Efficient Gene Targeting Mediated by Adeno-Associated Virus and DNA Double-Strand Breaks (original) (raw)
Abstract
Gene targeting is the in situ manipulation of the sequence of an endogenous gene by the introduction of homologous exogenous DNA. Presently, the rate of gene targeting is too low for it to be broadly used in mammalian somatic cell genetics or to cure genetic diseases. Recently, it has been demonstrated that infection with recombinant adeno-associated virus (rAAV) vectors can mediate gene targeting in somatic cells, but the mechanism is unclear. This paper explores the balance between random integration and gene targeting with rAAV. Both random integration and spontaneous gene targeting are dependent on the multiplicity of infection (MOI) of rAAV. It has previously been shown that the introduction of a DNA double-stranded break (DSB) in a target gene can stimulate gene targeting by several-thousand-fold in somatic cells. Creation of a DSB stimulates the frequency of rAAV-mediated gene targeting by over 100-fold, suggesting that the mechanism of rAAV-mediated gene targeting involves, at least in part, the repair of DSBs by homologous recombination. Absolute gene targeting frequencies reach 0.8% with a dual vector system in which one rAAV vector provides a gene targeting substrate and a second vector expresses the nuclease that creates a DSB in the target gene. The frequencies of gene targeting that we achieved with relatively low MOIs suggest that combining rAAV vectors with DSBs is a promising strategy to broaden the application of gene targeting.
Gene targeting is a process by which an exogenous gene replaces an endogenous gene by homologous recombination. It is used to create genetic change in murine embryonic stem cells and in certain other specialized cell types, such as the chicken B-cell line DT40 (5, 31). In general, however, the low spontaneous frequency of gene targeting has precluded its widespread experimental use in mammalian somatic cell genetics. In somatic cell lines, for example, gene targeting occurs at a frequency of approximately one event per million transfected cells (29). Moreover, while gene targeting would be a conceptually elegant way of performing gene therapy through gene correction, the low spontaneous frequency has also precluded its development for such an application. The regulation of gene targeting is intimately related to the metabolism of exogenous DNA. In particular, the relative frequency of gene targeting is directly related to the balance between the homologous and random integration of exogenous DNA and this balance has important consequences for the type of genetic change created. If, for example, exogenous DNA is integrated by gene targeting via homologous recombination, then subtle genomic changes can be created. If, on the other hand, exogenous DNA is integrated randomly, then insertional mutations are created, a different type of genetic alteration. Understanding whether cells passively or actively regulate the metabolism of exogenous DNA, therefore, has important consequences in understanding how genomes can be altered by the introduction of foreign DNA. Moreover, understanding the underlying mechanisms of gene targeting not only will give insight into how cells regulate genomic alterations but could also lead to its development as a tool for somatic cell genetics and gene therapy.
The creation of a DNA double-stranded break (DSB) in the target gene can increase the frequencies of both direct-repeat recombination and gene targeting several-thousand-fold (4, 7, 9, 23, 28, 30, 32). When conditions are optimized for DSB-mediated gene targeting, the frequency of gene targeting can reach 3 to 5% (22). Thus, DSBs seem to be a central element of the gene targeting mechanism. The powerful stimulatory role of DSBs in gene targeting was found by transfecting cells with naked DNA. For many cell types, however, transfection is inefficient. It is important, therefore, to find an efficient delivery system for the gene targeting components (a DNA substrate to direct the desired genetic change and a nuclease to create a gene-specific DSB).
Viral vectors, and in particular, recombinant adeno-associated virus (rAAV), have broad cellular tropism and may be preferred over transfection as a method for the delivery of gene targeting components (20, 25). AAV vectors possess a single-stranded DNA genome with inverted terminal repeats that form hairpin structures at each end and can integrate randomly by nonhomologous recombination (18, 26, 27, 34). The ability of rAAV vectors to stably transduce cells has led to their being used in human gene therapy trials for the treatment of hemophilia (16). Russell and Hirata, however, showed that rAAV could not only randomly integrate into the genome but could also mediate gene targeting (24). They found that using a multiplicity of infection (MOI) of 400,000 viral genomes/cell resulted in gene targeting frequencies of 0.1 to 0.3% in human cell lines (24). They have extended these findings to show that rAAV can mediate gene targeting in a variety of cell types, with a variety of small mutations, and in a locus-independent manner (12-14). While the use of rAAV for creating specific gene mutations by gene targeting for experimental purposes is promising (11), the extremely high MOIs required render the use of rAAV for gene correction for therapeutic purposes less promising. Not only are such high MOIs practically untenable, but the random integration of rAAV also has been associated with genomic rearrangements even at low MOIs (19). The mechanism by which rAAV mediates gene targeting also remains unclear, and understanding the mechanism may suggest ways to increase its efficiency.
