Default-mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI (original) (raw)
Abstract
Recent functional imaging studies have revealed coactivation in a distributed network of cortical regions that characterizes the resting state, or default mode, of the human brain. Among the brain regions implicated in this network, several, including the posterior cingulate cortex and inferior parietal lobes, have also shown decreased metabolism early in the course of Alzheimer's disease (AD). We reasoned that default-mode network activity might therefore be abnormal in AD. To test this hypothesis, we used independent component analysis to isolate the network in a group of 13 subjects with mild AD and in a group of 13 age-matched elderly controls as they performed a simple sensory-motor processing task. Three important findings are reported. Prominent coactivation of the hippocampus, detected in all groups, suggests that the default-mode network is closely involved with episodic memory processing. The AD group showed decreased resting-state activity in the posterior cingulate and hippocampus, suggesting that disrupted connectivity between these two regions accounts for the posterior cingulate hypometabolism commonly detected in positron emission tomography studies of early AD. Finally, a goodness-of-fit analysis applied at the individual subject level suggests that activity in the default-mode network may ultimately prove a sensitive and specific biomarker for incipient AD.
Reduced posterior cingulate cortex (PCC) activity is among the most common findings in positron emission tomography (PET) and single-photon emission computerized tomography (SPECT) studies of early Alzheimer's disease (AD) (1-3). Other studies have shown hypometabolism in the PCC in cognitively intact subjects with genetic susceptibility to AD (4-6). Most recently, functional deactivation profiles in the PCC were shown to differ between patients with AD and healthy controls in a semantic classification task (7). The consistency with which the PCC is implicated in these studies is puzzling in that this region is not among the first to show the neuropathological changes of AD (8). The leading hypothesis is that decreased PCC activity in incipient AD reflects decreased connectivity with medial temporal lobe (MTL) structures, such as the entorhinal cortex and hippocampus, which are among the first regions targeted by AD pathology (3, 9). This hypothesis is supported by data from the animal literature showing prominent connectivity between the PCC and the MTL (10-13) as well as hypometabolic changes in PCC after rhinal cortex ablation (14). Human studies have supported this hypothesis as well (15-17).
Although the PCC shows decreased resting-state activity in incipient AD, it tends to be among the most metabolically active regions in healthy subjects resting with their eyes closed (18). Furthermore, the PCC is among the most commonly “deactivated” brain regions, often showing increased activity during rest or a cognitively simple baseline task compared to a cognitively demanding experimental task (19). This combination of attributes led Raichle et al. (18) to propose that the PCC forms part of a “default mode” brain network. We subsequently used a functional connectivity analysis of functional MRI (fMRI) data to demonstrate significant resting-state coactivation of several regions within this putative network, including the PCC, bilateral inferior parietal cortex, left inferolateral temporal cortex, and ventral anterior cingulate cortex (20). We also demonstrated that the default-mode network persisted, virtually unchanged, during a sensory task with little cognitive demand (passive viewing of a flashing or still checkerboard). Although we detected a small cluster of coactivation within the most posterior aspect of the parahippocampal gyrus, we did not find conclusive evidence, at a field strength of 3 T, for connectivity between the PCC and other MTL structures such as the hippocampus or entorhinal cortex. Nonetheless, the prominent role of the PCC in this network led us to suspect that it might be abnormal in AD.
To test our hypothesis that the default-mode network might be abnormal in AD, we examined a dataset (#2-2000-1118W) from the fMRI data center (www.fmridc.org), an international repository of freely accessible raw fMRI data. These data were generously contributed by Buckner et al. (21), who studied healthy young subjects, healthy elderly subjects, and patients with very mild to mild AD during a simple sensory-motor task. Having previously detected the default-mode network not only during an eyes-closed resting condition but also during a simple visual processing task, we reasoned that the minimal cognitive requirements of this sensory-motor task would not disrupt the default-mode network, and that it would therefore be detectable in this data set.
In our previous study, we used a functional connectivity MRI analysis to detect coactivation in the network. This involves specifying a region-of-interest (ROI), averaging the time series of all of the voxels within it and using that time series as a covariate of interest in a whole-brain linear regression statistical parametric analysis. In the current study, we adapted independent component analysis (ICA) to derive the default-mode network in a more data-driven fashion (i.e., without requiring a priori specification of a seed region). Examination of the default-mode network in these groups revealed three critical findings: there is significant coactivation of the hippocampus in the default-mode network, the network is abnormal in the mildest stages of AD compared to healthy aging, and network activity holds potential as a noninvasive biomarker of incipient AD.
