Man9 nuclear magnetic resonance structures, this preferential binding is explained by a phenylalanine in DC-SIGN(R) that hinders potential binding to the inner trimannose branch point by sterically clashing with a core _N_-acetylglucosamine (GlcNAc) linked in a β1-4 bond to the first mannose. Binding can only occur when DC-SIGN(R) binds the outer trimannose branch point, which is only present in high-mannose _N_-glycans (14). Other studies have shown that DC-SIGN(R) have a higher affinity for Man9GlcNAc2 than for a single-mannose moiety and a higher affinity for glycopeptides with two rather than one Man9GlcNAc2 structure (14, 39).
Our conclusions with HIV differ from those reached by T. B. Geijtenbeek et al., who found that a gp120 lacking a signal sequence and produced in yeast cells, and thus lacking carbohydrate structures as well as native disulfide bonds and conformation, still bound DC-SIGN (20). Thus, Geitjtenbeek et al. proposed that binding of gp120 to DC-SIGN occurs independently of carbohydrate and is therefore fundamentally different than the ICAM-3-DC-SIGN interaction, which is dependent on carbohydrate (20). We feel that the denatured Envs used by Geijtenbeek et al. are not likely to bind DC-SIGN in a manner similar to a native, fully glycosylated Env, although further investigation is required. Our findings that DC-SIGN exhibits specificity for HIV and SIV Envs containing a greater proportion of high-mannose carbohydrates, along with the crystal structure of DC-SIGN are consistent with a model in which DC-SIGN-Env interactions are dependent on high-mannose _N_-glycans. Further, DC-SIGN(R) bind all HIV-1, HIV-2, and SIV Envs tested to date. All of these Envs are heavily glycosylated, and all contain high-mannose carbohydrate structures, but they do not all contain an absolutely conserved protein sequence that would likely be required if the direct protein-protein interaction model proposed by Geijtenbeek et al. is correct. Although direct protein-protein interactions may be involved, we feel that these are likely to be subsidiary in nature. The fact that other viral GPs can either bind DC-SIGN(R) or be induced to bind by altering _N_-glycan status to the high-mannose variety further diminishes the possibility that a primary protein sequence is the main determinant for efficient DC-SIGN(R) binding.
If DC-SIGN(R) selectively bind high-mannose _N_-glycans on viral Envs, then where exactly does DC-SIGN(R) bind? For HIV gp120, complex _N_-glycans are generally more N terminal, whereas high-mannose/hybrid _N_-glycans are more C terminal (32, 59). When located on the structure of the gp120 core, the high-mannose _N_-glycans cluster on one surface, whereas the complex _N_-glycans cluster on another surface with relatively little overlap (59). When an HIV Env trimer is modeled, complex _N_-glycans are on the external-lateral Env surface, whereas high-mannose _N_-glycans are on the Env surface facing the cell (Fig. 8). Thus, we believe it is likely that DC-SIGN(R) bind to high-mannose carbohydrates present on this area of HIV Env. For Ebola virus GP, the _N_-glycan type at particular N-linked sites has not been determined nor is there an available crystal structure. However, since the conversion of high-mannose _N_-glycans to complex is likely limited by the accessibility of the N-linked processing machinery to an oligosaccharide, we predict that a cluster of high-mannose _N_-glycans also exists on Ebola virus GP that accounts for DC-SIGN(R) binding keeping in mind that Ebola virus GP _N_-glycan composition can vary between isolates as well as within the same isolate depending on the cell type in which virus is produced, adding to the complexity of understanding potential DC-SIGN(R)-Ebola virus GP interactions.
FIG. 8.
Location of high-mannose or complex _N_-glycans on the HIV Env trimer. A model of the HIV Env trimer based on optimization of quantifiable surface parameters (30) is shown in three different views, each rotated 90° about a horizontal axis. The top panel is a view from the virus. The middle panel is a side view with the viral membrane above and the target cell below. The bottom panel is a view from the target cell. The left column is an α-carbon worm trace with gp120 in brown and CD4 in yellow. The protein proximal Man3GlcNAc2 pentasaccharide core conserved between high-mannose and complex _N_-glycans is shown in cyan and modeled as described previously (58). The right column depicts the solvent-accessible surface of gp120 with high-mannose _N_-glycans in blue, complex _N_-glycans in black (32), and the rest of the surface in white. (This figure was kindly provided by Peter D. Kwong.)
