Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors (original) (raw)
Abstract
Cerebellar granule cells are the most abundant neurons in the brain, and granule cell precursors (GCPs) are a common target of transformation in the pediatric brain tumor medulloblastoma. Proliferation of GCPs is regulated by the secreted signaling molecule Sonic hedgehog (Shh), but the mechanisms by which Shh controls proliferation of GCPs remain inadequately understood. We used DNA microarrays to identify targets of Shh in these cells and found that Shh activates a program of transcription that promotes cell cycle entry and DNA replication. Among the genes most robustly induced by Shh are cyclin D1 and N-myc. N-myc transcription is induced in the presence of the protein synthesis inhibitor cycloheximide, so it appears to be a direct target of Shh. Retroviral transduction of N-myc into GCPs induces expression of cyclin D1, E2F1, and E2F2, and promotes proliferation. Moreover, dominant-negative N-myc substantially reduces Shh-induced proliferation, indicating that N-myc is required for the Shh response. Finally, cyclin D1 and N-myc are overexpressed in murine medulloblastoma. These findings suggest that cyclin D1 and N-myc are important mediators of Shh-induced proliferation and tumorigenesis.
Keywords: neuron, cerebellum, granule cell
Granule cells are a critical element of the cerebellar circuitry that controls motor coordination (1). They are also targets of transformation in many cases of medulloblastoma, the most common malignant brain tumor in children (2, 3). However, the molecular mechanisms that regulate the growth and differentiation of granule cells are not fully understood.
One key regulator of granule cell development is Sonic hedgehog (Shh) (4–6). In the cerebellum, Shh is produced by Purkinje cells and granule cell precursors (GCPs) respond to it. Shh induces proliferation of GCPs in culture, and in vivo administration of Shh-blocking antibodies causes a dramatic reduction in the number of granule cells produced. These studies suggest that Shh is a critical mitogen for GCPs during normal development.
Shh signaling has also been implicated in medulloblastoma. Mutations in patched1, an antagonist of Shh signaling, result in Gorlin's syndrome, a disease characterized by skeletal abnormalities, skin tumors, and increased incidence of medulloblastoma (7). In addition, 20–30% of sporadic medulloblastomas harbor mutations in patched and other elements of the Shh pathway (8–10). Finally, mice in which one copy of the patched1 gene has been inactivated develop tumors that resemble medulloblastoma (11). Thus, Shh pathway-related proliferation is involved in both normal and malignant growth in the cerebellum.
The precise mechanisms by which Shh promotes proliferation and tumor formation are unknown. In most cells, the transmembrane protein encoded by patched1 represses transcription of Shh target genes (12). When Shh binds to Patched, the repression is relieved, and a protein called Smoothened becomes activated. Smoothened activation leads, through steps that are poorly understood, to posttranslational modification and nuclear translocation of Gli-family transcription factors. Once in the nucleus, Gli proteins bind to DNA and regulate target gene transcription.
Genes that mediate the effects of Shh on cell fate and differentiation have been identified in a number of tissues. In the embryonic spinal cord, Shh induces expression of homeodomain transcription factors that promote differentiation of progenitors into floor plate, motor neurons, or oligodendrocytes (13). In the limb, Shh regulates growth and digit identity by inducing transcription of Bmp2, Fgf4, and Hoxd13 (14). More recently, Shh targets have been identified by using microarrays. For example, in epithelial cells, transfection of gli1 results in increased transcription of genes encoding Cyclin D2, insulin-like growth factor binding protein 6, osteopontin, and plakoglobin (15). The significance of these genes for Shh responses during development and tumorigenesis is unknown.
Although Shh functions as a mitogen in a number of tissues, the genes that mediate this response remain poorly defined. To understand how Shh induces proliferation, we have performed microarray analysis on GCPs stimulated with Shh. Our analysis reveals that Shh induces critical regulators of cell cycle progression, DNA replication, and cell differentiation. One of the most robustly induced targets in these cells is the gene encoding the transcription factor N-myc. Here we show that N-myc is a direct target of the Shh pathway and plays a critical role in mediating the proliferative response.
Materials and Methods
Animals. Wild-type (C57BL/6 × CBA F1) and patched1 heterozygous mice were maintained in animal facilities at Stanford and Duke Universities.
Isolation of Granule Cell Precursors. GCPs were isolated from 7-day-old (P7) mice as described (4). Cerebella were digested in solution containing 10 units/ml papain (Worthington) and 250 units/ml DNase (Sigma) and triturated to obtain a cell suspension. This suspension was centrifuged through 35% and 65% Percoll (Pharmacia), and GCPs were harvested from the 35%/65% interface. Cells were resuspended in Neurobasal medium containing B27, sodium pyruvate, L-glutamine, and penicillin/streptomycin (all from Invitrogen) and transferred to dishes coated with poly(D-lysine) (Sigma).
Shh Stimulation and RNA Isolation. To identify Shh target genes, GCPs were cultured in medium with no growth factor or 3 μg/ml Shh-N (provided by Curis, Cambridge, MA, or purchased from R & D Systems). For some experiments, cycloheximide (10 μg/ml, Sigma) was added at the beginning of culture. After 1–24 h, cells were harvested and frozen. To isolate RNA for microarrays or Northern blotting, pellets were lysed in buffer containing 0.5% IGEPAL CA-630 (Sigma), digested with proteinase K, extracted with phenol:chloroform:isoamyl alcohol, and precipitated with ethanol. For microarrays, RNA was further purified by using RNeasy columns (Qiagen, Valencia, CA). RNA extraction from retrovirus-infected cells, adult cerebellum, and medulloblastoma cells was carried out by using TRIzol (Invitrogen).