Methods
As a proof of concept, we first applied ICA to our previously published resting-state data (referred to herein as Stanford University data), allowing us to compare the default-mode network detected with ICA to that detected with the ROI approach. An essentially identical ICA approach was then applied to the three subject groups from Washington University (referred to below as Washington University data). Further details regarding methods are available in the original publications (20, 21).
Subjects and Tasks. Stanford University data. Fourteen healthy right-handed subjects (seven females) participated in this study after giving informed consent in accordance with Stanford University's Institutional Review Board. Their ages ranged from 18 to 25 years, with a mean age of 21.2 years.
For the resting-state scan, subjects were instructed simply to keep their eyes closed and not to think of anything in particular. The scan lasted for 4 min.
Washington University data. Thirteen subjects with very mild to mild AD, 14 healthy elderly subjects, and 14 healthy young subjects were scanned during a simple sensory-motor paradigm (21). The AD subjects (six males) ranged in age from 68 to 83 (mean age 77.2). The elderly subjects (five males) ranged in age from 66 to 89 (mean age 74.9). The 14 healthy young subjects (five male) ranged in age from 18 to 24 (mean age 21.1). The subjects with dementia were scanned at a relatively early stage of their illness: 8 of the 13 AD patients had a Clinical Dementia Rating (CDR) of 0.5, placing them in the “very mild” category, whereas the other five had a CDR score of 1, placing them in the “mild” category (22).
The basic task required subjects to respond with a button press when a stimulus was presented. The stimulus, a flashing checkerboard, was presented for 1.5 sec either singly or in a paired sequential presentation with a 5.36-sec gap between presentations. Each run, A-D, consisted of 15 trials, pseudorandomly intermixed so that there were eight trials of one type (single or paired presentation) and seven trials of the other type per run. Subjects completed four runs. One AD subject had data only for three of four runs.
Imaging Methods. Stanford University data. Functional images were acquired on a 3-T General Electric Signa scanner using a standard whole-head coil. The following spiral pulse sequence parameters were used: repeat time, 2,000 ms; echo time, 30 ms; flip angle, 80°; and one interleave. To aid in the localization of functional data, a high-resolution T1-weighted spoiled grass gradient recalled 3D MRI sequence with the following parameters was used: 124 coronal slices; 1.5-mm thickness; no skip; repeat time, 11 ms; echo time, 2 ms; and flip angle, 15°. The images were reconstructed as a 124 × 256 × 256 matrix with a 1.5 × 0.9 × 0.9-mm spatial resolution. The structural scans were acquired on a 1.5-T General Electric Signa scanner.
Washington University data. Functional images were acquired on a Siemens (Iselin, NJ) 1.5-T scanner with an asymmetric spin-echo sequence sensitive to blood oxygenation level-dependent (BOLD) contrast. The following parameters were used: repeat time (TR), 2.68 sec; 3.75 × 3.75-mm in-plane resolution; T2* evolution time, 50 ms; α, 90°. Whole-brain volumes were acquired with 16 contiguous 8-mm-thick axial oblique slices (parallel to the plane connecting the anterior and posterior commisures). Each functional run lasted 5.5 min. High-resolution structural images were acquired in a series of three to four separate T1-weighted MP-RAGE anatomic images with the following parameters: resolution = 1 × 1 × 1.25 mm; TR, 9.7 msec; echo time, 4 msec; flip angle, 10°; TI, 20 msec; TD, 500 msec.
Data Processing. Stanford University data. Data were preprocessed and analyzed by using SPM99 (23) (www.fil.ion.ucl.ac.uk/spm). Images were corrected for movement by using least-squares minimization without higher-order corrections for spin history and normalized (24) to stereotaxic coordinates of Talairach and Tournoux (25). Images were then resampled every 2 mm by using sinc interpolation and smoothed with a 4-mm Gaussian kernel to decrease spatial noise.
Statistical maps were superimposed on a group average of the normalized high-resolution T1-weighted images and cluster locations interpreted by using known neuroanatomical landmarks.
Washington University data. Raw structural and functional data were received from the fMRI Data Center maintained at Dartmouth College (Hanover, NH). Functional data were then movement-corrected, normalized, and smoothed as described with the Stanford University data above. Examination of the smoothed images revealed a large spike artifact that contaminated each time point in one elderly subject's data. This subject's data were removed from subsequent analyses.
Statistical maps were superimposed on a group average of the normalized high-resolution T1-weighted images and cluster locations interpreted using known neuroanatomical landmarks.
Data Analysis. The following steps were done essentially identically, except where noted, on data from Stanford University and Washington University.
