Dimerization of Recombinant Tobacco Mosaic Virus Movement Protein (original) (raw)
Abstract
The p30 movement protein (MP) is essential for cell-to-cell spread of tobacco mosaic virus in planta. We used anion-exchange chromatography and preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to obtain highly purified 30-kDa MP, which migrated as a single band in native PAGE. Analytical ultracentrifugation suggested that the protein was monodisperse and dimeric in the nonionic detergent _n_-octyl-β-d-glucopyranoside. Circular dichroism (CD) spectroscopy showed that the detergent-solubilized protein contained significant α-helical secondary structure. Proteolysis of the C-tail generated a trypsin-resistant core that was a mixture of primarily monomers and some dimers. We propose that MP dimers are stabilized by electrostatic interactions in the C terminus as well as hydrophobic interactions between putative transmembrane α-helical coiled coils.
The 30-kDa movement protein (MP) is a nonstructural protein that is encoded by the single-stranded RNA genome of tobacco mosaic virus (TMV) (24). The MP is required for cell-to-cell spread of TMV infection (10, 26, 32) and participates in systemic spread of disease (2).
MPs of many plant viruses are associated with plasmodesmata, the pores that provide intercellular (symplastic) connections between plant cells (1, 14, 15, 25, 30). TMV infection results in a temporary increase in the size exclusion limit of plasmodesmata from ∼0.4 to ∼20 kDa in tobacco leaf epidermis and mesophyll tissues (29, 41). The MP is required for increased intercellular permeability, but the mechanisms responsible for increased size exclusion limits are unclear (32). Associated proteins such as pectin methylesterase may interact with MP to facilitate cell-to-cell spread of infection (8, 11).
Many plant and animal viruses form replication complexes in association with cellular membranes such as the endoplasmic reticulum (ER) (14-16, 22, 23, 25, 33, 36, 40). The MP appears to promote the aggregation of ER during the formation of virus replication “factories” (15, 18, 20, 22, 23). Consistent with this hypothesis, the MP behaves as an intrinsic membrane protein with a tendency to self-aggregate (6, 27, 33). However, molecular mechanisms of viral replication in association with membranes are unknown.
Recombinant viral MPs expressed in Escherichia coli typically form insoluble inclusion bodies. To obtain purified recombinant MP for biophysical analysis, we previously used anion-exchange chromatography to remove contaminating RNA. This one-step procedure yielded milligram quantities of MP contained in two peaks designated P1 and P2. Hydropathy analysis, circular dichroism (CD) spectroscopy, mass spectrometry, and proteolytic digestion experiments suggested that the MP solubilized in 0.1% sodium dodecyl sulfate (SDS) and 2 M urea is a polytopic, α-helical membrane protein (6). In the present study, biophysical analysis of highly purified TMV MP suggests that the full-length protein associates as a dimer in the nonionic detergent _n_-octyl-β-d-glucopyranoside (βOG).
MATERIALS AND METHODS
Preparation of full-length MP (MP-FL).
E. coli pET3MP was used to express recombinant MP in 4 liters of culture per preparation (6). In most experiments, lysozyme (1.0 mg/ml) was included in the buffer used to resuspend bacterial pellets. Inclusion bodies were isolated, solubilized, and subjected to anion-exchange chromatography (6). MP eluted in two peaks, P1 and P2, and the fractions containing purified MP were pooled and concentrated to ∼10 mg of protein per ml by ultrafiltration (6). Protein concentrations were estimated by comparison with bovine serum albumin standards in digital scans of Coomassie-stained proteins separated via SDS-polyacrylyamide gel electrophoresis (PAGE) gels (Adobe Systems, Inc., San Jose, Calif.).
