Transcription-dependent recombination and the role of fork collision in yeast rDNA (original) (raw)
Abstract
It is speculated that the function of the replication fork barrier (RFB) site is to avoid collision between the 35S rDNA transcription machinery and the DNA replication fork, because the RFB site is located near the 3′-end of the gene and inhibits progression of the replication fork moving in the opposite direction to the transcription machinery. However, the collision has never been observed in a blockless (fob1) mutant with 150 copies of rDNA. The gene FOB1 was shown previously to be required for replication fork blocking activity at the RFB site, and also for the rDNA copy number variation through unequal sister-chromatid recombination. This study documents the detection of fork collision in an fob1 derivative with reduced rDNA copy number (∼20) using two-dimensional agarose gel electrophoresis. This suggests that most of these reduced copies are actively transcribed. The collision was dependent on the transcription by RNA polymerase I. In addition, the transcription stimulated rDNA copy number variation, and the production of the extrachromosomal rDNA circles (ERCs), whose accumulation is thought to be a cause of aging. These results suggest that such a transcription-dependent fork collision induces recombination, and may function as a general recombination trigger for multiplication of highly transcribed single-copy genes.
Keywords: rDNA, gene amplification, double-strand break, sister-chromatid recombination, life span, transcription-mediated recombination
Replication fork barrier (RFB) sites have been identified in the genomes of many organisms. In eukaryotic cells, RFB sites are found in the ribosomal RNA gene repeats (rDNA). Among them, the RFB of the yeast Saccharomyces cerevisiae has been the most intensively studied. S. cerevisiae carries ∼150 copies of the rDNA unit that are tandemly repeated on Chromosome XII (Petes 1979). A single unit consists of two transcribed regions, 35S precursor rRNA and 5S rRNA coding regions, and two non-transcribed regions, NTS1 and NTS2 (Fig. 1). The DNA encoding 35S and 5S rRNA genes is transcribed by RNA polymerases I (Pol I) and III, respectively. The origin of replication (ARS) and the RFB are located in the NTS2 and NTS1, respectively (Skryabin et al. 1984; Brewer and Fangman 1988; Linskens and Huberman 1988). The RFB contains a specific nucleotide sequence of ∼100 bp that allows progression of the replication fork in the direction of 35S rRNA transcription, but not in the opposite direction (Brewer et al. 1992; Kobayashi et al. 1992).
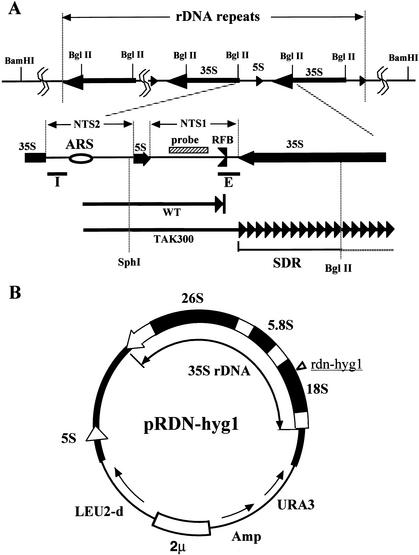
Figure 1.
(A) Structure of rDNA repeats in Saccharomyces cerevisiae. The locations of the 35S and 5S rRNA genes (the direction of transcription is indicated by arrows) are shown in the upper part. The recognition sites of _Bgl_II are also shown. The two nontranscribed spacer regions (NTS1 and NTS2), the ARS (replication origin), and its surrounding regions are expanded below. The RFB (replication fork barrier site) is indicated. The striped box is the region used for the rDNA-specific probe. The solid bars I and E are the components of HOT1. Two arrows in the bottom part show progression of one of the replication forks started from the ARS, bidirectionally. The upper fork (WT, wild type) is arrested at the RFB, and the lower one (TAK300; fob1, low rDNA copy number) is slowed down in the 35S rDNA. SDR is the region in which the replication fork is slowed down (Fig. 2C, panel c). The recognition sites of _Sph_I and _Bgl_II, which were used for the 2D gel analysis (Fig. 2C), are also shown. (B) Structure of the helper plasmid, pRDN-hyg1 (Chernoff et al. 1994). The filled box shows an rDNA unit. The hygromycin B-resistance mutation site is indicated in the 18S rDNA. This plasmid was used to isolate strains with low copy numbers of rDNA repeats.
The gene FOB1 was isolated as an essential gene for the RFB and HOT1 (see below) activities (Kobayashi and Horiuchi 1996). In _fob1_-defective mutants, replication fork blocking activity at the RFB is lost, and recombination in the rDNA repeats is much reduced (Kobayashi et al. 1998; Defossez et al. 1999; Kaeberlein et al. 1999; Johzuka and Horiuchi 2002; Merker and Klein 2002). Additionally, deletion of the RFB from the rDNA repeats resulted in defects in repeat expansion and contraction (Kobayashi et al. 2001). Therefore, inhibition of the replication fork seems to be required for the activation of recombination in the rDNA repeats.
It was previously shown that blockage of the replication fork at the terminus (Ter) site in Escherichia coli stimulates homologous recombination at nearby sister chromosomal regions (Horiuchi et al. 1994; Horiuchi and Fujimura 1995). The Ter site is a sequence that, together with the Tus protein, appears to function to ensure that two replication forks that have initiated bidirectionally from an origin of replication can enter the termination zone of the chromosome but cannot escape it (for review, see Hill 1996). Therefore, it was speculated that pausing of the DNA replication machinery at a replication fork blocking site would lead to the formation of double-strand breaks (DSBs) in the newly replicated DNA (Horiuchi et al. 1994: Kobayashi et al. 1998). These DSBs are then thought to be repaired by a recombination process via the formation of a new replication fork as shown in the model proposed in Horiuchi and Fujimura (1995), based on the DSB repair model of Szostak et al. (1983).
