Budding of PPxY-Containing Rhabdoviruses Is Not Dependent on Host Proteins TGS101 and VPS4A (original) (raw)
Abstract
Viral matrix proteins of several enveloped RNA viruses play important roles in virus assembly and budding and are by themselves able to bud from the cell surface in the form of lipid-enveloped, virus-like particles (VLPs). Three motifs (PT/SAP, PPxY, and YxxL) have been identified as late budding domains (L-domains) responsible for efficient budding. L-domains can functionally interact with cellular proteins involved in vacuolar sorting (VPS4A and TSG101) and endocytic pathways (Nedd4), suggesting involvement of these pathways in virus budding. Ebola virus VP40 has overlapping PTAP and PPEY motifs, which can functionally interact with TSG101 and Nedd4, respectively. As for vesicular stomatitis virus (VSV), a PPPY motif within M protein can interact with Nedd4. In addition, M protein has a PSAP sequence downstream of the PPPY motif, but the function of PSAP in budding is not clear. In this study, we compared L-domain functions between Ebola virus and VSV by constructing a chimeric M protein (M40), in which the PPPY motif of VSV M is replaced by the L domains of VP40. The budding efficiency of M40 was 10-fold higher than that of wild-type (wt) M protein. Overexpression of a dominant negative mutant of VPS4A or depletion of cellular TSG101 reduced the budding of only M40-containing VLPs but not that of wt M VLPs or live VSV. These findings suggest that the PSAP motif of M protein is not critical for budding and that there are fundamental differences between PTAP-containing viruses (Ebola virus and human immunodeficiency virus type 1) and PPPY-containing viruses (VSV and rabies virus) regarding their dependence on specific host factors for efficient budding.
Enveloped viruses require a membrane fusion event to release virions from the host cell at the final step of their life cycle. It has been shown that viral matrix proteins such as Gag for retroviruses, VP40 for Ebola virus, and M for rhabdoviruses including vesicular stomatitis virus (VSV) and rabies virus (RV) are by themselves able to bud from the cell surface in the form of lipid-enveloped virus-like particles (VLPs), suggesting that these proteins play important roles at this late-budding step (21, 23, 28, 43, 51). Subsequent investigations have shown that these proteins possess late-budding domains (L-domains), which are critical for efficient budding. To date, three L-domain motifs (PPxY, PT/SAP, and YxxL, where x is any amino acid) have been identified in these viral matrix proteins (12). The majority of retroviruses possess PPxY and/or PT/SAP motifs except for equine infectious anemia virus, which possesses a YxxL motif (15, 19, 26, 40, 49, 53, 57). Ebola virus VP40 possesses overlapping L-domain motifs (7PTAP10 and 10PPEY13), and a PPPY motif is present in M proteins of VSV and RV (10, 17, 18, 22). Different viral L-domains are functionally interchangeable between viruses belonging to heterologous families. For example, the PTAP motif of human immunodeficiency virus type 1 (HIV-1) Gag can be functionally replaced by the heterologous YPDL motif from EIAV Gag or the overlapping PTAPPEY motif from Ebola virus (27, 30).
Specific host proteins have been implicated in mediating the activity of each L-domain. For example, the YPDL motif of EIAV p9 was reported to interact with the AP-2 clathrin adaptor, which is involved in endocytosis of certain transmembrane proteins and which relocalizes AP-2 to the budding site (41). More recently, the YPDL motif was shown to interact with host protein AIP1, a central component of the MVB pathway (29a, 47a, 52a). The PPxY motifs of retroviruses, rhabdoviruses, and Ebola virus interact with the Nedd4-like E3 ubiquitin ligase via its WW domains (17, 18). Interactions between the PT/SAP motif of HIV-1 and Ebola virus and cellular tumor susceptibility gene 101 (TSG101) have been well characterized (13, 14, 29, 30, 39, 50, 52). For example, studies using a series of HIV-1 p6 mutants show good correlation between the ability of these mutants to interact with TSG101 and the efficiency of virus budding (13, 14, 39, 52), and depletion of TSG101 by small interfering RNA (siRNA) inhibits HIV-1 budding significantly (13). It has been suggested that the PTAP L-domains of HIV-1 and Ebola virus may function to recruit TSG101 to the sites of particle assembly in order to facilitate virus egress (30).
The ESCRT-I (endosomal sorting complex required for transport) complex consists of the class E vacuolar protein sorting proteins VPS23, VPS28, and VPS37 in yeast (24). ESCRT-I plays a conserved role in sorting proteins into the multivesicular body (MVB) in mammalian cells. Depletion of the human ortholog of VPS23p, TSG101, causes a defect of epidermal growth factor receptor trafficking consistent with the results observed in yeast (4). It has been suggested that ESCRT-I acts together with ESCRT-II and ESCRT-III complexes to regulate MVB sorting in a sequential manner (2, 3). In the final step of vacuolar sorting, an AAA-ATPase, VPS4, interacts with ESCRT-III to catalyze the release of the class E factors from the membrane (1, 5). Catalytically inactive VPS4A displays a dominant negative (DN) phenotype and induces the formation of aberrant enlarged endosomes lacking internal vesicle accumulation (1, 5, 7).
Dominant negative (DN) VPS4A proteins lacking the ability to bind or hydrolyze ATP were shown to inhibit budding of PTAP-containing HIV-1 as well as PPxY-containing murine leukemia virus (MuLV), suggesting that both types of retroviruses are dependent on VPS4 for efficient budding (13). In contrast, depletion of cellular TSG101 affected the budding of HIV-1 but not of MuLV (13). A dominant negative mutant of VPS4A has also been shown to reduce the budding of infectious Ebola virus by approximately 10-fold (29). The importance of host proteins VPS4A and TSG101 in the budding of rhabdoviruses remains to be determined.