Microarray Hybridization and Analysis. RNA from cells cultured with or without Shh-N for 6 h was converted to cDNA by using the Superscript Choice cDNA kit (Invitrogen) and a T7-dT(24) primer (Genset/Proligo, Boulder, CO). cRNA was generated by using a T7-transcription/labeling kit from Enzo Life Sciences and hybridized to Mu11K GeneChips (Affymetrix, Santa Clara, CA). Chips were scanned, and hybridization were data acquired by using AFFYMETRIX SUITE 5.0. Data were analyzed by using the R package, which carries out normalization, estimates gene expression levels, and determines statistical significance by using empirical Bayesian analysis (refs. 16 and 17; for details, see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). The identities of differentially expressed genes were verified by integrating data from the Affymetrix and Unigene databases.
Northern Analysis. RNA was run on agarose-formaldehyde gels and transferred to nylon membranes. Membranes were UV-crosslinked and incubated in QuikHyb (Stratagene) containing 32P-labeled probes for cyclin D1, cyclin D2, N-myc, or c-myc, each labeled by random priming. Membranes were washed and scanned by using a PhosphorImager (Molecular Dynamics). RNA amounts were normalized based on ethidium bromide staining of 28S rRNA.
Generation of Retroviruses. Genes were introduced into GCPs by using replication-defective retroviruses. cDNAs encoding wild-type and dominant-negative N-myc (ΔMBII, provided by Michael Cole, Princeton University, Princeton), cyclin D1 and D2 (from Charles Sherr, St. Jude Children's Research Hospital, Memphis, TN), and g_li1_ (from Ken Kinzler, Johns Hopkins University, Baltimore) were cloned into LZRS-IRES-GFP vectors (from Garry Nolan, Stanford University). Plasmids were transfected into 293T cells with helper plasmids encoding gag-pol and vesicular stomatitis virus envelope glycoprotein. Supernatants were harvested and concentrated by centrifugation. Retroviral stocks were tested on naive 293T cells, and equivalent titers were used for infection of GCPs.
Infection of Granule Cell Precursors and Measurement of BrdUrd Incorporation. To test effects of viruses on proliferation, supernatants were added to GCPs at the beginning of culture. Shh-N (3 μg/ml) was also added to some cultures at this time. Cells were maintained at 32°C for 18 h to maximize virus stability during infection and then transferred to 37°C. Cells were cultured for a total of 64 h, with BrdUrd (Roche) added during the last 16 h. At the end of culture, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, treated with DNase (50 units/ml) to expose BrdUrd epitopes, and stained with sheep anti-BrdUrd (BioDesign, Kennebunk, ME). Samples were observed by using a Nikon TE-200 microscope and OPENLAB software (Improvision, Lexington, MA). In most cultures, 20–50% of GCPs were infected. To quantitate proliferation of virus-infected cells, the percentage of green fluorescent protein-positive (GFP+) cells labeled with BrdUrd was determined for four randomly chosen fields.
Quantitation of Transcript Levels by Using Real-Time RT-PCR. To identify targets of N-myc in GCPs, cells were infected with control or N-myc retroviruses. After 24 h, cells were harvested and GFP+ cells were sorted by using a FACSVantage SE flow cytometer (BD Biosciences). RNA was isolated by using TRIzol and treated with DNase to eliminate genomic DNA. Reverse transcription was performed by using the Superscript II RNase H- kit (Invitrogen). cDNA was quantitated by using PicoGreen (Molecular Probes), and equal amounts of cDNA were used for analysis on a Roche LightCycler.
Results
Microarray Analysis of Genes Regulated by Shh in Granule Cell Precursors. To gain insight into the molecular mechanisms of Shh-induced proliferation, we analyzed RNA from GCPs cultured in the presence or absence of Shh-N (3 μg/ml) for 6 h. Five experiments were carried out, each using cells from an independent litter of mice. RNA was labeled and hybridized to Affymetrix Mu11K GeneChips. For each gene, the difference in expression level between untreated and Shh-treated cells (mbar) and the statistical significance of this difference [logarithm of odds ratio (lod)] were calculated. Of the 10,495 genes represented on the chips, we identified 134 (1.3%) whose expression changed significantly (lod > 0) in response to Shh. These included 126 genes whose expression increased and 8 genes whose expression decreased (see Table 3 and Fig. 6, which are published as supporting information on the PNAS web site). Representative genes from the most prevalent categories are shown in Table 1.
Table 1. Categories of genes regulated by Shh.