Fob1-dependent recombination in the rDNA repeats is required not only for expansion and contraction of the rDNA repeats (Kobayashi et al. 1998), but also for production of extrachromosomal rDNA circles (ERCs) whose accumulation enhances aging (Defossez et al. 1999). It was reported that in an fob1 mutant, ERCs are almost undetectable and the life span is 30%–60% longer than that of wild-type cells (Defossez et al. 1999). However, the mechanism for this longevity is still unknown.
The RFB and FOB1 are also required for HOT1 activity. HOT1 is a DNA element that stimulates mitotic intra- and interchromosomal recombination at nearby regions when inserted at a non-rDNA location (Keil and Roeder 1984). HOT1 consists of two elements: the E-element, which overlaps the “enhancer” for RNA polymerase I transcription (Elion and Warner 1984); and the I-element, which corresponds to the promoter region for Pol I transcription (Fig. 1; Keil and Roeder 1984; Voelkel-Meiman et al. 1987). Stimulation of Pol I transcription by the E-element was observed only for ectopic Pol I promoters and not for Pol I transcription of chromosomal rDNA (Wai et al. 2001). The same study also showed that FOB1 is required for transcription from ectopic Pol I promoters. Therefore, stimulation of recombination by HOT1 appears to be caused by strong Pol I transcription events from ectopic Pol I promoters (Huang and Keil 1995). It was also shown that the RFB overlaps the E-element, but the replication fork blocking activity itself is not required for HOT1 activity (Ward et al. 2000).
The location of the RFB site near the rDNA transcription termination site and its directionality in function led to suggestions that the RFB might have evolved to prevent collision between the transcription and replication machineries (Brewer et al. 1992; Kobayashi et al. 1992). However, in an fob1 mutant, whose replication blocking activity is deficient, collision was not observed at the RFB or any other sites (Kobayashi and Horiuchi 1996). Theoretically, in an fob1 cell, as the replication forks proceed bidirectionally, collision of the two machineries is expected. One possible reason for the apparent failure to detect the collision could be the presence of many transcriptionally inactive rDNA units. Sogo and collaborators estimated less than half of the rDNA repeat copies are transcriptionally active (Dammann et al. 1993). In addition, the active _ARS_s are also limited (∼20% of the _ARS_s initiate replication; Brewer and Fangman 1988; Linskens and Huberman 1988). Recently, Pasero et al. (2002) demonstrated that some of the inactive _ARS_s are repressed by the silencing machinery. However, in the silencing defective condition, more than half of the _ARS_s are still inactive. Therefore, even though the active _ARS_s tend to be located in the actively transcribed units (Muller et al. 2000), units whose transcription machinery collides with the replication fork would be many fewer than noncollision units, and therefore the collision was not detected.
In this paper, we isolated fob1 derivative strains whose rDNA copy number is ∼10–20. In these strains, most of the rDNA copies are expected to be transcribed (French et al. 2003). The results show that the rate of replication fork progression is slowed down in a 35S rDNA transcription-dependent manner. Moreover, transcription stimulated recombination, resulting in rDNA copy number variation and production of ERCs. These results suggest that collision of the replication fork with the transcription machinery takes place when RFB activity is lacking, and such a transcription-dependent collision is a trigger of recombination. This form of recombination may lead to gene duplication and/or chromosomal rearrangements.
Results
Isolation of low-copy-rDNA strains
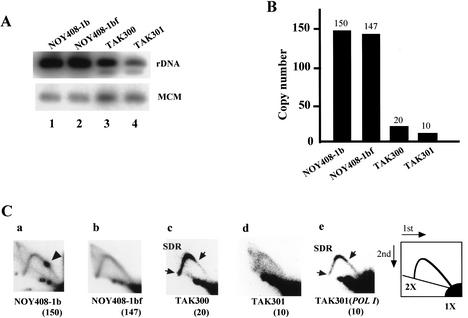
We used the following strategy to isolate a strain (TAK300) whose rDNA copy number is much reduced. A helper plasmid (pRDN-hyg1; Chernoff et al. 1994) that carries a copy of the rDNA repeats was transformed into the wild-type strain (Kobayashi et al. 2001). The 18S rRNA gene of the 35S precursor rRNA gene in this plasmid contains a mutation that makes the ribosomes resistant to hygromycin B (Fig. 1B). Because the mutation is recessive to the wild-type 18S rRNA gene, the original strain carrying this plasmid is sensitive to hygromycin B. Selection of hygromycin-resistant colonies produces strains in which chromosomal rDNA copy number is much reduced (Kobayashi et al. 2001). The FOB1 gene of these low-copy-rDNA strains was then disrupted to stabilize rDNA copy numbers. Plasmid pRDN-hyg1 was subsequently removed by using a plate including 5-FOA. To determine the copy number, the DNA was digested with _Bgl_II and subjected to electrophoresis followed by Southern analysis using a probe specific for chromosomal rDNA repeats (Fig. 1A). A single-copy gene, MCM2, was used as an internal control for normalization. As shown in Figure 2A, the intensities of the 4.6-kb _Bgl_II fragment indicate the rDNA copy number in TAK300 is significantly reduced relative to that in the control strains, NOY408-1b (wild-type strain), and the fob1 mutant, NOY408-1bf. Quantification after normalization by MCM2 intensity (Fig. 2B) indicated that the copy number of TAK300 was reduced to approximately one-eighth (∼20 copies) of that in the control strains. As we could not isolate any strain whose copy number was <20, we believe this is close to the minimum number of rDNA copies to allow cell growth. The doubling time of this strain was ∼20% longer than that of the wild type, and the rate of rRNA synthesis was similar to that of the wild type (data not shown). Recently, French et al. (2003) observed by Miller-spread electron microscopy that, in strains with ∼40 rDNA copies, most of the copies were transcribed to keep the rate of rRNA synthesis similar to that of the wild type. This suggests that, in our low-copy rDNA strain, most rDNA copies would be actively transcribed.
