Functional and biochemical dissection of the structure-specific nuclease ARTEMIS (original) (raw)
Abstract
During V(D)J recombination, the RAG1 and RAG2 proteins form a complex and initiate the process of rearrangement by cleaving between the coding and signal segments and generating hairpins at the coding ends. Prior to ligation of the coding ends by DNA ligase IV/XRCC4, these hairpins are opened by the ARTEMIS/DNA-PKcs complex. ARTEMIS, a member of the metallo-β-lactamase superfamily, shares several features with other family members that act on nucleic acids. ARTEMIS exhibits exonuclease and, in concert with DNA-PKcs, endonuclease activities. To characterize amino acids essential for its catalytic activities, we mutated nine evolutionary conserved histidine and aspartic acid residues within ARTEMIS. Biochemical analyses and a novel in vivo V(D)J recombination assay allowed the identification of eight mutants that were defective in both overhang endonucleolytic and hairpin-opening activities; the 5′ to 3′ exonuclease activity of ARTEMIS, however, was not impaired by these mutations. These results indicate that the hairpin-opening activity of ARTEMIS and/or its overhang endonucleolytic activity are necessary but its exonuclease activity is not sufficient for the process of V(D)J recombination.
Keywords: ARTEMIS, metallo-β-lactamase, SCID, V(D)J recombination
Introduction
The expression of T- and B-cell receptors on the surface of lymphoid progenitor cells is essential for the development of T and B cells. The process by which the exons encoding the antigen-binding domains of immunoglobulin and T-cell receptor proteins are assembled from individual V, (D), and J gene segments is termed V(D)J recombination (Tonegawa, 1983; Fugmann et al, 2000). This rearrangement of the genomic DNA is directed by the recombination signal sequences (RSSs) flanking each V, (D), or J gene segment. The RSSs consist of a palindromic heptamer and an AT-rich nonamer that are separated by spacer sequences of either 12 bp (12-RSS) or 23 bp (23-RSS) in length. V(D)J recombination occurs between one 12-RSS and one 23-RSS element (12/23 rule) and is initiated by the two lymphoid specific recombination-activating gene proteins 1 and 2 (RAG1 and RAG2). The RAG proteins specifically recognize the RSSs and initially introduce a nick in the DNA double strand between a coding subgenic V, (D), or J element and the heptamer of its flanking RSS (Schatz et al, 1989; Oettinger et al, 1990; Fugmann et al, 2000; Gellert, 2002). After the generation of this nick, the resulting free 3′-OH attacks the phosphodiester bond of the opposite strand. This cleavage occurs coordinately at the two coding element/heptamer borders of V, (D), or J elements and results in two hairpin-sealed coding ends. Thereafter, the corresponding coding end hairpins are opened and joined by the ubiquitously expressed factors of the nonhomologous end-joining DNA repair pathway (NHEJ). Before or during the joining process, nucleotides (N nucleotides) may be added by the lymphocyte-specific template-independent terminal deoxynucleotidyl transferase (TdT) or nucleotides may be deleted leading typically to imprecise coding joints (CJs). Furthermore, short palindromic sequences (P nucleotides) may appear at the CJs, presumably resulting from hairpin opening at positions other than the hairpin tip.
Recently, it was demonstrated that a protein called ARTEMIS opens the coding end hairpins (Ma et al, 2002). ARTEMIS associated with and phosphorylated by the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) possesses overhang endonucleolytic and DNA hairpin-opening activities in vitro (Ma et al, 2002). This hairpin-opening activity was confirmed in vivo in artemis knockout mice (Rooney et al, 2002). Without the activation by DNA-PKcs, ARTEMIS only exhibits its intrinsic DNA single-strand-specific 5′ to 3′ exonuclease activity in vitro.
Lack of ARTEMIS in humans leads to the phenotype of B-cell-negative, T-cell-negative severe combined immunodeficiency (B− T− SCID). Since fibroblasts and hematopoietic progenitor cells deficient in ARTEMIS showed increased sensitivity to ionizing radiation compared to wild-type cells, ARTEMIS, like several other components of the NHEJ pathway, seems to play a role in the general DNA double-strand break (dsb) repair pathway. Therefore, the type of SCID caused by a lack of ARTEMIS activity is called radio-sensitive SCID (RS-SCID) (Moshous et al, 2001).
ARTEMIS, as its homologs PSO2 (scSNM1) in yeast and muSNM1 in mouse, is a member of the large metallo-β-lactamase superfamily, because of the metallo-β-lactamase domain in the N-terminal region (amino acids 1–155) of ARTEMIS (Callebaut et al, 2002). In contrast to PSO2 and muSNM1, ARTEMIS may not be involved in the repair of DNA interstrand crosslinks (Rooney et al, 2002, 2003). Like all members of the metallo-β-lactamase superfamily, ARTEMIS needs divalent cations to be catalytically active. ARTEMIS requires Mg2+-, but is inactive in Mn2+- or Zn2+-containing buffers (Ma et al, 2002).
The protein domain fold characteristic of metallo-β-lactamases consists of a four-layered β-sandwich with two mixed β-sheets flanked by α-helices (Aravind, 1999; Wang et al, 1999). The sites for the coordination of the metal ions are located at one edge of the β-sandwich. Five sequence motifs of metallo-β-lactamases are highly conserved in enzymes of the superfamily and possibly participate in metal coordination, substrate binding, and enzymatic activities (Aravind, 1999; Wang et al, 1999). Motif one is typified by an aspartate located at the end of a β-strand, which might be important as the counterpart of an extensive positive charge of the second motif. Motif two consists of a highly conserved HxHxDH sequence. The first two histidines of motif two are speculated to take part in the coordination of the metal ions. The aspartate residue of motif two is positioned very closely to the catalytic centers of the enzymes and most likely participates in hydrolysis reactions, while the role of the third histidine is less obvious. Motifs three and five encompass single histidines, which might be important for the coordination of metal ions and binding of negatively charged substrates. In motif four, a single aspartate possibly participates in the hydrolysis reactions (Aravind, 1999; Wang et al, 1999). The essential amino acids of motifs one–four can be found in the human ARTEMIS protein; the position of motif five is less clear (Moshous et al, 2001). Additionally, amino acids 156–385 of ARTEMIS share several conserved features with other metallo-β-lactamases specifically acting on nucleic acids. This domain was named the β-CASP motif (metallo-β-lactamases-associated CPSF ARTEMIS SNM1 PSO2) and presumably includes motif 5 (Callebaut et al, 2002).
Having shown that ARTEMIS possesses intrinsic single-strand-specific 5′ to 3′ exonuclease activity and acquires endonucleolytic activities when complexed with DNA-PKcs, we characterized those residues of ARTEMIS that appear to be critical for its enzymatic properties. By testing in vitro mutated ARTEMIS proteins in biochemical analyses and in a novel plasmid-based in vivo V(D)J recombination assay, we identified eight residues of ARTEMIS that are essential for its endonucleolytic but not its exonucleolytic activity.
Results
Conserved residues in the metallo-_β_-lactamase/_β_-CASP domain of the ARTEMIS protein
Despite exhibiting considerable sequence differences, members of the metallo-β-lactamase superfamily share conserved motifs (Aravind, 1999; Wang et al, 1999). In particular, the metallo-β-lactamase/β-CASP domain of the human ARTEMIS protein, spanning amino acids 1–385, and presumably containing the active site of the enzyme, shows significant homologies to related proteins (Callebaut et al, 2002). By database investigation and alignment analysis, we selected four conserved aspartic acids (D17, D37, D136, D165) and five conserved histidines (H33, H35, H38, H115, H319) in the metallo-β-lactamase/β-CASP domain of human ARTEMIS. These residues are conserved in human ARTEMIS, in its rat and mouse orthologs, in its human paralogs SNM1 and SNM1B, in mouse SNM1B, in yeast SNM1 as well as in the L1 protein of Stenotrophomonas maltophilia (Supplementary Figure 1). Considering that aspartic acid and histidine residues are likely to be involved in coordinating metal ions of the active site as well as in binding and processing substrates in metallo-β-lactamases, we were interested in studying whether these conserved residues contribute to the active site of ARTEMIS.