| Unigene/GenBank no. | Statistical confidence, Iod | Expression difference, mbar | |
|---|---|---|---|
| Cell cycle/DNA replication | |||
| Cyclin D1 | Mm.22288 | 5.70 | 5.58 |
| Minichromosome maintenance-deficient 3 | Mm.4502 | 4.13 | 4.88 |
| Cyclin D2 | Mm.3141 | 4.11 | 3.33 |
| Proliferating cell nuclear antigen (PCNA) | Mm.7141 | 3.35 | 3.15 |
| Minichromosome maintenance-deficient 5 | Mm.5048 | 2.28 | 2.90 |
| Replication factor C3 | Mm.12553 | 1.91 | 2.49 |
| Cyclin G2 | Mm.3527 | 1.88 | -2.24 |
| Minichromosome maintenance-deficient 6 | Mm.4933 | 1.65 | 2.08 |
| Cell division cycle 25 homolog A (Cdc25A) | Mm.29800 | 1.63 | 2.04 |
| Replication protein A2 | Mm.2870 | 1.60 | 3.49 |
| Minichromosome maintenance deficient 2 | Mm.16711 | 1.26 | 2.84 |
| DNA binding/transcription | |||
| N-myc | Gb.X03919 | 4.49 | 6.52 |
| Tat-binding protein interacting protein | Mm.18344 | 1.89 | 2.09 |
| Dp-1 | Mm.925 | 1.89 | 2.09 |
| Cellular nucleic acid binding protein (CNBP) | Mm.7335 | 1.86 | 2.40 |
| High-mobility group AT-hook 1 | Mm.4438 | 0.66 | 1.81 |
| Sterol regulatory element-binding protein (SP) | Mm.214958 | 0.62 | 3.16 |
| Myb-binding protein (p160) | Mm.147946 | 0.51 | 2.08 |
| Chromatin structure/DNA repair | |||
| Uracil-DNA glycosylase | Mm.1393 | 4.01 | 3.90 |
| Helis helicase | Mm.57223 | 1.91 | 2.64 |
| Checkpoint kinase 1 (Chk1) | Mm.16753 | 1.52 | 2.24 |
| EST similar to DNA topoisomerase I | Gb.W10047 | 1.31 | 2.59 |
| Breast cancer 1 (Brca1) | Mm.1889 | 0.78 | 1.85 |
| Dihydrofolate reductase | Mm.23695 | 0.54 | 2.66 |
| Rad51/RecA homolog | Mm.231 | 0.14 | 2.32 |
| Metabolism/biosynthesis | |||
| Hexokinase II | Gb.Y11666 | 4.73 | 5.00 |
| Nucleoside transporter SLC29A1 | Mm.29744 | 4.06 | 6.04 |
| Spermidine synthase | Mm.10 | 3.78 | 3.35 |
| S-adenosylmethionine decarboxylase 1 | Mm.7880 | 3.62 | 3.48 |
| Delta-aminolevulinate dehydratase | Mm.6988 | 3.50 | 4.15 |
| IMP dehydrogenase 2 | Mm.6065 | 0.87 | 1.82 |
| Thymidylate synthase | Gb.M13352 | 0.84 | 2.72 |
| CTP synthase | Mm.1815 | 0.84 | 2.01 |
| Signaling/differentiation | |||
| Deltex 1 | Mm.1645 | 2.32 | 5.90 |
| Disintegrin-like metalloprotease (Adamts1) | Mm.1421 | 2.13 | 3.68 |
| Secreted frizzled-related protein 1 (sfrp1) | Mm.3171 | 1.74 | 2.45 |
| EST similar to hippocalcin A | Gb.W54905 | 1.74 | 2.40 |
| Glypican 1 | Mm.24193 | 0.93 | 2.48 |
| IGF-binding protein 5 (IGFBP-5) | Mm.218877 | 0.87 | -4.13 |
| v-crk-associated kinase substrate (p130cas) | Mm.3758 | 0.22 | 1.95 |
| Eph receptor B4 (EphB4) | Mm.34533 | 0.13 | 2.13 |
Consistent with the mitogenic effects of Shh, many of the differentially expressed genes encoded proteins involved in cell cycle regulation. These included Cyclin D1 and D2, the phosphatase Cdc25A, and members of the minichromosome maintenance (MCM) family that are involved in DNA replication. Shh also induced expression of transcription factors implicated in cell cycle progression, such as N-myc and Dp-1, and regulators of cholesterol metabolism, such as cellular nucleic acid binding protein (CNBP) and sterol regulatory element binding protein (SREBP-1). Genes involved in DNA repair and nucleotide and polyamine metabolism were also induced by Shh. Despite their diverse functions, many of these genes have previously been associated with cell proliferation. Finally, Shh-treated cells had increased RNA for proteins involved in preventing cell differentiation, such as Deltex, an activator of Notch signaling, and secreted frizzled-related protein 1 (sfrp1), an inhibitor of Wnt signaling. Thus, Shh may regulate growth by both increasing production of cell cycle activators and inhibiting differentiation signals.
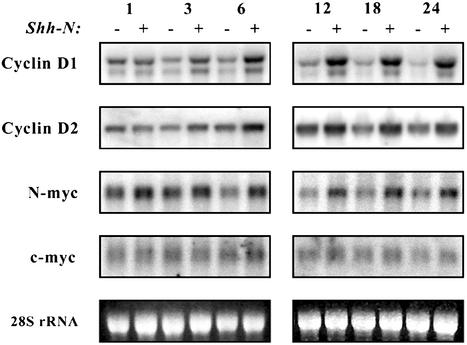
N-myc Is a Direct Target of Shh Signaling. Among the genes induced by Shh, the D-cyclins and N-myc are important regulators of the early stages of cell cycle progression and are therefore potential mediators of Shh-induced proliferation. To confirm that Shh treatment induces transcription of these genes, RNA from GCPs cultured with or without Shh for various time periods was analyzed by Northern blotting (Fig. 1). After 1 h of culture, untreated and Shh-treated cells showed no difference in expression of cyclin D1 or D2. By 3 h, the amount of cyclin D1 and D2 RNA began to decline in untreated cells and to increase in cells treated with Shh. This trend continued from 6–24 h, with levels of cyclin D1 and D2 RNA decreasing sharply in the absence of Shh and continuing to increase in its presence. Expression of cyclin D3 was not detected in GCPs at any time after Shh stimulation (data not shown).
Fig. 1.
Induction of D-cyclins and N-myc by Shh. GCPs were cultured in serum-free media containing no stimulus (-) or 3 μg/ml Shh-N (+) for the indicated times. Total RNA was separated by electrophoresis and transferred onto a nylon membrane. The membrane was hybridized with 32P-labeled cDNA probes for cyclin D1, cyclin D2, N-myc, or c-myc, and then exposed to a PhosphorImager. Images were processed by using photoshop (Adobe Systems, Mountain View, CA).
Whereas Shh induction of D-cyclins was detected only after 3 h of culture, increased N-myc expression was seen as early as 1 h after Shh treatment (Fig. 1). By 6 h of culture,significantly higher levels of N-myc RNA were observed in Shh-treated GCPs compared with unstimulated controls. Elevated levels of N-myc RNA persisted up to 24 h, the latest time period we investigated. In contrast, expression of c-myc was difficult to detect by Northern analysis and changed little in response to Shh.