Figure 2.
Analysis of low-copy-rDNA strains. (A) Southern hybridization analysis of rDNA copy numbers. DNA was digested with _Bgl_II and subjected to electrophoresis followed by Southern analysis using the rDNA probe (Fig. 1). A single-copy gene, MCM2, was used as an internal control for normalization. (B) Quantitation of the intensities of the bands. NOY408-1b (wild-type strain), NOY408-1bf (fob1), TAK300 (fob1; low-copy rDNA strain), TAK301 (fob1 pol1; low-copy rDNA strain). (C) Collision between the transcription and the replication machineries analyzed by 2D gel analysis. DNA was prepared from the strains indicated, digested with _Bgl_II and _Sph_I, and subjected to 2D agarose gel electrophoresis followed by Southern hybridization using the rDNA probe (see Fig. 1). A spot indicated by an arrowhead shows accumulation of Y-shaped DNA molecules at the RFB site (panel a). The slowdown region (SDR) is located between two arrows (panel c). The numbers in parentheses are copy numbers of rDNA in each strain. (Panel a) NOY408-1b (wild-type strain). (Panel b) NOY408-1bf (fob1). (Panel c) TAK300 (fob1; low-copy rDNA strain). (Panel d) TAK301 (fob1 pol1; low-copy rDNA strain). (Panel e) TAK301, complemented by a plasmid-borne RPA135 gene (fob1 POLI; low-copy rDNA strain).
Detection of collision between the transcription machinery and the replication fork
To monitor the progression of the replication fork in TAK300, DNA was isolated from logarithmically growing cells, subjected to two dimensional (2D) agarose gel electrophoresis (Brewer and Fangman 1987) after digestion with _Bgl_II and _Sph_I, and followed by Southern analysis using the rDNA probe (Fig. 1A). The results are shown in Figure 2C. As a control experiment, DNA from the wild-type strain (NOY408-1b) and the _fob1_-defective mutant strain (NOY408-1bf), which carry ∼150 copies of rDNA, were used (Fig. 2B; Kobayashi et al. 1998). As shown in Figure 2C, panel a, the spot corresponding to accumulated replication intermediates (RIs) at the RFB site was observed in the wild-type strain but not in the fob1 mutant strains NOY408-1bf and TAK300 (Kobayashi and Horiuchi 1996). However, there is a significant difference between NOY408-1bf and TAK300. In NOY408-1bf, the arc of Y-shaped RIs is largely uniform between 1× and 2× of the size of the region analyzed (Fig. 2C, panel b). This means that the replication fork progresses in this region at approximately the same rate. In contrast, in TAK300 there is an intense region of hybridization along the arc [indicated by SDR (slowdown region) bounded by arrows in Fig. 2C, panel c]. The SDR occupies the region between the RFB site and the position corresponding to 2× (the largest Y-shaped molecules just before their resolution into two linear molecules). As the RFB site is located near the 3′-end of the 35S rDNA (Fig. 1A), the results are consistent with a slow rightward progression of the replication fork within the 35S rRNA coding region (Fig. 1A, lower panel). This slowdown of progression may be caused by collision of the replication machinery with the 35S rDNA transcription machinery. To confirm this inference, we constructed another low-copy strain (TAK301) in which an essential Pol I subunit (RPA135) and FOB1 are defective and growth is supported by a multicopy helper plasmid (pNOY353) containing the 35S rRNA coding region fused to a strong Pol II promoter, the GAL7 promoter (Oakes et al. 1998). The rDNA copy number of rDNA in TAK301 was shown to be ∼10 (Fig. 2B). In this strain, no preferential slowdown of replication fork progression in the 35S rRNA coding region was observed (Fig. 2C, panel d), although the arc signal was weak presumably because of the low copy number of rDNA and the slower growth rate of this strain relative to the others. After complementation with a plasmid-borne RPA135 gene, the SDR was again observed (Fig. 2C, panel e), although the signal was a little weaker than that of TAK300. We suggest that the reason for the reduced signal is that the multicopy helper plasmid (pNOY353) compensates transcription of chromosomal rDNA. Therefore, the reduced level of chromosomal rDNA transcription results in a moderated level of collision. These results indicate that the slowdown of the replication fork observed in TAK300 is dependent on 35S rDNA transcription by Pol I.