In this paper, we explore whether the stimulation of gene targeting by DSBs can be combined with the use of rAAV vectors. We found that a DSB significantly stimulates rAAV-induced gene targeting, resulting in high gene targeting frequencies by using lower MOIs. These results suggest that combining AAV with the generation of DSBs is an intriguing strategy to perform gene targeting and has many characteristics that suggest it may be a powerful way to perform gene therapy through gene correction.
MATERIALS AND METHODS
DNA manipulations and cloning.
Standard molecular biology procedures were used to make all plasmids (1). rAAV.Subs was made by cloning a 2,700-bp _Xba_I fragment from plasmid RS2700 (22) into psub201. rAAV.Subs-Puro was made replacing the woodchuck posttranscriptional regulatory element of rAAV.Subs with a puromycin resistance cassette (the puromycin acetyltransferase gene driven by the simian virus 40 [SV40] promoter). rAAV.Sce was made by placing the I-_Sce_I (Sce) coding region downstream from the cytomegalovirus enhancer/chicken β-actin promoter into psub201. Plasmid RS2100 and plasmids that express the Sce endonuclease have been described elsewhere (22). Briefly, RS2100 is a repair substrate that contains the truncated green fluorescent protein (GFP) gene, internal ribosomal entry site, and CD8 portions but not the woodchuck posttranscriptional regulatory element of rAAV.Subs in a pBSII (Stratagene, La Jolla, Calif.) backbone. The Sce expression plasmid consisted of the cytomegalovirus promoter driving Sce expression in a pcDNA6 (Invitrogen, Carlsbad, Calif.) plasmid backbone.
Cell culture and cell lines.
293 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM)-10% calf serum-2 mM l-glutamine-100 U of penicllin/ml-100 μg of streptomycin/ml (10% DMEM) in a humidified chamber at 37°C and 5% CO2. Cells were split and passaged by following standard procedures (1). Cell line 293/A658 was made as previously described (22). Briefly, 2 million 293 cells were electroporated with 10 μg of linearized plasmid A658, and G418-resistant colonies were selected by culturing in 500 μg of G418 (Invitrogen)/ml from day 2 after electroporation until distinct colonies formed (14 days after electroporation). Individual colonies were picked and grown in 24-well plates. After the expansion of colonies in 24-well plates, individual colonies were analyzed for surface CD8α expression with a phycoerythrin-conjugated anti-CD8α antibody (BD Biosciences, San Diego, Calif.). We used a cell line that showed high, homogeneous surface expression of CD8α.
Transfection of 293 cells.
293 cells were transfected as previously described (21). Briefly, approximately 100,000 cells were plated in a 24-well plate the day prior to transfection. Just prior to infection, the medium was changed and, using a standard calcium phosphate technique (1), 200 ng of Sce expression plasmid and 200 ng of the repair substrate were cotransfected. Using this procedure, we routinely obtained transfection efficiencies of 20 to 30%. These efficiencies were lower than those usually obtained with 293T cells because we were using 293 cells that had not been SV40 T antigen transformed.
Production and purification of rAAV.
rAAV was produced and purified essentially as previously described (10). To produce rAAV, plates of 293T cells were transfected with three plasmids: pXX2, which supplied the Rep and Cap proteins of AAV2; pXX6, which contained the adenovirus helper functions; and a vector plasmid (33). For virus production, five 15-cm-diameter plates were transfected with 6 μg of pXX2, 25 μg of pXX6, and 19 μg of vector plasmid and incubated for 72 h. Virus was purified with iodixanol gradients as described previously (35). Titers of rAAV were determined by real-time PCR with SYBR Green I double-stranded DNA binding dye and an ABI Prism 7700 sequence detection system (PE Biosystems, Foster City, Calif.). Samples were prepared as previously described (8). Throughout this study, MOIs are referred to as the number of DNase-resistant genomic particles per cell.
Measurement of random integration.
The day prior to infection, 50 to 100,000 293-0 or 293/A658 cells were plated in parallel in a 24-well plate and allowed to adhere overnight in 10% DMEM. Just prior to infection, one well was counted to determine the number of cells in each well by following standard procedures. Just prior to infection, the medium in all wells was changed to DMEM-2% fetal bovine serum-2 mM l-glutamine-100 U of penicillin/ml-100 μg of streptomycin/ml (2% DMEM). rAAV.GFP or rAAV.Subs-Puro was then added to the desired MOI. 24 h after adding virus, the virus containing media was removed and replaced with 10% DMEM. For rAAV.GFP, at 48 h after infection, each well was passaged and an aliquot was analyzed by flow cytometry for the percentage of GFP-positive cells by a FACScan (settings, FSC E-1 6.06, SSC 350, FL-1 550, FL-2 550, FL-3 550; compensation, FL2-23.6%-FL-1 and FL3-18.1%-FL1) (BD Biosciences, San Jose, Calif.). Infected cells were then serially passaged and analyzed by flow cytometry until the percentage of GFP-positive cells stabilized. To determine the random integration frequency by using antibiotic resistance, we infected 293/A658 cells with various MOIs of rAAV.Subs-Puro as described above. Three days after infection, the wells were harvested and 250,000 cells were plated in a 10-cm-diameter plate. At day 4 or day 9 after infection, we added puromycin to a final concentration of 1 μg/ml and grew the cells until distinct colonies had formed (usually 14 days postinfection). Colonies were counted by first fixing them in 4% paraformaldehyde, followed by staining with 2% methylene blue in 70% ethanol and washing twice with distilled water. We found no difference in the number of puromycin-resistant colonies whether we began antibiotic selection at day 4 or at day 9 after infection. In general, we interpreted the number of GFP-positive cells at day 14 and the number of puromycin-resistant colonies as a measurement of the random integration frequency of rAAV in these proliferating 293 cells.