Preparative SDS-PAGE was used to remove the small fraction of contaminating fragments and aggregates of MP that remained in P1 and P2. Samples containing ∼10 to ∼25 mg of protein in ∼1 to ∼2.5 ml were mixed with 1/2 volume of 3× SDS-PAGE sample buffer (35) and warmed to 40°C for 8 min. The manufacturer's instructions were followed for preparative SDS-PAGE (model 491 Prep Cell; Bio-Rad Inc., Hercules, Calif.), except that the stacking gel volume was fourfold larger than the sample volume. Preparative separating gels were formed from 80 ml of 10% polyacrylamide, and stacking gels were formed from 4% polyacrylamide. (Separating and stacking gels were cast with 37.5:1 acrylamide-bisacrylamide.) Preparative electrophoresis was performed at 11.5 or 12.0 W. Immediately following elution of the tracking dye, 2.5-ml fractions were collected at an elution rate of 1 ml/min. Aliquots (12 μl) of the fractions were subjected to analytical (slab [described below]) SDS-PAGE to identify preparative SDS-PAGE fractions containing MP-FL. The MP-FL fractions were pooled, dialyzed extensively against TNEM2MU buffer (10 mM Tris [pH 9.0], 500 mM NaCl, 5 mM EDTA, 1 mM 2-mercaptoethanol, and 2 M urea) containing 0.1% SDS, and then concentrated to ∼10 mg of protein per ml by ultrafiltration as described above.
The protein was then dialyzed stepwise at 22°C from TNEM2MU buffer containing 0.1% SDS into TN buffer (10 mM Tris [pH 9.0], 400 mM NaCl) containing 0.05% (wt/vol) βOG detergent (Calbiochem, Inc., La Jolla, Calif.). The concentration of urea was halved every 2 h until the dialysis buffer contained 0.062 M urea-0.003% SDS. The protein was then dialyzed against TN buffer containing 0.05% βOG (lacking urea and SDS) overnight at 22°C. The contents of dialysis bags were subjected to centrifugation at 22°C for 15 min at 15,000 × g. The protein concentration of the supernatant was estimated as described above and was adjusted to ∼8 mg/ml by dilution with TN buffer containing 0.05% βOG. The purified MP-FL was divided into 100-μl aliquots, flash-frozen in liquid nitrogen, and stored at −80°C until needed. For biochemical studies, samples were thawed at 40°C for 5 min. MP was stable at 22°C but tended to aggregate with time when stored at 4°C.
Analytical SDS-PAGE, Western immunoblotting, and native PAGE.
Analytical SDS-PAGE and Western immunoblotting were performed as described previously (6). For native PAGE, we used 4 to 20% precast Novex Tris-glycine polyacrylamide minigels (Invitrogen, Carlsbad, Calif.). MP samples in TN buffer containing 0.05% βOG were mixed 1:1 (vol/vol) with 2× Novex native gel sample buffer. Native gel standards (Sigma Chemical Co., St. Louis, Mo.) were prepared according to the manufacturer's instructions. The electrode buffer contained 25 mM Tris base-192 mM glycine. Following electrophoresis, proteins were stained with Coomassie brilliant blue R-250.
Analytical ultracentrifugation.
Sedimentation velocity and equilibrium analytical ultracentrifugation of MP-FLP1 and MP-FLP2 were performed in TN buffer containing 0.05% βOG. Data were collected on a temperature-controlled Beckman XL-I analytical ultracentrifuge equipped with an An60 Ti rotor and photoelectric scanner (Beckman Instruments, Inc., Palo Alto, Calif.). A double-sector cell equipped with a 12-mm Epon centerpiece and sapphire windows was loaded with ∼400 μl of sample by using a Hamilton syringe. Rotor speeds were 3,000 to 50,000 rpm in continuous mode at 20°C, with a step size of 0.005 cm, employing an average of 1 scan per point. Analysis by a time derivative method using the DCDT+ program (31, 38) yielded the distribution of soluble species, which was represented by a range of s values.
Hydrodynamic molecular weights of the samples were confirmed by sedimentation equilibrium using the same analytical ultracentrifuge. The reference compartment was loaded with TN buffer containing 0.05% βOG (140 μl). Samples (100 μl) were monitored for _A_260 at 3,000 to 20,000 rpm (for MP-FLP1 and MP-FLP2) and 3,000 to 27,000 rpm (for MP-CP1) at 20°C. Nonlinear least-squares analysis of single species and two species models was performed using Origin software (Microcal Software, Inc., Northampton, Mass.).
Isolation of the core domain of MP.