Collision between the transcription machinery and the replication fork stimulates recombination in the rDNA repeats
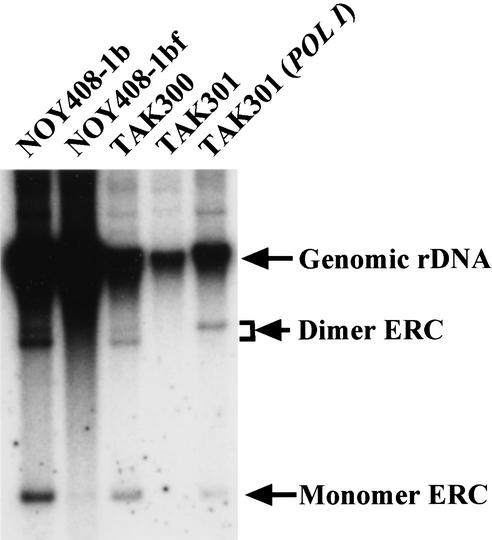
Fob1-dependent replication fork blocking activity at the RFB site is essential for the recombination in the rDNA repeats (Kobayashi et al. 1998; Defossez et al. 1999; Johzuka and Horiuchi 2002). Therefore, the inhibition of replication fork progression induced by transcription as observed in TAK300 could also stimulate the recombination. To examine this possibility, the level of ERCs, which are produced by intrachromosomal recombination from the rDNA repeats, was measured in the low-copy _fob1_-defective strain (TAK300) and in the other control strains. In the wild-type strain, ERCs are induced by Fob1-dependent recombination at the RFB (Defossez et al. 1999; Johzuka and Horiuchi. 2002). DNA was isolated from TAK300 and the other control strains, and the gel electrophoresis was performed. The results are shown in Figure 3. For the wild-type strain (NOY408-1b in Fig. 3), monomer and dimer ERCs were observed. As expected, these ERCs were nearly absent in the control fob1 strain (NOY408-1bf), which has ∼150 copies of rDNA. In contrast, in a low-copy _fob1_-defective strain, the ERCs could be detected (TAK300 in Fig. 3). Furthermore, in the low-copy _fob1_-defective strain whose RPA135 is deleted (TAK301; fob1, rpa135; Fig. 3), ERC molecules were not observed. And after complementation with a plasmid-borne RPA135 gene, ERCs were again observed. [Note: The dimer ERCs display a double band. We suggest the reason is because of torsional differences, because ERCs are known to be covalently closed circular molecules (Sinclair and Guarente 1997). We think the upper band might be an open circular molecule, resulting from DNA preparation.] These results suggest that the high level of transcription of rDNA in the low rDNA copy strain induces the production of ERCs by enhancing recombination, even in the _fob1_-defective background.
Figure 3.
Detection of the extrachromosomal rDNA circles (ERCs). DNA isolated from various strains was subjected to electrophoresis (0.6% agarose gel for 20 h at 1 V/cm) followed by Southern hybridization using the rDNA probe (see Fig. 1). The positions of monomer ERC, dimer ERC, and genomic rDNA are indicated by arrows. The strains are the same as used in Figure 2C. We used the same number of cells, which were determined by spectrometer to isolate the DNA.
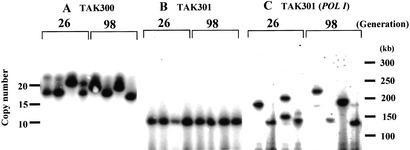
Next, we observed whether or not the recombination induced by transcription could change the copy number of rDNA. In wild-type cells, expansion and contraction of rDNA repeats is dependent on Fob1p and the RFB activity (Kobayashi et al. 1998, 2001). TAK300 cells from a single colony were streaked on a YPD plate followed by single-colony isolation, and this process was repeated five times. Eight colonies were picked, four from the first plate and four from the fifth plate, and DNA was isolated using agarose gel blocks to minimize DNA damage. The DNA was digested with the _Bam_HI restriction enzyme, whose recognition site does not exist in the rDNA repeats (Fig. 1A). Copy numbers of repeats were then estimated from the length of the fragments containing rDNA. As the _Bam_HI fragment includes the non-rDNA regions (39 kb) outside the repeats, the numbers of repeats were calculated after subtraction of 39 kb from the length of the _Bam_HI fragments. The results of the pulsed-field gel electrophoresis are shown in Figure 4A. The length of the repeats in the independent TAK300 colonies varied both in the first (∼26 generations from the starting single colony) and fifth (∼98) plates, and their rDNA copy numbers correspond to 17–23, suggesting that copy number can be altered in TAK300 within this range. In contrast, in the Pol I transcriptionless strain (TAK301; fob1 rpa135), the copy number of the rDNA repeats was not altered over 90 generations (Fig. 4B). When the wild-type RPA135 gene carried on a shuttle vector was introduced into this rpa135 strain, increases in the number of rDNA repeats were observed in some clones. As shown in Figure 4C, the rDNA copy number varied from 10 to 18. These results strongly suggest that transcription of rDNA by Pol I induces changes in the copy number even in the fob1 background, if the rDNA repeat number is kept small.
Figure 4.
Analysis of rDNA repeat size by pulsed-field gel electrophoresis (PFGE; CHEF, Bio-Rad). DNA was isolated using agarose gel blocks, digested with _Bam_HI, and subjected to PFGE, followed by hybridization with the rDNA probe (see Fig. 1). As there is no _Bam_HI recognition site in the repeats, the length of the fragment indicates the rDNA copy number. This fragment includes 39 kb of non-rDNA region. The numbers above the autoradiograph show the number of generations after purification from single colonies. (A) TAK300 (fob1; low-copy rDNA strain). (B) TAK301 (fob1 pol1; low-copy rDNA strain). (C) TAK301, complemented by a plasmid-borne RPA135 gene (fob1 POLI; low-copy rDNA strain).