Generation and expression of mutant ARTEMIS proteins
To examine the roles of the highly conserved amino acids D17, D37, D136, D165, H33, H35, H38, H115, and H319 of the ARTEMIS metallo-β-lactamase/β-CASP domain, these residues were mutated separately so that their individual role for the enzymatic activity of ARTEMIS could be investigated. We generated expression plasmids coding for the wild-type human ARTEMIS protein (ART-WT) and nine individual mutant ARTEMIS proteins (ARM11–19, where ARM designates ARTEMIS mutant) that carry asparagine residues instead of the conserved aspartic acids or alanines instead of the conserved histidines (Figure 1). In addition, we generated three plasmids coding for double-mutant ARTEMIS proteins (ARM33–35). The ARTEMIS cDNAs were cloned in frame with the coding sequence of a C-terminal myc-His tag. To test the stability of the encoded ARTEMIS-myc-His fusion proteins in eucaryotic cells, the corresponding expression plasmids were transiently transfected into HEK293T cells. Using an antibody directed against the myc tag, an immunoblot of cell lysates was performed (Figure 2A, Supplementary Figure 3A). As a loading control, the blot was stained with an antibody directed against SV40 large T. These experiments show that the wild-type and the mutant ARTEMIS proteins are stable in eucaryotic cells with some differences in expression levels. Since ARTEMIS is involved in V(D)J recombination and DNA dsb repair, both nuclear functions, it was important to establish that the wild-type and mutant ARTEMIS proteins are localized in the corresponding subcellular compartment (Moshous et al, 2001; Ma et al, 2002). Therefore, immunostaining of transfected HEK293T cells was performed, with an antibody directed against the myc epitope (Figure 2B, Supplementary Figure 3B). The results indicate that the majority of the wild-type ARTEMIS protein and the 12 mutant ARTEMIS proteins are homogeneously distributed in the nucleus (excluding nucleoli) and thus these proteins should be able to fulfill their roles in the processes of V(D)J recombination and DNA dsb repair.
Figure 1.
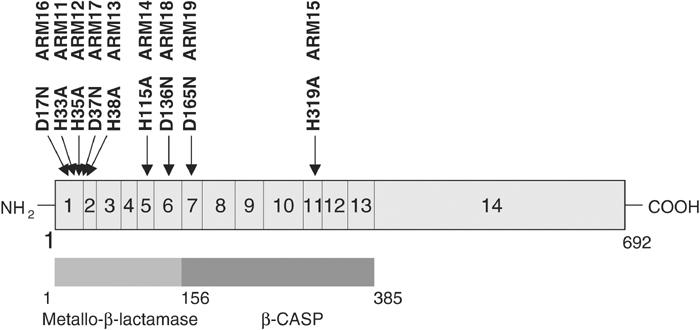
Mutagenesis of single conserved residues of the human ARTEMIS protein. The positions of the amino-acid changes of the mutant human ARTEMIS proteins (ARM11–19) are indicated; in addition, the exons of the Artemis gene as well as the amino-acid positions of known ARTEMIS domains are shown. In our analyses, the ARTEMIS protein consists of 692 amino acids with the first codon starting at nucleotide 41 of the GenBank sequence AW954867. This is in contrast to the translational start position nucleotide 62, which was assumed by Moshous et al and which leads to a protein of only 685 amino acids (Moshous et al, 2001; Ma et al, 2002). Furthermore, compared to the GenBank sequence AW954867, our cDNA exhibits a G1718T exchange leading to a leucine instead of a valine at codon 560 and a silent C to T exchange at nucleotide 1849. ARTEMIS cDNAs derived from human cell lines always contained a T at nucleotide 1718 as well as at nucleotide 1849.
Figure 2.
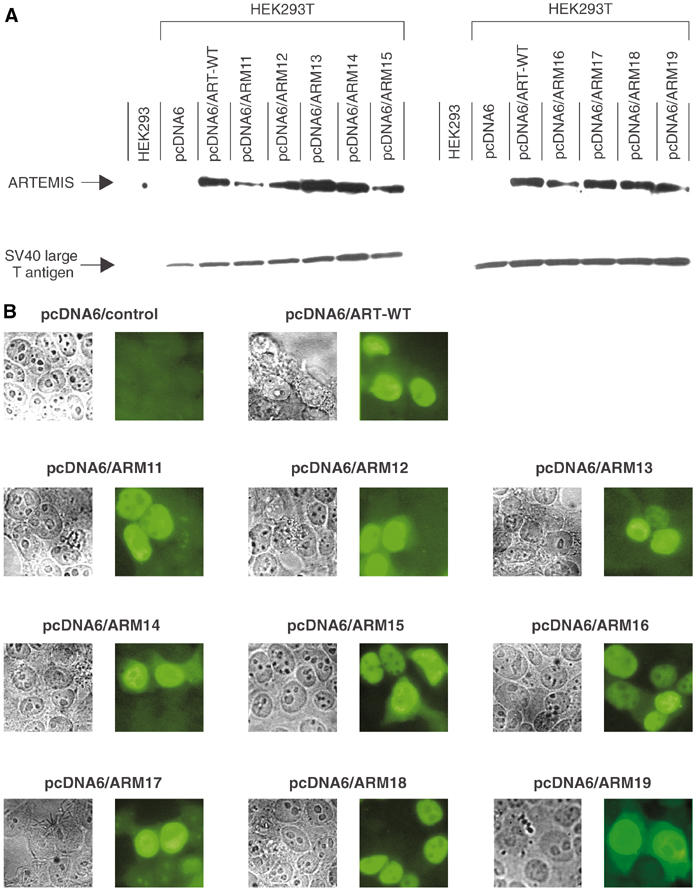
Expression of wild-type and mutant ARTEMIS proteins in human cells. (A) HEK293T cells were transfected with pcDNA6 expression plasmids coding for myc-His fusion proteins of either wild-type (ART-WT) or mutant (ARM11–19) ARTEMIS. As controls, HEK293 and HEK293T cells were transfected with the empty vector. The expression of the ARTEMIS-myc-His fusion proteins, as detected by immunoblot analysis, is shown in the upper panel; SV40 large T protein expression is shown below. (B) HEK293T cells were transfected with pcDNA6 expression plasmids coding for myc-His fusion proteins of wild-type (ART-WT) and mutant (ARM11–19) ARTEMIS proteins. The subcellular localization of the myc-His fusion proteins was detected by immunostaining using a mouse anti-human myc antibody. An empty vector transfection served as a control.