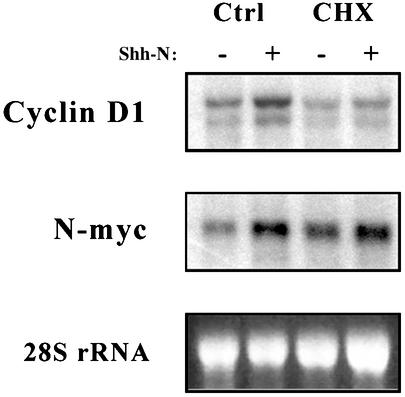
The kinetics of cyclin D1 and N-myc transcription suggested that they might be direct targets of Shh signaling. To test this hypothesis, Shh stimulation was carried out in the presence or absence of the protein synthesis inhibitor cycloheximide (Fig. 2). In the presence of cycloheximide, Shh induction of cyclin D1 was abolished but induction of N-myc was still observed. These data suggest that N-myc induction does not require protein synthesis and is likely to be a direct target of the Shh pathway.
Fig. 2.
N-myc induction does not require protein synthesis. GCPs were cultured for 6 h with no stimulus (-) or 3 μg/ml Shh-N (+), in the presence or absence of 10 μg/ml cycloheximide (CHX). RNA was separated by electrophoresis, transferred onto membranes, and hybridized with probes for cyclin D1 or N-myc. Membranes were exposed to a PhosphorImager, and images were processed by using photoshop.
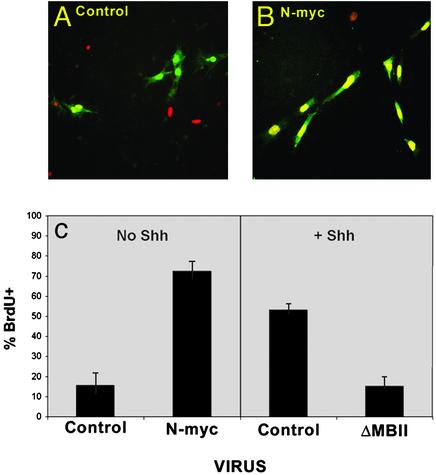
N-myc Is a Critical Mediator of Proliferation. The rapid increase in N-myc transcription stimulated by Shh suggested that N-myc might be an important mediator of Shh-induced proliferation. To test this possibility, we overexpressed N-myc in GCPs and examined the effects on proliferation. Cells were infected with retroviruses encoding no cDNA or N-myc followed by an internal ribosome entry site (IRES) and the coding sequence for GFP. After 48 h, cells were pulsed with BrdUrd and incubated for an additional 16 h before being fixed and stained with anti-BrdUrd antibodies. Cultures were examined for colocalization of GFP (infected cells) and BrdUrd (proliferating cells). Only 10–15% of cells infected with control retroviruses were positive for BrdUrd (Fig. 3). In contrast, 73% of the cells infected with viruses encoding N-myc were labeled with BrdUrd. These results indicate that overexpression of N-myc is sufficient to cause proliferation of GCPs.
Fig. 3.
Retroviral expression of N-myc promotes cell cycle entry. GCPs were infected with retroviruses carrying no cDNA (Control), N-myc, or dominant-negative N-myc (ΔMBII) and cultured in the presence or absence of Shh-N for 48 h. Cells were labeled with BrdUrd for 16 h and then stained with anti-BrdUrd antibodies. Representative images for control and N-myc-infected cells are shown in A and B. For each condition, the percentage of virus-infected cells (green staining) that had incorporated BrdUrd (red/yellow staining) was determined. Data in C represent mean ± SEM for four fields. Similar results were obtained in three independent experiments.
We also tested the ability of retroviruses encoding Cyclin D1 and D2, Gli-1, and Gli-2 to induce GCP proliferation (Table 2). Whereas overexpression of Gli-1 and Gli-2 caused strong proliferative responses (72% and 29% BrdUrd-positive, respectively, compared with 5% BrdUrd-positive for control virus-infected cells), cyclin D1 and D2 caused only small increases in proliferation (10–13% BrdUrd-positive). Under the conditions tested, overproduction of cyclins was not sufficient to induce cell cycle entry in the majority of cells. In contrast, Gli proteins, which activate a number of Shh target genes, are much more mitogenic for GCPs.
Table 2. Induction of proliferation by retroviruses encoding D-cyclins and Gli proteins.
| Virus | Stimulus | % BrdUrd+ | Fold induction |
|---|---|---|---|
| Control | — | 5±1 | 1 |
| Control | Shh | 33±3 | 6.6 |
| Gli-1 | — | 72±9 | 14.4 |
| Gli-2 | — | 29±8 | 5.8 |
| Cyclin D1 | — | 13±4 | 2.6 |
| Cyclin D2 | — | 10±1 | 2 |
Having found that overexpression of N-myc is sufficient to promote proliferation, we sought to determine whether N-myc activity is necessary for Shh-induced proliferation. To test this, GCPs were stimulated with Shh and infected with viruses encoding a mutant form of N-myc (ΔMBII) that has been shown to function as a dominant-negative inhibitor of N-myc-mediated cell transformation (18, 19). Expression of ΔMBII caused a dramatic reduction in BrdUrd incorporation in response to Shh (15% BrdUrd-positive, GFP-positive cells in ΔMBII-infected cultures compared with 53% BrdUrd-positive, GFP-positive cells in cultures infected with control vector; Fig. 3_C_). Thus, N-myc function is required for Shh-induced proliferation.