The requirements revealed here for efficient binding of HIV Env and Ebola virus GP to DC-SIGN(R) have interesting implications for viral interactions in vivo. Previous studies have shown that HIV gp120 from PBMC- versus MDM-grown virus differs in complex carbohydrate composition with MDM-derived gp120 containing _N_-acetyllactosamine repeats (33, 34, 55). Indeed, we found that the HIV-1 Ba-L virus grown in MDMs contained gp120 that was more heterogeneous and differed in complex carbohydrates than gp120 from the identical Ba-L virus grown in PBMCs. Consistent with our selective binding results with HIV gp120 produced in 293T cells, Env from HIV grown from MDMs did not interact with DC-SIGN(R) as well as Env from PBMC-derived virus, even though Endo H digestions showed that it contained some high-mannose carbohydrate chains. Having complex chains with extended _N_-acetyllactosamine repeats may prevent efficient interaction of DC-SIGN(R) with high-mannose chains, especially if such complex structures cluster on the outer regions of an Env trimer (Fig. 8). Alternatively, modification of a single N-linked site on gp120 in macrophages could impact binding if that site plays an important role in DC-SIGN interactions. DC-SIGN also did not efficiently enhance infectivity of MDM-derived Ebola GP pseudovirions even though macrophages are important early targets of Ebola virus infection in vivo (48). Perhaps the Ebola virus virion, by modulating _N_-glycan status, can more efficiently emerge from a macrophage by preventing reattachment by DC-SIGN, allowing subsequent infection of other cell types. We found that DC-SIGNR and ASGP-R were still able to enhance infectivity of MDM-derived EboZ-GP pseudovirions, which may allow for targeted infection of endothelial cells or hepatocytes.
Despite the inability of DC-SIGN to efficiently interact with MDM-derived HIV Env, DCs were still able to enhance the infectivity of MDM-derived HIV, suggesting additional unidentified mechanisms on DCs for HIV infectivity enhancement. Indeed, rhesus macaque DCs bind and transmit SIV independently of DC-SIGN (56). In addition, we and others have found that DC-SIGN accounts for only a fraction of the ability of DCs to bind HIV (3a, 52, 57) and that other C-type lectins on DC subsets and Langerhans cells can efficiently bind HIV Env (52a). It will be interesting to determine whether the carbohydrate structures on Env are selected for in part to prevent interactions with certain C-type lectins that have the additional role of antigen uptake and presentation to MHC (12, 27, 47) while balancing favorable interactions with other molecules that lead to efficient transmission. Since MDM-derived Ebola virus GP also interacted less well with DC-SIGN, we speculate that these mechanisms may also apply to Ebola virus in how it may evade capture, degradation, and immune activation by DCs. In addition, differences in N-linked glycosylation between Ebola virus GP subtypes may affect interactions with C-type lectins and lead to differences in replication dynamics in vivo.
In summary, differential glycosylation of viral Envs due to differences in producer cell type and virus strain can impact interactions with DC-SIGN(R) and possibly other C-type lectins, as we have demonstrated for HIV and Ebola virus. Therefore, this variable needs to be taken into account when virus-host interactions and the development of antiviral agents that target virus receptors and attachment factors are being assessed. It should be noted that ICAM-3 is also heavily glycosylated, with approximately half of its molecular weight consisting of various types of N-linked carbohydrate structures (13, 16). Thus, it is possible that modulation of ICAM-3 glycosylation may also affect its interactions with DC-SIGN in vivo. Finally, since DC-SIGN is potentially involved in antigen presentation by DCs (12), one may better target antigens to DC-SIGN by altering carbohydrate composition toward the high-mannose variety by expressing proteins in the presence of mannosidase inhibitors or in cell lines unable to process N-linked oligosaccharides to the hybrid or complex variety. Indeed, we could manipulate N-linked glycosylation of MLV Env and VSV-G such that these proteins could now interact efficiently with DC-SIGN. Such alterations may improve vaccine design for HIV and other pathogens that contain N-linked glycosylated proteins and may also prove useful in gene therapeutic strategies targeting cells expressing DC-SIGN(R).
Acknowledgments
We thank Genoveffa Franchini and Benhur Lee for plasmids, Navid Ahmad and Victor Holubowsky for technical support, and Tim Hart for sCD4. We especially thank Peter D. Kwong for providing a model of the HIV Env trimer delineating complex or high mannose _N_-glycans.