Phosphorylation of wild-type and mutant ARTEMIS proteins by DNA-PKcs
The human ARTEMIS protein alone possesses single-strand 5′ to 3′ exonuclease activity. After complex formation, DNA-PKcs phosphorylates ARTEMIS and ARTEMIS demonstrates endonucleolytic activities on 5′ and 3′ DNA overhangs as well as on hairpins (Ma et al, 2002). Since this latter activity is considered to be indispensable for V(D)J recombination, we examined whether the myc-His-tagged wild-type and mutant ARTEMIS proteins bind to and are phosphorylated by DNA-PKcs. Purified wild-type or mutant ARTEMIS proteins were used in an in vitro kinase assay, which as a prerequisite needs ARTEMIS:DNA-PKcs interaction. Wild-type ARTEMIS and the 12 mutant ARTEMIS proteins are phosphorylated by DNA-PKcs to similar levels (Figure 3, Supplementary Figure 3C). Together, these results demonstrate that the mutant ARM11–19 and ARM33–35 proteins are expressed in the correct subcellular compartment and adopt a conformation that allows DNA-PKcs to bind to and phosphorylate them comparably to the wild-type ARTEMIS protein.
Figure 3.
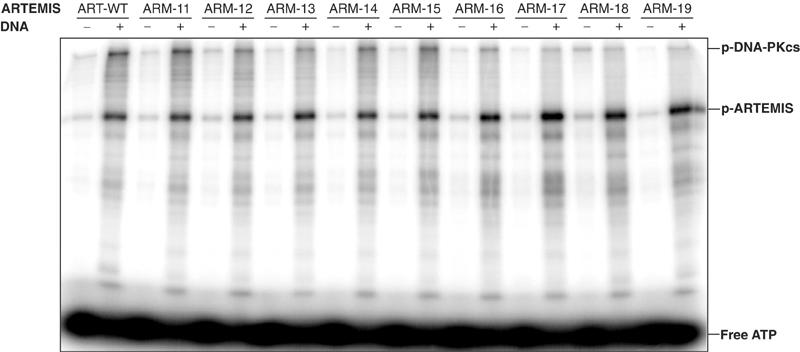
DNA-PKcs assay of ARTEMIS and ARTEMIS mutants. The ARTEMIS mutants ARM11–ARM19 are indistinguishable from the wild-type ARTEMIS in the ability of being phosphorylated by DNA-PKcs. Wild-type ARTEMIS and mutant ARTEMIS ARM11–ARM19 were subjected to a DNA-PKcs phosphorylation assay in the absence and presence of exogenous 35 bp DNA. The positions of phosphorylated DNA-PKcs, phosphorylated ARTEMIS (p-ARTEMIS), and free ATP are indicated.
In vitro cleavage activities of mutant ARTEMIS proteins
Two different substrates were used to test whether the mutant ARTEMIS proteins still exhibit both exonuclease and endonuclease activities in an in vitro cleavage assay. The first substrate is a DNA hairpin 20 bp in length with a 6-nt 5′ overhang (Figure 4, Supplementary Figure 3D). Only wild-type ARTEMIS (ART-WT) and ARM13 showed endonucleolytic activities. The ARM13 hairpin-opening activity was reduced about 50-fold and the overhang endonuclease activity was reduced about 4- to 6-fold as compared to ART-WT. The hairpin-opening and endonucleolytic cleavage activities of the other ARTEMIS mutants were undetectable. In the presence of DNA-PKcs, ART-WT and ARM13 are able to open the hairpin preferentially 3′ of the second nucleotide 3′ of the hairpin tip (the +2 position). The same two proteins removed the 5′ overhang by cutting at the positions blunt, blunt-1, blunt+1, and blunt+2, with the major product resulting from a cut at the blunt position. The second substrate was a 21 bp double-strand DNA with a 15 nt 3′ overhang (Figure 4). In this assay, again only ART-WT and ARM13 showed endonucleolytic activities. In a DNA-PKcs-dependent manner, both proteins preferentially cut the substrate 3′ of the positions blunt+4 and blunt+5. Importantly, in all of the experiments, the exonucleolytic activities of the tested ARTEMIS variants were not impaired.
Figure 4.
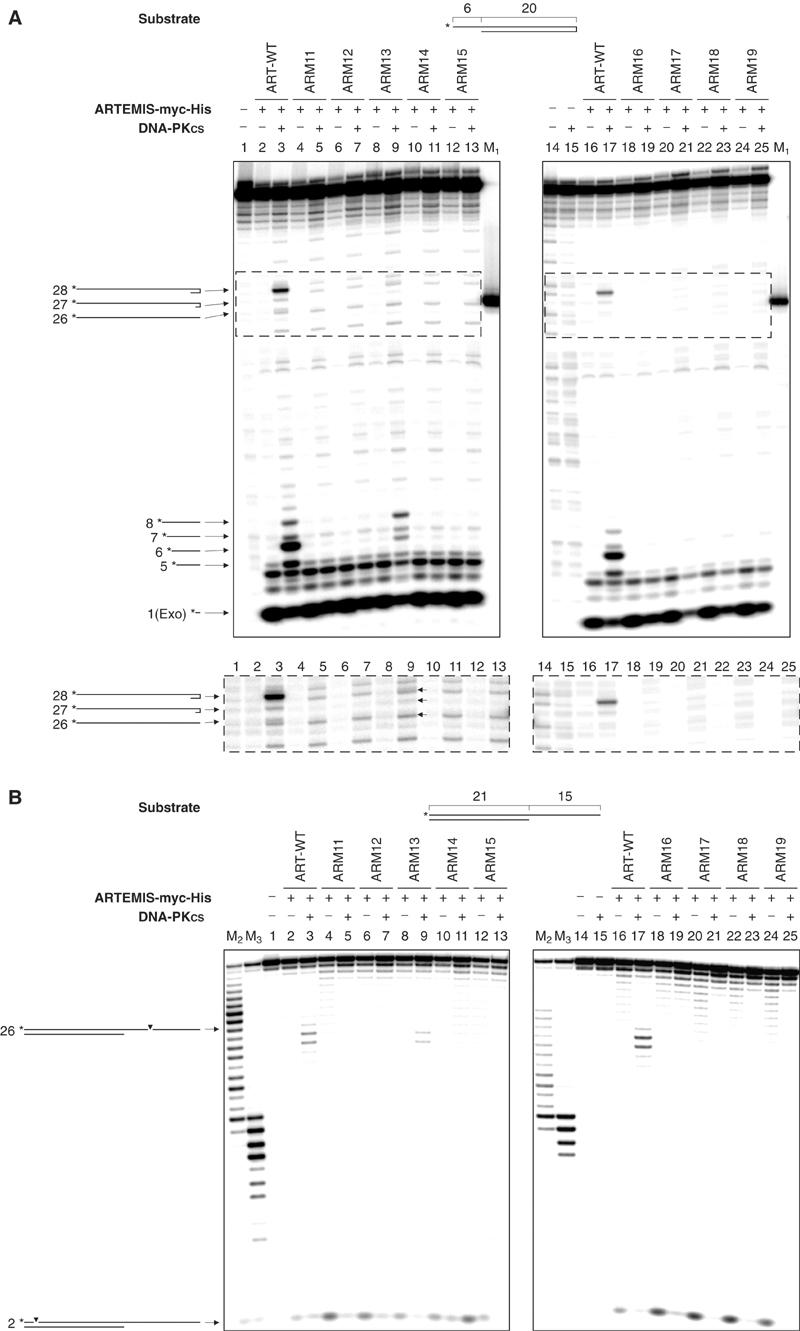
In vitro nuclease assay of ARTEMIS and ARTEMIS mutants. (A) The 5′ overhang processing and hairpin-opening activities of ARM11–ARM19. A 20-bp hairpin with 6-nt 5′ overhang was used as the substrate. The asterisks indicate the positions of the radioactive label. Wild-type ARTEMIS, histidine to alanine mutants ARM11–ARM15, and aspartic acid to asparagine mutants ARM16–ARM19 were incubated with the substrate indicated in the absence and presence of DNA-PKcs. Autoradiographs of sequencing gels are shown. M1 marks the position of the hairpin-opening product if the hairpin was opened at the tip. Positions and sizes of the major hairpin-opening and overhang processing products are indicated by arrows and numbers (in nt). The exonucleolytic product by ARTEMIS (1 nt) is indicated by ‘Exo' adjacent to the size of the product. Diagrams adjacent to the arrows are depicted to show the cleavage positions in the substrate that result in the corresponding products. The dotted boxes mark the parts of the gels overexposed and shown beneath. The arrows mark the hairpin products generated by the ARM13:DNA-PKcs complex. (B) A 21-bp DNA with 15-nt 3′ overhang was used as the substrate. The asterisks indicate the positions of the radioactive label. Diagrams adjacent to the arrows are depicted to show the cleavage positions in the substrate that result in the corresponding products. M2 and M3 were generated by incubating the labelled strand of the 3′ overhang substrate with Klenow enzyme at room temperature for 30 and 60 min, respectively.