Measurement of gene targeting.
Gene targeting frequencies were measured as followed. The day prior to infection, 293/A658 cells were plated at a density of approximately 50 to 100,000 cells per well in a 24-well plate. Just prior to infection, the cell number was determined by counting one well. The medium was then changed to 2% DMEM, and virus was added to the desired MOI. Twenty-four hours after infection, the virus-containing medium was replaced with 10% DMEM. At 48 to 72 h after infection, the cells were split into six-well plates and allowed to expand. At days 9 to 11 after infection, the percentage of GFP-positive cells was determined by flow cytometry as described above.
RESULTS
GFP gene targeting system and rAAV vectors.
To efficiently and quantitatively study gene targeting, we developed an assay based on the correction of a mutated GFP gene that has been integrated into the genome (Fig. 1A) (22). This assay was designed to mimic the correction of a small mutation in an endogenous gene. The mutation in the GFP gene is a 35-bp insertion consisting of a stop codon followed by the recognition site for the I-_Sce_I endonuclease (Sce) (an intron-encoded endonuclease from Saccharomyces cerevisiae) (15). In this assay, the frequency of gene targeting is measured by flow cytometry to quantitate the conversion of GFP-negative cells to GFP-positive cells. It was previously shown by this system that the 293/A658 cell line is GFP negative, GFP-positive cells are created only by gene targeting, the frequency of spontaneous gene targeting following transfection of a double-stranded DNA plasmid is 7.1 × 10−7, and the frequency increases over 2,000-fold when a DSB is introduced at the Sce site in the targeted GFP gene (22). In this paper, we report the results with a clonal 293 cell line harboring a single integrated copy of the GFP target gene but have obtained similar results with polyclonal cell lines in which the single insertion site of the target gene is at a variety of different locations. Throughout this paper, we report the frequencies without normalization to the efficiency of infection. The frequencies of gene targeting and random integration are reported as the number of events per million cells (a frequency of 10,000, for example, is equivalent to 1%).
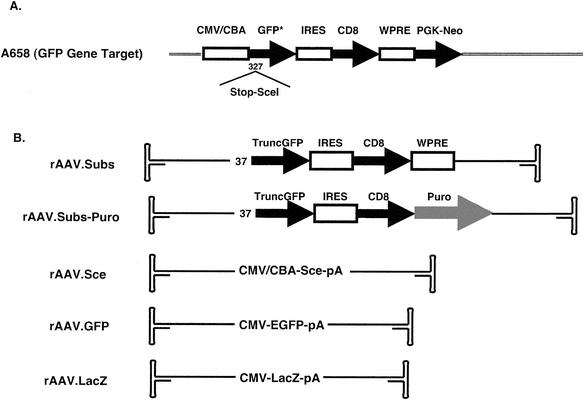
FIG. 1.
Schematics of the GFP gene targeting system components and rAAVs used. (A) Schematic depiction of artificial gene target (A658). A 35-bp insertion consisting of an in-frame stop codon followed by the recognition site for the Sce endonuclease was inserted at bp 327 of the GFP coding region. The entire insertion sequence is as follows: 5′ TAAGCTCTCGAGATTACCCTGTTATCCCTAAGCTT 3′. (B) Schematic representations of the rAAVs used in this paper. The viruses consist of a single-stranded DNA core with hairpin ends. Repair substrate viruses rAAV.Subs and rAAV.Subs-Puro are missing the first 36 nucleotides of the GFP coding region. Abbreviations: CMV/CBA, cytomegalovirus enhancer/chicken β-actin promoter; EGFP, enhanced GFP; IRES, internal ribosomal entry site; CD8, coding region for the human CD8α coding region; WPRE, woodchuck posttranscriptional regulatory element (36); PGK-Neo, neomycin phosphotransferase gene driven by the phosphoglycerate kinase promoter; TruncGFP, GFP coding region that begins at bp 37 of the coding region; Sce, coding region for the I-_Sce_I endonuclease; Puro, puromycin acetyltransferase gene driven by the SV40 promoter and containing a polyadenylation signal sequence; CMV, cytomegalovirus promoter and enhancer; pA, polyadenylation signal sequence; LacZ, coding region for the β-galactosidase gene.