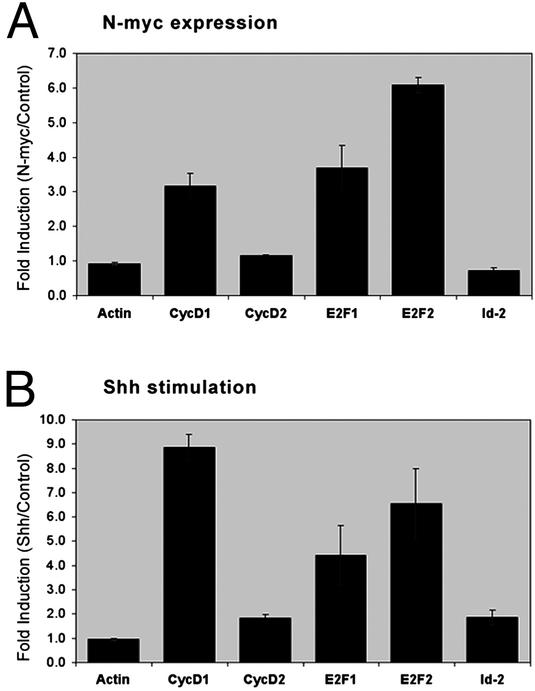
N-myc Promotes Expression of Cell Cycle Genes. Myc proteins can promote cell cycle progression by a variety of mechanisms, including repressing expression of cyclin-dependent kinase inhibitors (CKIs) and inducing expression of D-cyclins, E2F transcription factors, and the inhibitor of differentiation Id2 (20–24). To determine how N-myc promotes cell cycle progression in GCPs, we examined expression of these genes in cells infected with N-myc retroviruses. Cells were incubated with control or N-myc-encoding viruses for 24 h, and infected cells were isolated by fluorescence-activated cell sorting. Expression of cell cycle genes was measured by using real-time RT-PCR. Compared with control virus-infected cells, N-myc virus-infected cells had significantly increased levels of cyclin D1 (3.2-fold), E2F1 (3.7-fold), and E2F2 (6.1-fold) RNA (Fig. 4_A_). In contrast, expression of cyclin D2 and Id2 did not differ between control and N-myc virus-infected cells. Increased expression of cyclin D1 (8.9-fold), E2F1 (4.4-fold), and E2F2 (6.6-fold) was also observed in cells treated with Shh for 24 h (Fig. 4_B_). Shh stimulation caused small (1.8-fold) increases in expression of cyclin D2 and Id2. Expression of genes encoding the cdk inhibitors p15, p18, p19, p21, and p27 did not change significantly in response to N-myc or Shh (data not shown). These data suggest that N-myc may promote proliferation by inducing transcription of cyclin D1 and _E2F_s.
Fig. 4.
N-myc promotes expression of cell cycle genes. GCPs were infected with retroviruses carrying no cDNA (control) or wild-type N-myc (A), or cultured in medium containing no stimulus (control) or 3 μg/ml Shh-N (B). After 24 h, cells were harvested (and for virus-infected cultures, cells were fluorescence-activated cell sorted to isolate GFP-positive cells), and total RNA was isolated and reverse transcribed. Equal amounts of cDNA were analyzed by real-time PCR with primers for the indicated genes. Data represent mean ± SEM for experiments performed in triplicate.
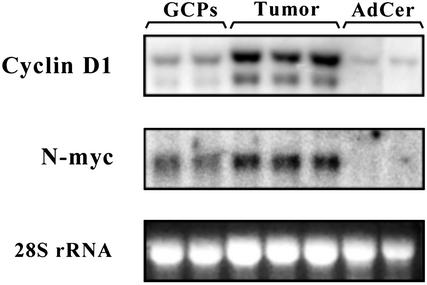
Expression of cyclin D1 and N-myc Is Elevated in Medulloblastoma. Aberrant Shh signaling contributes to medulloblastoma in both humans and mice. To determine whether N-myc and cyclin D1 are targets of the Shh pathway in medulloblastoma, we examined their expression in tumors from patched1 mutant mice. Cerebellar tumors were dissected from ptc1+/- mice, and total RNA was subjected to Northern analysis. Ptc1 tumors contained substantially higher levels of cyclin D1 and N-myc RNA than GCPs and normal adult cerebellum (Fig. 5). These data suggest that normal targets of the Shh signaling pathway are also overexpressed in tumors from ptc1 mutant mice and may contribute to tumor initiation or maintenance.
Fig. 5.
cyclin D1 and N-myc are expressed in medulloblastomas from patched1 mutant mice. Total RNA from GCPs, tumor cells from patched1 mutant mice, and normal adult cerebellum were separated by electrophoresis, transferred onto membranes, hybridized with probes specific for cyclin D1 or N-myc, and then exposed to a PhosphorImager screen. Images were processed by using photoshop.
Discussion
The Transcriptional Response to Shh in Granule Cell Precursors. Shh is an important regulator of growth and differentiation in developing tissues. We have examined the genes induced by Shh in GCPs, where its known effects are primarily mitogenic. Our results indicate that Shh induces a number of genes involved in cell cycle progression and DNA replication, including D-cyclins, Cdc25A, and members of the minichromosome maintenance (MCM) family. Several of these are known targets of Myc-family transcription factors (21, 22, 25, 26), so their induction in GCPs may be a consequence of N-myc expression. Shh also represses transcription of Cyclin G2, which acts as an inhibitor of growth in a variety of cell types (27, 28). Thus, Shh may regulate growth by increasing production of cell cycle activators, as well as by inhibiting production of cell cycle inhibitors.
