Rds3p Is Required for Stable U2 snRNP Recruitment to the Splicing Apparatus (original) (raw)
Abstract
Rds3p is a well-conserved 12-kDa protein with five CxxC zinc fingers that has been implicated in the activation of certain drug transport genes and in the pre-mRNA splicing pathway. Here we show that Rds3p resides in the yeast spliceosome and is essential for splicing in vitro. Rds3p purified from yeast stably associates with at least five U2 snRNP proteins, Cus1p, Hsh49p, Hsh155p, Rse1p, and Ist3p/Snu17p, and with the Yra1p RNA export factor. A mutation upstream of the first Rds3p zinc finger causes the conditional release of the putative branchpoint nucleotide binding protein, Ist3p/Snu17p, and weakens Rse1p interaction with the Rds3p complex. The resultant U2 snRNP particle migrates exceptionally slowly in polyacrylamide gels, suggestive of a disorganized structure. U2 snRNPs depleted of Rds3p fail to form stable prespliceosomes, although U2 snRNA stability is not affected. Metabolic depletion of Yra1p blocks cell growth but not splicing, suggesting that Yra1p association with Rds3p relates to Yra1p's role in RNA trafficking. Together these data establish Rds3p as an essential component of the U2 snRNP SF3b complex and suggest a new link between the nuclear processes of pre-mRNA splicing and RNA export.
The cellular pre-mRNA splicing apparatus, or spliceosome, is composed of five small nuclear RNAs (snRNAs) and over 70 proteins (reviewed in references 9, 12, and 14). Recent advances in the biochemical purification and analysis of spliceosomal complexes have greatly facilitated the identification of splicing factors (23, 32, 35, 53, 54, 62). Complementary genetic studies have provided proof of function for many of these proteins and have led to the identification of other splicing-relevant factors, some of which show weak or transient association with the spliceosome (10). In addition to contributing to our understanding of spliceosomal organization and dynamics, such work has uncovered intriguing links between the cellular machineries that function in the transcription, processing, transport, and decay of mRNA (reviewed in references 19, 21, 51, and 64).
Cellular mRNAs destined for transport to the cytoplasm are recruited by specific components of the RNA export machinery (recently reviewed in references 17 and 38). The presence of introns enhances gene expression in mammals (11, 18, 34, 67) and may greatly facilitate RNA export under certain conditions (47). In mammals, a set of approximately eight proteins (including Y14, Mago, DEK, RNPS1, SRm160, Upf3, UAP56, and REF/Aly) are deposited 20 to 24 nucleotides upstream of mRNA exon junctions in a splicing-dependent manner (37, 42). These exon-junction complex (EJC) proteins are implicated in several cellular functions, including pre-mRNA splicing, mRNA 3′-end formation, RNA export, conventional RNA stability, and nonsense-mediated decay. Recently, however, the general importance of splicing to RNA export has been questioned (57, 65), and a possible new role for EJC-associated proteins in translational loading has been suggested (46, 55). Although much remains to be learned about the multiple roles of EJC proteins, it is clear that the splicing-dependent recruitment of proteins to mRNA is an important conserved feature of eukaryotic gene expression.
The mammalian UAP56 protein was first identified as a DExD/H-box protein that interacts with the U2AF65 splicing factor during the recruitment of the U2 snRNP particle to the prespliceosome (22). The Saccharomyces cerevisiae UAP56 homolog, Sub2p, is an essential splicing factor that appears to function at an equivalent step in yeast spliceosome assembly. The requirement for Sub2p in splicing is relieved if the nonessential U2AF65 homolog, Mud2p, is first removed (39), suggesting that Sub2p acts, at least in part, to dislodge Mud2p from the pre-mRNP and thereby allow U2 snRNP access to the branchpoint sequence. The yeast REF/Aly homolog, Yra1p, is an RNA binding protein that interacts with Sub2p and functions in the nuclear export of both spliced and unspliced mRNAs (73; see also references in references 17 and 38). UAP56/Sub2p may promote REF/Aly/Yra1p recruitment to spliced RNA (48), and Yra1p recruitment to mRNA depends upon (or is enhanced by) proper mRNA 3′-end formation and, at least for certain RNAs, pre-mRNA splicing (44). Yra1p interacts with the Abf1p transcription factor (31), and both UAP56/Sub2p and REF/Aly/Yra1p are present in the THO transcription elongation complex (74), providing possible splicing-independent means for RNA association.
RDS3 was recently identified in a screen for yeast mutants that show synthetic growth defects when combined with a nonlethal mutation in the CLF1 splicing factor gene (78). Clf1p is a component of the nineteen complex (NTC), a salt-stable collection of 8 to 12 splicing factors (28, 50, 70, 82; J. Woolford, personal communication), the penta-snRNP (71), and the U2,U5,U6-RNP complex that likely represents an endogenous yeast spliceosome (56, 81). Considerable overlap exists in the genes identified by genetic studies with LSR1/U2 snRNA (86), PRP17 (5), and CLF1 (78). These include (in at least two of the three studies) genes encoding the U2 snRNA, multiple NTC-associated proteins (Clf1p, Syf2, Brr2p, and perhaps /Ecm2p and Prp8p), and second-step (or later) splicing factors (Prp16p, Prp17p, Prp22p, and Slu7p). The NTC-U2 snRNP connection is of particular interest as present spliceosome models place U2 snRNA at or near the enzyme's active site. Ben-Yehuda et al. (5) report that overexpression of U2 snRNA suppresses the growth defects of certain SYF1, SYF2, and CLF1/SYF3 mutations, suggesting that increased abundance of this snRNA might stabilize a sensitive interaction between the NTC and the U2 snRNP particle. Consistent with this, heat inactivation of the temperature-sensitive Clf1Δ2p mutant derivative reduces Rse1p and Hsh155p (U2 snRNP protein) association with the U2,U5,U6-RNP and causes the release of Prp19p and Cef1p from the NTC (81).
The 12-kDa Rds3 protein has five CxxC zinc finger repeats and is highly conserved, with the human Rds3p homolog showing remarkable 95 and 56% levels of sequence identity with the Drosophila melanogaster and yeast counterparts, respectively. Curiously, while depletion of Rds3p from yeast blocks pre-mRNA splicing (78), extensive proteomic analyses have not identified Rds3p as a component of the yeast splicing apparatus (56, 70, 71, 81). Based on changes in drug transporter mRNA levels and drug sensitivity observed in the absence of Rds3p function, Turcotte and colleagues suggested that Rds3p acts as a transcriptional activator (3, 4). If so, the splicing defect observed with the synthetic lethal, rds3-1/slc6-1 mutant (78) might be indirect and caused by the impaired biosynthesis of one or more essential pre-mRNA splicing factors. Here we show that Rds3p is a critical pre-mRNA splicing factor and an integral component of the yeast spliceosome. Extracts that lack Rds3p activity are splicing defective and arrest spliceosome assembly prior to stable U2 snRNP recruitment. In addition, Rds3p interacts with a set of at least five U2 snRNP proteins present in the SF3b subcomplex (Cus1p, Hsh155p, Hsh49p, Its3p/Snu17p, and Rse1p) and with the Yra1p RNA export protein. At the restrictive temperature, the single amino acid change in Rds3-1p blocks interaction with the Ist3p/Snu17p branchpoint binding protein and weakens Rds3p association with the Clf1Δ2p-sensitive protein Rse1p. These and related data provide evidence for an Rds3p-dependent step in prespliceosome formation and suggest that NTC-U2 snRNP interactions contribute to later stages of the spliceosome cycle. In addition, the recovery of Yra1p with the Rds3p complex suggests a possible additional link between the pre-mRNA splicing and RNA export pathways.