Residues of ARTEMIS critical for its in vivo enzymatic activity
A novel plasmid-based cellular test system was established that measures the ability of wild-type and mutant ARTEMIS proteins for inversional V(D)J recombination in vivo on an extrachromosomal substrate (Supplementary Figures 2A and B). Primary dermal ARTEMIS-negative fibroblasts from a patient with a homozygous deletion of ARTEMIS exons 1–3 (unpublished data) were cotransfected with RAG1 and RAG2 expression plasmids, a V(D)J recombination substrate plasmid, and expression plasmids for either wild-type or mutant ARTEMIS proteins. After 48 h, the transfection efficiency was measured by staining cells with a PerCP-conjugated antibody against the murine MHC class I gene H-2Kk, which is encoded by the substrate plasmid and is constitutively expressed in transfected primary fibroblasts (Supplementary Figures 2C and D). PerCP-positive cells were detected by FACS analysis. V(D)J recombination of the substrate plasmid leads to an inversion of an EGFP-ORF. Successful recombination places the EGFP-ORF in the correct orientation with respect to the SV40 promoter. Consequently, cells with recombined substrate plasmid exhibit green fluorescence (Supplementary Figures 2A and B). The percentage of green cells in the subpopulation of transfected cells was quantified for cells transfected with different ARTEMIS protein-expressing plasmids (Table I). As a control, we tested our system with a pre-recombined plasmid (pMACS11-19Flip). As additional controls, similar experiments were performed in ARTEMIS-positive fibroblasts (Table I). Transfection efficiencies for ARTEMIS-positive control fibroblasts ranged from 18 to 37%; only 56–60% of these transfected cells expressed the pre-recombined EGFP, indicating that we may underestimate true recombination frequencies by approximately two-fold. In the control fibroblasts, V(D)J recombination efficiency was RAG2 dependent, but was independent of the addition of an ARTEMIS expression plasmid (Supplementary Figures 2C, Table I). In ARTEMIS-deficient cells, transfection efficiencies ranged from 14 to 49%, with expression of a pre-recombined EGFP in 49–60% of the transfected cells. Without ARTEMIS, the ARTEMIS-deficient cells did not carry out significant detectable numbers of recombination events (0.0–0.1% of transfected cells exhibited green fluorescence), while ARTEMIS rescued the V(D)J recombination to a level of 2.8–4.1% in transfected cells. Omitting the RAG2 expression plasmid during transfection yielded only up to 0.1% positive cells, indicating that the appearance of positive signals is also RAG-dependent in the ARTEMIS-negative cells. We conclude that in an ARTEMIS-deficient cell line, there is functional complementation of V(D)J recombination after transfection of an ARTEMIS expression vector.
Table 1.
V(D)J recombination assays in human primary dermal fibroblasts
| | Positive control | −RAG2 | −ART | +ART | ARM11 | ARM12 | ARM13 | ARM14 | ARM15 | ARM16 | ARM17 | ARM18 | ARM19 | ARM33 | ARM34 | ARM35 | | | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | ----- | ---- | ---- | ----- | ----- | ----- | ----- | ----- | ----- | ----- | ----- | ---------- | ----- | ----- | ----- | ---------- | | ARTEMIS-positive fibroblasts | | | | | | | | | | | | | | | | | | Exper. 1 | 55.70 | 0.12 | 3.12 | 3.32 | | | | | | | | | | | | | | Exper. 2 | 57.48 | 0.05 | 3.88 | 4.33 | | | | | | | | | | | | | | Exper. 3 | 59.74 | 0.05 | 2.59 | 3.22 | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | ARTEMIS-deficient fibroblasts | | | | | | | | | | | | | | | | | | Exper. 1 | 49.11 | 0.09 | 0.09 | 2.79 | 0.48 | 0.21 | 1.88 | 0.09 | 0.03 | 0.09 | 0.07 | 0.16 | 0.63 | 0.09 | 0.14 | NA | | Exper. 2 | 58.63 | 0.05 | 0.01 | 3.65 | 0.27 | 0.10 | 1.50 | 0.03 | 0.01 | 0.02 | 0.00 | NA | 0.25 | 0.02 | 0.01 | 0.03 | | Exper. 3 | 60.46 | 0.08 | 0.05 | 4.06 | 0.41 | 0.17 | 2.21 | 0.04 | 0.07 | 0.03 | 0.14 | 0.09/0.04a | 0.46 | 0.03 | 0.05 | 0.08/0.06a | | ARTEMIS-positive fibroblasts and ARTEMIS-negative fibroblasts were tested in three independent experiments. Human primary dermal fibroblasts were cotransfected with V(D)J recombination substrate vector and expression plasmids coding for RAG1, RAG2, wild-type ARTEMIS (+ART), or mutant ARTEMIS proteins (ARM11–19, ARM33–35). As controls, the same cotransfections without the expression plasmids coding for RAG2 (−RAG2) or wild-type ARTEMIS (−ART) were performed. As a positive control, a pre-recombined substrate vector was transfected. In each assay, 3–5 × 104 fibroblasts were analyzed by FACS. The percentages of recombination-positive cells out of the subpopulation of transfected fibroblasts are depicted. | | | | | | | | | | | | | | | | | | a Two independent values were ascertained. NA=not available. | | | | | | | | | | | | | | | | |
When expression plasmids for single amino-acid mutant ARTEMIS proteins (ARM11–19) instead of wild-type ARTEMIS were introduced into the ARTEMIS-negative fibroblasts, five of the nine mutant ARTEMIS variants (ARM14, 15, 16, 17, 18) were unable to complement the defects in V(D)J recombination at all (Table I). Three mutants (ARM11, 12, 19) showed a V(D)J recombination efficiency on average of about 11, 4.5, and 13% respectively when compared to ART-WT-expressing cells. When the expression plasmid encoding ARM13 was cotransfected, a 1.5–2.2% efficiency was measured, which was half of wild-type levels (2.8–4.1% of transfected cells). For the double-mutant proteins ARM33–35, no V(D)J recombination activities above background were observed. Taken together, the in vitro and in vivo data indicate that the conserved amino acids D17, D37, D136, D165, H33, H35, H115, and H319 of ARTEMIS are critical for its hairpin-opening activity, whereas H38 is dispensable for this feature of the protein.
The analysis of the CJs of recombined substrate plasmids isolated from ARTEMIS-negative fibroblasts revealed V(D)J recombination products when wild-type ARTEMIS or the mutant ARTEMIS proteins with in vivo activities above background values (ARM11, 12, 13, 19) (Supplementary Figure 4) were introduced. Importantly, even without an ARTEMIS-encoding plasmid, a low level of near-normal recombination products was isolated from ARTEMIS-deficient fibroblasts. The corresponding CJs show an increased extent of coding end deletions and longer P nucleotides compared to CJs from ARTEMIS-positive cells. CJs from ARTEMIS-deficient cells with addition of either wild-type ARTEMIS or ARM11, 12, 13, and 19 are similar to CJs from ARTEMIS-positive cells.