The transcription-dependent ERCs shorten the life span
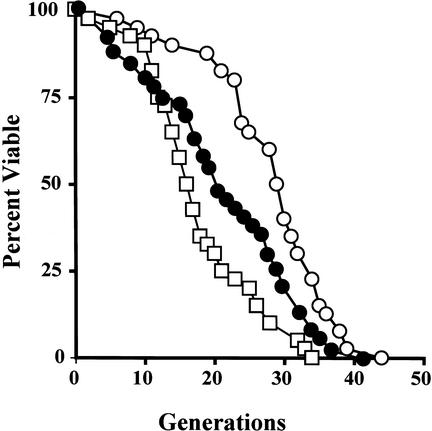
It was originally reported that a cause of aging in S. cerevisiae is the accumulation of ERCs (Sinclair and Guarente 1997). Defossez et al. (1999) observed that an _fob1_-defective mutant had an extended life span compared with wild-type cells, and that the ERC level in the mutant was much less than that of the wild type (Fig. 3). We repeated the experiment with our _fob1_-defective strains and obtained similar results. Daughter cells that bud off from a virgin mother cell during her life were counted. As shown in Figure 5, the life span of the fob1 mutant (shown by empty circles) measured in this way was 28 generations, and 18 generations for the wild-type strain (shown by empty squares); that is, the life span of the fob1 mutant was ∼56% longer than that of the wild-type strain (P < 0.001). We then measured the life span of the low-copy _fob1_-defective strain (TAK300) whose ERCs were observed in Figure 3. The life span (22 generations; Fig. 5, indicated by solid circles) was ∼21% shorter than that of the control normal-copy-number fob1 strain (P < 0.001), but it was still longer than that of the wild type strain (P = 0.03).
Figure 5.
Life span analysis. The analysis was performed on NOY408-1b (white box), NOY408-1bf (white circle), and TAK300 (filled circle) as described previously (Kennedy et al. 1994). Life span was determined by scoring the number of daughter cells produced by each mother cell before cessation of all division. The average life spans were as follows: NOY408-1b = 18; NOY408-1bf = 28; TAK300 = 22.
Discussion
We report here that in an _fob1_-defective strain whose rDNA copy number is reduced to ∼20, slowdown of replication fork procession in the 35S rDNA was observed by using 2D gel electrophoresis. As the slowdown activity was dependent on 35S rDNA transcription, we propose that the activity is the result of collision between the replication and transcription machineries. Furthermore, recombination was induced by transcription, as detected by ERC production and rDNA copy number variation. These results suggest that collision of the two polymerases may induce this recombination in the rDNA.
Stimulation of mitotic recombination by transcription is known in yeast cells (Thomas and Rothstein 1989; White et al. 1992; Nevo-Caspi and Kupiec 1994; Huang and Keil 1995; Saxe et al. 2000). Although the mechanism of transcription-associated recombination is still unclear, the torsional stress created by high levels of transcription is thought to lead to recombination-initiating lesions (Lui and Wang 1987). Mutations of structural genes for topoisomerases TOP1, TOP2, and TOP3 confer hyperrecombination in the rDNA (Christman et al. 1988; Wallis et al. 1989). Recently, Olavarrieta et al. (2002) reported that transcription proceeding in the opposite direction to replication also creates torsional stress. It is possible that the slowdown of the replication fork reported in this paper may be a result of the torsional stress, as well as the physical collision of both DNA and RNA polymerases. Inhibition of replication is known to be a cause of recombination as mentioned above. Some mutations in DNA polymerase and helicase genes enhance recombination in the rDNA (Ivessa et al. 2000). In some cases, fork arrests were observed along the genome, and they could be triggers of recombination via double-strand breaks. The slowdown region (SDR; Fig. 2C, panel c) may have similar recombination effects as fork arrest, as discussed below (Fig. 6B).
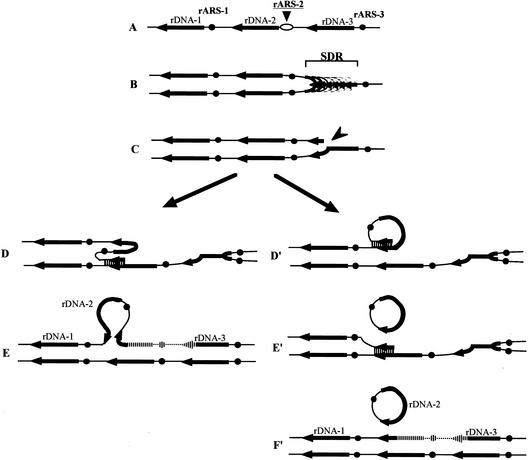
Figure 6.
The transcription-dependent recombination model for rDNA repeat expansion, contraction, and production of ERCs in TAK300 (low rDNA copy number, fob1). The positions of the ARS and the 35S rDNA are shown as filled dots and arrows, respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the _ARS_s (ARS-2) bidirectionally (A). As all of the 35S rDNA units are transcribed fiercely in this strain, the rightward replication fork is slowed down in the 35S rDNA (SDR; B), and we propose that it stimulates a DNA double-strand break (indicated by an arrowhead in C). As the strain is fob1, the replication fork barrier (RFB) site at the 3′-end of the 35S rDNA is not functioning. (D) A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place, and a new replication fork is formed. (E) The new replication fork meets with the leftward replication fork, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as a striped line. If the strand invasion is at a site in an upstream repeat (e.g., right side of ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. If the strand invasion is at a site in the same chromatid (_D_′), an ERC is produced (_E_′). Then another invasion takes place (_E_′, lower part) and two sister chromatids are formed (_F_′). The region that is replicated twice is indicated as a striped line in _F_′. A similar model that does not involve transcription-related ERC production is presented by Rothstein and Gangloff (1999).
In Escherichia coli and Bacillus subtilis, all of the rRNA genes are transcribed in the same direction as replication proceeds. As transcription of the genes is highly active in these cells, the direction of the transcription may be the same as the direction of replication to avoid collision. In our laboratory, a recombination hot spot at the end of the rRNA gene was recently identified in E. coli (K. Ohsumi and T. Horiuchi, in prep.). The hot spot activity can be observed when the replication initiates from a site other than the origin (oriC) and some replication forks proceed in the direction opposite to rDNA transcription. This hot spot activity was dependent on the transcription of the rRNA gene, and the slowdown of the replication fork also occurred in the transcribed region (K. Ohsumi and T. Horiuchi, in prep.). Therefore, transcription-dependent recombination in rRNA genes seems to be a general phenomenon from bacterial to eukaryotic cells.