In this study, we compared the functions of L-domains derived from viruses belonging to either the Filoviridae (Ebola virus), or the Rhabdoviridae (VSV and RV) families. Our results not only demonstrate that the L-domain of VSV M protein can be functionally replaced by that of Ebola virus VP40 but also that the VP40 L-domain enhances the budding of M protein by 11-fold. Interestingly, overexpression of DN VPS4A or depletion of TSG101 by siRNA reduced the budding of proteins having the Ebola virus L-domain but did not significantly affect the budding of proteins having a rhabdovirus L-domain. Furthermore, DN VPS4A expression or depletion of TSG101 failed to inhibit the budding of infectious VSV and RV. Taken together, these results suggest that although filoviruses and rhabdoviruses bud in an L-domain-dependent manner, there are key differences in their dependence on specific host proteins for efficient budding.
MATERIALS AND METHODS
Cells and viruses.
BHK-21, COS-1, and human 293T cells were maintained in Dulbecco's minimum essential medium (Life Technologies, Rockville, Md.) supplemented with 10% fetal calf serum (HyClone) and 1× penicillin-streptomycin (Life Technologies). VSV (Indiana serotype) and a recombinant vaccinia virus (VvT7), which expresses a bacteriophage T7 RNA polymerase, were propagated in BHK-21 cells. Virus stocks were subjected to titer determination by plaque assay on BHK-21 cells.
Plasmid construction.
A plasmid encoding VP40-WT has been described previously (29). The VSV M gene was inserted into the _Eco_RI and _Xho_I restriction sites of the pcDNA3.1(+) vector (Invitrogen) to yield pT7-Mwt by using a standard PCR and cloning techniques. A chimeric M gene (M40; see Fig. 1A) was constructed using primers 5VSVMEcoRI (TATATGAATTCACCATGAGTTCCTTAAAGAAGAT), 3VSVMXhoI (TATATCTCGAGTCATTTGAAGTGGCTGATAGAATC), 5VSVMVP40L (TATATGGATCCGGAATATATGGAGGCCATAAGCATGGAGTATGCTCCG), and 3VSVMVP40L (TATATTCCGGAGGAGCAGTAGGCAATATAACCCGTTTCTTAGATTTCTTACC) to contain the L-domains of Ebola virus VP40. M40 was inserted into the _Eco_RI and _Xho_I sites of the pcDNA3.1(+) vector. The expression plasmid encoding hemagglutinin (HA)-tagged TSG101 was kindly provided by P. Bates (University of Pennsylvania), and plasmids encoding enhanced green fluorescent protein (EGFP)-fused VPS4A-WT and DN mutants VPS4A-E228Q and VPS4A-K173Q were kindly provided by W. Sundquist (University of Utah). These EGFP-fused VPS4A genes were also subcloned into the _Nhe_I and _Bam_HI sites of the pcDNA3.1(+) vector for expression of these genes by bacteriophage T7 polymerase in the VvT7 expression system.
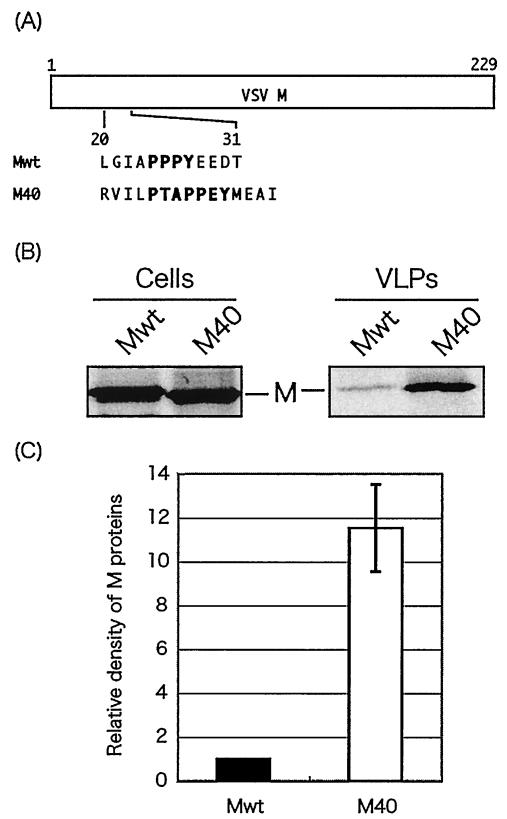
FIG. 1.
Budding assay of VSV M proteins. (A) Schematic representation of expression plasmids encoding Mwt and chimeric M40. The amino acid sequences of the L-domains are indicated by their single-letter code. (B) Radiolabeled cell lysates and VLPs were immunoprecipitated with anti-M MAb, and immunoprecipitates were analyzed by SDS-PAGE. (C) M proteins in VLPs were quantitated by densitometry with the ImageJ 1.24 software, and the bars represent an average of six independent experiments. The level of Mwt was normalized to 1.0, and the relative value of M40 is indicated.