G.L. was supported by an NIH MSTP grant, G.S. was supported by a long-term EMBO fellowship, S.P. was supported by a Deutsche Forschungsgemeinschaft fellowship, P.B. was supported by NIH grants R01 AI43455 and CA76256, D.W. was supported by NIH grant HL 62060-04 and a Pediatric AIDS Foundation grant, J.A.H. was supported by NIH grant R01 AI45378, and R.W.D. was supported by NIH grants R01 40880 and R01 35383, a Burroughs Wellcome Fund translational research award, and an Elizabeth Glaser Scientist award from the Pediatric AIDS Foundation. Support was also provided by the Penn Center for AIDS Research (NIH grant P30 AI45008).
REFERENCES
- 1.Alexander, W. A., B. Moss, and T. R. Fuerst. 1992. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 66**:**2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, C. P., F. Lasala, J. Carrillo, O. Muniz, A. L. Corbi, and R. Delgado. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76**:**6841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baribaud, F., S. Pöhlmann, T. Sparwasser, M. T. Kimata, Y. K. Choi, B. S. Haggarty, N. Ahmad, T. Macfarlan, T. G. Edwards, G. J. Leslie, J. Arnason, T. A. Reinhart, J. T. Kimata, D. R. Littman, J. A. Hoxie, and R. W. Doms. 2001. Functional and antigenic characterization of human, rhesus macaque, pigtailed macaque, and murine DC-SIGN. J. Virol. 75**:**10281-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Baribaud, F., S. Pöhlmann, G. Leslie, F. Mortari, and R. W. Doms. 2002. Quantitative expression and virus transmission analysis of DC-SIGN on monocyte-derived dendritic cells. J. Virol. 76**:**9135-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashirova, A. A., T. B. Geijtenbeek, G. C. van Duijnhoven, S. J. van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193**:**671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, S., M. Spiess, and H. D. Klenk. 1995. The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J. Gen. Virol. 76**:**393-399. [DOI] [PubMed] [Google Scholar]
- 6.Cameron, P. U., P. S. Freudenthal, J. M. Barker, S. Gezelter, K. Inaba, and R. M. Steinman. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257**:**383-387. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206**:**935-944. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89**:**8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doranz, B. J., S. S. Baik, and R. W. Doms. 1999. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J. Virol. 73**:**10346-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74**:**7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87**:**745-756. [DOI] [PubMed] [Google Scholar]
- 12.Engering, A., T. B. Geijtenbeek, S. J. van Vliet, M. Wijers, E. van Liempt, N. Demaurex, A. Lanzavecchia, J. Fransen, C. G. Figdor, V. Piguet, and Y. van Kooyk. 2002. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 168**:**2118-2126. [DOI] [PubMed] [Google Scholar]
- 13.Fawcett, J., C. L. Holness, L. A. Needham, H. Turley, K. C. Gatter, D. Y. Mason, and D. L. Simmons. 1992. Molecular cloning of ICAM-3, a third ligand for LFA-1, constitutively expressed on resting leukocytes. Nature 360**:**481-484. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg, H., D. A. Mitchell, K. Drickamer, and W. I. Weis. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294**:**2163-2166. [DOI] [PubMed] [Google Scholar]
- 15.Feldmann, H., S. T. Nichol, H. D. Klenk, C. J. Peters, and A. Sanchez. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199**:**469-473. [DOI] [PubMed] [Google Scholar]
- 16.Funatsu, O., T. Sato, P. Kotovuori, C. G. Gahmberg, M. Ikekita, and K. Furukawa. 2001. Structural study of N-linked oligosaccharides of human intercellular adhesion molecule-3 (CD50). Eur. J. Biochem. 268**:**1020-1029. [DOI] [PubMed] [Google Scholar]
- 17.Geijtenbeek, T. B., D. J. Krooshoop, D. A. Bleijs, S. J. van Vliet, G. C. van Duijnhoven, V. Grabovsky, R. Alon, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 1**:**353-357. [DOI] [PubMed] [Google Scholar]
- 18.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100**:**587-597. [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek, T. B., R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, G. J. Adema, Y. van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100**:**575-585. [DOI] [PubMed] [Google Scholar]