Discussion
In this study, several conserved amino acids in the metallo-β-lactamase/β-CASP domain of the human ARTEMIS were mutated individually or in combination by site-directed mutagenesis. Among all of the single point mutants, we identified eight that exhibited markedly decreased DNA endonucleolytic activities (hairpin opening and endonucleolytic processing of overhangs) in vitro and reduced capability to support inversional V(D)J recombination in vivo. The results of the in vivo and in vitro assays are in accordance. Thus, the corresponding residues D17, H33, H35, D37, H115, D136, D165, and H319 are critical for the DNA endonucleolytic activities of ARTEMIS, most likely due to their metal coordinating ability. Only the ARTEMIS variant H38A exhibited an activitiy that is about half of wild-type ARTEMIS in the in vivo recombination assay. Thus, H38, although highly conserved in the HxHxDH motif of metallo-β-lactamases, seems not to be essential for the activity of the enzyme. Interestingly, none of the mutations influenced the exonucleolytic activity of ARTEMIS.
Mutant ARTEMIS proteins specifically lose endonucleolytic activities
To rule out the possibility that the wild-type and mutant ARTEMIS-myc-His fusion proteins tested in our system are not stably expressed in eucaryotic cells, we performed Western blot and immunostaining analyses (Figures 2A and B, Supplementary Figures 3A and B). All mutant ARTEMIS proteins, as well as wild-type ARTEMIS, were detected in the lysates of transfected cells and were localized in the nucleus. Furthermore, ARM11–19 and ARM33–35 were phosphorylated by DNA-PKcs to levels comparable to wild-type ARTEMIS (Figure 3, Supplementary Figure 3C). Thus, the mutations and the myc-His tags do not interfere with the binding to DNA-PKcs and the mutant ARTEMIS myc-His fusion proteins do not yield gross conformational changes compared to the wild type. Based on this, the plasmid-encoded ARTEMIS variants should have the same overall ability to support V(D)J recombination as the endogenous ARTEMIS.
When the mutant ARTEMIS proteins were tested in in vivo V(D)J recombination assays, only ARM13 (H38A) was able to complement V(D)J recombination in vivo in ARTEMIS-deficient cells comparable to wild-type ARTEMIS. For all other mutants (D17N, D37N, D136N, D165N, H33A, H35A, H115A, H319A, H35A/H115A, H35A/H319A, D17N/H319A), the ability to support V(D)J recombination was abolished or markedly decreased. The corresponding in vitro assays showed that only wild-type ARTEMIS and ARM13 exhibit endonucleolytic and hairpin-opening activities, whereas all other mutants do not. These results underline the significance of the conserved residues D17, D37, D136, D165, H33, H35, H115, and H319 of the human ARTEMIS protein in the process of V(D)J recombination. Amino acid H38, on the other hand, despite its conservation, is not essential for endonucleolytic activities of ARTEMIS in vitro and the capability of ARTEMIS to facilitate V(D)J recombination in vivo. However, in the in vitro assays, ARM13 showed a decreased endonucleolytic activity on hairpins (about 50-fold reduced compared to ART-WT) and on overhangs (about 4- to 6-fold reduction); one could envision that three-dimensional constraints for hairpin placement in the active site are more restricted than those for DNA overhangs. On the other hand, in the in vivo recombination assay, cells with ARM13 achieved V(D)J recombination frequencies comparable to cells with ART-WT, indicating that the in vivo assay is more sensitive for detecting residual activity than the in vitro assay. This is further supported by our observation that ARM11, 12, and 19 exhibit V(D)J recombination activities in vivo slightly above background, whereas in vitro, no endonuclease activities were detected. These observations are in accordance with the finding of Kienker et al (2000) that residual amounts of NHEJ activities, DNA-PKcs in their case, are sufficient for in vivo V(D)J recombination. Moreover, compared to wild-type ARTEMIS, the relative amounts of the different products of the 5′ overhang endonucleolytic activity of ARM13 in vitro were altered. This result could give a hint that H38 is important for the orientation and/or configuration of the substrate in the active center of ARTEMIS.
There are several possible explanations for the fact that the 5′ to 3′ DNA exonuclease activity is not impaired in any of the ARTEMIS single or double conserved residue mutants. One possibility is that the β-lactamase fold represents the portion of the active site most critical for binding of the DNA substrates for endonucleolytic cleavage, but is not important for substrate binding for exonucleolytic cleavage. There are other nucleases that have both exo- and endonucleolytic properties, which can be separated, including FEN-1 and recBCD (Kowalczykowski and Eggleston, 1994; Lieber, 1997). In both of these other enzymes, one active site is assumed, despite the disparate nature of the exo- and endonucleolytic activities. A second possibility is that there is some degree of redundancy within the active site, such that a mutation of any one residue is insufficient to destabilize a divalent metal ion cofactor, especially in an active site where there may be two divalent metal cofactor ions. This scenario seems unlikely since double mutants concerning either one of the two metal coordinating sites or both sites still preserve exonuclease activity. A third possibility is that the mutated residues are directly involved in the binding of ARTEMIS to DNA-PKcs. This argument may be ruled out, because all mutants were shown to be phosphorylated by DNA-PKcs in vitro. A fourth possibility is that there is a second independent active site within the protein. Only co-crystals with various substrates can rule this out definitively. A fifth possibility is that the exonucleolytic activity is contributed by another protein that co-purifies with ARTEMIS. However, no second protein has been identified in careful biochemical and genetic searches thus far. These included purifications from mammalian cells as well as insect cells and purifications that included variations in the monovalent salt concentrations over a range from 50 to 1000 mM monovalent salt. We also purified FLAG-tagged ARTEMIS from 293T cells by immunoprecipitation and elution with FLAG peptide. The ratio of the exo- and endonuclease activities did not vary despite the different purification methods, cell types used, or salt concentrations. This suggests, but by no means proves, that the exonuclease is intrinsic to ARTEMIS. We favor the first explanation, namely that the exonucleolytic activity relies on the same active site as the endonucleolytic activity and that the β-lactamase fold is important for the substrate binding for the endo- but not the exonucleolytic activity.
The residual hairpin-opening activity in ARTEMIS-deficient cells
Quantitatively, the V(D)J recombination activity in ARTEMIS null human cells is about 100-fold reduced. Of the small numbers of CJs formed, about 70% have unusually long P nucleotides or unusually long deletions from one or both coding ends. These unusual features suggest that some low-level mechanism of opening the hairpins exists. This low-level mechanism could be due to an alternative process, such as other nucleases opening the hairpins at a 100-fold lower efficiency than the ARTEMIS:DNA-PKcs complex.
How does the residual level of hairpin opening observed in ARTEMIS null human cells correlate with the numbers of B and T cells seen in ARTEMIS null patients or mice? In the ARTEMIS null patients, no B or T cells of patients' origin are observed in peripheral blood. Perhaps the 100-fold reduction in V(D)J recombination efficiency plus the fact that only about one-third of these appear to be normal CJs may result in a 300-fold reduction of each coding end joining. This reduction may be sufficient to block Ig or TCR production. Interestingly, a similar level of reduction in RAG function leads to a B- and T-cell-negative SCID phenotype in humans (Schwarz et al, 1996). Importantly, more than half of the ARTEMIS-deficient SCID patients analyzed in our laboratory have T lymphocytes received transplacentally from their mothers (data not shown). Could this explain the level of lymphocytes seen in the ARTEMIS null mice? Recent evidence indicates that there is more transplacental transfer in mice than was previously thought (Marleau et al, 2003). In contrast, it may be that a very low level of alternative hairpin opening in mice is sufficient to permit some lymphocytes to survive.