The genes encoding VPS4A-WT and the two DN mutants VPS4A-K173Q and VPS4A-E228Q were PCR amplified from pEGFP-C1-VPS4-WT, pEGFP-C1-VPS4-K, or pEGFP-C1-VPS4-E, respectively, using the forward primer 5′-TTTCGTACGACATGACAACGTCAACCCTCC-3′, containing a _Bsi_WI site (boldface), and the reverse primer 5′-AAAGCTAGCTTAACTCTCTTGCCCAAAGTC-3′, containing an _Nhe_I site (boldface). The PCR products were digested with _Bsi_WI and _Nhe_I and cloned into the previously described recombinant vaccine strain-based rabies virus cDNA (pSPBN) (32). The resulting plasmids were designated pSPBN-VPS4-WT, pSPBN-VPS4-K, and pSPBN-VPS4-E. These recombinant viruses express only wild-type or mutant VPS4A proteins and not green fluorescent protein (GFP)-VPS4A chimeric proteins.
Antibodies.
Monoclonal antibody (MAb) 23H12 specific for the M protein of VSV was kindly provided by D. S. Lyles (Wake Forest University School of Medicine, Winston-Salem, N.C.). Antibody against the VP40 of Ebola virus was kindly provided by J. Paragas (U.S. Army Medical Research Institute for Infectious Diseases, Fort Detrick, Md.). Monoclonal antibodies against the HA epitope tag (Roche Biochemicals, Indianapolis, Ind.), GFP (Torrey Pines Biolabs Inc.), and human TSG101 (4A10; Gene Tex) were used as specified by the suppliers.
Budding assay.
A functional budding assay for VP40 has been described previously (29). The VvT7-based budding assay for VSV M has been described previously (16). Briefly, cells cultured in six-well plates were infected with VvT7 at a multiplicity of infection (MOI) of 10 and then transfected with the indicated plasmids using Lipofectamine (Invitrogen). At 2 h posttransfection (p.t.), the cells were metabolically labeled with [35S]Met-Cys (Perkin-Elmer) (100 μCi/ml) for 8 h. Culture medium was harvested and clarified by centrifugation at 2,500 rpm for 10 min. The supernatant was then centrifuged at 36,000 rpm (Beckman SW41 rotor) for 2.5 h through a 10% sucrose cushion. The pellet was suspended in 400 μl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]), immunoprecipitated with anti-M MAb, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Cells were washed with 1× phosphate-buffered saline and lysed in 400 μl of RIPA buffer. The cell lysates were also immunoprecipitated with appropriate antibodies and analyzed by SDS-PAGE. Protein bands were visualized by autoradiography and quantitated by densitometry with the ImageJ 1.2.4 program or PhosphorImager.
Recovery of infectious RV from cDNA.
Recovery of recombinant RV using reverse genetics has been described previously (45). Briefly, BSR-T7 cells, which stably express T7 RNA polymerase, were transfected with 5 μg of full-length RV cDNA in addition to plasmids encoding RV N, P, L, and G proteins by using a calcium phosphate transfection kit (Stratagene), as instructed by the vendor. At 3 days posttransfection, supernatants were transferred onto fresh BSR cells; infectious RV was detected 3 days later by immunostaining with fluorescein isothiocynate against the RV N protein (Centocor, Inc.). The VPS4 inserts were confirmed by reverse transcription-PCR, restriction endonuclease digestion, and automated DNA sequencing.
VPS4A transfection and VSV infection.
Cells cultured in six-well plates were transfected with the appropriate amount of VPS4-WT or VPS4-EQ by using Lipofectamine. At 24 h p.t., the cells were infected with VSV at an MOI of 0.1 and metabolically labeled with [35S]Met-Cys. At 0, 4, 6, 8, 10, and 12 h postinfection (p.i.), culture medium was harvested and clarified at 2,500 rpm for 5 min. The supernatant was then centrifuged at 36,000 rpm (Beckman SW41 rotor) for 2.5 h through a 20% sucrose cushion. Virion pellets were suspended in 400 μl of RIPA buffer, immunoprecipitated with anti-M MAb, and analyzed by SDS-PAGE as described above. Cells were also harvested at 12 h p.i. and analyzed in the same way as described above. Nonradiolabeled samples were also prepared, and the virus yield at each time point was determined by a plaque assay using BHK-21 cells.
Multicycle growth curve of recombinant RVs.
BSR cells were plated in 60-mm-diameter dishes and infected at an MOI of 0.01 with SPBN, SPBN-VPS4-WT, SPBN-VPS4-K, or SPBN-VPS4-E 16 h later. After incubation at 37°C for 1 h, inocula were removed and cells were washed four times with phosphate-buffered saline to remove any unabsorbed virus. A 3-ml volume of complete medium was added back, and 100 μl of supernatant was removed at various time points after infection. Virus aliquots were subjected to titer determination in duplicate on BSR cells.
siRNA transfection.
Human 293T cells cultured in six-well plates were transfected with a combination of 0.2 μg of TSG101-specific or nonspecific siRNA (Dharmacon Inc.) and 2.0 μg of pCAGGS.MCS vector by using Lipofectamine 2000 (Invitrogen). At 24 h p.t., the cells were transfected a second time with the same combination of siRNA and plasmid. After 12 h the cells were used in the budding assay as described above or infected with VSV at an MOI of 0.1. At 10 h p.i., culture medium and cells were harvested and analyzed as described above. The virus yield was determined by a plaque assay using BHK-21 cells.
RESULTS
Comparison of budding efficiency between L-domains of VSV M and Ebola virus VP40.