To isolate the protease-resistant core of MP (MP-C), chromatographic peak P1 was prepared in TNEM2MU buffer containing 0.1% SDS. The protein was gradually dialyzed against an increasing percentage of TN buffer containing 0.05% βOG as described above. After a preliminary study to establish digestion conditions, modified trypsin (Promega, Inc., Madison, Wis.) was added to 10 to 20 mg of MP in 1 to 2 ml with a 1:500 (wt/wt) ratio of trypsin to MP. Proteolysis was stopped after 10 to 20 min by reactions with 1/2 volume of 3× SDS-PAGE sample buffer. Samples were heated to 40°C for 8 min, and preparative SDS-PAGE was performed as described above. Fractions containing MP-CP1 domain were identified, pooled, and processed as described above for MP-FL.
CD spectroscopy.
CD spectra were recorded with an AVIV model 202SF CD spectrometer equipped with a Peltier temperature-controlled cell holder. MP samples at 1 mg/ml in TN buffer containing 0.05% βOG were placed in a Suprasil quartz cell with a path length of 0.1 cm (Hellma, Forest Hills, N.Y.). CD spectra were recorded at wavelengths from 200 to 250 nm at 25°C in 0.5-nm steps with an averaging time of 2.0 s per step. Molar ellipticity was calculated by the formula [θ]λ = (θ × MW)/(10_cl_), where θ is the measured ellipticity in degrees at a given wavelength (λ), MW is the molecular mass of MP (30 kDa for MP-FL monomer and 28 kDa for MP-C), c is the protein concentration in milligrams per milliliter, and l is the light path length in centimeters.
RESULTS
Isolation of MP-FL.
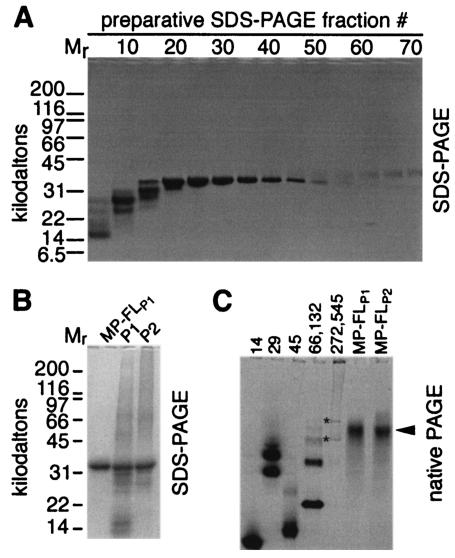
Anion-exchange chromatography of recombinant MP yields peaks designated P1 and P2 that are free of detectable RNA and are highly purified (6). However, they do contain a small fraction of proteolytic fragments and aggregates of MP (Fig. 1B, lanes P1 and P2). Proteolytic degradation was minimized (but not eliminated) by inclusion of 1 mg of lysozyme per ml in the buffer used for resuspension of the bacterial pellet (data not shown). Because analytical SDS-PAGE is capable of separating the MP-FL from fragments and aggregates (Fig. 1) and SDS apparently does not denature the protein (6), we used preparative SDS-PAGE to isolate highly purified MP-FL (Fig. 1A and B). Analytical SDS-PAGE (Fig. 1A) was used to identify fractions containing isolated MP-FL. Pooled samples that contained concentrated MP-FLP1 (Fig. 1B) and MP-FLP2 (not shown) displayed a single band of the predicted mobility. Although not seen in Fig. 1B, discrete bands with reduced mobility were frequently detected in MP-FLP1 and MP-FLP2.
FIG. 1.
Preparative SDS-PAGE was used to isolate soluble MP-FL. (A) SDS-PAGE of every fifth fraction. (B) MP-FLP1 contained isolated MP monomer, in contrast with chromatographic peaks P1 and P2 (6), which contained MP aggregates, monomer, and fragments. Mr, molecular mass standards in kilodaltons (A and B). (C) Migration of MP-FLP1 and MP-FLP2 in native PAGE suggested that the MP was essentially monodisperse. Numbers above the first five lanes denote the molecular masses of protein standards (in kilodaltons): 14, α-lactoglobulin; 29, carbonic anhydrase; 45, egg white albumin; 66 and 132, BSA monomers and dimers (and some higher-order oligomers); 272 and 545, urease trimers and hexamers. Lanes containing MP-FLP1 or MP-FLP2 are indicated. Asterisks mark urease trimer and hexamer bands. An arrowhead points to the MP. Note that migration was not necessarily proportional to the molecular mass in native PAGE (panel C only). Gels were stained with Coomassie brilliant blue R-250.