Shh stimulation of GCPs also causes transcription of genes indirectly involved in cell growth and proliferation, including transcription factors (N-myc and Dp-1), regulators of chromatin structure and DNA repair (Hells helicase, Chk1, and Brca1), and enzymes required for polyamine and nucleotide synthesis (spermidine and thymidylate synthases). Among the transcription factors induced by Shh are also two that have been implicated in cholesterol metabolism: SREBP-1 and CNBP. Induction of these genes might reflect a requirement for increased cholesterol synthesis in proliferating cells (29). On the other hand, the links between Shh signaling and cholesterol metabolism [ref. 30; cholesterol modification of Shh proteins, structural homology between Patched and NPC-1 (a regulator of cholesterol trafficking), and common effects of cholesterol deficiency and Shh pathway mutations on embryogenesis] suggest that induction of these transcription factors may have direct consequences for transduction of the Shh signal.
Shh also causes increased expression of genes involved in other signaling pathways. Among the most potently induced of these are Deltex and Sfrp1, regulators of Notch and Wnt signaling, respectively (31, 32). In the cerebellum, Notch signaling promotes proliferation and prevents differentiation of GCPs (33). Thus, induction of Deltex could contribute to the ability of Shh to maintain cells in an undifferentiated state. A similar role might be played by Sfrp1, which antagonizes Wnt signaling. In granule cells, Wnts have been implicated in several aspects of differentiation, including axonal remodeling and synapse formation (34). Further studies will be necessary to determine the roles of these genes in granule cell development.
Targets of Shh were also described in a recent microarray screen by using cells from P5 cerebellum (35). A number of genes identified in that study overlap with those described here. In particular, genes associated with the cell cycle (D-cyclins, MCM genes, and N-myc) were strongly induced by Shh in both studies. However, there were also many targets unique to each study. For example, several transcription factors and signaling molecules identified as targets in our study (e.g., SREBP-1, Deltex-1, and Sfrp-1) were not reported to be induced by Zhao et al. (35); similarly, these investigators found increased expression of kinases (STK1 and AYK1) and cell surface molecules (integrin alpha-X and CXCR4) that we did not detect in our analysis. One difference between the studies is our use of highly enriched GCPs (90–95% cells expressing Math1, a marker of GCPs), in contrast to the previous study, in which Math1-positive cells were 20% of the population. In addition, we examined the effects of Shh on freshly isolated cells, rather than on cells that had been rested in serum-containing medium for 12–16 h. These conditions may have allowed us to detect less abundant transcripts and more rapidly induced genes. The overlap in gene expression between the two studies highlights the genes that are most robustly induced by Shh in GCPs.
N-myc and D-Cyclins as Targets of Shh. Among the most dramatically induced genes in our screen were cyclin D1 and N-myc. D-cyclins were previously reported to be targets of Shh in GCPs (36). As in that study, we observed increased cyclin D1 RNA levels within 3 h of Shh stimulation and found that induction was inhibited by cycloheximide. We also detected increased expression of cyclin D2, but the degree of induction was markedly less than that of cyclin D1. Importantly, relatively high levels of cyclin D2 persisted in GCPs cultured in the absence of Shh for 24 h, even though the majority of such cells had exited the cell cycle (R.J.W.-R., unpublished observations). This suggests that Cyclin D2 may have a function in postmitotic granule cells in addition to its role in GCPs. Consistent with this notion, cyclin D2 knockout mice not only have defects in GCP proliferation but also in granule cell survival and differentiation (37). Although _cyclin D1_-deficient mice have no obvious cerebellar abnormalities, mice lacking both cyclin D1 and D2 are much more severely affected than those lacking only D2 (38). Thus, cyclins D1 and D2 play critical but partially redundant roles in granule cell development.
N-myc transcription increases within 1 h of Shh stimulation, considerably sooner than cyclin D1 transcription. Induction occurs in the presence of cycloheximide, suggesting that N-myc may be a direct target of Shh signaling. Similar findings were recently reported by Kenney et al. (39). These investigators also showed that N-myc is expressed in Shh-responsive cells in the developing spinal cord, suggesting it may be a general target of Shh signaling in neural tissue. Although examination of sequences in and around the N-myc gene (e.g., refs. 40–42) does not reveal any consensus Gli-binding sites, it is possible that such sites are present in other parts of the gene, or that Shh regulates N-myc expression through Gli-independent mechanisms.
N-myc Plays a Critical Role in Proliferation of Granule Cell Precursors. Infection with a retrovirus encoding N-myc was sufficient to promote BrdUrd incorporation in a large percentage of infected GCPs. In fact, _N-myc_-induced proliferation was comparable to that induced by Gli-1, which mimics a potent Shh signal. Thus, N-myc might account for much of the proliferation caused by Shh. Further evidence for the importance of N-myc comes from our observation that dominant-negative N-myc completely blocks the proliferative response to Shh. Together these findings suggest that N-myc is a critical mediator of Shh-induced proliferation.
Among the transcriptional targets that have been shown to be important in Myc-induced cell cycle entry are D-cyclins, (21, 22), E2Fs (23, 43), Id2 (24), and cdk inhibitors (20). In GCPs, we found that retroviral transduction of N-myc causes robust increases in cyclin D1 RNA. N-myc has also been reported to induce expression of Cyclin D1 and D2 proteins and to repress expression of cyclindependent kinase inhibitor (CKI) proteins in these cells (38, 39, 44). Because we see no effect of N-myc on cyclin D2 and CKI mRNA levels, it is possible that N-myc regulates the levels of these proteins indirectly, perhaps by modulating translation or stability. In addition to cyclin D1, we found that N-myc overexpression results in increased levels of E2F1 and E2F2 RNA. In the presence of D-cyclins, which promote Rb phosphorylation and dissociation from E2Fs, E2F proteins directly regulate expression of a host of genes that are essential for cell cycle progression (45). Thus, by promoting expression of N-myc, Shh initiates a cascade of gene expression that results in a potent proliferative response.