It has been reported that the integrity of amino acid sequence of not only the L-domains themselves but also their flanking amino acids are important for their functions (30, 53). To compare the functions of L-domains found in VSV M and Ebola virus VP40, a chimeric M protein (M40) was constructed in which the PPPY motif and flanking amino acids of M protein were replaced by PTAPPEY and flanking amino acids of VP40 (Fig. 1A). The ability of both Mwt and M40 to bud as VLPs was examined using a functional budding assay. Cells were first infected with VvT7 and then transfected with the Mwt or M40 plasmids. VLPs were harvested from the culture medium of Mwt- or M40-transfected BHK-21 cells by ultracentrifugation through a sucrose cushion followed by immunoprecipitation with anti-M MAb. A striking difference in budding efficiency was observed between Mwt and M40, although equivalent amounts of the two proteins were detected in cell lysates (Fig. 1B). Average quantitation data from six independent experiments revealed that the budding efficiency of M40 VLPs was approximately 11-fold greater than that of Mwt VLPs (Fig. 1C). These results indicate that the PPPY motif and flanking amino acids of VSV M can be replaced with the PTAPPEY motif and flanking amino acids of VP40 to generate a more efficient budding protein.
DN VPS4A inhibits budding of VP40 VLPs.
VPS4A is an ATPase and is one of the final effectors in the MVB sorting pathway in cells (1, 5). Recent studies using DN VPS4A proteins indicate that the enzymatic activity of VPS4A is required for efficient budding of HIV-1, MuLV, and Ebola virus (13, 29). We sought to determine whether DN VPS4A would inhibit the budding of viral matrix proteins independent of their L-domain or whether inhibition of budding by DN VPS4A is L-domain specific. We first transfected cells with either VP40-WT or VP40-dPTA mutant, in which the 7PTA9 amino acids have been deleted to disrupt only the PTAP motif. At the same time, we cotransfected cells with either VPS4-WT or DN VPS4-EQ (Fig. 2). VLPs were pelleted from the culture medium, and VP40 was detected by immunoprecipitation. Protein expression controls demonstrate that equivalent amounts of VP40-WT and VP40-dPTA were expressed in transfected cells (Fig. 2A). In contrast, budding of VP40-WT VLPs was reduced by an average of 6.3-fold in the presence of VPS4-WT and more dramatically (670-fold) in the presence of VPS4-EQ (Fig. 2B, lanes 1 to 3). As expected, budding of VP40-dPTA in the absence of VPS4A proteins was reduced compared to that of VP40-WT (compare lanes 1 and 4). The level of VP40-dPTA budding observed is probably due to the remaining presence of the PPEY motif. Interestingly, budding of VP40-dPTA was reduced by an average of fivefold in the presence either VPS4-WT or VPS4-EQ (compare lanes 5 and 6). These results suggest that the enzymatic activity of VPS4A is clearly required for efficient budding of a PTAP-containing VP40 protein (VP40-WT) but is not required for budding of a PTAP-deficient VP40 protein (VP40-dPTA).
FIG. 2.
Effect of DN VPS4A on budding of VP40-WT and VP40-dPTA VLPs. Human 293T cells were cotransfected with a combination of VP40-WT or VP40-dPTA and the indicated VPS4A plasmid (wt or DN EQ mutant). Cell lysates (A) and VLPs (B) were immunoprecipitated with the appropriate antibodies to detect VP40 or VPS4A as indicated. VP40 in VLPs was quantitated by PhosphorImager analysis.
DN VPS4A inhibits the budding of M40 but not of Mwt.
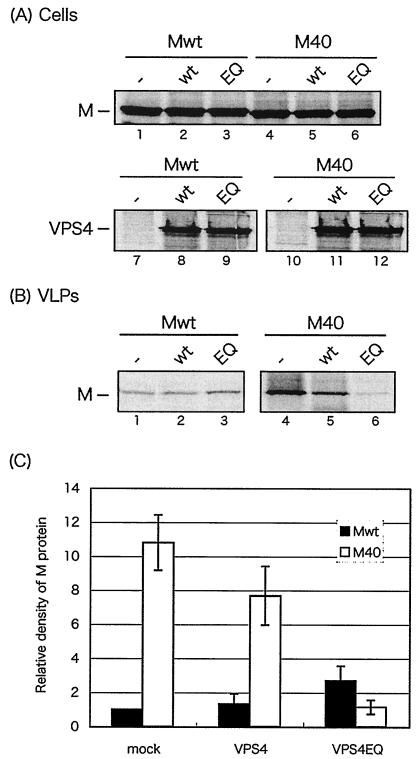
To determine further that inhibition of budding by DN VPS4A occurs in an L-domain-specific manner, we compared the effect of DN VPS4A expression on budding of Mwt and M40 (Fig. 3). BHK-21 cells were first infected with VvT7 and then cotransfected with a combination of Mwt or M40 plus either VPS4-WT or VPS4-EQ. Expression of equivalent amounts of VPS4-WT and VPS4-EQ in cells was readily observed as a control (Fig. 3A, lanes 7 to 12), as were equivalent amounts of Mwt and M40 (lanes 1 to 6). Interestingly, budding of Mwt was unaffected by coexpression of either VPS4-WT or VPS4-EQ (Fig. 3B, lanes 1 to 3). In contrast, budding of M40, although slightly reduced in the presence of VPS4-WT, was more significantly reduced in the presence of VPS4-EQ (lanes 4 to 6). Quantitation of protein expression from three independent experiments revealed that the amount of Mwt in VLPs was virtually unchanged in the absence or presence of VPS4 proteins whereas the amount of M40 in VLPs was reduced approximately 10-fold in cells coexpressing VPS4-EQ (Fig. 3C). These results indicate that budding of M40 was inhibited by DN VPS4A whereas budding of Mwt was not. Taken together, these results suggest that dependence on host VPS4A for efficient budding is regulated, at least in part, by characteristics of the viral L-domain motif.