Soluble MP-FL is primarily dimeric.
We used dialysis to remove urea and SDS from the sample and to solubilize MP-FLP1 and MP-FLP2 in the nonionic detergent βOG (0.05% [wt/vol]). The dialysis began at ∼50% of the SDS critical micelle concentration and ended with dialysis against a buffer lacking SDS, so we expect that the SDS had been largely replaced by βOG. Nevertheless, there may still be some SDS molecules that are tightly bound to the protein. We note that other membrane proteins, such as aquaporin AqpZ from E. coli (4), the Streptomyces lividans potassium channel homolog (9), phospholamban from cardiac sarcoplasmic reticulum (3), and glycophorin A from human erythrocyte membranes (12, 21), retain their oligomeric state and secondary structure in SDS. Indeed, MP displayed a remarkable degree of α-helical secondary structure in the presence of 2 M urea and 0.1% SDS (6). Therefore, even if some tightly bound SDS molecules remain with the protein in βOG buffer, we expect that our physicochemical measurements are relevant to understanding the structure of MP. Native PAGE showed single bands of similar mobility for MP-FLP1 and MP-FLP2 (Fig. 1C). Thus, native gel electrophoresis demonstrated that MP-FLP1 and MP-FLP2 are monodisperse in TN buffer containing 0.05% βOG. However, native PAGE does not provide reliable estimates of molecular mass, especially with highly basic proteins such as the TMV MP.
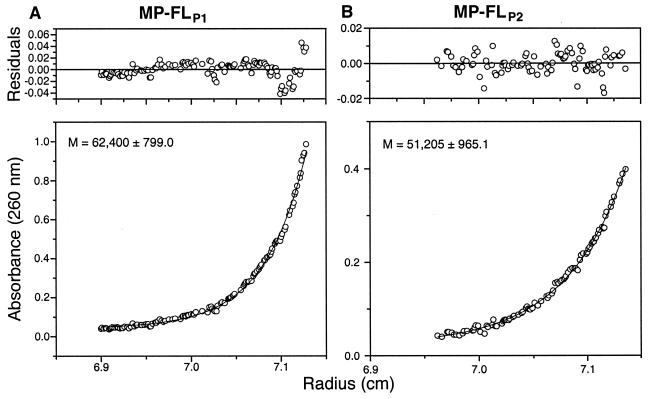
Analytical ultracentrifugation can provide good estimates of molecular mass and dispersity. Equilibrium analytical ultracentrifugation suggested that MP-FLP1 and MP-FLP2 were monodisperse and had mean molecular masses of ∼62 and ∼51 kDa, respectively (Fig. 2). Because TMV MP monomers have a molecular mass of 30 kDa (6, 13), MP-FLP1 and MP-FLP2 presumably associate as dimers in TN buffer containing 0.05% βOG. The modest difference in molecular mass estimated from analytical ultracentrifugation may be due to slight conformational differences between MP-FLP1 and MP-FLP2 and/or differences in bound detergent. Sedimentation velocity analytical ultracentrifugation also suggested that MP-FLP1 and MP-FLP2 are dimeric (data not shown).
FIG. 2.
Equilibrium analytical ultracentrifugation of MP-FLP1 and MP-FLP2. Residuals (upper panels) and _A_260 (lower panels) versus radius are shown. Molecular mass estimates are also shown for MP-FLP1 (A) and MP-FLP2 (B). The data are fitted very well by a one-state model, suggesting that the 30-kDa MP-FL is monodisperse and dimeric in TN buffer containing 0.05% βOG.
A trypsin-resistant core domain lacks the C-tail and is primarily monomeric.