MATERIALS AND METHODS
Yeast strains and plasmid constructions.
We previously published details on the construction of the GAL1::rds3-1 fusion gene and its expression in a yeast rds3::Kanr knockout strain (78). The RDS3-TAP fusion construct was made by insertion of the TAP coding sequence (as described in reference 81) into a Sma_I site introduced at the 3′ end of RDS3 by inverse PCR with oligonucleotides Rds3-3 (5′ TTCGAATTCCCTTGGCTCAACTCCTTGCA 3′) and Rds3-4 (CATGGATCCCGGGTACCTTTTTCTTTTTCTCGAAATG) on plasmid backbone YIplac111 (25). The amplified DNA includes 170 bp of 5′-flanking sequence. The Ycplac111-RDS3-TAP plasmid was transformed into yeast heterozygous for an rds3::Kanr deletion obtained from the American Type Culture Collection. A haploid strain that exclusively expresses the plasmid-borne RDS3-TAP allele (i.e., QWY101, matahis3 lys2 ura3 rds3::Kan, Ycplac111_-RDS3-TAP) was isolated from the meiotic offspring of this diploid parent. The rds3-1-TAP strain (i.e., QWY102, matahis3 lys2 ura3 rds3::Kan, Ycplac111_-rds3-1-TAP_) was prepared in a like manner except that genomic DNA from the rds3-1 mutant (78) was substituted for that of the wild-type DNA (here strain MGD454-46D, α Cyhr leu2-3,112 his trp1-289 ura3-52).
Plasmids bearing the GAL1::GFP-YRA1 and the YRA1-ProtA fusion genes have been described in detail elsewhere (73). The plasmids were introduced into the YRA1 shuffle yeast strain (73) (_MAT_α ade2 his3 leu2 trp1 ura3 yra1::HIS3, pRS316-YRA1) by transformation, and the resident pRS316-YRA1 plasmid was removed by fluoroorotic acid selection (8) prior to analysis.
Protein purification and analysis.
The Rds3-TAP complex was prepared as described previously (62) with minor modifications (81) from 20 liters of yeast (for proteomic analysis) or from 10 ml of metabolically labeled yeast. Yeast proteins were 35S labeled by the addition of 1 mCi of Trans-35S (ICN) to 1 ml of yeast concentrated from 10 ml of culture (at an optical density at 600 nm of 1.0). The cells were incubated with the labeled amino acids at 30°C for 3 h. For the temperature-inactivation experiments, the cultures were shifted to 37°C for the final hour of metabolic labeling. The cell pellets were collected by centrifugation, washed once with water, and broken by vortexing the pellet with glass beads for 4 min in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 200 mM NaCl, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 0.5% NP-40). TAP purification was then performed scaled down for use with the reduced volumes as recently described (81). Where indicated, the NaCl concentration was adjusted to 150, 300, or 500 mM during the binding and wash steps of protein A agarose chromatography. RNase-treated samples were incubated for 15 min at 23°C with 22 μg of RNase A/ml and 444 U of RNase T1 (Ambion)/ml before TAP purification. To reduce contamination by background proteins, where indicated (see figure legends) the released complexes were resolved on a 15-ml 10 to 30% glycerol gradient in 50 mM Tris-HCl (pH 7.4)-20 mM NaCl-5 mM MgCl2 after calmodulin release. Proteins present in the bottom two-thirds of the gradient were pooled and precipitated with 6% trichloroacetic acid. The samples were then resolved on a sodium dodecyl sulfate-5 to 10% gradient polyacrylamide gel with Benchmark molecular weight markers (Invitrogen). The labeled bands were visualized with a Typhoon phosphorimager (Molecular Dynamics). Western blots of the Rds3-TAP, Clf1-TAP, and Yra1-ProtA were incubated with a 1:1,000 dilution of rabbit anti-peroxidase primary antibody (Sigma) followed by incubation with a 1:1,000 dilution of goat anti-rabbit immunoglobulin G (IgG) (heavy-plus-light chain)-alkaline phosphatase secondary antibody (Gibco/BRL) prior to development with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium according to the supplier's recommendation.
Mass analysis.
The tandem affinity-purified protein complexes were size fractionated on a 5 to 10% polyacrylamide gel, stained with silver, digested with trypsin, and assayed by mass analysis after fractionation with a Luna C18 column (Phenomenex, Torrance, Calif.) as previously described (81). Alternatively, the proteins released from calmodulin agarose were assayed by direct analysis of large protein complexes without gel fractionation by two-dimensional liquid chromatography of the tryptic fragments with strong ion-exchange and C18 packing materials (Whatman) on a Deca mass spectrometer (ThermoFinnigan, San Jose, Calif.). Mass analysis was performed on TAP-purified samples with and without the added step of gradient fractionation. The tandem mass spectra were converted to mass-intensity lists and searched against the nonredundant Owl database with SEQUEST software and the nonredundant National Center for Biotechnology database with MASCOT software. The sums of unique and overlapping peptides obtained in three mass analyses were as follows: Rse1p, 25; Hsh155p, 15; Cus1p, 36; Hsh49p, 10; Yra1p, 11; and Rds3p, 2.
RNA isolation and in vitro splicing.
To deplete Rds3p, GAL1::rds3-1 yeast was grown at 30°C in YP broth with 2% galactose (36) to an optical density at 600 nm of approximately 0.20. The yeast was then harvested by centrifugation and resuspended in 2× the original culture volume with YP broth containing 2% glucose. The culture was incubated at 37°C for 8 h prior to extraction of RNA (66) or extract preparation as described by Umen and Guthrie (76).
To biochemically deplete Rds3-TAP, 0.5 ml of extract was bound at 4°C for 2 h with 100 μl of calmodulin resin (Stratagene) made up to a 2 mM concentration with CaCl2. The beads were then removed by centrifugation at 4,000 × g for 5 min. We found that considerable tagged protein persisted in the extract after this treatment but that much of the residual Rds3-TAP could be depleted if the extract was then immune precipitated with 100 μl of rabbit IgG agarose (Sigma). The Rds3-TAP-depleted extract was dialyzed against 2 liters of buffer D (10 mM HEPES [pH 7.9], 50 mM KCl, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 20% glycerol) for 15 h prior to use in the splicing assays. For Yra1-ProtA and Rds3-TAP purification, 0.5 ml of extract was bound to 100 μl of calmodulin resin as described above. After extensive washing, the bound protein was released with 3 column volumes of IPP150 [150 mM NaCl, 10 mM Tris-HCl (pH 8.0), 1 mM Mg (OAc)2, 1 mM imidazole, 2 mM EGTA, and 10 mM β-mercaptoethanol) and then assayed by Western blotting.
For the Rds3p reconstitution assay, 1.5 μl (∼150 ng) of Rds3-TAP protein purified by double-affinity selection (or buffer D only) was mixed with 4 μl of the depleted or inactivated splicing extract and incubated under splicing conditions but without ATP or pre-mRNA substrate for 5 min at 23°C. Pre-mRNA prepared from _Dde_I-cleaved SPRP51A DNA (renamed RPS17A to reflect the change in the ribosome nomenclature) and ATP was then added, and the reaction continued for 2, 15, or 45 min. For chase experiments, the pre-mRNA was preincubated with the Rds3-inactive extract under splicing conditions for 5 min followed by the simultaneous addition of 1.5 μl (∼500 ng) of released protein (or buffer D) and a 50 M excess (150 ng) of cold substrate in a 10-μl splicing reaction mixture. U2 snRNA was targeted for degradation with RNase H (69), and spliceosomes were selected by streptavidin agarose chromatography (85) as previously described. Spliceosomes and snRNP complexes were resolved on 4% polyacrylamide (80:1 acrylamide:bisacrylamide)-0.5% agarose gels run in 50 mM Tris-glycine buffer (pH 8.8) overnight at a 30-V constant voltage. Details of the in vitro splicing, spliceosome assembly, and affinity purification assays are published elsewhere (7, 66, 85).