FIG. 3.
Effect of DN VPS4A on budding of VSV Mwt and M40 VLPs. BHK-21 cells were infected with VvT7 and then cotransfected with the indicated plasmids. Cell lysates (A) and VLPs (B) were immunoprecipitated with the appropriate antibodies to detect VSV M or VPS4A. (C) Mwt and M40 present in VLPs were quantitated, and the bars represent an average of three independent experiments. The level of Mwt in VLP samples from cells not receiving VPS4A (−) was normalized to 1.0.
DN VPS4A does not inhibit budding of VSV or RV.
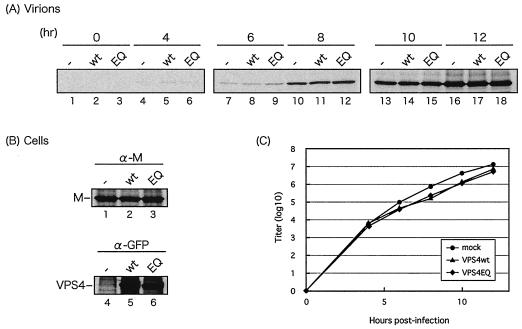
We next sought to determine whether expression of DN VPS4A would inhibit the budding of live virus (Fig. 4). COS-1 cells were first transfected with either VPS4-WT or VPS4-EQ for 24 h and then infected with VSV at an MOI of 0.1 PFU/ml. Medium samples were harvested at 0, 4, 6, 8, 10, and 12 h p.i., and VSV M protein present in the medium was detected by immunoprecipitation (Fig. 4A). M protein was first detected at 4 h p.i. (Fig. 4A, lanes 4 to 6), and its level increased steadily through 12 h p.i. (lanes 16 to 18). Interestingly, identical amounts of M protein were detected at all times p.i. in the absence of VPS4A (lanes 1, 4, 7, 10, 13, and 16), in the presence of VPS4-WT (lanes 2, 5, 8, 11, 14, and 17), and in the presence of VPS4-EQ (lanes 3, 6, 9, 12, 15, and 18). As a control, expression of VPS4-WT and VPS4-EQ was shown to be equivalent in cell lysates, as was expression of M protein following infection with VSV (Fig. 4B). It should be noted that identical results were obtained when human 293T cells were used rather than COS-1 cells (data not shown). In addition to detecting M protein in medium samples by immunoprecipitation, titers of infectious VSV were determined in an identical but separate experiment. COS-1 cells were transfected with the appropriate VPS4A expression plasmid as described above and then infected with VSV at an MOI of 0.1 PFU/ml. Virus yields from both VPS4-WT- and VPS4-EQ-expressing cells were virtually identical to those from mock-transfected cells at all time points examined (Fig. 4C). These results suggest that enzymatic activity of VPS4A is not crucial for budding of infectious VSV, which is in contrast to findings obtained with infectious Ebola virus, HIV-1, and MuLV (12, 27). Whether VSV budding is dependent on VPS4B (a human ortholog of VPS4A) or occurs independently of both VPS4A and VPS4B remains to be determined.
FIG. 4.
Effect of DN VPS4A on VSV budding. (A and B) COS-1 cells were transfected with vector alone (−), VPS4-WT (wt), or VPS4-EQ (EQ). At 24 h p.t., the cells were infected with VSV at an MOI of 0.1 PFU/ml, and culture medium was harvested at 0, 4, 6, 8, 10, and 12 h p.i. Virions (A) and cell lysates (B) were harvested at 12 h p.i. M protein was immunoprecipitated from virions (A), and both M and VPS4A proteins (B) were immunoprecipitated from cell extracts. (C) The virus titer was determined by plaque assay, and titers were plotted against time p.i.
As further proof that VPS4A is not critical for budding of rhabdoviruses, recombinant rabies viruses expressing VPS4A proteins were recovered using reverse genetics and growth curves of recombinant and WT rabies viruses were compared. VPS4-WT, VPS4-EQ, or VPS4-KQ was inserted between the G and L genes in the full-length cDNA clone of rabies virus SPBN (Fig. 5A). These viruses were successfully recovered from DNA by using reverse genetics, and their multistep growth kinetics were examined (Fig. 5B). No significant differences were observed among WT SPBN and the three recombinant viruses, since they exhibited virtually identical (<1.0 log unit difference) growth curves over a 3-day period (Fig. 5B). Thus, results using three different experimental approaches indicate that expression of DN mutants of VPS4A does not significantly affect the replication and budding of VSV and RV.
FIG. 5.
Growth kinetics of recombinant RV expressing VPS4A proteins. (A) Schematic representation of the genome of WT SPBN and recombinant genomes containing VPS4-WT, VPS4-EQ, and VPS4-KQ. All inserts and 5′ and 3′ promoter regions were sequenced. (B) BSR-T7 cells were infected with the indicated viruses at an MOI of 0.01 PFU/ml, and culture medium was harvested at 8, 24, 48, and 72 h p.i. Virus titer at each time point was determined in duplicate on BSR cells and is plotted against time p.i.
Effect of TSG101 depletion on budding of Mwt, M40, and infectious VSV.