To study gene targeting by rAAV with the GFP system, we generated the rAAV vectors shown schematically in Fig. 1B. rAAV.Subs and rAAV.Subs-Puro can both serve as repair substrates in the gene targeting reaction. Both have short, inactivating 5′ truncations in the GFP gene and do not express GFP. rAAV.Subs has 2,700 bp of homology to the GFP gene target, and rAAV.Subs-Puro has 2,100 bp of homology. rAAV.Subs-Puro also contains a puromycin resistance cassette. It can be used to measure both gene targeting through the measurement of GFP-positive cells and random integration by counting puromycin-resistant colonies. rAAV.Sce expresses the Sce endonuclease through a strong, ubiquitous promoter. We used rAAV-GFP to measure the frequencies of transduction and random integration. rAAV.LacZ has no homology to the target GFP gene, and we used it to control for nonspecific effects from rAAV infection. When rAAV.Subs, rAAV.Subs-Puro, and rAAV.Sce were infected into 293 cells that do not contain the GFP target gene (293-0 cells), we detected no GFP-positive cells (less than two per million) (data not shown). Furthermore, we also did not detect GFP-positive cells when rAAV.Subs and rAAV.Sce were coinfected into 293-0 cells (data not shown). Thus, the rAAV repair substrate and Sce viruses do not recombine to form functional GFP on their own. As a further control, when rAAV.Sce was infected into 293/A658 target cells, no GFP-positive cells were generated because an in-frame stop codon precedes the site of the Sce-induced DSB and repair of the DSB by nonhomologous end-joining does not remove this stop codon.
Random integration by using rAAV.
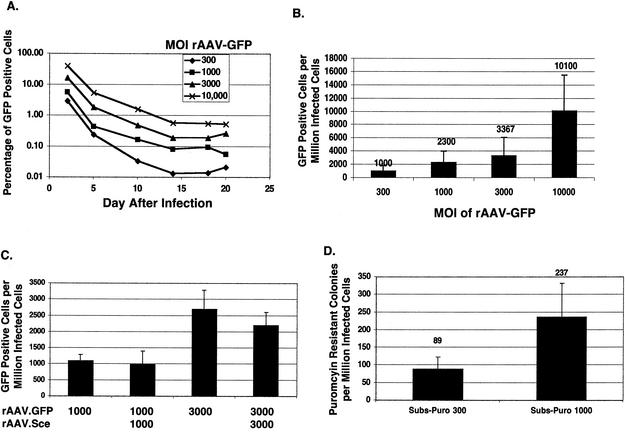
We examined the frequency of random integration for rAAV by assessing the random integration frequency with either rAAV.GFP or rAAV.Subs-Puro over a range of MOIs (Fig. 2). The kinetics of generating stably transduced cells was similar at different MOIs, and after day 14, the number of transduced cells remained stable (Fig. 2A). The percentage of GFP-positive cells remained stable until at least day 24 after infection (data not shown). The stability of GFP expression after random integration beyond 24 days was not examined. The frequency of random integration was dependent on the MOI, and by using an MOI of 10,000, approximately 1% of the cells were stably GFP positive (Fig. 2B). The frequency of random integration was not affected by the generation of a simultaneous DSB by coinfection of rAAV.GFP with rAAV.Sce (Fig. 2C). The target gene was being cleaved after infection with rAAV.Sce because there were small mutations in the target after rAAV.Sce infection, consistent with the repair of a DSB by a mutagenic DSB repair mechanism (data not shown). These mutations were not found when rAAV.Sce was not infected. We found that the random integration frequency was 10-fold lower when we used antibiotic resistance as a marker rather than GFP expression (compare random integration frequency by using GFP [Fig. 2B] with random integration frequency by using puromycin resistance [Fig. 2D]), suggesting that GFP is a more sensitive measure of random integration than antibiotic selection.
FIG. 2.
Random integration with rAAV. (A) The time course of rAAV transduction is illustrated. This graph shows representative examples of the time course of transduction of 293 cells with rAAV.GFP at different MOIs. (B) The random integration frequency of rAAV.GFP at various MOIs is shown. The random integration frequency at each MOI was determined by measuring the percentage of GFP-positive cells by flow cytometry at day 14 after infection from at least four different infections. (C) The random integration frequency of rAAV.GFP is not affected by the presence of a DSB. 293/A658 cells were infected with rAAV.GFP ± rAAV.Sce at the MOIs indicated, and the percentage of stably transduced cells was measured by flow cytometry. (D) The random integration frequency with rAAV.Subs-Puro is shown. 293/A658 cells were infected with rAAV.Subs-Puro at an MOI of either 300 or 1,000, and the numbers of puromycin-resistant colonies were counted at day 21 after infection.
Gene targeting using rAAV.