RESULTS
Rds3p is a yeast pre-mRNA splicing factor.
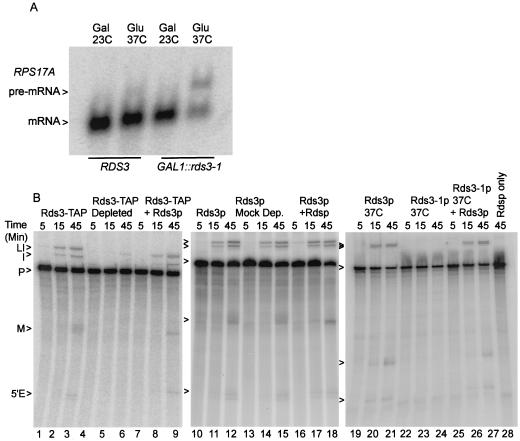
The GAL1::rds3-1 gene consists of the galactose-regulated GAL1 promoter (33) fused to the coding sequence of the temperature-sensitive rds3-1 allele (78). The removal of Rds3-1p activity from yeast by simultaneous transcriptional repression and temperature inactivation blocks cellular pre-mRNA splicing (Fig. 1A) (see also reference 78). Wild-type yeast cells are not splicing impaired under the same conditions. The decreased splicing competence observed with Rds3-1p depletion might result from the direct loss of an essential splicing factor or from the removal of a protein required for the expression of splicing factor genes. To distinguish between these alternatives, yeast whole-cell extracts were assayed for splicing activity in the presence or absence of Rds3p.
FIG. 1.
Rds3p activity is required for pre-mRNA splicing. (A) Northern blot analysis of RNA isolated from yeast strains that express a wild-type RDS3 gene and from yeast strains that express a glucose-repressible GAL1::rds3-1 allele. Samples were taken from yeast grown constitutively on galactose at room temperature (Gal, 23°C) and after shift to glucose-based medium (Glu) for 10 h at 37°C. (B) In vitro assay of splicing conducted in the presence and absence of active Rds3p or Rds3-TAP. The extract used in lanes 4 to 9 and 12 to 18 was preincubated with the TAP-specific calmodulin and IgG affinity resins prior to the assay. In lanes 22 to 27, extract was prepared from the glucose-repressed, heat-inactivated GAL1::rds3-1 mutant. For lanes 7 to 9, 16 to 18, and 25 to 28 the double-affinity-purified Rds3-TAP complex was included in the splicing reaction mixture. The positions of the RPS17A pre-mRNA (P), lariat intermediate (LI), 5′ exon (5′E), mRNA (M), and excised intron (I) are indicated.
Splicing extract was prepared from a strain which expresses a TAP-tagged Rds3 protein that is fully functional in vivo (data not shown) and supports pre-mRNA splicing in vitro (Fig. 1B, lanes 1 to 3). Approximately 70% of the Rds3p-TAP could be removed from this extract by sequential calmodulin agarose and protein A agarose chromatography (data not shown). This two-step depletion protocol greatly inhibits splicing in the Rds3-TAP extract (lanes 4 to 6) but not in a mock-depleted extract prepared from a strain that lacks a TAP-tagged protein (lanes 10 to 12 and 13 to 15). Splicing is completely abolished if extracts are prepared from metabolically depleted GAL1::rds3-1 yeast (lanes 22 to 24; see also Materials and Methods). The medium and temperature shifts used for metabolic depletion do not appreciably impair splicing when a wild-type (i.e., RDS3) yeast strain is used (lanes 19 to 21). While the immune depletion procedure removes Rds3p and any stably bound proteins (see below), the GAL1::rds3-1 metabolic depletion protocol should be specific for Rds3-1p (although secondary effects resulting from Rds3-1p removal cannot be ruled out). Independent of whether Rds3p activity is reduced by biochemical or genetic means, splicing is impaired prior to 5′-splice-site cleavage, consistent with the splicing block observed in vivo (78). Splicing can be restored in the Rds3p-depleted extracts by the addition of an affinity-purified Rds3p complex (lanes 7 to 9 and 25 to 27; see below). As expected, the purified Rds3p complex has no splicing activity in the absence of cell extract (lane 28). These data suggest an essential, Rds3p-sensitive step in the assembly or function of the yeast spliceosome.
Rds3p interacts directly with the spliceosome.
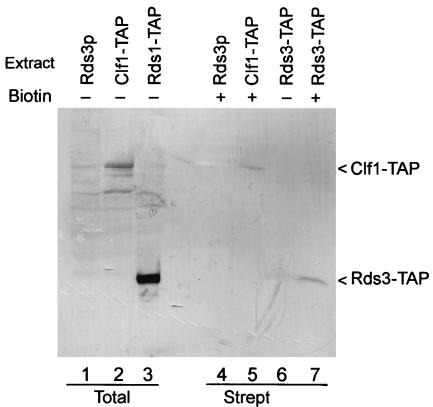
While the spliceosome contains many stably bound proteins, some factors interact transiently with this complex to facilitate conformational changes required during assembly (e.g., Prp16p [68]). Spliceosomes assemble efficiently on biotin-substituted pre-mRNA (e.g., see references 6, 30, and 81). To learn if Rds3p is a stable spliceosomal constituent, spliceosomes were assembled in vitro on biotin-substituted pre-mRNA, recovered by streptavidin agarose chromatography, and then assayed for the presence of Rds3-TAP (Fig. 2). In addition to background bands (lane 1), the anti-TAP antibody recognizes prominent bands of approximately 100 and 32 kDa in total unfractionated extract from yeast cells that express Clf1-TAP (lane 2) and Rds3-TAP (lane 3), respectively. As expected, the TAP epitope increases the mass of each protein by approximately 20 kDa. Clf1-TAP is recovered in spliceosomes purified by streptavidin selection (lane 5) (81) while few or no background bands are present in this sample or in a negative control sample prepared from extract that lacks a TAP-tagged protein (lane 4). Rds3-TAP recovery is greatly enriched in the assembled spliceosome (lane 7) compared with a negative-control sample prepared with a pre-mRNA substrate that lacks biotin (lane 6). Thus, Rds3p stably interacts with the yeast spliceosome under moderate-salt conditions (i.e., 200 mM NaCl).
FIG. 2.
Rds3p is present in the yeast spliceosome. (A) Western blot of whole-cell yeast extracts (lanes 1 to 3) and affinity-purified splicing complexes (lanes 4 to 7) probed with a TAP-specific antibody. Spliceosomes were assembled for 45 min on biotin-substituted (+) or unmodified (−) RPS17A pre-mRNA in wild-type extracts (Rds3p) and in extracts where the epitope-tagged Rds3-TAP or Clf1-TAP served as the only source of these essential proteins. The positions of the relevant tagged proteins are indicated on the right. Strept, streptavidin.
Diminished Rds3p protein level or activity blocks prespliceosome formation.