Independent component analysis. For each subject, the smoothed normalized fMRI images were concatenated across time to form a single 4D image. For the Washington University data, the first two time points were eliminated to allow for equilibration of the magnetic field (this is done at the acquisition stage at Stanford University). For each subject, the 4D dataset was then analyzed with FSL melodic ICA software (www.fmrib.ox.ac.uk/fsl/melodic2/index.html). ICA is a statistical technique that separates a set of signals into independent uncorrelated and non-Gaussian spatiotemporal components (26). When applied to the T2* signal of fMRI, ICA allows not only for the removal of high- and low-frequency artifacts (27, 28), but also for isolation of task-activated neural networks (27, 29). Most recently, ICA has been used to identify low-frequency neural networks that are active during resting-state (visual fixation) fMRI data (30). There is no consensus, as yet, on how to choose the optimal number of components, although methods to do so are in development (31). We chose to have the analysis output 24 components for the Stanford University data and 31 components for the Washington University data (approximately one-fourth to one-fifth the number of timepoints in the respective scans). The ICA software could not converge on 31 components in a total of 11 runs from nine subjects (two AD, two elderly, five young). For these runs, the number of components generated ranged from 22 to 42.
Selection of the best-fit component. An automated two-step process was then used to select the component in each subject that most closely matched the default-mode network. First, because functional connectivity networks have been detected in low-frequency ranges (32), a frequency filter was applied to remove any components in which high-frequency signal (>0.1 Hz) constituted 50% or more of the total power in the Fourier spectrum. Next, a template of the default-mode network was used to select the “best-fit” of the remaining low-frequency components in each subject. To do this, we developed a nonlinear template-matching procedure that involved taking the average z score of voxels falling within the template minus the average z score of voxels outside the template and selecting the component in which this difference (the goodness of fit) was the greatest. z scores here reflect the degree to which a given voxel's time series correlates with the time series corresponding to the specific ICA component.
A different template was used for selecting the best-fit components in the two data sets. Because the PCC appears to be a critical node in the default-mode network (20), we used the PCC cluster from our previous ROI-based study as the template in selecting the best-fit component for the Stanford University data. This also allowed us to demonstrate that the entire network can be derived with ICA by using only one of the nodes as a template. In an effort to better distinguish AD subjects from elderly subjects, a broader template was chosen for the Washington University data consisting of all brain regions in the default-mode network, rather than the PCC cluster alone. Because AD subjects show metabolic deficits in the parietal lobes (3) as well as in the PCC, we reasoned that incorporating other regions into the goodness-of-fit calculation would allow a better separation of the AD patients from the healthy elderly controls. Thus, in selecting the best-fit components and their associated goodness-of-fit scores for the Washington University data, we used the entire default-mode network, as defined by applying ICA to the Stanford University data (Fig. 1_B_), as the template.
Fig. 1.
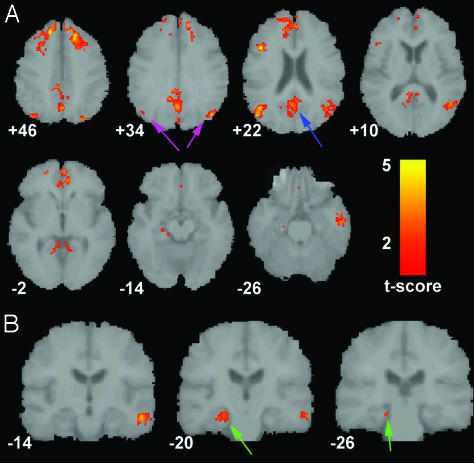
Validation of the ICA approach (Stanford University data). Axial images showing the default-mode network as detected with ROI-based (A) and ICA-based (B) approaches in a group of healthy young adults scanned on a 3-T magnet at Stanford University. The blue arrows indicate the PCC. The left side of the image corresponds to the left side of the brain. The numbers beneath each image refer to the z coordinate in Talairach space. T score bars are shown at right. Functional images were overlaid on the group-averaged structural image. Joint height and extent thresholds of P < 0.001 were used to determine significant clusters.
Group statistical maps. For the Stanford University data, the best-fit components from each subject were then combined in a second-level random-effects analysis to generate group statistical maps for the default-mode network. Significant clusters of activation were determined by using the joint expected probability distribution (33) with height (P < 0.001) and extent (P < 0.001) thresholds, corrected at the whole-brain level.
For the Washington University data, where 39 of 40 subjects had four scans processed, a single median image was generated from each subject's four best-fit components (i.e., the highest and lowest best-fit component images were discarded, and the two middle best-fit components were averaged). In the AD subject who underwent only three runs, the midvalue best-fit component became the median image. Because of the increased power associated with using median images, clusters of activation for the young, elderly, and AD groups from Washington University were determined by using more stringent height and extent thresholds of P < 0.0001, corrected at the whole-brain level.