In our experimental system using a low-copy-rDNA strain, most of the rDNA units should be transcriptionally active (French et al. 2003). However, this system may have the opposite effect on the activity of _ARS_s. Pasero et al. (2002) reported that ARS activity in the rDNA is partially repressed by the silencing machinery. Therefore, in a low-copy-rDNA strain, the silencing machinery will be in excess relative to the copy number and may reduce the number of active _ARS_s in the rDNA. This may explain the weak signal of replication intermediates in the 2D analysis (Fig. 2C, panel d), in addition to the poor growth rate of the cells.
In wild-type strains of E. coli and yeast, collision is not observed because the replication forks are protected against the rDNA transcription machinery by the replication fork barrier site, Ter in E. coli and RFB in yeast. Therefore, one of the physiological functions of the replication fork barrier sites could be prevention of such a collision (Brewer 1988). This seems to suggest that the collision, or the resulting recombination, should impose a burden on the cells. One possible reason is that the collision may lead to damage of the gene, and the repair may induce mutation. The damage may cause double-strand breaks (DSBs), which are known as a substrate for recombination enzymes (Fig. 6C; for review, see Rothstein et al. 2000). This kind of recombination repair, which includes new DNA synthesis, may result in errors more frequently than normal DNA replication (Strathern et al. 1995). The collision may result in not only replication pausing but also in inhibition of transcription elongation, and the inhibition would be disadvantageous for the cells. Aguilera and collaborators isolated genes, HPR1 and THO2, which are responsible for transcript elongation (Chavez and Aguilera 1997; Piruat and Aguilera 1998). In the mutants defective in these genes, the rate of recombination between tandemly repeated sequences is extremely high. The authors speculated that the transcriptional block is a source of recombination in these mutants. A similar block of elongation might be induced by collision in the rDNA repeats and cause instability of the repeats. Additionally, high levels of nonrDNA transcription are known to increase spontaneous mutation rates in yeast (Datta and Jinks-Robertson 1995). Such a high level of transcription may also induce similar effects to the collision and lead to damage of the gene, although the mechanism is unknown.
In eukaryotic cells, which have multireplicon genomes, the collision of DNA replication machinery with RNA transcription machinery may not be avoidable. In fact, in S. cerevisiae, most replication origins have been mapped on Chromosome VI, and the direction of the replication is not always coincident with that of transcription (Friedman et al. 1997: Yamashita et al. 1997). Deshpande and Newlon (1996) reported that there are replication pausing sites at transfer RNA (tRNA) genes. The sites are polar, stalling replication forks only when they are opposite to the direction of the transcription. It was shown that the pausing activities are dependent on RNA polymerase III transcription. The authors suggested that head-on collisions between transcription and replication machineries result in the pausing activity. Of course, collision was not only caused by the directionality of the polymerases. In the case of low transcription of a gene, even if the direction of the polymerases is opposite, collision may be rare. In contrast, actively transcribed genes, like tRNA and rRNA genes, can actually inhibit replication if the replication fork proceeds in the opposite direction (France 1992). When the collision takes place and it functions as a recombination hot spot, the gene may be duplicated (or amplified) to increase the level of transcripts. We present a model that shows how the collision induces ERC production and rDNA copy number variation in Figure 6 (for details, see the legend).
The life span of TAK300 was significantly shorter (P < 0.001) than that of a control fob1 mutant whose rDNA copy number is ∼150 (Fig. 5). As the only difference between the two strains is the rDNA copy number, the change in life span seems to be related to copy number. As shown in Figure 3, in TAK300 the accumulation of ERCs, which is thought to be a trigger of aging, was observed, but it was not seen in the control fob1 mutant. Therefore, it could be speculated that the reduction in ERC in _fob1_-defective strains is partially ameliorated by recombination triggered by fork collision in the low-copy-number strain, suggesting that ERC accumulation decreases life span as previously described (Sinclair and Guarente 1997). However, the relationship between ERC accumulation and aging is still controversial (Hoe et al. 1999), and whether ERC accumulation reduces TAK300's life span, or whether another factor(s) related to rDNA copy number does, is not known. Further analysis is required to resolve this problem. [Note: TAK301 contains the rDNA helper plasmid (pNOY353), which may also affect life span, independently of rDNA copy number. Therefore, we did not measure life span in this strain (Sinclair and Guarente 1997).]
Our experimental system is an artificial one. As we speculate that collision is rare in wild-type strains because of the large number of transcriptionally inactive repeats, we had to delete FOB1 and keep the rDNA copy number low to observe collision. However, in any region except rDNA repeats, genes exist as a single copy and also RFB sites are not located at the end of the genes. Therefore, our artificial system mimics general highly transcribed gene structures in the genome.
Materials and methods
Strains, plasmids, and media
The yeast strains and plasmids used are listed in Table 1. SD is a synthetic glucose medium (Kaiser et al. 1994). SD supplemented with required amino acids and adenine was used to select transformants or strains carrying unstable plasmids. Isolation of low-copy-rDNA strains is described in the text. For the selection, YPD medium containing 0.3 μg/mL hygromycin B (Sigma) was used. SG and YEP-galactose are the same as SD and YEPD (Yano and Nomura 1991), respectively, except that 2% glucose is substituted by 2% galactose. SG supplemented with required amino acids and adenine was used to grow strains carrying a helper plasmid pNOY353, whose 35S rDNA is under the control of a galactose-inducible promoter. The incubation temperature was 30°C.
Table 1.