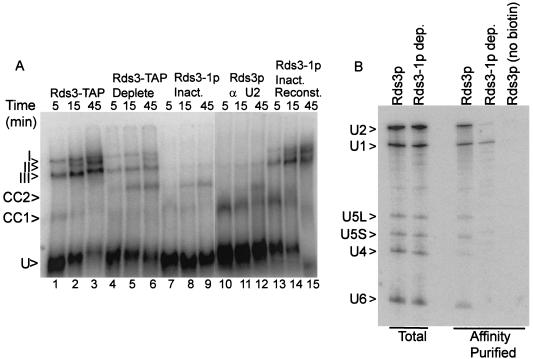
To investigate what role Rds3p may serve in splicing, spliceosome assembly was monitored in the presence and absence of Rds3p activity. With the splicing-competent Rds3-TAP extract, three major bands corresponding to the prespliceosome (III), the snRNP-complete spliceosome (I), and the mature spliceosome after U4 snRNA release (II) are resolved by native gel electrophoresis (Fig. 3A, lanes 1 to 3) (59). The abundance of the mature spliceosome complex increases throughout the 45-min time course of assembly. When the Rds3p is 70% depleted by affinity selection, spliceosome assembly is greatly reduced, with much of the pre-mRNA arresting in a pair of bands that migrate more quickly than the U1-U2-dependent prespliceosome (lanes 4 to 6). Extracts from cells metabolically depleted of Rds3-1p accumulate these novel bands almost exclusively (lanes 7 to 9), indicating a failure in prespliceosome assembly. When a wild-type extract or the Rds3p-depleted extracts are preincubated with RNase H and an oligonucleotide targeted to the U2 snRNA, only the U1-dependent commitment complex forms (69) (lanes 10 to 12 and data not shown). The Rds3p-arrested bands precisely comigrate with these commitment complex bands. Under all conditions, the branchpoint-dependent CC2 band predominates at later time points. The Rds3p-defective complexes can be chased through the assembly pathway with Rds3p purified from yeast, even in the presence of saturating amounts of cold competing substrate (to block de novo spliceosome assembly) (lanes 13 to 15) (see Materials and Methods).
FIG. 3.
Rds3p is required for the commitment complex to prespliceosome transition. (A) Native gel electrophoresis of splicing complexes assembled on RPS17A pre-mRNA. Rds3-TAP splicing extract was assayed without additional manipulation (lanes 1 to 3) and after preabsorption with calmodulin agarose and protein A agarose affinity resins (lanes 4 to 6). Extracts were also prepared from glucose-grown, temperature-inactivated GAL1::rds3-1 yeast (lanes 7 to 9 and 13 to 15) and from wild-type yeast (lanes 10 to 12). The wild-type extract was preincubated with RNase H and an anti-U2 oligonucleotide prior to assay in order to block assembly at the commitment complex stage. In lanes 13 to 15, the extract was preincubated with pre-mRNA for 5 min prior to the addition of a 50-fold excess of unlabeled substrate and the complementing Rds3-TAP complex. The positions of the unassembled pre-mRNA (U), commitment complex bands (CC1 and CC2), prespliceosome (III), snRNP complete splicing complex (I), and catalytically active spliceosome (II) are indicated at the left. (B) Northern analysis of splicing complex snRNAs. Shown is a membrane transfer of RNA present in wild-type extract (Rds3p) and extract prepared from the glucose-grown, temperature-inactivated GAL1::rds3-1 culture (Rds3-1p dep.) probed for the spliceosomal snRNAs. RNA in the unfractionated extract (Total) is compared with that released from affinity-purified splicing complexes assembled on biotin-substituted or unmodified (no biotin) pre-mRNA. The positions of the spliceosomal snRNAs are noted at the left.
To further characterize the Rds3-1p inactivated splicing complexes, spliceosomes were assembled on biotin-substituted pre-mRNA, affinity purified, and assayed for snRNA content (Fig. 3B). Spliceosomes recovered after 30 min of assembly in a splicing-competent extract contain each of the spliceosomal species found in total, unfractionated extract. U4 snRNA levels are underrepresented relative to the total extract due to release from the complex with spliceosome activation (59). In contrast, U1 snRNA is recovered almost exclusively from complexes assembled in the Rds3-1p heat-inactivated extract. Only trace levels of snRNA are present when an identical but non-biotin-substituted pre-mRNA is used. These results are consistent with the native gel assembly pattern and document an Rds3p-sensitive step in the recruitment of the U2 snRNP to the U1-containing pre-mRNP.
Rds3p interacts with protein components of the U2 snRNP and the RNA nuclear export pathway.
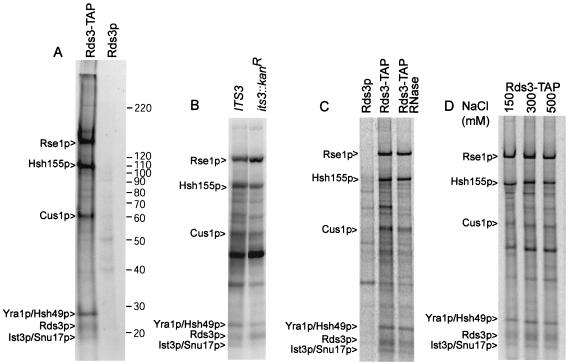
The impaired prespliceosome assembly observed after Rds3p removal might be explained if Rds3p functions as an integral U2 snRNP protein or as a remodeling factor required for stable U2 snRNP recruitment (for instance, similar to Sub2p). To address this issue, Rds3-TAP was purified from metabolically labeled yeast and assayed for the presence of associated proteins. Five prominent protein bands greater than 20 kDa reproducibly copurify with 35S-labeled Rds3-TAP (Fig. 4A). Four proteins were identified by mass spectroscopy as known components of the U2 snRNP SF3b complex, namely, Cus1p, Hsh49p, Hsh155p, and Rse1p. A fifth protein, the Yra1p RNA export factor, was found to comigrate with Hsh49 (60, 73, 75, 87). No other RNA export factors were found in our analysis of the Rds3-TAP complex, although loss during purification cannot be excluded.
FIG. 4.
Protein characterization of the Rds3-TAP complex. (A) Profile of Rds3-TAP-associated proteins. 35S-labeled proteins from Rds3-TAP or the wild-type control (Rds3p) cultures were isolated by the two-step TAP affinity purification and glycerol gradient separation (see Materials and Methods) and resolved on a sodium dodecyl sulfate-5 to 10% gradient polyacrylamide gel with Benchmark molecular mass markers (indicated on right in kilodaltons). Bands present in the Rds3p control are background proteins. The protein assignments in the Rds3-TAP lane were made based on mass analysis of tryptic digests of gel bands and of the entire complex without prior gel fractionation (see Materials and Methods). (B) Ist3p/Snu17p is present in the Rds3p complex. The bands correspond to proteins recovered by Rds3p-TAP affinity purification in a wild-type (IST3/SNU17) and null allele (ist3::Kanr) background. The background bands are somewhat greater here than in panel A since the Rds3-TAP complex was not gradient fractionated prior to analysis. A minor background band migrates with or just above the position of Ist3p/Snu17p. (C) The prominent Rds3p complex bands are RNase resistant. Rds3-TAP yeast extract was used for affinity purification (without gradient separation) before and after extensive digestion with RNase A and RNase T1. The first lane (Rds3p) shows the positions of background proteins. (D) The Rds3p complex is salt stable. Rds3-TAP extracts were adjusted to the indicated NaCl level during the binding and wash steps of protein A-IgG selection. Subsequent calmodulin agarose chromatography was conducted under standard (150 mM) salt conditions (62).