The role of ARTEMIS exonuclease activity for DNA repair
Having shown that the ARTEMIS mutants specifically lost their endonucleolytic activities but still retained the 5′ to 3′ DNA exonuclease activity raises the question of whether the exonuclease of ARTEMIS is sufficient for the DNA dsb repair in cells. In the genomic DNA of an RS-SCID patient, we detected a compound heterozygosity of an H35D and an M1T mutation (unpublished data). Both alleles are expressed at the RNA level in primary skin fibroblasts of the patient. No matter whether the M1T mutant exhibits endo- and/or exonuclease activities, knowing that in vitro the H35D mutation leads to a specific loss of the endonuclease activity and retention of the exonuclease activity, in analogy to the H35A (ARM12) mutation, the observation that the patient's fibroblasts are radiosensitive may suggest that the 5′ to 3′ exonuclease activity of ARTEMIS alone is not sufficient for efficient DNA dsb repair (data not shown). Additional experiments that may clarify the roles of the ARTEMIS endonuclease and exonuclease activities in DNA dsb repair will be performed using the mutant ARTEMIS proteins described in this study.
Interestingly, a recent study on the ARTEMIS-related metallo-β-lactamase SNM1 of Saccharomyces cerevisiae, which is specific for the repair of DNA interstrand crosslinks, investigated a mutant form of the protein presenting a D to A mutation in the HxHxDH motif (metallo-β-lactamase motif 2) (Li and Moses, 2003). This mutation corresponds to the D37N (ARM17) mutation in ARTEMIS. The mutant SNM1 protein was deficient in repairing DNA dsb caused by interstrand crosslinks. Although the function of SNM1 in the process of DNA dsb repair remains unclear, this result agrees with our finding that the aspartate in the HxHxDH motif of ARTEMIS is essential for enzymatic activity. Furthermore, this report supports our hypothesis that an intact motif 2, and therefore the endonucleolytic activity of ARTEMIS, is essential for the repair of a DNA dsb.
Residues involved in the active site of ARTEMIS
To see how the mutations may affect the active site of ARTEMIS and to determine possible roles for the mutated residues in the endonuclease/hairpin-opening activity, we modeled the active center of human ARTEMIS using crystal structures of the related metallo-β-lactamases from S. maltophilia and Bacillus cereus (Protein Data Bank entries 1SML and 1BVT, respectively). All mutated residues in this analysis are conserved between ARTEMIS and the metallo-β-lactamases (Supplementary Figure 1), and thus should adopt similar positions in the three-dimensional structure. The majority of the mutated residues of ARTEMIS in this analysis evidently participate in the formation of the metal-binding site in the active center (Figure 5). The high conservation of the active site histidines and aspartates between ARTEMIS and metallo-β-lactamases suggests that the active site of ARTEMIS binds two metal cofactors, consistent with the requirement of the ARTEMIS nuclease activity for magnesium ions (Ma et al, 2002). In addition, the participation of the mutated residues in metal coordination indicates that the mechanism for disruption of nuclease activity results from defects in the formation of the metal-containing active site of ARTEMIS. D37 and histidines 33, 35, 38, 115, and 319 directly coordinate the two proposed active site metals. Thus, mutations in these residues would lead to reduced or abolished binding of one or both metals. Aspartic acids 17, 136, and/or 165, which do not directly bind to the metals, may form salt bridges to the HxHxDH motif backbone and the side chain of H38 and H33, respectively, and probably stabilize the conformation of the HxHxDH motif to coordinate optimally the metal cofactors.
Figure 5.
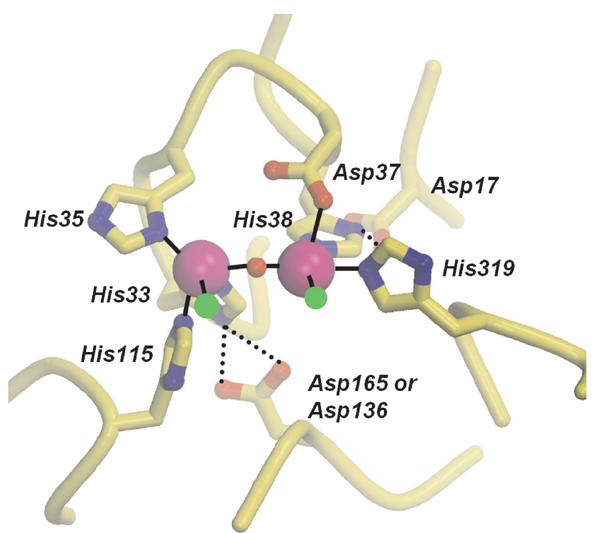
Model of the active center of the metallo-β-lactamase/β-CASP domain of the human ARTEMIS protein. Active site residues between ARTEMIS and metallo-β-lactamases, which have been mutated in this analysis, are modeled on the crystal structures of metallo-β-lactamases from S. maltophilia and B. cereus (Protein Data Bank entries 1SML and 1BVT, respectively). The mutated residues are conserved between ARTEMIS and metallo-β-lactamases and are shown in a ball-and-stick representation (with yellow carbons, blue nitrogen, and red oxygen atoms). The conserved clustering of histidine and aspartate residues in ARTEMIS suggests that two metal ions (magenta spheres) are positioned in the active site as has been observed in metallo-β-lactamase crystal structures. Asp37 and histidines 33, 35, 38, 115, and 319 directly coordinate the two active site metals in 1BVT1 (solid lines). Aspartic acids 17, 136, and/or 165 do not directly bind to the metals but likely orient histidines 38 and 33 for efficient metal coordination (dashed lines). A putative water molecule might complete the coordination sphere of the active site metal ions and was modeled (red sphere). Two additional coordination sites on the active site metals (green spheres) might bind the DNA phosphate backbone at the scissile bond and the attacking hydroxyl ion, as observed in the two-metal active site of the nuclease Mre11. The mutations in this analysis therefore likely perturb metal binding to ARTEMIS and correct formation of the active site, suggesting that the endonuclease activity and hairpin-opening activity but not the exonuclease activity are strongly dependent on correct formation of the active site environment.
The corresponding loop harboring either ARTEMIS D136 or D165 (i.e. the aspartic acid not stabilizing H33) is not found in the sequences of available metallo-β-lactamase crystal structures. The severe effect of a mutation of this residue on endonuclease activity suggests that it is also located in the active site of ARTEMIS and may participate in metal binding. Participation of this aspartic acid oxygen atom in metal coordination could explain the preference of ARTEMIS for magnesium cofactors over zinc cofactors. The latter, preferred by other metallo-β-lactamases, preferentially binds to soft ligands, whereas the former prefers hard ligands such as oxygen atoms.
The mutation in H38 still retains V(D)J recombination in vivo and possesses some residual endonucleolytic activity in vitro. In the absence of H38, one metal (the right in Figure 5) will probably bind with less affinity and this likely results in a decreased substrate binding. But the retention of the second metal, which is coordinated by different ligands, may still ensure nucleophilic attack of the metal-activated water on bound substrate DNA, resulting in a decreased but not abolished nuclease activity.
Taken together, the participation of all mutated residues in forming the metal-binding environment suggests that the endonuclease activity and the hairpin-opening activity, but probably not the exonuclease activity, are strongly dependent on the correct formation of the active site environment of ARTEMIS, in particular, on the binding of metal cofactors.