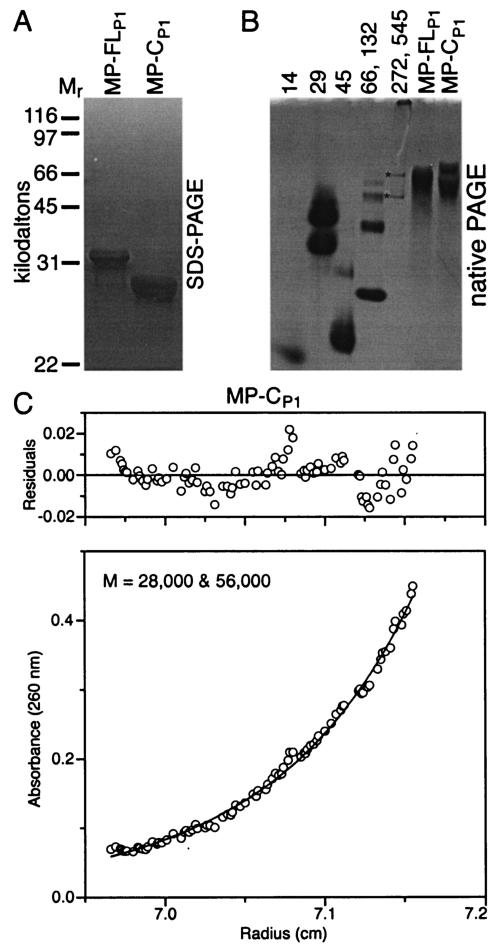
Proteolysis of chromatographic peaks P1 and P2 generates a trypsin-resistant core domain (MP-C) (6). MP-CP1 was isolated by preparative SDS-PAGE and displayed the expected mobility (∼28 kDa) by analytical SDS-PAGE (Fig. 3A). In native PAGE, MP-CP1 exhibited two dominant bands (Fig. 3B). Equilibrium analytical ultracentrifugation experiments strongly supported a two-species monomer-dimer model with molecular masses of 28 and 56 kDa (Fig. 3C) in which the monomer was the dominant molecular species. Sedimentation velocity analytical ultracentrifugation also strongly supported the two-species model (data not shown).
FIG. 3.
Isolated core domain of MP from P1 (MP-CP1) was composed of monomers and dimers. Migration of preparations of MP-FLP1 and MP-CP1 in analytical SDS-PAGE (A) and native PAGE (B). Protein standards and asterisks are as described in the legend to Fig. 1. (C) Equilibrium analytical ultracentrifugation of MP-CP1. The results are fitted very well by a two-species model, suggesting that the MP core exists primarily as monomers and some dimers.
MP in βOG has α-helical secondary structure.
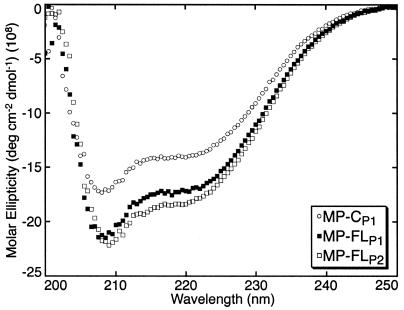
CD spectroscopy was used to compare the secondary structure content of MP-CP1, MP-FLP1, and MP-FLP2 in the detergent βOG. The minima at 208 and 222 nm suggested that the MP preparations exhibited a high content of α-helix (Fig. 4) (7). The CD spectra of MP-FLP1 and MP-FLP2 were similar to those of P1 and P2 in 0.1% SDS and 2 M urea (6). Although CD spectroscopy is not a quantitative technique, the α-helical content of MP-CP1 appeared to be slightly lower than that of MP-FLP1 and MP-FLP2 (Fig. 4).
FIG. 4.
CD spectroscopy suggested that the α-helical content of MP-CP1 was slightly lower than that of MP-FLP1 and MP-FLP2. MP was at 1 mg/ml in TN buffer containing 0.05% βOG. Molar ellipticity is shown from 200 to 250 nm; shorter-wavelength data were unreliable due to UV absorption by the buffer.
DISCUSSION
When TMV-MP is produced in E. coli, it accumulates in inclusion bodies along with RNA. However, the RNA was removed by anion-exchange chromatography, revealing two peaks, designated P1 and P2, both of which contained highly purified MP (6). Characterization of the protein by hydropathy analysis, CD spectroscopy, mass spectrometry, and proteolytic digestion experiments supported a model in which MP is an integral membrane protein with two transmembrane α-helices, a flexible, proteolytically sensitive C-terminal tail and a tightly folded “core” that is resistant to proteolysis. In the present study, we used preparative SDS-PAGE to obtain highly purified, homogenous preparations of MP-FLP1, MP-FLP2, and a trypsin-resistant core of MP-FLP1, named MP-CP1. The estimated molecular mass of the MP-FLP2 dimer (∼51 kDa) was lower than that of MP-FLP1 (∼62 kDa) (Fig. 2), possibly due to tighter folding or a change in bound detergent.