Although the final identified SF3b subunit, Ist3p/Snu17p, is 17.1 kDa (20, 27), we were unable to get a reliable mass analysis for the 17-kDa Rds3p-associated band. Confirmation of this assignment was sought with a strain having the IST3/SNU17 gene deleted (20, 27). The RDS3-TAP gene was transformed on a centromeric plasmid into the ist3::Kanr mutant strain, and Rds3-TAP complexes were isolated as before. The 17-kDa band is reproducibly reduced or absent in the ist3::Kanr mutant (Fig. 4B). A faint band can sometimes be detected at or just above the position of Ist3p/Snu17p, but this is likely one of two 16- to 18-kDa background proteins that occasionally copurify during TAP selection (i.e., see Fig. 1A in reference 81). A Cus1p protein tagged with TAP was found to produce an equivalent set of proteins (Q. Wang and B. C. Rymond, unpublished data). Additional evidence for the Ist3p/Snu17p assignment is presented below in the descriptions of U2 snRNP complexes assembled in the absence of Ist3p/Snu17p or Rds3p.
CxxC domains are present in DNA methyltransferases and a number of proteins that bind nucleic acids (e.g., see references 16 and 40 and references within). Nevertheless, the Rds3p complex is stable after addition of RNases A and T1 under conditions that degrade all detectable RNA, consistent with the presence of only trace amounts of snRNA present in this structure (Fig. 4C and data not shown). This does not rule out the possibility that Rds3p weakly or transiently binds RNA, however. The Rds3-TAP complex appears intact up to at least 500 mM NaCl (Fig. 4D). With the exception of Ist3p/Snu17p, each of the Rds3p-associated proteins is essential for cell viability.
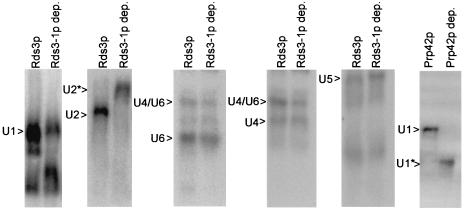
No reproducible differences were observed between wild-type and Rds3-1p-depleted extracts in the levels or electrophoretic mobilities of U1, U4, U5, or U6 snRNA-bearing snRNP complexes (Fig. 5). In contrast, the mobility of the U2 snRNP particle is greatly retarded after the removal of Rds3-1p (U2*, Fig. 5). This upward shift contrasts with the more commonly observed increased mobility seen with the removal of other snRNP proteins, for instance, the U1 proteins Prp39p (45) and Prp42p (U1*, Fig. 5) (52). The altered U2 snRNP mobility is similar to what was recently reported for Ist3p/Snu17-deficient U2 snRNP particles (27) and is consistent with an Rds3p-dependent contribution to U2 snRNA structure or snRNP protein composition.
FIG. 5.
Rds3-1p depletion specifically alters the U2 snRNP particle. Yeast snRNP particles prepared from wild-type yeast (Rds3p) and GAL1::rds3-1 yeast (Rds3-1p dep.) were resolved by native gel electrophoresis and hybridized with snRNA-specific probes. Both cultures were grown in glucose-based medium at 30°C and shifted to 37°C for 1 h prior to extract preparation. The positions of the various snRNP complexes resolved are indicated.
Previously it was shown that, when normal RNA 3′-end formation is blocked, yeast U2 snRNA accumulates in an unexpected, polyadenylated form (1). The electrophoretic mobility of U2 snRNA on denaturing gels does not change in the presence or absence of Rds3p (data not shown), showing that the slower U2 snRNP mobility observed with Rds3p removal is not due to lengthened U2 snRNA. RNase H does not cleave U2 snRNA when oligo(dT) is added, reinforcing the view that this RNA is not modified by a poly(A) tract. Based on this, we believe that the U2 snRNP mobility shift seem with Rds3p depletion results from a conformational change associated with protein dissociation. Presumably the removal of Rds3p (and any Rds3p-dependent proteins) makes the U2 snRNP particle less compact and hence more sensitive to the frictional forces encountered during electrophoresis.
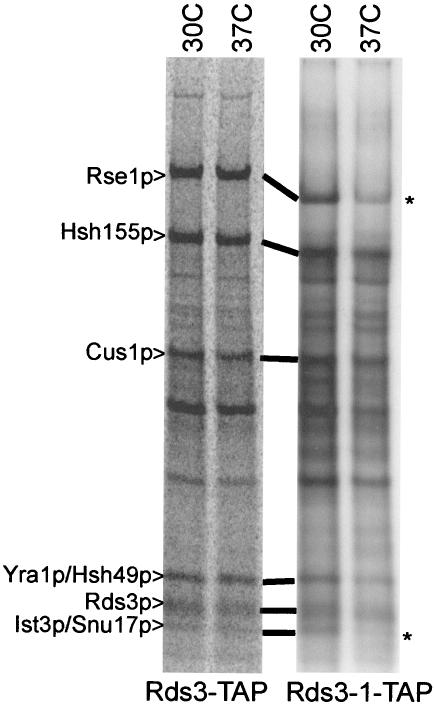
Mutation upstream of the first zinc finger weakens Ist3p/Snu17p and Rse1p association with the U2 SF3b complex.
The Rds3-1 protein contains an aspartic acid for glycine substitution at residue 20 (i.e., GLLCEKC→DLLCEKC; the first zinc finger of Rds3p is underlined). To learn if this lesion influences U2 SF3b integrity, we repeated the Rds3p complex purification with yeast bearing the rds3-1 point mutation. When corrected for minor loading differences, an equivalent pattern of proteins is recovered from Rds3-TAP and Rds3-1-TAP cultures grown at 30°C (Fig. 6). In contrast, when the yeast strains are shifted to 37°C prior to extract preparation, Ist3p/Snu17p is reproducibly released and Rse1p is reduced two- to threefold in the Rds3-1-TAP-purified SF3b complex. SF3b remains intact when the wild-type control strain is treated similarly. Thus, the _rds3-1_-induced splicing block is correlated with an altered U2 snRNP structure with reduced SF3b protein complexity.
FIG. 6.
Rds3-1-TAP inactivation impairs Ist3p/Snu17p and Rse1p association with the Rds3p complex. Radiolabeled proteins were recovered by TAP purification (without gradient separation) from wild-type (Rds3-TAP) and mutant (Rds3-1-TAP) cultures grown constitutively at 30°C or shifted to 37°C for 1 h prior to harvest. The asterisk indicates the position of reproducible bands lost or greatly reduced with temperature shift.
Yra1p is dispensable for yeast pre-mRNA splicing.
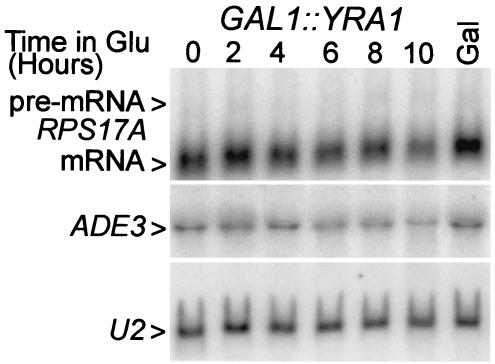
The recovery of Yra1p with Rds3-TAP raises the question of whether Yra1p plays any role in pre-mRNA splicing. Yra1p is known to bind RNA and to autoregulate its own expression by a negative feedback loop that acts at the level of splicing (61). To learn whether reduced Yra1p levels impair general cellular splicing, we made use of a yeast strain that expresses a biologically active GFP-YRA1 fusion gene regulated by the GAL1 promoter (73) (referred to here as GAL1::YRA1). Growth of this strain is galactose dependent; GAL1::YRA1 yeast strains do not form colonies on 5-fluoroorotic acid medium, which selects against the plasmid-linked URA3 gene (reference 73 and data not shown). When the GAL1::YRA1 strain is grown on galactose-based medium, its splicing efficiency is equivalent to that observed with wild-type yeast (i.e., an RPS17A pre-mRNA/RNA ratio of approximately 20 [Fig. 7 and 1A ]). When the GAL1::YRA1 gene is transcriptionally repressed, logarithmic growth continues for approximately 6 h and then becomes progressively impaired over the next 4 h. At late time points RPS17A mRNA abundance decreases slightly but with no accumulation of pre-mRNA. A slight mRNA reduction was also observed with the intronless ADE3 mRNA, suggesting a modest decrease in snRNA stability, possibly due to mislocalization. U2 snRNA levels did not change, consistent with the fact that Yra1p does not function in the export of snRNA. Thus, when YRA1 expression is limiting, pre-mRNA splicing continues unabated.