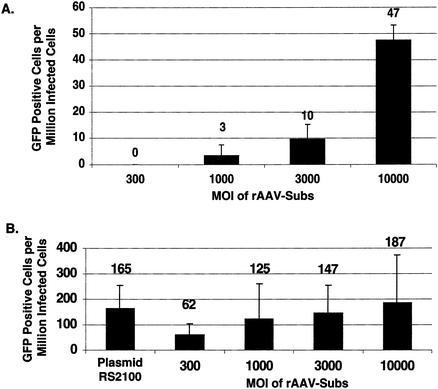
To explore whether rAAV could increase the frequency of spontaneous gene targeting by the GFP system, we infected 293/A658 target cells with rAAV.Subs. We found that the frequency of spontaneous gene targeting increased as the MOI increased (Fig. 3A). The background frequency of gene targeting with transfected DNA by this system is 0.71 events/million transfected cells (22); thus, we found a significant increase only in the frequency of spontaneous gene targeting when we used MOIs of 3,000 or greater. These experiments demonstrated that rAAV can mediate gene targeting in somatic mammalian cells in a dose-dependent fashion and confirmed the results of Russell and Hirata (24).
FIG. 3.
Gene targeting with rAAV. (A) The spontaneous frequency of gene targeting with rAAV-Subs is dependent on the MOI. (B) Transfection of the Sce expression plasmid stimulates rAAV-mediated gene targeting. 293/A658 cells were either cotransfected with an Sce expression plasmid (Sce driven by the PGK promoter) and repair substrate plasmid (RS2100) or simultaneously transfected with the Sce expression plasmid and infected with rAAV.Subs. Plasmid RS2100 contains the 2,100-bp repair substrate in rAAV.Subs-Puro, but in the pBS SK(+) (Stratagene) backbone (22). The targeting frequencies in this figure are not adjusted for transfection or infection efficiency.
Studies of plasmid-mediated gene targeting suggest that a DSB in the target gene stimulates gene targeting by activating the homology-directed repair of the DSB. To examine whether rAAV-mediated gene targeting can proceed through a similar mechanism, we measured the frequency of gene targeting by rAAV in the setting of a DSB (Fig. 3B). We simultaneously infected 293/A658 cells with rAAV.Subs and transfected a Sce expression plasmid. We found that at all MOIs of rAAV.Subs, the transfection of the Sce expression plasmid increased the frequency of gene targeting. The stimulation was most significant at low MOIs (stimulation of greater than 60-fold at an MOI of 300) and reached a plateau at higher MOIs (4-fold stimulation at an MOI of 10,000). With the transfection of an Sce expression plasmid, the introduction of the repair substrate by infection with rAAV did not significantly stimulate gene targeting more than that by the transfection of supercoiled plasmid DNA (RS2100) (Fig. 3B).
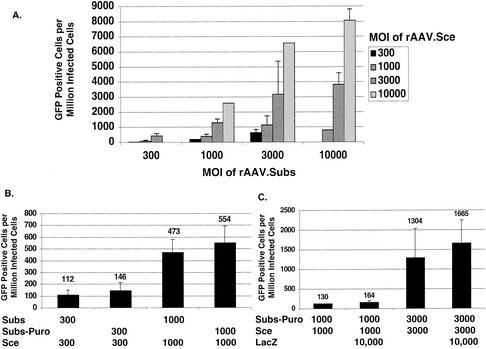
To investigate whether gene targeting could be stimulated with only rAAV vectors, we developed a two-virus system by using coinfection of rAAV.Subs and rAAV.Sce (Fig. 4). We found that the frequency of gene targeting was directly proportional to the MOI of both viruses (Fig. 4A). Thus, when the MOI of rAAV.Subs was held constant, the gene targeting frequency increased as the MOI of rAAV.Sce increased. Conversely, when the MOI of rAAV.Sce was held constant, the frequency of gene targeting increased as the MOI of rAAV.Subs increased. The gene targeting frequency was highest (8,000 events/million cells or 0.8%) when we coinfected rAAV.Subs and rAAV.Sce at an MOI of 10,000 (the highest MOI tested). Using a combination of allele-specific PCR and sequencing, we molecularly analyzed GFP-positive cells and showed that gene targeting had occurred (data not shown). Substituting rAAV.Subs-Puro for rAAV.Subs as the repair substrate virus did not change the frequency of gene targeting (Fig. 4B). The frequency of gene targeting was also not changed by coinfection with a high MOI of a nonspecific rAAV (rAAV.LacZ) (Fig. 4C). If the Sce expression cassette and repair substrate were on the same virus (one-virus system), the frequency of gene targeting was identical to that by using the two-virus system (data not shown).
FIG. 4.
DSB-mediated gene targeting can be stimulated by coinfection of rAAV.Sce and rAAV.Subs. (A) Gene targeting frequencies were obtained after coinfecting different MOIs of rAAV.Subs (along the x axis) and rAAV.Sce (in the figure key). The data for MOIs between 300 and 3,000 are the averages of four to six different samples from two or three different experiments. The data when the MOI was 10,000 for either virus are the result of one or two samples from a single experiment. (B) rAAV DSB-mediated gene targeting frequency was the same for rAAV.Subs and rAAV.Subs-Puro. (C) rAAV DSB-mediated gene targeting frequency was not changed by coinfection with a nonspecific rAAV.