CD spectroscopy showed that MP retains ordered α-helical structure in the presence of chaotropes such as urea and SDS (6). Such resistance to denaturation has been observed for some soluble proteins such as staphylococcal nuclease, which retains “native-like” structure in the presence of 8 M urea (37). Some membrane proteins, including MP, also resist denaturation by chaotropes (4, 6, 9, 12, 17). Nevertheless, for further biophysical characterization, we transferred the purified MP from chaotropic buffers containing 0.1% SDS and 2 M urea into a buffer containing the nonionic detergent βOG.
Similar to our previous experiments with buffer containing 1% Tween 20 (in which MP demonstrated lower solubility and increased polydispersity), trypsin treatment of MP-FL in TN buffer containing 0.05% βOG released a core polypeptide with a molecular mass of ∼28 kDa by SDS-PAGE. Resistance to trypsin implies structural stability of this core. We believe that the core, which contains abundant lysine and arginine residues at its C terminus, is compact. Acidic regions near the carboxy terminus of the MP have been designated regions A and C, while a basic region was designated region B (Fig. 5) (34). Trypsin removed region C from the monomer to yield the core (6).
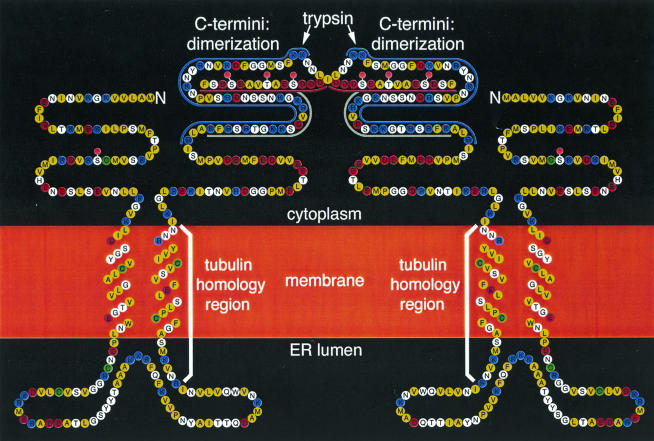
FIG. 5.
Topological model of the MP dimer refined from that of Brill et al. (6). Hydrophobic amino acid residues are yellow, basic residues are blue, acidic residues are red, and Cys residues are green. The trypsin-resistant core contains the first 249 or 250 amino acid residues, including the peptide (gray bar) that was used to produce anti-MP antibodies used previously. The acidic C terminus (residues 249 to 268) was rapidly removed by trypsin (6) to yield the trypsin-resistant core domain (Fig. 3 and 4). As defined by Saito et al. (34), domain C (residues 252 to 268) is acidic (red bar) and domain B (residues 206 to 250) is basic (blue bar). Cytoplasmic, transmembrane, ER lumenal, and core domains were previously proposed (6). Transmembrane domains are presumed to be α-helical. Ser37 (19), Ser258, Thr261, and Ser265 (39) were reported to be phosphorylation sites (pink circles). Because dimerization (Fig. 3C) and α-helicity (Fig. 4) decrease as the full-length protein is converted to the core lacking the C terminus, we propose that the acidic C terminus participates in dimerization. Dimerization could be mediated in part by charge-charge interactions between acidic region C and basic region B of another monomer.
Analytical ultracentrifugation experiments showed that MP-FLP1 and MP-FLP2 solubilized in 0.05% βOG were dimeric (Fig. 2), whereas MP-CP1 was predominantly monomeric, with a small fraction of dimers (Fig. 3C). MP-CP1 also had a somewhat lower α-helical content than did MP-FL (Fig. 4). Taken together, these results support a model in which interactions between positively charged residues in region B and negatively charged residues in region C may participate in the stabilization of dimers and α-helices of the MP (Fig. 5). Residual dimers in MP-CP1 suggest that the transmembrane domains may participate in stabilizing MP dimers, possibly as a dimer of antiparallel α-helical coiled coils, analogous to the hepatitis core capsid protein (42).
We proposed previously that MP residues 150 to 169 span ER membranes (6). In contrast, Boyko et al. (5) proposed that residues 144 to 169 include a region of homology with tubulin and may participate in lateral contacts with or within microtubules. It is possible that residues ∼144 to ∼169 of the MP interact with microtubules and membranes at different stages of TMV infection.