A two-sample t test was used to compare the healthy elderly and AD group maps. To avoid detection of clusters that did not appear in either group map, we used a masking procedure in which we generated a group map for all 26 subjects (significant clusters defined as in the single-group analyses above) and limited the search for clusters that differed between the two groups to this combined group map. Significant clusters of activation for the two-sample t test were determined by using combined height (P < 0.05) and extent (P < 0.05) thresholds, corrected at the whole-brain level.
Goodness-of-fit analysis. In addition to the statistical maps, we explored differences between the AD and elderly groups in the goodness-of-fit metric. Each subject had a score for each of their four best-fit components (one from each run) reflecting the degree to which their best-fit component network matched the default-mode template defined by the Stanford University data. One AD subjects had scores for only three runs. We calculated the median goodness-of-fit score for each subject. A two-sample t test with a significance level of P < 0.05 was used to determine whether the group means were significantly different. The Mann-Whitney test was also used as a confirmatory test of significance given the nonparametric nature of the data. The individual subject data were also plotted as a scattergram, which was used to select a cutoff point in the median goodness-of-fit scores that provided the optimal sensitivity and specificity in distinguishing AD subjects from healthy elderly controls.
Results
Behavioral Data. Mean response times across all trials were available for all 14 elderly subjects and 11 of 13 AD subjects. The group mean response time was 575.9 msec (SD 525.1) for the elderly subjects and 630.3 msec (SD 534.4) for the AD subjects. The means were not significantly different (P = 0.8, two-sample t test). The numbers of correct responses across all trials were available for all 14 elderly subjects and 12 of 13 AD subjects. Of 90 possible responses, the mean number of missed responses per subject was 5.5 (SD 12.7) in the elderly group and 3.25 (SD 5.0) in the AD group. The means were not significantly different (P = 0.6, two-sample t test). Please refer to the original paper by Buckner et al. (21) for behavioral data in young subjects.
Validation of ICA Approach (Stanford University Data). Fig. 1 compares the default-mode network as detected in our previous publication by the ROI approach (Fig. 1 A) and as detected here by the ICA approach (Fig. 1_B_). Significant overlap between the two approaches is demonstrated across the majority of clusters including the PCC, bilateral inferior parietal cortex, left inferolateral temporal cortex, medial prefrontal cortex, and ventral anterior cingulate cortex. Specific cluster locations for the two approaches are available as supporting information, which is published on the PNAS web site.
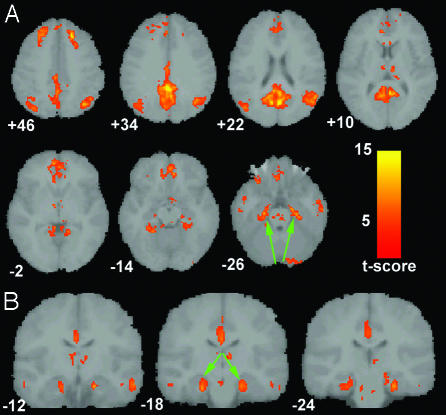
Default-Mode Network in Healthy Young Subjects (Washington University Data). Fig. 2 demonstrates the default-mode network in 14 healthy young subjects from Washington University. Coronal sections (Fig. 2_B_) highlight the prominent coactivation of the MTL and hippocampus not detected in the 3-T data from Stanford University. Specific cluster locations are available as supporting information.
Fig. 2.
Hippocampal coactivation in the default-mode network (Washington University data). Axial (A) and coronal (B) images showing the default-mode network for 14 healthy young subjects scanned on a 1.5-T magnet at Washington University. The green arrows highlight the prominent bilateral coactivation in the hippocampus and underlying entorhinal cortex. The numbers beneath each coronal image refer to the y coordinate in Talairach space. Joint height and extent thresholds of P < 0.0001 were used to determine significant clusters. Other details are as in Fig. 1.
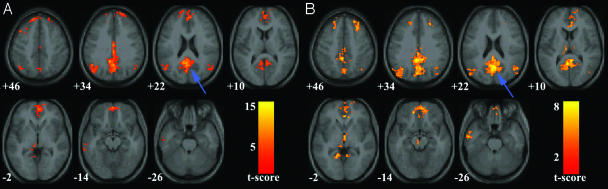
Default-Mode Network in Healthy Aging and AD Subjects (Washington University Data). Fig. 3 compares the default-mode network in the healthy elderly (Fig. 3_A_) and AD groups (Fig. 3_B_). Specific cluster locations are available as supporting information.