From the measurements of the random integration and gene targeting frequencies, we calculated the relative frequency of gene targeting by using rAAV (Table 1). The relative frequency of gene targeting is the ratio of the number of targeted events compared to the total number of integration events (targeted events plus random integration events). A higher relative frequency of gene targeting means that the number of desired targeted events is increased compared to the number of undesired insertional mutations. The relative frequency of spontaneous gene targeting with rAAV was significantly better (1 in 215 at an MOI of 10,000) than that obtained with plasmid DNA (1 in 14,000) (Table 1). The relative frequency of gene targeting increased significantly with the introduction of a DSB because the frequency of gene targeting increased dramatically and the frequency of random integration did not change. The relative frequency of gene targeting at an MOI of 10,000 was 1 in 2 (or 50% of all events were targeted) (Table 1). Importantly, increasing the MOI from 300 to 3,000 increased both the absolute and relative frequencies of gene targeting. Increasing the MOI from 3,000 to 10,000, however, increased the absolute number of gene targeting events but did not further increase the relative frequency of gene targeting. While most cells had undergone either gene targeting or random integration, there were occasionally cells in which both events occurred (data not shown).
TABLE 1.
Random integration and gene targeting frequencies with rAAV
| DNA | Random integration frequencya (GFP) | Gene targeting frequency | _n_-Fold stimulation by Sce | Relative gene targeting frequency (substrate + Sce) | ||
|---|---|---|---|---|---|---|
| Absolutea (substrate) | Relative (substrate) | Absolutea (substrate + Sce) | ||||
| Plasmid | 10,000 | 0.71 | 7.1 × 10−5 (1/14,000) | 1,600 | 2,200 | 1.4 × 10−1 (1/7) |
| MOI of rAAV | ||||||
| 300 | 1,000 | <2 | <2 × 10−3 (<1/500) | 15 | >7 | 1.5 × 10−2 (1/68) |
| 1,000 | 2,300 | 3 | 1.3 × 10−3 (1/750) | 393 | 130 | 1.5 × 10−1 (1/7) |
| 3,000 | 3,300 | 10 | 2.9 × 10−3 (1/340) | 3180 | 320 | 4.9 × 10−1 (1/2) |
| 10,000 | 10,100 | 47 | 4.6 × 10−3 (1/215) | 8,070 | 170 | 4.4 × 10−1 (1/2) |
DISCUSSION
Gene correction would be the safest mode of gene therapy for genetic diseases. It has significant advantages over gene addition approaches because it does not require knowledge of the regulatory mechanisms that control gene expression, it does not suffer from the problem of transgene silencing, and it does not generate insertional mutations, which are a risk for any gene addition approach. The problem of insertional mutations has been highlighted by patients developing leukemia from insertional mutations in the gene therapy trial to treat X-linked SCID (17). But the low spontaneous frequency of gene targeting has precluded its use as a therapeutic tool. In this paper, we describe progress towards solving this problem by showing that combining rAAV infection with the generation of DSBs in the target gene can significantly increase both the absolute and relative frequencies of gene targeting.
rAAV is an efficient, seemingly nonpathogenic virus for the delivery of DNA to a wide range of cell types (20, 25). Prior work by Russell and Hirata (24) showed that rAAV could increase the gene targeting frequency in human somatic cells. Our results provide the first independent confirmation of their work. We found that rAAV increased both the absolute and relative frequencies of spontaneous gene targeting compared to the transfection of plasmid DNA (Table 1). The absolute and relative frequencies of spontaneous gene targeting mediated by rAAV are of an order to consider for use in experimental purposes but are still too low to consider for therapeutic purposes.
DNA DSBs in the target gene can stimulate the absolute gene targeting frequency by several-thousand-fold and concomitantly improve the relative frequency of gene targeting by using plasmid DNA (4, 7, 9, 22, 23, 28, 30, 32). Our work shows that combining rAAV infection with the introduction of a DSB also improves both the absolute and relative frequencies of gene targeting. The stimulation of rAAV gene targeting by a DSB is evidence that one mechanism of rAAV-mediated gene targeting is through the homology-directed repair of a DSB. Thus, rAAV can mediate gene targeting through a mechanism common to that of naked DNA targeting. Whether rAAV-mediated spontaneous gene targeting is also initiated by DNA damage remains to be determined. We found that by coinfecting with rAAVs that contain the repair substrate and an Sce expression cassette, we could obtain an absolute gene targeting frequency of 8,000 per million cells (0.8%) and a relative frequency of 1 in 2. The use of a rAAV that expresses Sce thus stimulated the absolute and relative frequency of gene targeting by 100- to 300-fold. In comparison with that determined by Russell and Hirata (24), the combined use of rAAV.Subs and rAAV.Sce increased the frequency of gene targeting by 4- to 8-fold while using 40-fold less virus. Using lower MOIs to achieve the same or greater gene targeting frequencies has several advantages. First, it makes the use of rAAV to mediate gene targeting practically tenable. Second, it increases the safety of using rAAV. Miller et al. showed that infection with rAAV is associated with genomic rearrangements (19). Whether rAAV is an active part of creating the rearrangements or just a passive marker remains to be determined. Moreover, we have demonstrated that the number of insertional mutations (another form of genomic instability) is directly related to the MOI of rAAV. Thus, being able to use lower MOIs makes rAAV significantly safer to use.