While our paper was under revision, Poinsignon et al (2004) reported the same residues of ARTEMIS to be essential for V(D)J recombination as described here. The agreement between their in vivo cellular assay and ours is quite strong. Our study has the following additional aspects: First, we have carried out biochemical studies in addition to the cellular ones. This permits us to determine effects on endo- and exonuclease catalytic activities. Only with biochemical assays can the mode of ARTEMIS action be determined to be nucleolytic; otherwise, the ‘active site' defined could be for any other type of enzymatic or even non-enzymatic activity. Second, from biochemistry, we know that the metal ions required in the active site of ARTEMIS are almost certain to be magnesium ions, rather than the zinc ions assumed by Poinsignon et al. Third, we have determined that the exonuclease activity remains even though the endonuclease has been ablated. Fourth, we have been able to show that the amount of in vitro enzymatic activity can be markedly reduced and still be adequate for a significant level of V(D)J recombination as discussed above.
In summary, the identification of eight residues critical for the endonucleolytic activity of human ARTEMIS on DNA provides new insights into the catalytic mechanism and sheds light on the structures of the active sites of other members of the metallo-β-lactamase superfamily. Furthermore, the understanding of the organization of the active site of ARTEMIS might lead to the development of specific inhibitors for the ARTEMIS DNA endonucleolytic and/or exonucleolytic activities, which could be of therapeutic significance. The in vivo V(D)J recombination assay described here is a powerful tool for diagnostic applications as well as for the characterization of proteins involved in the process of V(D)J recombination. Recently, Dai et al (2003) speculated on the existence of additional factors essential for V(D)J recombination. The system presented here would be useful in identifying such possible components in recombination-deficient cells.
Materials and methods
Alignment of ARTEMIS protein sequences
A comparison of the amino-acid sequence of the metallo-β-lactamase/β-CASP domains of human ARTEMIS with sequences of homologous and orthologous proteins was performed using the CLUSTALW software (http://npsa-pbil.ibcp.fr/cgi- bin/npsa_automat.pl?page=npsa _clustalw.html) with the following parameters: end gap penalty: 1; gap distance penalty: 8; gap extension penalty: 0.2; gap open penalty: 0.2; k-tuple size: 1; matrix: pam; pairwise alignment gap penalty: 3; scoring method: percent; top diagonals: 5; window size: 5 (Thompson et al, 1994).
Construction of ARTEMIS-myc-His and mutated ARTEMIS-myc-His expression plasmids
Full-length human ARTEMIS cDNA was amplified by recombinant Pfu DNA polymerase (Stratagene, La Jolla), from a human thymus matchmaker cDNA library (BD Biosciences, Palo Alto). The ARTEMIS cDNA was amplified using primers Kpn1ARTcDNA5′N (5′-GGGTACCGCTATGAGTTCTTTCGAGGG-3′) and Not1ARTcDNA3′w/oSTOP (5′-ATAAGAATGCGGCCGCCAGGTATCTAAGAGTGAG-C-3′). A plasmid encoding the fusion protein ARTEMIS-myc-His (pcDNA6/ArtWT-myc-His) was constructed by cloning the wild-type ARTEMIS cDNA (ART-WT) into the pcDNA6/myc-His vector Version A (Invitrogen, Breda, Netherlands) after a _Kpn_I/_Not_I digest. The point mutated ARTEMIS-expressing vectors

were generated using the QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla). Primers for mutagenesis are available on request (ulrich.pannicke@medizin.uni-ulm.de). Subsequent to cloning, the wild-type and mutant ARTEMIS ORFs of the corresponding plasmids were confirmed by sequencing.
Construction of the RAG1 and RAG2 expression plasmids
For construction of pcWTRAG1, the blunted _Kpn_I/_Sau_3A fragment of the human RAG1 cDNA (GenBank entry M29474) was inserted into the blunted _Hin_dIII site of pCDM8 (Invitrogen, Breda, Netherlands). Subsequently, a _Sna_BI/_Not_I fragment of pcWTRAG1 was subcloned into _Sna_BI/_Not_I-digested pcDNA6/myc-His Version A to generate the RAG1-expressing plasmid pcWTRAG1.
For construction of pcWTRAG2, the _Sna_BI/_Dra_I fragment of the human RAG2 cDNA (GenBank entry M94633) was inserted into the blunted _Hin_dIII site of pCDM8. Subsequently, a _Sna_BI/_Not_I fragment of pcWTRAG2 was subcloned into _Sna_BI/_Not_I-digested pcDNA6/myc-His Version A to generate the RAG2-expressing plasmid pcWTRAG2. The RAG1 and RAG2 ORFs were confirmed by sequencing.
Construction of a V(D)J recombination substrate
For cloning of the V(D)J recombination inversion substrate pMACS11-19VDJ, an EGFP cDNA containing fragment, isolated by an _Age_I/_Mlu_I digest of pEGFP-C1 (BD Biosciences, Palo Alto), was inserted into the inversional V(D)J recombination cassette of pGG52 (Gauss and Lieber, 1996). Accordingly, the EGFP cDNA is flanked by a 12 bp spacer RSS at the 5′ end and a 23 bp spacer RSS at the 3′ end. The RSSs have an identical orientation. This whole recombination cassette, that is, EGFP ORF flanked by the two RSSs, was liberated by a _Spe_I/_Not_I digest, was blunt ended and then inserted into the _Sma_I site of the pMACS KK.II vector (Miltenyi Biotec, Auburn). The resulting pMACS11-19VDJ substrate carries the EGFP cDNA in the inverted orientation with respect to the SV40 promoter. The control plasmid pMACS11-19Flip is equivalent to pMACS11-19VDJ after V(D)J recombination has taken place. By the process of V(D)J recombination, the EGFP cDNA was inverted. Therefore, the ORF is in the correct orientation to the SV40 promoter. The corresponding signal joint is located 3′ of the EGFP ORF. The pMACS11-19VDJ and the pMACS11-19Flip plasmids both carry a truncated MHC class I cDNA H-2KK controlled by an H-2KK promoter. The EGFP ORFs of these plasmids were confirmed by sequencing.
Immunoblotting
HEK293 and HEK293T cells were transfected by the calcium phosphate precipitation method (Wigler et al, 1979) with the pcDNA6/myc-His Version A plasmids, coding for either wild type or mutated versions of ARTEMIS fused to the myc-His tag. After 48 h, cells were harvested and lysed in the lysis buffer (50 mM Tris–HCl pH 8.0, 62.5 mM EDTA, 1% (w/v) NP-40, 0.4% (w/v) sodium deoxycholate). Lysates were spun down in a microcentrifuge at 14 000 g to pellet insoluble debris. Protein concentrations of the lysates were determined by the Bio-Rad Dc Protein assay (Bio-Rad, Hercules). A 10 μg portion of each lysate was run on a 10% SDS–polyacrylamide gel. The proteins were transferred to PVDF membranes (Millipore, Billerica) and the blots were developed using mouse anti-myc (Ma et al, 2002) and mouse anti-SV40 large T (Santa Cruz Biotechnology, Santa Cruz) antibodies.