FIG. 7.
Metabolic depletion of Yra1p does not impair pre-mRNA splicing. RNA was isolated from the GAL1::_YRA1_-dependent yeast culture grown constitutively in galactose (Gal) and after shift to glucose medium for the indicated periods of time. The RNA was fractionated on a denaturing formaldehyde gel and hybridized with probes for the intron-bearing RPS17A (formerly known as RP51A) transcripts, the intronless ADE3 mRNA, and the U2 snRNA.
DISCUSSION
Rds3p is shown here to be an essential and stable component of the yeast spliceosome. In the absence of Rds3p activity, spliceosome assembly arrests at the U1-dependent commitment complex stage, and U2 snRNP addition is blocked. Several lines of evidence point to a basic defect in the U2 snRNP as being causative. First, the commitment complexes formed in the absence of Rds3p can be chased through the splicing pathway when complemented with exogenous Rds3p complex. Thus, the early pre-mRNP appears intact. Second, U2 snRNPs assembled in the absence of Rds3p activity show retarded electrophoretic mobility on native gels, indicative of altered structure. Finally, Rds3p complexes purified from yeast contain each of the previously defined U2 snRNP SF3b proteins, namely, Cus1p, Hsh49p, Hsh155p, Rse1p, and Ist3p/Snu17p, establishing Rds3p as a previously unrecognized component of the yeast U2 snRNP SF3b complex. The Rds3p-SF3b assignment is bolstered by the fact that its human counterpart, SF3b14b, was recently reported as one of eight SF3b proteins expressed in HeLa cells (84). And while the function of SF3b14b was not evaluated, the results presented here predict that SF3b14b-mediated interactions will be important in mammalian spliceosome assembly. Two other proteins, approximately 10 kDa (SF3b10) and 125 kDa (SF3b125) in size, were also identified in the HeLa cell SF3b complex. Database comparisons indicate that neither one has an obvious counterpart in S. cerevisiae. On high-percentage polyacrylamide gels we observed a 10-kDa protein band enriched in the Rds3-TAP complex (Wang and Rymond, unpublished), but since no protein of this size was identified by our mass spectroscopic analyses, we cannot rule out the possibility that this is a breakdown product of a larger SF3b protein. In either case, the absence of clear SF3b10 and SF3b125 homologs suggests either that yeast does not require the associated activities or that it has incorporated SF3b10 and SF3b125 function into structurally diverse proteins.
Rds3p was identified as SLC6 (synthetic lethal with _clf1_Δ2) based on a synthetic lethal interaction between slc6-1/rds3-1 and a deletion mutant of CLF1 that lacks the second of 15 consecutive TPR protein motifs (78). While Clf1p interacts genetically or biochemically with early splicing factors (e.g., Prp40p and Mud2p), it is critical in assembly only after prespliceosome formation, during and after stable recruitment of the U4/U5.U6 tri-snRNP particle (13, 81). Clf1-TAP isolation copurifies the 8- to 12-protein-member NTC complex and a more elaborate complex that contains the U2, U5, and U6 snRNAs and at least 20 additional splicing factors (81). The latter complex (i.e., the Clf1p-U2,U5,U6 RNP) has a protein and snRNA composition characteristic of what might be expected in a late-stage or postcatalytic spliceosome. It is unclear why Rds3p was not found in the native Clf1p complexes or in other recently described yeast splicing complexes (e.g., see references 56 and 71), although automated analysis filters are known to occasionally exclude authentic protein components from large mass spectrum data sets (2). Small proteins, such as Rds3p (and, as seen here, Ist3p/Snu17p), generate few proteolytic fragments, adding to the challenge of proteomic identification.
The biochemical analyses of the Rds3p and Clf1p complexes provide a possible explanation for the synthetic lethal defect observed between slc6-1/rds3-1 and _clf1_Δ2 (78). Two U2 snRNP SF3b proteins, Rse1p and Hsh155p, show reduced abundance in the Clf1Δ2p RNP after temperature inactivation (81). Similar treatment reduces Rse1p and Ist3p/Snu17p association with the Rds3-1-TAP complex. Thus, for both the rds3-1 and _clf1_Δ2 mutations, the loss of splicing activity is correlated with the loss of U2 snRNP proteins, with Rse1p release being the common factor. It seems likely that the rds3-1/_clf1_Δ2 synthetic lethality results from a failure to properly integrate a defective U2 snRNP particle into the spliceosome. While the stage of assembly impaired by the rds3-1/_clf1_Δ2 synthetic defect has not been determined, the first known NTC-dependent step occurs after prespliceosome formation (references 13 and 81) and references within). Thus, either Clf1p acts earlier in assembly than previously shown or NTC structure plays a significant role in U2 snRNP reorganization during U4/U6.U5 tri-snRNP addition or spliceosome activation.
While U2 snRNP particles migrate abnormally slowly in the absence of either Ist3p/Snu17p or Rds3p (this study and reference 27), spliceosome assembly differs considerably in each case. In an Ist3p/Snu17p-deficient extract, an snRNP-complete spliceosome is formed that also shows retarded electrophoretic mobility. In contrast, spliceosome assembly is blocked much earlier, at the commitment complex CC2 stage, in the absence of Rds3p. The late arrest point for the _ist3/snu17_-null mutant is intriguing, as the mammalian homolog of Ist3p/Snu17p, p14 (also called SF3b14a), cross-links to the branchpoint adenosine in the spliceosome and therefore may play a role in positioning the nucleophile used for the first catalytic step of splicing (83). In support of this, p14 also binds Hsh155, a protein that contacts the pre-mRNA on both sides of the branchpoint and remains bound through the catalytic steps of splicing (29, 49, 63, 80). Curiously, p14 appears to be located at the center of a deep pocket within SF3b, seemingly inaccessible to RNA (26, 77). The yeast homolog, Its3p/Snu17p, is absent from SF3b complexes after Rds3-1 protein inactivation, and this might contribute to the splicing defect observed in the rds3-1 mutant. However, while haploid yeast strains bearing the ist3::Kanr null are viable (24), the rds3::Kanr mutation is lethal under standard laboratory conditions (24, 78). Presumably the rds3::Kanr mutation presents a more egregious insult to U2 snRNP function. Plainly, Rds3p is no longer present, and based on the results obtained with Rds3-1p, Its3p and perhaps Rse1p association with SF3b is likely impaired. The result is an Rds3p-deficient U2 snRNP particle that can no longer stably bind the commitment complex.