Yeast strains and plasmids used
| Designation | Genotypes and comments |
|---|---|
| Strain | |
| NOY408-1b | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3, 112 can1-100 (Nogi et al. 1991) |
| NOY408-1bf | Same as NOY408-1b except for fob1Δ::HIS3 (Kobayashi et al. 1998) |
| TAK300 | Same as NOY408-1bf except for the low copy number of rDNA |
| TAK301 | Same as TAK300 except for rpa135Δ::LEU2, pNOY353 |
| Plasmid | |
| pRDN-hygl | Multicopy plasmid carrying a single rDNA repeat with a hygromycin-B-resistance mutation (Chernoff et al. 1994) |
| pNOY353 | Multicopy plasmid carrying _GAL7_-35S rDNA, 5S, _2_μ, TRP1 (Oakes et al. 1998) |
| YCplac33 | Monocopy plasmid vector, ARS1, URA3 (1Gietz and Sugino 1988) |
| YCp-RPA135 | YCplac33 carrying RPA135 |
Determination of rDNA copy number
DNA digested with _Bgl_II was subjected to 1% agarose gel electrophoresis and analyzed by Southern hybridization using rDNA probes (0.5-kb fragment prepared by PCR in Fig. 1). This probe hybridizes only to the chromosomal rDNA, not to plasmid pNOY353. For MCM2 normalization, an MCM2 1.4-kb fragment prepared by PCR was used (Kobayashi et al. 1998). The level of hybridization was determined by PhosphorImager analysis (BAS2000; Fuji Film). To determine absolute copy numbers of rDNA, the copy number of the wild-type strain NOY408-1b was determined by a competitive PCR assay (Diviacco et al. 1992) as previously described (Kobayashi et al. 1998). The copy numbers of other strains were determined by comparison with the NOY408-1b.
Detection of the expansion and contraction of rDNA repeats
The approximate number of generations of yeast cells used for rDNA copy determination was first estimated from the size of colonies formed from single transformants. Single colonies with 1-mm diameter contain approximately 2 × 105 cells, and we assume that this corresponds to 18 generations starting from an individual ancestor cell. After repeated streaking, liquid cultures were inoculated using colonies with 1-mm diameter and the number of generations grown in liquid medium was calculated by measuring the absorbance at 600 nm.
For pulsed-field gel electrophoresis (PFGE), chromosomal DNA isolated as previously described (Smith et al. 1988) was digested with _Bam_HI and subjected to gel electrophoresis in a 1% agarose gel, 0.5× TBE buffer, using the CHEF-DRII (Bio-Rad) with a pulse time of 10–20 sec, 200 CV, at 14°C for 21 h. The gel was then stained with 0.5 μg/mL ethidium bromide at room temperature for 30 min, photographed, and subjected to Southern hybridization analysis (Sambrook et al. 1989) using the rDNA specific probe.
Other methods
Life span analysis was performed as described previously (Kennedy et al. 1994). The Mann-Whitney nonparametric test was used to determine statistical significance between the different average life spans using StatView software (SAS Institute Inc.). Replication fork blocking and slowdown activities were analyzed using 2D gel electrophoresis as described previously (Brewer and Fangman 1987).
Acknowledgments
The authors thank Dr. M. Nomura (University of California-Irvine) for critical reading of the manuscript, S.-J. Hoe (Tokyo University Eigene Co. Ltd) and H. Ikeda (Tokyo University Kitazato Institute) for technical advice on life span analysis, and A. Ganley in our lab for preparation of the manuscript. This work was supported in part by grants-in-aid for Scientific Research 13141205, 13480234 (to T.H. and T.K.), and 14380332 (to T.K.) from the Ministry of Education, Science and Culture, Japan; a grant from the Ministry of Health and Welfare, Japan (to T.K.); and a grant from the Human Frontier Science Organization, France (to T.K.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1085403.
References
- Brewer B.J. 1988. When polymerases collide: Replication and the transcriptional organization of the E. coli chromosome. Cell 53**:** 679–686. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51**:** 463–471. [DOI] [PubMed] [Google Scholar]
- ____. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55**:** 637–643. [DOI] [PubMed] [Google Scholar]
- Brewer B.J., Lockshon, D., and Fangman, W.L. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71**:** 267–276. [DOI] [PubMed] [Google Scholar]
- Chavez S. and Aguilera, A. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes & Dev. 11**:** 3459–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Vincent, A., and Liebman, S.W. 1994. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 13**:** 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M.F., Dietrich, F.S., and Fink, G.R. 1988. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55**:** 413–425. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini, R., Koller, T., and Sogo. J.M. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21**:** 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A. and Jinks-Robertson, S. 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268**:** 1616–1619. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Prusty, R., Kaeberlein, M., Lin, S.J., Ferrigno, P., Silver, P.A., Keil, R.L., and Guarente. L. 1999. Elimination of replication fork block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3**:** 447–455. [DOI] [PubMed] [Google Scholar]
- Deshpande A.M. and Newlon, C.S. 1996. DNA replication fork pause sites dependent on transcription. Science 272**:** 1030–1033. [DOI] [PubMed] [Google Scholar]
- Diviacco S., Norio, P., Zentilin, L., Menzo, S., Clementi, M., Biamonti, G., Riva, S., Falaschi, A., and Giacca. M. 1992. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene 122**:** 313–320. [DOI] [PubMed] [Google Scholar]
- Elion E.A. and Warner, J.R. 1984. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39**:** 663–673. [DOI] [PubMed] [Google Scholar]
- France S. 1992. Consequences of replication fork movement through transcription units in vivo. Science 258**:** 1362–1365. [DOI] [PubMed] [Google Scholar]
- French S.L., Osheim, Y.N., Cioci, F., Nomura, M., and Beyer, A.L. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23**:** 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K.L., Brewer, B.J., and Fangman, W.L. 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2**:** 667–678. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino, A. 1988. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74**:** 527–534. [DOI] [PubMed] [Google Scholar]
- Hill T.M. 1996. Features of the chromosomal terminus region. In Escherichia coli and Salmonella (ed. F.C. Neidhardt), Vol. 2, pp. 1602–1614. ASM Press, Washington, DC. [Google Scholar]
- Hoe S.-J., Tatebayashi, K., Ohsugi, I., Shimamoto, A., Furuichi, Y., and Ikeda, H. 1999. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells 4**:** 619–625. [DOI] [PubMed] [Google Scholar]