Fig. 3.
Default-mode network in healthy elderly and AD subjects (Washington University data). Axial images showing the default-mode network for the healthy elderly (A) and AD (B) groups. The blue arrows indicate the PCC. The hippocampus and underlying entorhinal cortex (green arrows) were detected bilaterally in healthy elderly subjects (A) but only in the right hemisphere in the AD group (B). Joint height and extent thresholds of P < 0.0001 were used to determine significant clusters. Other details are as in Fig. 1.
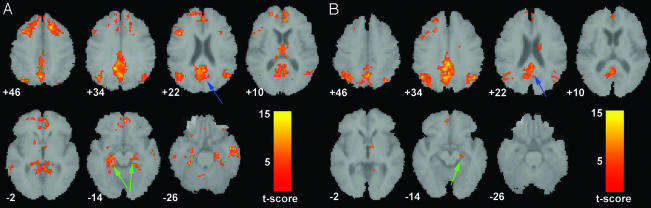
Fig. 4_A_ shows the statistical map resulting from the two-sample t test comparing the default-mode network in the healthy elderly vs. AD groups. Fig. 4_B_ shows a coronal section of the same contrast to highlight a 112-voxel cluster in the left hippocampus that survived the height but not the extent threshold. The reverse contrast (AD vs. healthy elderly) did not yield any significant clusters.
Fig. 4.
Increased default-mode network activity in healthy elderly. (A) Axial images showing the results of a two-sample t test contrasting the default-mode network in the healthy elderly group vs. the AD group. The blue arrow indicates the PCC. The magenta arrows indicate the inferior parietal lobes. (B) Coronal images showing a 112-voxel cluster in the left hippocampus (green arrows) that survived the height but not the extent threshold. Joint height and extent thresholds of P < 0.05 were used to determine significant clusters. Other details are as in Fig. 1.
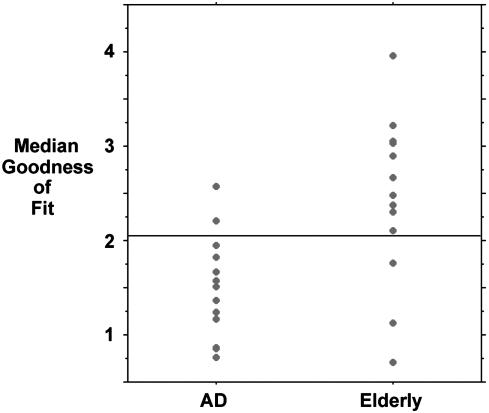
Fig. 5 is a scattergram showing the median goodness-of-fit metric for each subject in the two groups. The means were 2.4 (SD 0.9) for the healthy elderly group and 1.5 (SD 0.5) for the AD group. The means were significantly different using both a two-sample t test (P = 0.003) and a Mann-Whitney test (P = 0.007). Using a cutoff of 2.1, the test yields a sensitivity of 85% and a specificity of 77% in distinguishing AD subjects from healthy elderly subjects.
Fig. 5.
Individual scores for goodness of fit to standard default-mode network. A scattergram shows the median goodness of fit for each subject in the AD and healthy elderly groups using the Stanford University ICA-derived default-mode template. The group means were significantly different in a two-sample t test (P < 0.01). The horizontal line indicates a cutoff point of 2.1 where 11 of 13 AD subjects and 10 of 13 elderly subjects are correctly categorized, yielding a sensitivity of 85% and a specificity of 77%.
Discussion
Our study examining the default-mode network in early AD has generated several important results pertaining both to AD specifically and the default-mode network more generally. The main findings were that the hippocampus appears to play a prominent role in the default-mode network, network activity is deficient in AD compared to healthy elderly controls, and a metric of network activity shows promise as a clinical marker of AD.
Currently, the behavioral correlates of the default-mode network remain uncharacterized, although there are several potentially inclusive hypotheses. Some investigators have suggested that the network has a role in attending to environmental stimuli, both internally and externally generated (18, 34). Others have suggested that it mediates processes such as reviewing past knowledge in preparing for future actions (35). The current findings offer further support for our hypothesis that the default-mode network is closely involved in episodic memory processing. In our previous study, we hypothesized that the network might have some role in memory processing based on the prominent role of the PCC within it and evidence linking the PCC to memory functions (16, 36, 37). Although we detected a cluster of posterior parahippocampal coactivation in that 3-T study, there was little else to implicate medial temporal memory regions in the default-mode network. By contrast, we have now detected significant bilateral hippocampal/entorhinal cortex coactivation in the default-mode network in healthy young and elderly subjects as well as unilateral MTL coactivation in the AD group. We believe the discrepancy in hippocampal coactivation between our initial study and the current study can be traced to the different field strengths in the two studies. That is, the hippocampal coactivation in the default-mode network detected across all three groups here at 1.5-T was most likely lost to susceptibility artifact at 3-T (38). One of the most consistent species-independent findings in neuroscience is that the hippocampus is integral to episodic memory processing (39). Further, episodic memory loss is the cardinal feature of AD and the most common presenting symptom (40). Combining these two facts with our findings of (i) significant bilateral hippocampal coactivation in the two healthy Washington University groups and (ii) deficient hippocampal activity in the AD group makes a compelling, albeit indirect, case that the network plays a critical role in episodic memory processing. In future studies, we hope to gain more insight into the putative memory functions of the default-mode network by exploring correlations between network activity and neuropsychological measures.