Using the two-virus system, we found that the frequency of gene targeting was dependent on the amounts of both Sce virus and substrate virus. When, for example, the amount of Sce virus was held constant, the targeting frequency increased dramatically as the amount of substrate virus was increased. Conversely, when the amount of substrate virus was held constant, the targeting frequency increased dramatically as the amount of Sce virus was increased. Thus, by increasing the number of DSBs, the frequency of gene targeting for a given MOI of substrate virus increased dramatically. The dual dependence on substrate virus and Sce virus suggests that both the repair substrate and the DSB were limiting in the targeting reaction. Overall, the results we obtained are in agreement and complement the results obtained by D. Miller et al. (18a). Despite using different markers as target genes, different cell lines, and different ways to introduce the Sce endonuclease, we both found that the frequency of rAAV-mediated gene targeting was increased significantly by the introduction of a DSB and that the mechanism of rAAV-mediated gene targeting can proceed through the homology-directed repair of a DSB.
Although the frequency of spontaneous gene targeting with rAAV is superior to that with plasmids, we found that the frequencies of DSB-mediated gene targeting are similar. We obtained gene targeting frequencies of 1% when transfecting plasmid DNA (which increases to 3 to 5% when we normalize for transfection efficiency), which is of the same magnitude of 0.8% that we obtained by using rAAV (22). The use of rAAV to introduce both components necessary for gene targeting (a repair substrate and an endonuclease to create a DSB) represents a major advance because it significantly broadens the range of cell types that can be considered for gene targeting. The data of Miller et al. (18a) provide direct evidence that rAAV and DSBs can be used to stimulate gene targeting in primary cells.
While an absolute gene targeting frequency of 0.8% and a relative frequency of 50% approach levels of therapeutic utility and safety, important problems remain. The first problem is to develop methods to create sequence-specific breaks at any site in the genome in a safe and efficient manner. Chimeric nucleases, fusions between a zinc finger DNA binding domain and an endonuclease domain, have been shown to cleave extrachromosomal DNA in Xenopus oocytes and genomic DNA in Drosophila melanogaster (2, 3, 6). It was recently shown that chimeric nucleases can stimulate gene targeting in human somatic cells by several-thousand-fold (22). Performing gene targeting by designing chimeric nucleases to stimulate gene targeting at any gene and by using rAAV to introduce the nucleases is an experimental approach that should be investigated and has a strong probability of working. The second problem is to understand further the parameters of rAAV that make it an efficient substrate for gene targeting. Our data suggest that a mechanism of rAAV-mediated gene targeting, as for other forms of gene targeting, is through the repair of DNA DSBs. It was previously shown that the frequency of DSB-mediated gene targeting by using plasmid DNA is dependent on the amount of repair substrate introduced (22). The dose response of gene targeting with rAAV suggests that mass action effects, i.e., simply the ability of rAAV to bring DNA efficiently into cells, are an important aspect of the mechanism of rAAV-mediated gene targeting. But other features, including the possibilities that the single-stranded DNA structure of rAAV is preferred by the gene targeting machinery and that rAAV infection activates the machinery of gene targeting may also contribute. The third problem is to further reduce the number of random insertions. While a relative frequency of gene targeting of 1 in 2 is excellent, it still means that for each targeted event, there is an insertional mutation. In fact, occasionally both events can occur in the same cell. It may be possible to shift the balance of integration events further towards targeted events and away from random events and thus continue to reduce the risk of insertional mutagenesis. Nonetheless, we believe that our study and those of others support the promise of AAV as a vector for gene correction type gene therapy, particularly for genetic diseases that result from small mutations.
Acknowledgments
We thank Elissa Denney for excellent technical support over the course of this work. We thank Dan Miller and David Russell for sharing unpublished results.
This work was supported by the Howard Hughes Medical Institute (Physician Postdoctoral Fellow Award), Burroughs-Wellcome Fund (Career Development Award), and NIH grant KO8 HL70268-01 to M.H.P.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology. John Wiley and Sons, Inc., Boston, Mass.