It was surprising to recover Yra1p with Rds3p, as no previous report revealed REF/Aly/Yra1p association with U2 snRNP components. We note, however, that REF/Aly/Yra1p does copurify with mammalian spliceosomes (35, 54) and has been found by proteomic analysis in yeast complexes that contain, among numerous other proteins, the spliceosome assembly factor, Sub2p, and the intron release protein, Prp43p (32). Metabolic depletion of Yra1p, while lethal, does not inhibit pre-mRNA splicing, showing that Yra1p association with the U2 snRNP is not a critical feature of the basal splicing apparatus (although a subtle or gene-specific contribution to splicing cannot be ruled out). Since the majority of yeast genes are intron free, we expect only a limited amount of Yra1p to be snRNP associated. Consistent with this, we find that the banding pattern of proteins recovered with Yra1p is very different than what is observed with Rds3p (Wang and Rymond, unpublished). In addition, while approximately 50% of Rds3-TAP can be recovered from extracts by one-step calmodulin affinity chromatography, only 5 to 10% of the Yra1p copurifies (Fig. 8) (here, Yra1p is expressed as an Yra1-ProtA fusion [73]; IgG agarose was not used in this selection). The Yra1p recovered appears specific, however, as it is much greater than the background observed when Rds3-TAP is not coexpressed (Fig. 8, Yra1p-ProtA, bound). Furthermore, Yra1p has not been reported as a “sticky” contaminant of TAP-purified complexes (e.g., see references 23, 32, and 81). Presumably much or all of the unbound Yra1p is associated with mRNA from intronless genes and from spliced mRNAs after spliceosomal release. Additional work is needed to determine if Yra1p resides within SF3b, binds independently to Rds3p, or associates with undetected components (e.g., trace amounts of RNA) that may reside in the Rds3-TAP sample. Also, the results presented here, while intriguing, do not address the important issue of whether the Yra1p/Rds3p complex interaction is significant in the nuclear export of mRNA from intron-bearing genes.
FIG. 8.
Not all Yra1p is associated with Rds3p. A Western blot of extracts prepared from yeast that express Yra1-ProtA only or Yra1-ProtA and Rds3-TAP simultaneously was probed with antibodies to the common protein A component. Total, unfractionated extract; Bound, proteins released from calmodulin agarose. No protein A agarose was used as this would select for both tagged proteins. Approximately 10 extract equivalents were loaded in the Bound lane compared to the Total lane.
A model for Rds3p function in splicing is presented in Fig. 9. Here, Sub2p stimulates the displacement of Mud2p from the branchpoint region of the commitment complex (39). This event permits the Rds3p-dependent incorporation of the U2 snRNP into the commitment complex. Prp5p-induced changes to the U2 snRNP (58) allow U2 snRNA to base pair with the pre-mRNA in the prespliceosome. Also within the prespliceosome, Ist3p/Snu17p replaces Bbp at the branchpoint sequence, although the process by which this occurs is unknown. Yra1p may be recruited with the U2 snRNP or bound after prespliceosome formation through its affinity for one or more SF3b proteins and Sub2p (72, 88). The tri-snRNP is then added and likely stabilized in part through NTC-fostered contacts with the U5 snRNP (e.g., Prp8p, Brr2p, and Snu114p) and the U2 snRNP (e.g., Rse1p, Hsh155p, and possibly Rds3p and Ist3p/Snu17p) (78, 81; this study). After catalysis, Yra1p, Sub2p, and other proteins (e.g., the transcriptionally loaded RNA export factor, Npl3p [41, 43]) are then released from the spliceosome with the mRNA in a Prp22p-dependent step (15, 79). Rds3p likely remains bound to the U2 snRNP, although it is conceivable that this protein and perhaps other SnRNP factors associated with the mRNP through the point of nuclear export.
FIG. 9.
Model for Rds3p function and Yra1p recruitment in splicing. Assembly of the splicing apparatus from the branchpoint-dependent commitment complex (CC2) through the point of mRNA release is diagrammed. For clarity's sake only a subset of the splicing-related snRNP and non-snRNP proteins are indicated. Rds3p is shown as essential for the stable addition of the U2 snRNP particle to the commitment complex in prespliceosome formation. Yra1p is presented as being either associated with the U2 snRNP or recruited independently to the spliceosome through contacts that might include both Sub2p and one or more components of the Rds3p-SF3b complex. Sometime prior to or concurrent with the Prp22p-dependent mRNA release step, components of the RNA export pathway are deposited on the processed mRNA. Rds3p likely remains stably associated with the snRNP components of the splicing apparatus, although its release with the RNA export factors cannot be ruled out.
Acknowledgments
We are grateful to Bert Lynn and the UK Mass Spectroscopy Facility for proteomic analysis of the Rds3p complex. We thank the Hurt lab for generously providing the yra1::HIS3 knockout strain and plasmids containing the GAL1 and protein A fusions to YRA1. Bernard Turcotte is acknowledged for discussions while this work was in progress. We thank Martha Peterson for her helpful comments on the manuscript and Kevin Vincent and Georgia Zeigler for reagents and technical assistance, respectively.
This work was sponsored by NIH award GM42476 to B.C.R. Proteomic analysis of the Rds3p complex was supported by the Kentucky NSF EPS-0132295 award.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17**:**3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebersold, R., and M. Mann. 2003. Mass spectrometry-based proteomics. Nature 422**:**198-207. [DOI] [PubMed] [Google Scholar]
- 3.Akache, B., and B. Turcotte. 2002. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J. Biol. Chem. 277**:**21254-21260. [DOI] [PubMed] [Google Scholar]
- 4.Akache, B., K. Wu, and B. Turcotte. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29**:**2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Yehuda, S., I. Dix, C. S. Russell, M. McGarvey, J. D. Beggs, and M. Kupiec. 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156**:**1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bindereif, A., and M. R. Green. 1987. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 6**:**2415-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanton, S., A. Srinivasan, and B. C. Rymond. 1992. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell. Biol. 12**:**3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154**:**164-175. [DOI] [PubMed] [Google Scholar]
- 9.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36**:**333-360. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. D., M. Plumpton, and J. D. Beggs. 1992. The genetics of nuclear pre-mRNA splicing: a complex story. Antonie Leeuwenhoek 62**:**35-46. [DOI] [PubMed] [Google Scholar]
- 11.Buchman, A. R., and P. Berg. 1988. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 8**:**4395-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Chung, S., M. R. McLean, and B. C. Rymond. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5**:**1042-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, C. A., and C. Guthrie. 2000. The question remains: is the spliceosome a ribozyme? Nat. Struct. Biol. 7**:**850-854. [DOI] [PubMed] [Google Scholar]
- 15.Company, M., J. Arenas, and J. Abelson. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349**:**487-493. [DOI] [PubMed] [Google Scholar]
- 16.Cross, S. H., R. R. Meehan, X. Nan, and A. Bird. 1997. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 16**:**256-259. [DOI] [PubMed] [Google Scholar]
- 17.Cullen, B. R. 2003. Nuclear RNA export. J. Cell Sci. 116**:**587-597. [DOI] [PubMed] [Google Scholar]
- 18.Deng, T. L., Y. Li, and L. F. Johnson. 1989. Thymidylate synthase gene expression is stimulated by some (but not all) introns. Nucleic Acids Res. 17**:**645-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3**:**195-205. [DOI] [PubMed] [Google Scholar]
- 20.Entian, K. D., T. Schuster, J. H. Hegemann, D. Becher, H. Feldmann, U. Guldener, R. Gotz, M. Hansen, C. P. Hollenberg, G. Jansen, W. Kramer, S. Klein, P. Kotter, J. Kricke, H. Launhardt, G. Mannhaupt, A. Maierl, P. Meyer, W. Mewes, T. Munder, R. K. Niedenthal, M. Ramezani Rad, A. Rohmer, A. Romer, A. Hinnen, et al. 1999. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262**:**683-702. [DOI] [PubMed] [Google Scholar]
- 21.Farina, K. L., and R. H. Singer. 2002. The nuclear connection in RNA transport and localization. Trends Cell Biol. 12**:**466-472. [DOI] [PubMed] [Google Scholar]
- 22.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11**:**1864-1872. [DOI] [PubMed] [Google Scholar]
- 23.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415**:**141-147. [DOI] [PubMed] [Google Scholar]
- 24.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418**:**387-391. [DOI] [PubMed] [Google Scholar]
- 25.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74**:**527-534. [DOI] [PubMed] [Google Scholar]