- 20.Geijtenbeek, T. B., G. C. van Duijnhoven, S. J. van Vliet, E. Krieger, G. Vriend, C. G. Figdor, and Y. van Kooyk. 2002. Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for intercellular adhesion molecule 3 and HIV-1. J. Biol. Chem. 277**:**11314-11320. [DOI] [PubMed] [Google Scholar]
- 21.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus: structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263**:**11760-11767. [PubMed] [Google Scholar]
- 22.Geyer, H., R. Kempf, H. H. Schott, and R. Geyer. 1990. Glycosylation of the envelope glycoprotein from a polytropic murine retrovirus in two different host cells. Eur. J. Biochem. 193**:**855-862. [DOI] [PubMed] [Google Scholar]
- 23.Hattori, N., F. Michaels, K. Fargnoli, L. Marcon, R. C. Gallo, and G. Franchini. 1990. The human immunodeficiency virus type 2 vpr gene is essential for productive infection of human macrophages. Proc. Natl. Acad. Sci. USA 87**:**8080-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hioe, C. E., L. Bastiani, J. E. Hildreth, and S. Zolla-Pazner. 1998. Role of cellular adhesion molecules in HIV type 1 infection and their impact on virus neutralization. AIDS Res. Hum. Retrovir. 14(Suppl. 3)**:**S247-S254. [PubMed] [Google Scholar]
- 25.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96**:**6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jameson, B., F. Baribaud, S. Pöhlmann, D. Ghavimi, F. Mortari, R. W. Doms, and A. Iwasaki. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 76**:**1866-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, W., W. J. Swiggard, C. Heufler, M. Peng, A. Mirza, R. M. Steinman, and M. C. Nussenzweig. 1995. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature 375**:**151-155. [DOI] [PubMed] [Google Scholar]
- 28.Kaushal, G. P., and A. D. Elbein. 1994. Glycosidase inhibitors in study of glycoconjugates. Methods Enzymol. 230**:**316-329. [DOI] [PubMed] [Google Scholar]
- 29.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T-cell infection. Immunity 16**:**135-144. [DOI] [PubMed] [Google Scholar]
- 30.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74**:**1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, B., G. Leslie, E. Soilleux, U. O'Doherty, S. Baik, E. Levroney, K. Flummerfelt, W. Swiggard, N. Coleman, M. Malim, and R. W. Doms. 2001. cis expression of DC-SIGN allows for more efficient entry of human and simian immunodeficiency viruses via CD4 and a coreceptor. J. Virol. 75**:**12028-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265**:**10373-10382. [PubMed] [Google Scholar]
- 33.Liedtke, S., M. Adamski, R. Geyer, A. Pfutzner, H. Rubsamen-Waigmann, and H. Geyer. 1994. Oligosaccharide profiles of HIV-2 external envelope glycoprotein: dependence on host cells and virus isolates. Glycobiology 4**:**477-484. [DOI] [PubMed] [Google Scholar]
- 34.Liedtke, S., R. Geyer, and H. Geyer. 1997. Host-cell-specific glycosylation of HIV-2 envelope glycoprotein. Glycoconj. J. 14**:**785-793. [DOI] [PubMed] [Google Scholar]
- 35.Lin, G., B. Lee, B. S. Haggarty, R. W. Doms, and J. A. Hoxie. 2001. CD4-independent use of rhesus CCR5 by human immunodeficiency virus type 2 implicates an electrostatic interaction between the CCR5 N terminus and the gp120 C4 domain. J. Virol. 75**:**10766-10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Lin, G., F. Baribaud, J. Romano, R. W. Doms, and J. A. Hoxie. Identification of gp120 binding sites on CXCR4 using CD4-independent human immunodeficiency virus type 2 Env proteins. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 36.Martin, K. A., R. Wyatt, M. Farzan, H. Choe, L. Marcon, E. Desjardins, J. Robinson, J. Sodroski, C. Gerard, and N. P. Gerard. 1997. CD4-independent binding of SIV gp120 to rhesus CCR5. Science 278**:**1470-1473. [DOI] [PubMed] [Google Scholar]
- 37.Means, R. E., T. Matthews, J. A. Hoxie, M. H. Malim, T. Kodama, and R. C. Desrosiers. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75**:**3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier, M., M. D. Bider, V. N. Malashkevich, M. Spiess, and P. Burkhard. 2000. Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor. J. Mol. Biol. 300**:**857-865. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR: subunit organization and binding to multivalent ligands. J. Biol. Chem. 276**:**28939-28945. [DOI] [PubMed] [Google Scholar]