- 2.Bibikova, M., D. Carroll, D. J. Segal, J. K. Trautman, J. Smith, Y. G. Kim, and S. Chandrasegaran. 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 21**:**289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikova, M., M. Golic, K. G. Golic, and D. Carroll. 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161**:**1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenneman, M., F. S. Gimble, and J. H. Wilson. 1996. Stimulation of intrachromosomal homologous recombination in human cells by electroporation with site-specific endonucleases. Proc. Natl. Acad. Sci. USA 93**:**3608-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capecchi, M. 1989. Altering the genome by homologous recombination. Science 244**:**1288-1292. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasegaran, S., and J. Smith. 1999. Chimeric restriction enzymes: what is next? Biol. Chem. 380**:**841-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choulika, A., A. Perrin, B. Dujon, and J.-F. Nicolas. 1995. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccaromyces cerevisiae. Mol. Cell. Biol. 15**:**1968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, K. R., X. Liu, J. P. McGrath, and P. R. Johnson. 1999. Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum. Gene Ther. 10**:**1031-1039. [DOI] [PubMed] [Google Scholar]
- 9.Donoho, G., M. Jasin, and P. Berg. 1998. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol. 18**:**4070-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grifman, M., M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini, and M. D. Weitzman. 2001. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3**:**964-975. [DOI] [PubMed] [Google Scholar]
- 11.Hirata, R., J. Chamberlain, R. Dong, and D. W. Russell. 2002. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 20**:**735-738. [DOI] [PubMed] [Google Scholar]
- 12.Hirata, R. K., and D. W. Russell. 2000. Design and packaging of adeno-associated virus gene targeting vectors. J. Virol. 74**:**4612-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue, N., R. Dong, R. K. Hirata, and D. W. Russell. 2001. Introduction of single base substitutions at homologous chromosomal sequences by adeno-associated virus vectors. Mol. Ther. 3**:**526-530. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, N., R. K. Hirata, and D. W. Russell. 1999. High-fidelity correction of mutations at multiple chromosomal positions by adeno-associated virus vectors. J. Virol. 73**:**7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jasin, M. 1996. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 12**:**224-228. [DOI] [PubMed] [Google Scholar]
- 16.Kay, M. A., C. S. Manno, M. V. Ragni, P. J. Larson, L. B. Couto, A. McClelland, B. Glader, A. J. Chew, S. J. Tai, R. W. Herzog, V. Arruda, F. Johnson, C. Scallan, E. Skarsgard, A. W. Flake, and K. A. High. 2000. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 24**:**257-261. [DOI] [PubMed] [Google Scholar]
- 17.Marshall, E. 2002. Gene therapy a suspect in leukemia-like disease. Science 298**:**34-35. [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin, S. K., P. Collis, P. L. Hermonat, and N. Muzyczka. 1988. Adeno-associated virus general transduction vectors: analysis of proviral structures. J. Virol. 62**:**1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Miller, D. G., L. M. Petek, and D. W. Russell. 2003. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol. Cell. Biol. **23:**3550-3557. [DOI] [PMC free article] [PubMed]
- 19.Miller, D. G., E. A. Rutledge, and D. W. Russell. 2002. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 30**:**147-148. [DOI] [PubMed] [Google Scholar]
- 20.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158**:**97-129. [DOI] [PubMed] [Google Scholar]
- 21.Pomerantz, J. L., E. M. Denny, and D. Baltimore. 2002. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 21**:**5184-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porteus, M. H., and D. Baltimore. Chimeric nucleases stimulate gene targeting in human cells. Science, in press. [DOI] [PubMed]
- 23.Rouet, P., F. Smih, and M. Jasin. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14**:**8096-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, D. W., and R. K. Hirata. 1998. Human gene targeting by viral vectors. Nat. Genet. 18**:**325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell, D. W., and M. A. Kay. 1999. Adeno-associated virus vectors and hematology. Blood 94**:**864-874. [PMC free article] [PubMed] [Google Scholar]
- 26.Rutledge, E. A., and D. W. Russell. 1997. Adeno-associated virus vector integration junctions. J. Virol. 71**:**8429-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samulski, R. J., L. S. Chang, and T. Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63**:**3822-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sargent, R. G., M. A. Brenneman, and J. H. Wilson. 1997. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 17**:**267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sedivy, J. M., and P. A. Sharp. 1989. Positive genetic selection for gene disruption in mammalian cells by homologous recombination. Proc. Natl. Acad. Sci. USA 86**:**227-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smih, F., P. Rouet, P. J. Romanienko, and M. Jasin. 1995. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 23**:**5012-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonoda, E., C. Morrison, Y. M. Yamashita, M. Takata, and S. Takeda. 2001. Reverse genetic studies of homologous DNA recombination using the chicken B-lymphocyte line, DT40. Philos. Trans. R. Soc. Lond. Sec. B Biol. Sci. 356**:**111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taghian, D. G., and J. A. Nickoloff. 1997. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 17**:**6386-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72**:**2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71**:**9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6**:**973-985. [DOI] [PubMed] [Google Scholar]
- 36.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73**:**2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]