Plant virus MPs may generally interact with cellular macromolecules through hydrophobic interactions (28). The TMV MP is known to be associated with membranes in vivo (22, 27, 33). Oligomerization of the MP may play a role in the aggregation of ER-containing TMV replication complexes. The MP may also interact with and anchor other proteins associated with replication complexes, such as TMV replicase and coat protein (15, 22).
Acknowledgments
We thank Anchi Cheng and Masaaki Fujiki for help with some of the experiments, James R. Williamson for use of a Biocad perfusion chromatography workstation, and Jeffrey Kelley for use of analytical ultracentrifugation equipment.
Support was provided by an NIH-NRSA postdoctoral fellowship (L.M.B.), a Skaggs Foundation Fellowship (S.D.), an HHMI postdoctoral fellowship (B.D.), NIH grant GM066087 (M.Y.), NSF grant MCB 9631124, NIH grant GM62979 (R.N.B.), and the Donald Danforth Plant Science Center (R.N.B.). During the course of this work, M.Y. was an Established Investigator of the American Heart Association sponsored by Bristol-Myers Squibb and is now the recipient of a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund.
REFERENCES
- 1.Aaziz, R., S. Dinant, and B. L. Epel. 2001. Plasmodesmata and plant cytoskeleton. Trends Plant Sci. 6**:**326-330. [DOI] [PubMed] [Google Scholar]
- 2.Arce-Johnson, P., U. Reimann-Philipp, H. S. Padgett, R. Rivera-Bustamante, and R. N. Beachy. 1997. Requirement of the movement protein for long distance spread of tobacco mosaic virus in grafted plants. Mol. Plant-Microbe Interact. 10**:**691-699. [Google Scholar]
- 3.Arkin, I. T., P. D. Adams, K. R. MacKenzie, M. A. Lemmon, A. T. Brünger, and D. M. Engelman. 1994. Structural organization of the pentameric transmembrane α-helices of phospholamban, a cardiac ion channel. EMBO J. 13**:**4757-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borgnia, M. J., D. Kozono, G. Calamita, P. C. Maloney, and P. Agre. 1999. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J. Mol. Biol. 291**:**1169-1179. [DOI] [PubMed] [Google Scholar]
- 5.Boyko, V., J. Ferralli, J. Ashby, P. Schellenbaum, and M. Heinlein. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2**:**826-832. [DOI] [PubMed] [Google Scholar]
- 6.Brill, L. M., R. S. Nunn, T. W. Kahn, M. Yeager, and R. N. Beachy. 2000. Recombinant tobacco mosaic virus movement protein is an RNA-binding, α-helical membrane protein. Proc. Natl. Acad. Sci. USA 97**:**7112-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C. T., C.-S. C. Wu, and J. T. Yang. 1978. Circular dichroic analysis of protein conformation: inclusion of the β-turns. Anal. Biochem. 91**:**13-31. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M.-H., J. Sheng, G. Hind, A. K. Handa, and V. Citovsky. 2000. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19**:**913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes, D. M., and E. Perozo. 1997. Structural dynamics of the Streptomyces lividans K+ channel (SKC1): oligomeric stoichiometry and stability. Biochemistry 36**:**10343-10352. [DOI] [PubMed] [Google Scholar]
- 10.Deom, C. M., M. J. Oliver, and R. N. Beachy. 1987. The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science 237**:**389-394. [DOI] [PubMed] [Google Scholar]
- 11.Dorokhov, Y. L., K. Mäkinen, O. Y. Frolova, A. Merits, J. Saarinen, N. Kalkkinen, J. G. Atabekov, and M. Saarma. 1999. A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett. 461**:**223-228. [DOI] [PubMed] [Google Scholar]
- 12.Furthmayr, H., and V. T. Marchesi. 1976. Subunit structure of human erythrocyte glycophorin A. Biochemistry 15**:**1137-1144. [DOI] [PubMed] [Google Scholar]
- 13.Goelet, P., G. P. Lomonossoff, P. J. G. Butler, M. E. Akam, M. J. Gait, and J. Karn. 1982. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. USA 79**:**5818-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinlein, M., B. L. Epel, H. S. Padgett, and R. N. Beachy. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270**:**1983-1985. [DOI] [PubMed] [Google Scholar]
- 15.Heinlein, M., H. S. Padgett, J. S. Gens, B. G. Pickard, S. J. Casper, B. L. Epel, and R. N. Beachy. 1998. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10**:**1107-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, M., and L. Zhang. 1999. Association of the movement protein of alfalfa mosaic virus with the endoplasmic reticulum and its trafficking in epidermal cells of onion bulb scales. Mol. Plant-Microbe Interact. 12**:**680-690. [Google Scholar]