- 40.Mizuochi, T., T. J. Matthews, M. Kato, J. Hamako, K. Titani, J. Solomon, and T. Feizi. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9: presence of complex-type oligosaccharides with bisecting _N_-acetylglucosamine residues. J. Biol. Chem. 265**:**8519-8524. [PubMed] [Google Scholar]
- 41.Mizuochi, T., M. W. Spellman, M. Larkin, J. Solomon, L. J. Basa, and T. Feizi. 1988. Carbohydrate structures of the human immunodeficiency virus recombinant envelope glycoprotein gp120 produced in Chinese hamster ovary cells. Biochem. J. 254**:**599-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68**:**469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pöhlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75**:**4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pöhlmann, S., G. J. Leslie, T. G. Edwards, T. Macfarlan, J. D. Reeves, K. Hiebenthal-Millow, F. Kirchhoff, F. Baribaud, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus: virus binding and transfer are dissociable functions. J. Virol. 75**:**10523-10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pöhlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 98**:**2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves, J. D., and T. F. Schulz. 1997. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J. Virol. 71**:**1453-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallusto, F., M. Cella, C. Danieli, and A. Lanzavecchia. 1995. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J. Exp. Med. 182**:**389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnittler, H. J., and H. Feldmann. 1999. Molecular pathogenesis of filovirus infections: role of macrophages and endothelial cells. Curr. Top. Microbiol. Immunol. 235**:**175-204. [DOI] [PubMed] [Google Scholar]
- 49.Simmons, G., J. D. Reeves, S. Hibbitts, J. T. Stine, P. W. Gray, A. E. Proudfoot, and P. R. Clapham. 2000. Co-receptor use by HIV and inhibition of HIV infection by chemokine receptor ligands. Immunol. Rev. 177**:**112-126. [DOI] [PubMed] [Google Scholar]
- 50.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein downmodulation and loss of cell adherence. J. Virol. 76**:**2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Simmons, G., J. D. Reeves, L. H. Vandenberghe, F. Baribaud, R. C. Netter, J. L. Riley, R. W. Doms, P. Bates, and S. Pöhlmann. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology, in press. [DOI] [PubMed]
- 51.Soilleux, E. J., R. Barten, and J. Trowsdale. 2000. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 165**:**2937-2942. [DOI] [PubMed] [Google Scholar]
- 52.Turville, S. G., J. Arthos, K. Mac Donald, G. Lynch, H. Naif, G. Clark, D. Hart, and A. L. Cunningham. 2001. HIV gp120 receptors on human dendritic cells. Blood 98**:**2482-2488. [DOI] [PubMed] [Google Scholar]
- 52a.Turville, S. G., P. U. Cameron, A. Handley, G. Lin, S. Pöhlmann, R. W. Doms, and A. L. Cunningham. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3**:**975-983. [DOI] [PubMed] [Google Scholar]
- 53.Ugolini, S., I. Mondor, and Q. J. Sattentau. 1999. HIV-1 attachment: another look. Trends Microbiol. 7**:**144-149. [DOI] [PubMed] [Google Scholar]
- 54.Weissman, D., H. Ni, D. Scales, A. Dude, J. Capodici, K. McGibney, A. Abdool, S. N. Isaacs, G. Cannon, and K. Kariko. 2000. HIV gag mRNA transfection of dendritic cells (DC) delivers encoded antigen to MHC class I and II molecules, causes DC maturation, and induces a potent human in vitro primary immune response. J. Immunol. 165**:**4710-4717. [DOI] [PubMed] [Google Scholar]
- 55.Willey, R. L., R. Shibata, E. O. Freed, M. W. Cho, and M. A. Martin. 1996. Differential glycosylation, virion incorporation, and sensitivity to neutralizing antibodies of human immunodeficiency virus type 1 envelope produced from infected primary T-lymphocyte and macrophage cultures. J. Virol. 70**:**6431-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, L., A. A. Bashirova, T. D. Martin, L. Villamide, E. Mehlhop, A. O. Chertov, D. Unutmaz, M. Pope, M. Carrington, and V. N. KewalRamani. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. USA 99**:**1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, L., T. D. Martin, R. Vazeux, D. Unutmaz, and V. N. KewalRamani. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76**:**5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393**:**705-711. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39**:**11194-11204. [DOI] [PubMed] [Google Scholar]