The Role of N-myc in Cerebellar Development and Tumorigenesis. Our studies demonstrate an important role for N-myc in proliferation of GCPs. A role for N-myc in cerebellar development is also suggested by studies of mice in which the N-myc gene has been ablated in neuronal progenitors (44). These mice have a significant reduction in the size of the cerebellum and cerebral cortex, and develop ataxia, behavioral abnormalities, and tremors. The cerebellar defects stem from impairment of multiple cell types in the embryonic cerebellum, including GCPs and progenitors that give rise to other cerebellar neurons and glia. Although these studies support a role for N-myc in early cerebellar development, they do not address its function in postnatal GCPs. Most of the growth of the mouse cerebellum occurs in the first 3 weeks after birth. Because the mechanisms that control embryonic and postnatal proliferation of GCPs may be distinct, it will be important to examine the effects of N-myc inactivation during postnatal stages.
Elucidating the mechanisms by which Shh induces proliferation of GCPs also has significant implications for understanding tumorigenesis. We found robust expression of N-myc in medulloblastoma cells from patched1 mutant mice, compared with both proliferating GCPs and normal adult cerebellum. Elevated N-myc expression has also been reported in human medulloblastomas, particularly those of the desmoplastic type, which result from Shh pathway mutations (3). These findings support a model in which aberrant activation of Shh signaling leads to increased expression of N-myc and predisposes to medulloblastoma. Overexpression of N-myc in B lymphocytes can cause lymphomas, albeit slow-growing ones (46). Whether aberrant N-myc expression is sufficient to cause tumors in GCPs remains unknown. However, the role of N-myc in proliferation of normal GCPs, and its overexpression in both mouse and human medulloblastomas, suggests that it may be a critical mediator of tumor initiation or maintenance. Therefore, therapies designed to inhibit N-myc expression or function in vivo may be valuable approaches to treating medulloblastoma.
Supplementary Material
Supporting Information
Acknowledgments
We thank the Stanford Protein and Nucleic Acid Facility for microarray hybridization, Curis for Shh-N, Michael Cole for wild-type and dominant-negative N-myc, Mike Cook of the Duke Cancer Center for assistance with fluorescence-activated cell sorting, Greg Riggins for help with real-time PCR, and Barb Helfrich, Chris Counter, Webster Cavenee, and Cynthia Wetmore for helpful comments on the manuscript. We thank David Rowitch for communication of his work on cerebellar development. This work was supported by grants from the Geyer Foundation (to M.P.S.), the North Carolina Supercomputing Center (to S.M.L.), and the James S. McDonnell Foundation (to R.J.W.-R.). M.P.S. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: GCP, granule cell precursor; Shh, Sonic hedgehog.
References
- 1.Altman, J. & Bayer, S. A. (1997) Development of the Cerebellar System: In Relation to Its Evolution, Structure and Functions (CRC, Boca Raton, FL).
- 2.Katsetos, C. D. & Burger, P. C. (1994) Semin. Diagn. Pathol. 11**,** 85-97. [PubMed] [Google Scholar]
- 3.Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415**,** 436-442. [DOI] [PubMed] [Google Scholar]
- 4.Wechsler-Reya, R. J. & Scott, M. P. (1999) Neuron 22**,** 103-114. [DOI] [PubMed] [Google Scholar]
- 5.Dahmane, N. & Ruiz-i-Altaba, A. (1999) Development (Cambridge, U.K.) 126**,** 3089-3100. [DOI] [PubMed] [Google Scholar]
- 6.Wallace, V. A. (1999) Curr. Biol. 9**,** 445-448. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, R. L., Rothman, A. L., Xie, J., Goodrich, L. V., Bare, J. W., Bonifas, J. M., Quinn, A. G., Myers, R. M., Cox, D. R., Epstein, E. H., Jr., & Scott, M. P. (1996) Science 272**,** 1668-1671. [DOI] [PubMed] [Google Scholar]
- 8.Raffel, C., Jenkins, R. B., Frederick, L., Hebrink, D., Alderete, B., Fults, D. W. & James, C. D. (1997) Cancer Res. 57**,** 842-845. [PubMed] [Google Scholar]
- 9.Pietsch, T., Waha, A., Koch, A., Kraus, J., Albrecht, S., Tonn, J., Sorensen, N., Berthold, F., Henk, B., Schmandt, N., et al. (1997) Cancer Res. 57**,** 2085-2088. [PubMed] [Google Scholar]
- 10.Taylor, M. D., Liu, L., Raffel, C., Hui, C. C., Mainprize, T. G., Zhang, X., Agatep, R., Chiappa, S., Gao, L., Lowrance, A., et al. (2002) Nat. Genet. 31**,** 306-310. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich, L. V., Milenkovic, L., Higgins, K. M. & Scott, M. P. (1997) Science 277**,** 1109-1113. [DOI] [PubMed] [Google Scholar]
- 12.Ingham, P. W. & McMahon, A. P. (2001) Genes Dev. 15**,** 3059-3087. [DOI] [PubMed] [Google Scholar]
- 13.Marti, E. & Bovolenta, P. (2002) Trends Neurosci. 25**,** 89-96. [DOI] [PubMed] [Google Scholar]
- 14.Hayes, C., Brown, J. M., Lyon, M. F. & Morriss-Kay, G. M. (1998) Development (Cambridge, U.K.) 125**,** 351-357. [DOI] [PubMed] [Google Scholar]
- 15.Yoon, J. W., Kita, Y., Frank, D. J., Majewski, R. R., Konicek, B. A., Nobrega, M. A., Jacob, H., Walterhouse, D. & Iannaccone, P. (2002) J. Biol. Chem. 277**,** 5548-5555. [DOI] [PubMed] [Google Scholar]
- 16.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98**,** 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irizarry, R. A., Gautier, L. & Cope, L. (2002) in The Analysis of Gene Expression Data: Methods and Software, eds. Parmigiani, G., Garrett, E. S., Irizarry, R. A. & Zeger, S. L. (Springer, New York).