TSG101 is the mammalian ortholog of yeast VPS23, which is required for MVB formation in yeast. Mammalian TSG101 functionally interacts with HIV-1 Gag and Ebola virus VP40 in a PTAP-dependent manner (30). We sought to determine whether depletion of TSG101 using siRNAs would affect the budding of Mwt and M40 VLPs and infectious VSV. To ensure that TSG101 siRNA was functioning properly, TSG101-specific siRNA or a nonspecific (NS) siRNA control was transfected into human 293T cells together with a TSG101-expressing plasmid. At 24 h p.t., the cells were transfected a second time with the appropriate siRNA in a similar manner. After an additional 12 h of incubation, the cells were radiolabeled and expression of TSG101 was examined by immunoprecipitation with anti-TSG101 antibody (Fig. 6A). As expected, expression of TSG101 was virtually absent in cells receiving TSG101-specific siRNA but TSG101 was readily detected in cells receiving NS siRNA (Fig. 6A). Thus, efficient shutoff of plasmid-expressed TSG101 was evident, indicating that this siRNA transfection protocol should be sufficient to suppress synthesis of endogenously expressed TSG101.
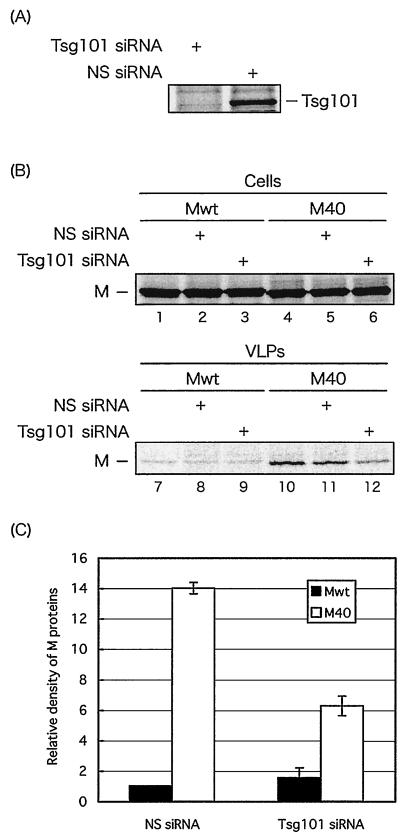
FIG. 6.
Effect of TSG101 depletion on budding of Mwt and M40 VLPs. (A) Human 293T cells were transfected with TSG101-specific or nonspecific (NS) siRNAs together with a TSG101-encoding plasmid (pCAGGS.MCS vector). TSG101 was detected by immunoprecipitation with anti-TSG101 antibody. (B) Immunoprecipitation of Mwt and M40 proteins from cell extracts (lanes 1 to 6) and VLPs (lanes 7 to 12) in the absence of siRNA, in the presence of NS siRNA, or in the presence of TSG101 siRNA as indicated. (C) Quantitation of Mwt and M40 in VLPs from three independent experiments. The level of Mwt protein in VLP samples from NS siRNA-transfected cells was normalized to 1.0.
We compared the budding efficiency of Mwt and M40 VLPs in human 293T cells depleted of TSG101 as described above. As a control, equivalent amounts of Mwt and M40 proteins were detected in the cell extracts (Fig. 6B, lanes 1 to 6). The amount of Mwt detected in VLPs was constant for cells receiving no siRNA, NS siRNA, and TSG101-specific siRNA (Fig. 6B, lanes 7 to 9). In contrast, the number of M40 VLPs budding from cells receiving TSG101-specific siRNA was reduced an average of 2.5-fold compared to that budding from cells receiving NS siRNA (compare lanes 10 and 12). Budding of M40 VLPs from cells receiving either no siRNA or NS siRNA was identical (compare lanes 10 and 11). PhosphorImager analysis was used to quantitate the Mwt and M40 VLPs, and the results of three independent experiments are shown in Fig. 6C. It should be noted that TSG101-specific siRNA inhibited the budding of VP40-WT VLPs by a similar amount (two- to threefold) in a separate experiment (J. M. Licata and R. N. Harty, unpublished data). These results suggest that depletion of TSG101 affects the budding of M40 but does not affect the budding of Mwt in this assay.
We next sought to extend these findings by determining whether depletion of TSG101 would affect the budding of live VSV. Human 293T cells were first transfected with the appropriate siRNAs as described above and then infected with VSV at an MOI of 0.1 PFU/ml. At 10 h p.i., M protein present in the supernatant as well as in cells was detected by immunoprecipitation. In addition, the yield of infectious VSV budding from siRNA-transfected cells was determined by a duplicate plaque assay. Equivalent amounts of M protein were present in siRNA-transfected cells, and no difference was observed in the amount of M protein present in virions budding from cells receiving NS siRNA or TSG101 siRNA (Fig. 7). Moreover, VSV was able to bud equally well from cells receiving TSG101 siRNA (2.6 × 108 PFU/ml), or NS siRNA (2.5 × 108 PFU/ml). Taken together, these results demonstrate that host TSG101 and VPS4A are not critical for budding of rhabdoviruses or rhabdoviral matrix proteins that possess PPxY L-domains.
FIG. 7.
Effect of TSG101 depletion on VSV budding. Human 293T cells were transfected with TSG101-specific or NS siRNAs as indicated and then infected with VSV at an MOI of 0.1 PFU/ml. M protein present in cell lysates and virions was detected by immunoprecipitation. Virus production (10 h p.i.) from cells receiving TSG101-specific or NS siRNA was determined by a duplicate plaque assay on BHK-21 cells.