Although the MTL is the initial site of histopathological changes in AD (8), the PCC is the most common site of early metabolic and perfusion abnormalities. Disrupted connectivity between the hippocampus/entorhinal cortex and the PCC has been invoked as the mechanism behind PCC hypometabolism and hypoperfusion in early AD (3, 9). There is evidence from a number of human studies supporting prominent connectivity between the PCC and MTL. PET studies have demonstrated task-driven PCC and MTL interactions across groups of subjects (15, 16). Studies of “retrosplenial amnesia” (41) and PCC hypometabolism in amnestic patients without AD (17) also provide support for PCC-MTL interactions. At the neuronal level, connectivity between these two regions has been demonstrated in animal studies (10-13). The ICA approach used here extracts the network en bloc and does not provide direct measures of interregional connectivity. Nonetheless, based on burgeoning evidence in animal and human studies, a strong case can be made that the coactivation of PCC and MTL detected here reflects connectivity between these two regions. Activity in these two regions was deficient in the AD group compared to the elderly controls (Fig. 4). Although reduced connectivity with parietal or other cortical regions could account for decreased PCC activity, given the relatively focal MTL pathology in early AD and the converging evidence for MTL-PCC connectivity, we believe our findings support the hypothesis that impaired MTL-PCC connectivity accounts for the decreased PCC metabolism/perfusion detected in PET and SPECT studies (1, 2, 9).
A unique advantage provided by the method used here is that it allows one to examine task-independent network activity in individual subjects. It is this critical distinction that allowed us to demonstrate the clinical potential of our approach in the diagnosis of AD. By using a standard template to select the best-fit component for each subject, we have developed an automated ICA technique for detecting the default-mode network. The relative stability of the network across laboratories, field strengths, and healthy subject populations also speaks to its universality. It appears that the default-mode network is a readily and reproducibly detectable neural network operating in the resting state and in tasks with low cognitive demand. Detection of the network at 1.5 T is important in that the vast majority of clinical scans are done at this field strength. A number of attributes make ICA-based detection of the default-mode network a promising candidate in the ongoing quest to find a safe noninvasive biomarker of incipient AD. Unlike most structural imaging methods, the process can be automated, minimizing manpower requirements and the potential for investigator bias. The absence of a task eliminates issues such as performance differences among groups and practice effects with repeated scanning. The enhanced spatial resolution and reliance on endogenous signal rather than radionuclide tracers also make this approach preferable to PET and SPECT methods.
We have reported a metric here, reflecting the goodness of fit of a subject's default-mode network to a standard default-mode template, which distinguishes individual AD subjects from healthy elderly subjects with a sensitivity of 85% and a specificity of 77%. These sensitivity and specificity values are in the range considered clinically relevant by a recent Working Group on biomarkers in AD (42). The results are particularly encouraging in light of the fact that 8 of 13 AD patients were in the earliest stages of the disease (Clinical Dementia Rating score of 0.5) (22). In subsequent studies, this approach may be optimized by using a template that includes only regions where network activity differs between AD subjects and elderly controls (Fig. 4). It will also be important to determine whether better sensitivity and specificity can be achieved by examining network activity during a true resting paradigm, as opposed to during a sensorymotor task, as was done here. Ideally, such an optimized network metric might prove efficacious in making the critical clinical distinction between which at-risk subjects will convert to AD and which will not.