- 18.MacGregor, D., Li, L. H. & Ziff, E. B. (1996) J. Cell Physiol. 167**,** 95-105. [DOI] [PubMed] [Google Scholar]
- 19.McMahon, S. B., Wood, M. A. & Cole, M. D. (2000) Mol. Cell. Biol. 20**,** 556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gartel, A. L., Ye, X., Goufman, E., Shianov, P., Hay, N., Najmabadi, F. & Tyner, A. L. (2001) Proc. Natl. Acad. Sci. USA 98**,** 4510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daksis, J. I., Lu, R. Y., Facchini, L. M., Marhin, W. W. & Penn, L. J. (1994) Oncogene 9**,** 3635-3645. [PubMed] [Google Scholar]
- 22.Bouchard, C., Thieke, K., Maier, A., Saffrich, R., Hanley-Hyde, J., Ansorge, W., Reed, S., Sicinski, P., Bartek, J. & Eilers, M. (1999) EMBO J. 18**,** 5321-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sears, R., Ohtani, K. & Nevins, J. R. (1997) Mol. Cell. Biol. 17**,** 5227-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasorella, A., Noseda, M., Beyna, M., Yokota, Y. & Iavarone, A. (2000) Nature 407**,** 592-598. [DOI] [PubMed] [Google Scholar]
- 25.Galaktionov, K., Chen, X. & Beach, D. (1996) Nature 382**,** 511-517. [DOI] [PubMed] [Google Scholar]
- 26.Shohet, J. M., Hicks, M. J., Plon, S. E., Burlingame, S. M., Stuart, S., Chen, S. Y., Brenner, M. K. & Nuchtern, J. G. (2002) Cancer Res. 62**,** 1123-1128. [PubMed] [Google Scholar]
- 27.Bennin, D. A., Don, A. S., Brake, T., McKenzie, J. L., Rosenbaum, H., Ortiz, L., DePaoli-Roach, A. A. & Horne, M. C. (2002) J. Biol. Chem. 277**,** 27449-27467. [DOI] [PubMed] [Google Scholar]
- 28.Horne, M. C., Donaldson, K. L., Goolsby, G. L., Tran, D., Mulheisen, M., Hell, J. W. & Wahl, A. F. (1997) J. Biol. Chem. 272**,** 12650-12661. [DOI] [PubMed] [Google Scholar]
- 29.Heiniger, H. J., Chen, H. W., Boissonneault, G. A., Hess, M., Cottier, H. & Stoner, R. D. (1985) Ann. N.Y. Acad. Sci. 459**,** 111-128. [DOI] [PubMed] [Google Scholar]
- 30.Incardona, J. P. & Eaton, S. (2000) Curr. Opin. Cell Biol. 12**,** 193-203. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto, N., Yamamoto, S., Inagaki, F., Kawaichi, M., Fukamizu, A., Kishi, N., Matsuno, K., Nakamura, K., Weinmaster, G., Okano, H. & Nakafuku, M. (2001) J. Biol. Chem. 276**,** 45031-45040. [DOI] [PubMed] [Google Scholar]
- 32.Jones, S. E. & Jomary, C. (2002) BioEssays 24**,** 811-820. [DOI] [PubMed] [Google Scholar]
- 33.Solecki, D. J., Liu, X. L., Tomoda, T., Fang, Y. & Hatten, M. E. (2001) Neuron 31**,** 557-568. [DOI] [PubMed] [Google Scholar]
- 34.Hall, A. C., Lucas, F. R. & Salinas, P. C. (2000) Cell 100**,** 525-535. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, Q., Kho, A., Kenney, A. M., Yuk Di, D. I., Kohane, I. & Rowitch, D. H. (2002) Proc. Natl. Acad. Sci. USA 99**,** 5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenney, A. M. & Rowitch, D. H. (2000) Mol. Cell. Biol. 20**,** 9055-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huard, J. M., Forster, C. C., Carter, M. L., Sicinski, P. & Ross, M. E. (1999) Development (Cambridge, U.K.) 126**,** 1927-1935. [DOI] [PubMed] [Google Scholar]
- 38.Ciemerych, M. A., Kenney, A. M., Sicinska, E., Kalaszczynska, I., Bronson, R. T., Rowitch, D. H., Gardner, H. & Sicinski, P. (2002) Genes Dev. 16**,** 3277-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenney, A. M., Cole, M. D. & Rowitch, D. H. (2003) Development (Cambridge, U.K.) 130**,** 15-28. [DOI] [PubMed] [Google Scholar]
- 40.Katoh, K., Sawai, S., Ueno, K. & Kondoh, H. (1988) Nucleic Acids Res. 16**,** 3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takehana, K., Nakada, S., Hara, E., Taya, Y., Sekiya, S. & Oda, K. (1991) Gene 103**,** 219-225. [DOI] [PubMed] [Google Scholar]
- 42.Ibson, J. M. & Rabbitts, P. H. (1988) Oncogene 2**,** 399-402. [PubMed] [Google Scholar]
- 43.Schuldiner, O. & Benvenisty, N. (2001) Oncogene 20**,** 4984-4994. [DOI] [PubMed] [Google Scholar]
- 44.Knoepfler, P. S., Cheng, P. F. & Eisenman, R. N. (2002) Genes Dev. 16**,** 2699-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trimarchi, J. M. & Lees, J. A. (2002) Nat. Rev. Mol. Cell Biol. 3**,** 11-20. [DOI] [PubMed] [Google Scholar]
- 46.Sheppard, R. D., Samant, S. A., Rosenberg, M., Silver, L. M. & Cole, M. D. (1998) Oncogene 17**,** 2073-2085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information