- Horiuchi T. and Fujimura, Y. 1995. Recombinational rescue of the stalled DNA replication fork: A model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J. Bacteriol. 177**:** 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Fujimura, Y., Nishitani, H., Kobayashi, T., and Hidaka, M. 1994. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J. Bacteriol. 176**:** 4656–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.S. and Keil, R.L. 1995. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple _cis_-acting sequences. Genetics 141**:** 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou, J.-Q., and Zakian, V.A. 2000. The Saccharomyces Pif1pDNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100**:** 479–489. [DOI] [PubMed] [Google Scholar]
- Johzuka K. and Horiuchi, T. 2002. Replication fork block protein, Fob1, act as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7**:** 99–113. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey, M., and Guarente, L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 13**:** 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis, S., and Mitchell, A. 1994. Methods in yeast genetics, pp. 208–209. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Keil R.L. and Roeder, G.S. 1984. _Cis_-acting recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39**:** 377–386. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Austriaco Jr., N.R., and Guarente, L. 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell. Biol. 127**:** 1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayaski T. and Horiuchi, T. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1**:** 465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka, M., Nishizawa, M., and Horiuchi, T. 1992. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol. Gen. Genet. 233**:** 355–362. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck, D.J., Nomura, M., and Horiuchi, T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes & Dev. 12**:** 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nomura, M., and Horiuchi, T. 2001. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 21**:** 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M.H.K. and Huberman, J.A. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8**:** 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui L.F. and Wang, J.C. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. 84**:** 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker R.J. and Klein, H.L. 2002. _hpr1_Δ affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 22**:** 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Lucchini, R., and Sogo, J.M. 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5**:** 767–777. [DOI] [PubMed] [Google Scholar]
- Nevo-Caspi Y. and Kupiec, M. 1994. Transcriptional induction of Ty recombination in yeast. Proc. Natl. Acad. Sci. 91**:** 12711–12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano, R., and Nomura, M. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. 88**:** 3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M.L., Aris, J.P., Brockenbrough, J.S., Wai, H., Vu, L., and Nomura, M. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 143**:** 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olavarrieta L., Hernández, P., Krimer, D.B., and Schvartzman, J.B. 2002. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 322**:** 1–6. [DOI] [PubMed] [Google Scholar]
- Pasero P., Bensimon, A., and Schwob, E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & Dev. 16**:** 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T.D. 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. 76**:** 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat J.I. and Aguilera, A. 1998. A novel yeast gene, THO2, is involved in RNA Pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17**:** 4859–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. and Gangloff, S. 1999. The shuffling of a mortal coil. Nat. Genet. 22**:** 4–6. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Michel, B., and Gangloff, S. 2000. Replication fork pausing and recombination or “gimme a break.” Genes & Dev. 14**:** 1–10. [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. In Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saxe D., Datta, A., and Jinks-Robertson, S. 2000. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol. Cell. Biol. 20**:** 5404–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente, L. 1997. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 91**:** 1033–1042. [DOI] [PubMed] [Google Scholar]
- Skryabin K.G., Eldarov, M.A., Larionov, V.L., Bayev, A.A, Klootwijk, J. de Regt, V.C., Veldman, G.M., Planta, R.J., Georgiev, O.I., and Hadjiolov, A.A. 1984. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 12**:** 2955–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.L., Klco, S.R., and Cantor, C.R. 1988. Pulse-field gel electrophoresis and the technology of large DNA molecules. In Genome analysis (ed. K.E. Dacis), pp. 41–112. IRL Press, Oxford, UK.
- Strathern J.N, Shafer, B.K., and McGill, C.B. 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140**:** 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver, T.L., Rothstein, R.J., and Stahl, F.W. 1983. The double-strand-break repair model for recombination. Cell 33**:** 25–35. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothtein, R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56**:** 619–630. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil, R.L., and Roeder, G.S. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48**:** 1071–1079. [DOI] [PubMed] [Google Scholar]
- Wai H., Johzuka, K., Vu, L., Eliason, K., Kobayashi, T., Horiuchi, T., and Nomura, M. 2001. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 21**:** 5541–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J.W., Chrevet, G., Brodsky, G., Rolf, M., and Rothstein, R. 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58**:** 409–419. [DOI] [PubMed] [Google Scholar]
- Ward T.R., Hoang, M.L., Prusty, R., Lau, C.K., Keil, R.L., Fangman, W.L., and Brewer, B.J. 2000. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: Shared sequences but independent activities. Mol. Cell. Biol. 20**:** 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.A., Detloff, P., Strand, M., and Petes, T.D. 1992. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr. Genet. 21**:** 109–116. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Hori, Y., Shinomiya, T., Obuse, C., Tsurimoto, T., Yoshikawa, H., and Shirahige, K. 1997. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells 2**:** 655–665. [DOI] [PubMed] [Google Scholar]
- Yano R. and Nomura, M. 1991. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: A suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol. Cell. Biol. 11**:** 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]