DISCUSSION
In this study, we performed a functional comparison of the L-domains from Ebola virus VP40 and the VSV M protein. We sought to determine whether these L-domains were functionally interchangeable and whether host proteins VPS4A and TSG101 were functionally relevant to the budding of proteins or viruses containing these different L-domains. It has been reported that viral L-domains are interchangeable and are able to maintain their function in a heterologous background (10, 30). Our results demonstrate that the PTAPPEY motif of Ebola virus VP40 not only functions as an L-domain in the context of the VSV M protein but also results in a 10-fold increase in budding efficiency of the chimeric M40 protein. This result may reflect the fact that the PTAPPEY motif of Ebola virus can utilize the cellular machinery more efficiently than can that of VSV, perhaps due to the presence of overlapping PTAP and PPEY motifs. A similar enhancement of budding was observed by Martin-Serrano et al. after inserting the VP40 motif into the HIV-1 Gag protein (30). Ebola virus may have evolved in such a way as to maintain a remarkably efficient L-domain that functions to promote the rapid release and spread of these very large virus particles. While PTAP and PPxY motifs may ultimately direct viral matrix proteins into the same general pathway for budding, host proteins responsive to PTAP motifs may not necessarily be responsive to PPPY motifs.
A number of cellular factors have been shown to functionally interact with viral L-domains. For example, the PPxY-, PT/SAP-, and YxxL-type L-domains can interact with the WW domains of the Nedd4-like E3 ubiquitin ligases, TSG101, and the clathrin adapter AP-2 and AIP1, respectively (17, 18, 25, 29, 29a, 30, 41, 47, 47a, 52, 52a, 54). Functional interactions between TSG101 and the PTAP motifs present in HIV-1 Gag and Ebola virus VP40 have now been well documented (13, 14, 29, 30, 31, 39, 50, 52). In addition to TSG101, VPS4A is an ATPase that functions downstream in the MVB sorting pathway and also modulates the budding of PTAP-containing HIV-1 and Ebola virus (13, 29). Our results using three different approaches indicate that TSG101 and VPS4A are not crucial for efficient budding of M protein alone or for budding of VSV and RV. However, we cannot rule out a role for the VPS4B protein in rhabdovirus budding. The dependence on TSG101 and VPS4A for efficient budding maps to the viral L-domain present, since budding of the M40 chimeric protein containing the PTAPPEY motif from VP40 in a VSV M background was responsive to the presence or absence of TSG101 and VPS4A. As expected, budding of full-length VP40-WT was also sensitive to inhibition by depletion of TSG101 (data not shown) and by DN VPS4A mutants (Fig. 2). Interestingly, the decreased level of budding of the VP40-dPTA mutant was not affected by coexpression of DN VPS4A (Fig. 2). Hence like the rhabdoviral M proteins, VP40-dPTA contains only a PPxY motif and was insensitive to coexpression of DN VPS4A. This may suggest that the overlapping PTAP and PPEY motifs of VP40 are utilized to recruit different cellular proteins and machinery for budding. Ebola virus may be capable of utilizing more than one cellular pathway for budding due to the overlapping nature of its L-domains, whereas the rhabdoviruses would probably be restricted to a PPxY-dependent pathway. These findings suggest that replacement of the PPxY motifs of either VSV or RV with the PTAPPEY motif of Ebola virus may result in recombinant VSV or RV that buds more efficiently than WT viruses do. It would be of interest to determine whether such recombinant rhabdoviruses would become dependent on or remain independent of host TSG101 and VPS4A proteins for budding. These experiments using VSV and RV reverse-genetic systems are currently in progress. Conversely, replacement of the PTAPPEY motif of Ebola virus with a simple rhabdovirus-like PPxY motif may result in attenuation of Ebola virus budding. A PPxY-containing Ebola virus may no longer be capable of interacting efficiently with host proteins TSG101 and/or VPS4A, leading to a reduction in virus yield. Such findings may have important implications for future vaccine and antiviral strategies for Ebola virus.
It is known that PPxY-type motifs can interact with WW domains of cellular proteins such as Nedd4-like E3 ubiquitin ligases (17, 18, 25, 50, 54, 55). A role for ubiquitin and ubiquitin ligases in virus budding and in protein-sorting pathways involving TSG101 and VPS4A has been documented (9, 17, 18, 24, 25, 34-38, 42, 46, 50, 55). PPxY-containing viruses such as VSV and RV may preferentially depend on host proteins such as Nedd4 for efficient budding, rather than on TSG101 and VPS4A. More subtle mutations of the PTAPPEY sequence of VP40 are being generated to elucidate further the function of the PTAP sequence versus the PPEY sequence in mediating interactions with host proteins to promote budding (Licata and Harty, unpublished). Since many of the matrix proteins of RNA viruses contain more than one L-domain motif (15, 26, 29, 48), it is important to determine whether one or multiple L-domains are necessary for virus budding. VSV M protein contains a PSAP motif downstream of the PPPY motif; however, a role for the PSAP motif in budding remains unclear. Since depletion of TSG101 did not affect the budding of Mwt or VSV, a biologically relevant interaction between VSV PSAP and host TSG101 is unlikely. In addition, we have recently recovered a PSAP mutant of VSV by reverse genetics, and preliminary findings indicate that the PSAP mutant virus replicates and buds just as well as WT VSV from BHK-21 cells (T. Irie and R. N. Harty, unpublished data).