- 17.Imagawa, T., T. Watanabe, and T. Nakamura. 1986. Subunit structure and multiple phosphorylation sites of phospholamban. J. Biochem. (Tokyo) 99**:**41-53. [DOI] [PubMed] [Google Scholar]
- 18.Kahn, T. W., M. Lapidot, M. Heinlein, C. Reichel, B. Cooper, R. Gafny, and R. N. Beachy. 1998. Domains of the TMV movement protein involved in subcellular localization. Plant J. 15**:**15-25. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami, S., H. S. Padgett, D. Hosokawa, Y. Okada, R. N. Beachy, and Y. Watanabe. 1999. Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J. Virol. 73**:**6831-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11**:**535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science 276**:**131-133. [DOI] [PubMed] [Google Scholar]
- 22.Más, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147**:**945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Más, P., and R. N. Beachy. 2000. Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc. Natl. Acad. Sci. USA 97**:**12345-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews, R. E. F. 1991. Plant virology, 3rd ed. Academic Press, New York, N.Y.
- 25.McLean, B. G., J. Zupan, and P. C. Zambryski. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7**:**2101-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meshi, T., Y. Watanabe, T. Saito, A. Sugimoto, T. Maeda, and Y. Okada. 1987. Function of the 30 kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J. 6**:**2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, P. J., C. A. Fenczik, C. M. Deom, and R. N. Beachy. 1992. Developmental changes in plasmodesmata in transgenic tobacco expressing the movement protein of tobacco mosaic virus. Protoplasma 170**:**115-127. [Google Scholar]
- 28.Mushegian, A. R., and E. V. Koonin. 1993. Cell-to-cell movement of plant viruses. Insights from amino acid sequence comparisons of movement proteins and from analogies with cellular transport systems. Arch. Virol. 133**:**239-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oparka, K. J., D. A. M. Prior, S. Santa Cruz, H. S. Padgett, and R. N. Beachy. 1997. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J. 12**:**781-789. [DOI] [PubMed] [Google Scholar]
- 30.Padgett, H. S., B. L. Epel, T. W. Kahn, M. Heinlein, Y. Watanabe, and R. N. Beachy. 1996. Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J. 10**:**1079-1088. [DOI] [PubMed] [Google Scholar]
- 31.Philo, J. S. 2000. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279**:**151-163. [DOI] [PubMed] [Google Scholar]
- 32.Pickard, B. G., and R. N. Beachy. 1999. Intercellular connections are developmentally controlled to help move molecules through the plant. Cell 98**:**5-8. [DOI] [PubMed] [Google Scholar]
- 33.Reichel, C., and R. N. Beachy. 1998. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 95**:**11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito, T., Y. Imai, T. Meshi, and Y. Okada. 1988. Interviral homologies of the 30K proteins of tobamoviruses. Virology 167**:**653-656. [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16**:**4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shortle, D., and M. S. Ackerman. 2001. Persistence of native-like topology in a denatured protein in 8 M urea. Science 293**:**487-489. [DOI] [PubMed] [Google Scholar]
- 38.Stafford, W. F., III. 1992. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 203**:**295-301. [DOI] [PubMed] [Google Scholar]
- 39.Waigmann, E., M.-H. Chen, R. Bachmaier, S. Ghoshroy, and V. Citovsky. 2000.. Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19**:**4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, B. M., R. Medville, S. G. Lazarowitz, and R. Turgeon. 1997. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71**:**3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf, S., C. M. Deom, R. N. Beachy, and W. J. Lucas. 1989. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246**:**377-379. [DOI] [PubMed] [Google Scholar]
- 42.Wynne, S. A., R. A. Crowther, and A. G. W. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3**:**771-780. [DOI] [PubMed] [Google Scholar]