Supplementary Material
Supporting Tables
Acknowledgments
We thank Gary Glover for his insight into resting-state connectivity, Randy Buckner and colleagues for making their data available to us via the fMRI Data Center, Jack Van Horn for help with accessing and converting the raw data from the fMRI Data Center, Christian Beckmann and Stephen Smith for making the FSL ICA software available, and two anonymous reviewers for constructive comments. This work was made possible through the following sources: National Institutes of Health Grants MH19938, MH01142, HD31715, HD40761, and MH62430 and a grant from the Ruth K. Broad Biomedical Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AD, Alzheimer's disease; DLPFC, dorsolateral prefrontal cortex; ICA, independent component analysis; PCC, posterior cingulate cortex; ROI, region of interest; fMRI, functional MRI; MTL, medial temporal lobe; SPECT, single-photon emission computerized tomography; PET, positron-emission tomography.
References
- 1.Minoshima, S., Giordani, B., Berent, S., Frey, K. A., Foster, N. L. & Kuhl, D. E. (1997) Ann. Neurol. 42**,** 85-94. [DOI] [PubMed] [Google Scholar]
- 2.Johnson, K. A., Jones, K., Holman, B. L., Becker, J. A., Spiers, P. A., Satlin, A. & Albert, M. S. (1998) Neurology 50**,** 1563-1571. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda, H. (2001) Ann. Nucl. Med. 15**,** 85-92. [DOI] [PubMed] [Google Scholar]
- 4.Reiman, E. M., Caselli, R. J., Yun, L. S., Chen, K., Bandy, D., Minoshima, S., Thibodeau, S. N. & Osborne, D. (1996) N. Engl. J. Med. 334**,** 752-758. [DOI] [PubMed] [Google Scholar]
- 5.Small, G. W., Ercoli, L. M., Silverman, D. H., Huang, S. C., Komo, S., Bookheimer, S. Y., Lavretsky, H., Miller, K., Siddarth, P., Rasgon, N. L., et al. (2000) Proc. Natl. Acad. Sci. USA 97**,** 6037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiman, E. M., Caselli, R. J., Chen, K., Alexander, G. E., Bandy, D. & Frost, J. (2001) Proc. Natl. Acad. Sci. USA 98**,** 3334-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lustig, C., Snyder, A. Z., Bhakta, M., O'Brien, K. C., McAvoy, M., Raichle, M. E., Morris, J. C. & Buckner, R. L. (2003) Proc. Natl. Acad. Sci. USA 100**,** 14504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braak, H. & Braak, E. (1991) Acta Neuropathol. 82**,** 239-259. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, K. M., O'Sullivan, V. T., Soper, N. D., Nagy, Z., King, E. M., Smith, A. D. & Shepstone, B. J. (2002) Brain 125**,** 1772-1781. [DOI] [PubMed] [Google Scholar]
- 10.Vogt, B. A., Finch, D. M. & Olson, C. R. (1992) Cereb. Cortex 2**,** 435-443. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki, W. A. & Amaral, D. G. (1994) J. Comp. Neurol. 350**,** 497-533. [DOI] [PubMed] [Google Scholar]
- 12.Morris, R., Petrides, M. & Pandya, D. N. (1999) Eur. J. Neurosci. 11**,** 2506-2518. [DOI] [PubMed] [Google Scholar]
- 13.Lavenex, P., Suzuki, W. A. & Amaral, D. G. (2002) J. Comp. Neurol. 447**,** 394-420. [DOI] [PubMed] [Google Scholar]
- 14.Meguro, K., Blaizot, X., Kondoh, Y., Le Mestric, C., Baron, J. C. & Chavoix, C. (1999) Brain 122**,** 1519-1531. [DOI] [PubMed] [Google Scholar]
- 15.McIntosh, A. R., Grady, C. L., Haxby, J. V., Ungerleider, L. G. & Horwitz, B. (1996) Cereb. Cortex 6**,** 571-584. [DOI] [PubMed] [Google Scholar]
- 16.Della-Maggiore, V., Sekuler, A. B., Grady, C. L., Bennett, P. J., Sekuler, R. & McIntosh, A. R. (2000) J. Neurosci. 20**,** 8410-8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aupee, A. M., Desgranges, B., Eustache, F., Lalevee, C., de la Sayette, V., Viader, F. & Baron, J. C. (2001) NeuroImage 13**,** 1164-1173. [DOI] [PubMed] [Google Scholar]
- 18.Raichle, M. E., MacLeod, A. M., Snyder, A. Z., Powers, W. J., Gusnard, D. A. & Shulman, G. L. (2001) Proc. Natl. Acad. Sci. USA 98**,** 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shulman, G. L., Fiez, J. A., Corbetta, M., Buckner, R. L., Miezin, F. M., Raichle, M. E. & Petersen, S. E. (1997) J. Cognit. Neurosci. 9**,** 648-663. [DOI] [PubMed] [Google Scholar]
- 20.Greicius, M. D., Krasnow, B., Reiss, A. L. & Menon, V. (2003) Proc. Natl. Acad. Sci. USA 100**,** 253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckner, R. L., Snyder, A. Z., Sanders, A. L., Raichle, M. E. & Morris, J. C. (2000) J. Cognit. Neurosci. 12**,** 24-34. [DOI] [PubMed] [Google Scholar]
- 22.Morris, J. C. (1993) Neurology 43**,** 2412-2414. [DOI] [PubMed] [Google Scholar]
- 23.Friston, K. J., Holmes, A. P., Poline, J. B., Grasby, P. J., Williams, S. C., Frackowiak, R. S. & Turner, R. (1995) NeuroImage 2**,** 45-53. [DOI] [PubMed] [Google Scholar]
- 24.Friston, K. J., Ashburner, J., Frith, C. D., Poline, J. B., Heather, J. D. & Frackowiak, R. S. D. (1995) Hum. Brain Mapp. 2**,** 165-189. [Google Scholar]
- 25.Talairach, J. & Tournoux, P. (1988) Co-Planar Stereotaxic Atlas of the Human Brain (Thieme, Stuttgart).