- 26.Golas, M. M., B. Sander, C. L. Will, R. Luhrmann, and H. Stark. 2003. Molecular architecture of the multiprotein splicing factor SF3b. Science 300**:**980-984. [DOI] [PubMed] [Google Scholar]
- 27.Gottschalk, A., C. Bartels, G. Neubauer, R. Luhrmann, and P. Fabrizio. 2001. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol. Cell. Biol. 21**:**3037-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottschalk, A., G. Neubauer, J. Banroques, M. Mann, R. Luhrmann, and P. Fabrizio. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18**:**4535-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18**:**4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabowski, P. J., and P. A. Sharp. 1986. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science 233**:**1294-1299. [DOI] [PubMed] [Google Scholar]
- 31.Hieronymus, H., and P. A. Silver. 2003. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33**:**155-161. [DOI] [PubMed] [Google Scholar]
- 32.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415**:**180-183. [DOI] [PubMed] [Google Scholar]
- 33.Johnston, M., and R. W. Davis. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4**:**1440-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonsson, J. J., M. D. Foresman, N. Wilson, and R. S. McIvor. 1992. Intron requirement for expression of the human purine nucleoside phosphorylase gene. Nucleic Acids Res. 20**:**3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8**:**426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiser, C., S. Michaelis, and A. Mitchell (ed.). 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6**:**673-682. [DOI] [PubMed] [Google Scholar]
- 38.Keene, J. D. 2003. Organizing mRNA export. Nat. Genet. 33**:**111-112. [DOI] [PubMed] [Google Scholar]
- 39.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 15**:**42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J. H., K. S. Voo, and D. G. Skalnik. 2001. Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 276**:**44669-44676. [DOI] [PubMed] [Google Scholar]
- 41.Lee, M. S., M. Henry, and P. A. Silver. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 10**:**1233-1246. [DOI] [PubMed] [Google Scholar]
- 42.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19**:**6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15**:**1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei, E. P., and P. A. Silver. 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 16**:**2761-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lockhart, S. R., and B. C. Rymond. 1994. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 14**:**3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu, S., and B. R. Cullen. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9**:**618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96**:**14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413**:**644-647. [DOI] [PubMed] [Google Scholar]
- 49.MacMillan, A. M., C. C. Query, C. R. Allerson, S. Chen, G. L. Verdine, and P. A. Sharp. 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 8**:**3008-3020. [DOI] [PubMed] [Google Scholar]
- 50.Makarova, O. V., E. M. Makarov, S. Liu, H. P. Vornlocher, and R. Luhrmann. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21**:**1148-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416**:**499-506. [DOI] [PubMed] [Google Scholar]
- 52.McLean, M. R., and B. C. Rymond. 1998. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 18**:**353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neubauer, G., A. Gottschalk, P. Fabrizio, B. Seraphin, R. Luhrmann, and M. Mann. 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA 94**:**385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20**:**46-50. [DOI] [PubMed] [Google Scholar]
- 55.Nott, A., S. H. Meislin, and M. J. Moore. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9**:**607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald, and K. L. Gould. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22**:**2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohno, M., A. Segref, S. Kuersten, and I. W. Mattaj. 2002. Identity elements used in export of mRNAs. Mol. Cell 9**:**659-671. [DOI] [PubMed] [Google Scholar]
- 58.Perriman, R., and M. Ares, Jr. 2000. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 14**:**97-107. [PMC free article] [PubMed] [Google Scholar]
- 59.Pikielny, C. W., B. C. Rymond, and M. Rosbash. 1986. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature 324**:**341-345. [DOI] [PubMed] [Google Scholar]
- 60.Portman, D. S., J. P. O'Connor, and G. Dreyfuss. 1997. YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA 3**:**527-537. [PMC free article] [PubMed] [Google Scholar]
- 61.Preker, P. J., K. S. Kim, and C. Guthrie. 2002. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8**:**969-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24**:**218-229. [DOI] [PubMed] [Google Scholar]
- 63.Query, C. C., S. A. Strobel, and P. A. Sharp. 1996. Three recognition events at the branch-site adenine. EMBO J. 15**:**1392-1402. [PMC free article] [PubMed] [Google Scholar]
- 64.Reed, R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15**:**326-331. [DOI] [PubMed] [Google Scholar]
- 65.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98**:**1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rymond, B. C., C. Pikielny, B. Seraphin, P. Legrain, and M. Rosbash. 1990. Measurement and analysis of yeast pre-mRNA sequence contribution to splicing efficiency. Methods Enzymol. 181**:**122-147. [DOI] [PubMed] [Google Scholar]
- 67.Ryu, W. S., and J. E. Mertz. 1989. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 63**:**4386-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwer, B., and C. Guthrie. 1991. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 349**:**494-499. [DOI] [PubMed] [Google Scholar]
- 69.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59**:**349-358. [DOI] [PubMed] [Google Scholar]
- 70.Stevens, S. W., I. Barta, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6 × U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7**:**1543-1553. [PMC free article] [PubMed] [Google Scholar]
- 71.Stevens, S. W., D. E. Ryan, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9**:**31-44. [DOI] [PubMed] [Google Scholar]
- 72.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413**:**648-652. [DOI] [PubMed] [Google Scholar]
- 73.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19**:**410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417**:**304-308. [DOI] [PubMed] [Google Scholar]
- 75.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Seraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6**:**638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Umen, J. G., and C. Guthrie. 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 9**:**855-868. [DOI] [PubMed] [Google Scholar]
- 77.Varani, G., and A. Ramos. 2003. Splicing factor 1 in the pocket. Structure (Cambridge) 11**:**481-482. [DOI] [PubMed] [Google Scholar]
- 78.Vincent, K., Q. Wang, S. Jay, K. Hobbs, and B. C. Rymond. 2003. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics 164**:**895-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner, J. D., E. Jankowsky, M. Company, A. M. Pyle, and J. N. Abelson. 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17**:**2926-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang, C., K. Chua, W. Seghezzi, E. Lees, O. Gozani, and R. Reed. 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 12**:**1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang, Q., K. Hobbs, B. Lynn, and B. C. Rymond. 2003. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 278**:**7875-7883. [DOI] [PubMed] [Google Scholar]
- 82.Weidenhammer, E. M., M. Singh, M. Ruiz-Noriega, and J. L. Woolford, Jr. 1996. The PRP31 gene encodes a novel protein required for pre-mRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 24**:**1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Will, C. L., C. Schneider, A. M. MacMillan, N. F. Katopodis, G. Neubauer, M. Wilm, R. Luhrmann, and C. C. Query. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20**:**4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Will, C. L., H. Urlaub, T. Achsel, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21**:**4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie, J., K. Beickman, E. Otte, and B. C. Rymond. 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17**:**2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu, D., D. J. Field, S. J. Tang, A. Moris, B. P. Bobechko, and J. D. Friesen. 1998. Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol. 18**:**2055-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zenklusen, D., P. Vinciguerra, Y. Strahm, and F. Stutz. 2001. The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 21**:**4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22**:**8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]