Although many published reports have focused on the functional similarities of viral L-domains and mechanisms of RNA virus budding, it is not surprising that important differences have also been noted (11, 53, 56; also see above). For example, the L-domain of HIV-1 Gag functions in a cell type-dependent manner (11), and not all L-domains are functionally interchangeable (53, 56). In addition, certain L-domains can functionally replace the positively charged region within influenza virus M1 protein, whereas others cannot (20). Finally, viruses such as Ebola virus, HIV-1, and influenza A virus appear to bud preferentially from lipid-raft microdomains whereas rhabdoviruses appear to bud from nonraft membrane domains (6, 8, 33, 44). Such differences in the budding sites of RNA viruses may correlate with the need for different cellular proteins and/or pathways required for virus budding.
In summary, our findings suggest that in addition to many similarities, there are fundamental differences in the budding mechanisms of rhabdoviruses and filoviruses. The composition of the viral L-domain and perhaps flanking sequences appears to dictate which host proteins and machinery will be important for efficient budding. Experiments to identify proteins other than TSG101 and VPS4A (e.g., Nedd4-like ubiquitin ligases and ubiquitin itself) that may be important for the budding of PPxY-containing rhabdoviruses are under way.
Acknowledgments
We thank members of the Harty laboratory for support, Shiho Irie for technical assistance, and J. Paragas for critical reading of the manuscript.
J.M.L. is supported by NIH training grant T32-AI007324. M.J.S. is supported by NIH grant AI49153. R.N.H. is supported by NIH grants AI46499 and AI53393. R.N.H. and T.I. are supported by a Health Research Faculty development block grant.
REFERENCES
- 1.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17**:**2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3**:**271-282. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3**:**283-289. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1**:**248-258. [DOI] [PubMed] [Google Scholar]
- 5.Babst, M., T. K. Sato, L. M. Banta, and S. D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16**:**1820-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195**:**593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, N., and P. Woodman. 2000. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276**:**11735-11742. [DOI] [PubMed] [Google Scholar]
- 8.Brown, E. L., and D. S. Lyles. 2003. Organization of the vesicular stomatitis virus glycoprotein into membrane microdomains occurs independently of intracellular viral components. J. Virol. 77**:**3985-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conibear, E. 2002. An ESCRT into the endosome. Mol. Cell 10**:**215-216. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73**:**3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76**:**105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76**:**4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107**:**55-65. [DOI] [PubMed] [Google Scholar]
- 14.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77**:**6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77**:**9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73**:**2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implication for filovirus budding. Proc. Natl. Acad. Sci. USA 97**:**13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75**:**10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expression protease. J. Virol. 69**:**6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui, E. K., S. Barman, T. Y. Yang, and D. P. Nayak. 2003. Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motif. J. Virol. 77**:**7078-7092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75**:**5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74**:**9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Justice, P. A., W. Sun, Y. Li, Z. Ye, P. R. Grigera, and R. R. Wagner. 1995. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J. Virol. 69**:**3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106**:**145-155. [DOI] [PubMed] [Google Scholar]
- 25.Kikonyogo, A.,F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98**:**11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76**:**10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76**:**1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., L. Luo, M. Schubert, R. R. Wagner, and C. Y. Kang. 1993. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J. Virol. 67**:**4415-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Licata J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77**:**1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Martin-Serrano, J., A. Yaravoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor protein. Proc. Natl. Acad. Sci. USA 100**:**12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7**:**1313-1319. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77**:**4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGettigan, J. P., S. Sarma, J. M. Orenstein, R. J. Pomerantz, and M. J. Schnell. 2001. Expression and immunogenicity of human immunodeficiency virus type 1 Gag expressed by a replication-competent rhabdovirus-based vaccine vector. J. Virol. 75**:**8724-8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74**:**3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278**:**111-121. [DOI] [PubMed] [Google Scholar]
- 35.Ott, D. E., L. V. Coren, R. C. Sowder, Jr., J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76**:**3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, R. C. Sowder, Jr., J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77**:**3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, Jr., Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72**:**2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97**:**13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9**:**812-817. [DOI] [PubMed] [Google Scholar]
- 40.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71**:**6541-6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72**:**10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15**:**446-455. [DOI] [PubMed] [Google Scholar]
- 43.Sakalian, M., and E. Hunter. 1998. Molecular events in the assembly of retrovirus particles. Adv. Exp. Med. Biol. 440**:**329-339. [DOI] [PubMed] [Google Scholar]
- 44.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274**:**2038-2044. [DOI] [PubMed] [Google Scholar]
- 45.Schnell, M. J., H. D. Foley, C. A. Siler, J. P. McGettigan, B. Dietzschold, and R. J. Pomerantz. 2000. Recombinant rabies virus as potential live-viral vaccines for HIV-1. Proc. Natl. Acad. Sci. USA 97**:**3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97**:**13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97**:**13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114**:**689-699. [DOI] [PubMed] [Google Scholar]
- 48.Strecker, T., R. Eichler, J. ter Meulen, W. Weissenhorn, H. Dieter Klenk, W. Garten, and O. Lenz. 2003. Lassa virus z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 77**:**10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanzi, G. O., A. J. Piefer, and P. Bates. 2003. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J. Virol. 77**:**8440-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326**:**493-502. [DOI] [PubMed] [Google Scholar]
- 51.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of Ebola virus matrix protein VP40. Virology 283**:**1-6. [DOI] [PubMed] [Google Scholar]
- 52.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98**:**7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H.-Y. Chung, E. Morita, H. E. Wang, T. Davis, G.-P. He, D. M. Cimbora, A. Scott, H.-G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114**:**701-713. [DOI] [PubMed] [Google Scholar]
- 53.Xiang, Y., C. E. Cameron, and J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76 gag late assembly domain. J. Virol. 70**:**5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3**:**636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77**:**9987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74**:**7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18**:**4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]