- 26.Beckmann, C. F. & Smith, S. M. (2004) IEEE Trans. Med. Imaging 23**,** 137-152. [DOI] [PubMed] [Google Scholar]
- 27.McKeown, M. J., Jung, T. P., Makeig, S., Brown, G., Kindermann, S. S., Lee, T. W. & Sejnowski, T. J. (1998) Proc. Natl. Acad. Sci. USA 95**,** 803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley, M. A., Haughton, V. M., Carew, J., Cordes, D., Moritz, C. H. & Meyerand, M. E. (2002) Am. J. Neuroradiol. 23**,** 49-58.11827875 [Google Scholar]
- 29.Gu, H., Engelien, W., Feng, H., Silbersweig, D. A., Stern, E. & Yang, Y. (2001) NeuroImage 14**,** 1432-1443. [DOI] [PubMed] [Google Scholar]
- 30.Calhoun, V. D., Pekar, J. J., McGinty, V. B., Adali, T., Watson, T. D. & Pearlson, G. D. (2002) Hum. Brain Mapp. 16**,** 158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calhoun, V. D., Adali, T., McGinty, V. B., Pekar, J. J., Watson, T. D. & Pearlson, G. D. (2001) NeuroImage 14**,** 1080-1088. [DOI] [PubMed] [Google Scholar]
- 32.Cordes, D., Haughton, V. M., Arfanakis, K., Carew, J. D., Turski, P. A., Moritz, C. H., Quigley, M. A. & Meyerand, M. E. (2001) Am. J. Neuroradiol. 22**,** 1326-1333. [PMC free article] [PubMed] [Google Scholar]
- 33.Poline, J. B., Worsley, K. J., Evans, A. C. & Friston, K. J. (1997) NeuroImage 5**,** 83-96. [DOI] [PubMed] [Google Scholar]
- 34.Gusnard, D. A., Akbudak, E., Shulman, G. L. & Raichle, M. E. (2001) Proc. Natl. Acad. Sci. USA 98**,** 4259-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binder, J. R., Frost, J. A., Hammeke, T. A., Bellgowan, P. S., Rao, S. M. & Cox, R. W. (1999) J. Cognit. Neurosci. 11**,** 80-95. [DOI] [PubMed] [Google Scholar]
- 36.Maguire, E. A. & Mummery, C. J. (1999) Hippocampus 9**,** 54-61. [DOI] [PubMed] [Google Scholar]
- 37.Maddock, R. J., Garrett, A. S. & Buonocore, M. H. (2001) Neuroscience 104**,** 667-676. [DOI] [PubMed] [Google Scholar]
- 38.Krasnow, B., Tamm, L., Greicius, M. D., Yang, T. T., Glover, G. H., Reiss, A. L. & Menon, V. (2003) NeuroImage 18**,** 813-826. [DOI] [PubMed] [Google Scholar]
- 39.Eichenbaum, H. (2001) Behav. Brain Res. 127**,** 199-207. [DOI] [PubMed] [Google Scholar]
- 40.McKhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D. & Stadlan, E. M. (1984) Neurology 34**,** 939-944. [DOI] [PubMed] [Google Scholar]
- 41.Valenstein, E., Bowers, D., Verfaellie, M., Heilman, K. M., Day, A. & Watson, R. T. (1987) Brain 110**,** 1631-1646. [DOI] [PubMed] [Google Scholar]
- 42.The Ronald and Nancy Reagan Research Institute of the Alzheimer's Association and the National Institute on Aging Working Group (1998) Neurobiol. Aging 19**,** 109-116. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Tables