Immunostaining of HEK293T cells
HEK293T cells placed on sterile coverslips were transfected by the calcium phosphate precipitation method (Wigler et al, 1979) with pcDNA6/myc-His Version A, coding for wild type or mutant forms of ARTEMIS. After 48 h, cells were washed twice in 1 × PBS (without Ca2+ and Mg2+). The cells were fixed in 1.5% paraformaldehyde, washed twice in 1 × PBS (without Ca2+ and Mg2+), and treated with 50 mM NH4Cl. Afterwards, the cells were permeabilized in buffer A (0.25% gelatin and 0.01% saponin in 1 × PBS (without Ca2+ and Mg2+)) supplemented with 0.1% NP-40. The cells were washed 3 × in buffer A and incubated with mouse anti-myc antibody (diluted 1:2000 in buffer A) for 1 h at 4°C. After three washes in buffer A, the cells were incubated with goat anti-mouse IgG-FITC (Santa Cruz Biotechnology, Santa Cruz) diluted 1:2000 in buffer A) for 1 h at 4°C followed by two washes in buffer A and two washes in 1 × PBS (without Ca2+ and Mg2+). The coverslips were mounted with slide mounting solution (90% glycerol and 2.3% DABCO). Subsequently, cells were analyzed by fluorescence microscopy.
V(D)J recombination assay in fibroblasts
Human primary ARTEMIS-negative or ARTEMIS-positive fibroblasts were transfected using the AMAXA NHDF-Neo Nucleofector Kit (AMAXA Biosystems, Cologne, Germany). Transfections were performed with 1.2 μg pcWTRAG1, 1.8 μg pcWTRAG2, 8.0 μg pMACS11-19VDJ or pMACS11-19Flip and 1.0 μg pcDNA6/myc-His Version A, pcDNA6/ArtWT-myc-His or pcDNA6 vectors coding for mutant ARTEMIS proteins (ARM11–19, ARM33–35). After 48 h, cells were harvested and analyzed by FACS.
FACS analysis
Human primary skin fibroblasts transfected with pMACS11-19Flip or pMACS11-19VDJ (after V(D)J recombination) were stained with a biotin-anti mouse H-2KK antibody (BD Biosciences, San Diego) against the truncated MHC class I protein H-2KK encoded by the plasmids. Subsequently, the biotin-labeled antibodies were detected by streptavidin–peridin chlorophyll protein (PerCP) staining (BD Biosciences, San Diego). The excitation wavelength for the FACS analysis was 488 nm. In the first step, healthy fibroblasts were grouped according to their morphology by a forward scatter/sideward scatter measurement. The fibroblasts transfected with either pMACS11-19VDJ or pMACS11-19Flip (PerCP-positive cells) were detected in fluorescence channel 3 (650 nm long pass) and were distinguishable from fibroblasts showing only autofluorescence when a fluorescence channel 2 (FL2) (585±21 nm)/fluorescence channel 3 (PerCP) analysis was performed. In the third step, EGFP-positive fibroblasts were specified in channel 1 (EGFP) (530±15 nm) out of the subpopulation of PerCP-positive cells by a fluorescence channel 1 (EGFP)/fluorescence channel 2 (FL2) analysis (Supplementary Figure 2B).
Analysis of coding joints
Plasmid DNA was isolated from fibroblasts after V(D)J recombination by the QIAprep 8 Miniprep Kit (QIAGEN, Hilden, Germany). CJs from rearranged substrate plasmids were amplified using primers binding to the pMACS-SV40 promoter (5′-AAG CTC CTC GAG GAA CTG AAA AACC-3′) and the EGFP-ORF (5′-CTT GCC GTA GGT GGC ATC GCC-3′) and subcloned into the pCR2.1-TOPO vector (Invitrogen, Breda, Netherlands). The sequences of individual CJs were analyzed using the primer binding to the pMACS-SV40 promoter.
Protein purification
Proteins were purified as specified by Ma et al (2002).
DNA-PKcs kinase assay and in vitro nuclease assay
DNA-PKcs kinase assay and in vitro nuclease assay were carried out as described (Ma et al 2002). The sequence of the hairpin used for Figure 4 is 5′-TTT TTG ATT ACT ACG GTA GTA GCT ACG TAG CTA CTA CCG TAG TAA T-3′ (YM-164). The 3′ overhang substrate used in Figure 4 was generated by annealing YM-149 with YM-68 (sequences were described in Ma et al, 2002).
Supplementary Material
Figure Sup_1. The Human ARTEMIS Protein
Figure Sup_2. In Vivo V(D)J Recombination Assay in Human Primary Dermal Fibroblasts
Figure Sup_3. In Vitro Nuclease Assay of ARTEMIS Double Mutants
Figure Sup_4. Sequence Analysis of V(D)J Recombination Coding Joints
Acknowledgments
We thank the members of the Hopfner, Lieber, and Schwarz laboratories for helpful discussions. We thank I Brackmann, I Janz, FM Koch, and EM Rump for excellent technical assistance. We are grateful to R Sailer and W Strauss for advice on immunostaining as well as to S Radecke for invaluable help in FACS analyses. We thank W Friedrich of the Department of Children and Adolescent Medicine, University of Ulm, for discussion and generous support. This work was funded by the Institute for Clinical Transfusion Medicine and Immunogenetics, Ulm, and by NIH grants to MRL. KPH acknowledges support by the DFG.
References
- Aravind L (1999) An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol 1: 69–91 [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon JP, de Villartay JP (2002) Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res 30: 3592–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA (2003) Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci USA 100: 2462–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG (2000) The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 18: 495–527 [DOI] [PubMed] [Google Scholar]
- Gauss GH, Lieber MR (1996) Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol 16: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71: 101–132 [DOI] [PubMed] [Google Scholar]
- Kienker LJ, Shin EK, Meek K (2000) Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res 28: 2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Eggleston AK (1994) Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem 63: 991–1043 [DOI] [PubMed] [Google Scholar]
- Li X, Moses RE (2003) The beta-lactamase motif in Snm1 is required for repair of DNA double-breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair 2: 121–129 [DOI] [PubMed] [Google Scholar]
- Lieber MR (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays 3: 233–240 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR (2002) Hairpin opening and overhang processing by an ARTEMIS/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Marleau AM, Greenwood JD, Wei Q, Singh B, Croy BA (2003) Chimerism of murine fetal bone marrow by maternal cells occurs in late gestation and persists into adulthood. Lab Invest 83: 673–681 [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP (2001) ARTEMIS, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105: 177–186 [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Poinsignon C, Moshous D, Callebaut I, de Chasseval R, Villey I, de Villartay JP (2004) The metallo-β-lactamase/β-CASP domain of ARTEMIS constitutes the catalytic core for V(D)J recombination. J Exp Med 199: 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J (2003) Defective DNA repair and increased genomic instability in ARTEMIS-deficient murine cells. J Exp Med 197: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, De Vido J, Foy D, Chaudhuri J, Lombard D, Alt FW (2002) Leaky scid phenotype associated with defective V(D)J coding end processing in ARTEMIS-deficient mice. Mol Cell 10: 1379–1390 [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D (1989) The V(D)J recombination activating gene, RAG-1. Cell 59: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger RA, Hansen-Hagge TE, Desiderio S, Lieber MR, Bartram CR (1996) RAG mutations in human B cell-negative SCID. Science 274: 97–99 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302: 575–581 [DOI] [PubMed] [Google Scholar]
- Wang Z, Fast W, Valentine AM, Benkovic SJ (1999) Metallo-beta-lactamase: structure and mechanism. Curr Opin Chem Biol 3: 614–622 [DOI] [PubMed] [Google Scholar]
- Wigler M, Sweet R, Sim GK, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R (1979) Transformation of mammalian cells with genes from prokaryotes and eukaryotes. Cell 16: 777–785222468 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure Sup_1. The Human ARTEMIS Protein
Figure Sup_2. In Vivo V(D)J Recombination Assay in Human Primary Dermal Fibroblasts
Figure Sup_3. In Vitro Nuclease Assay of ARTEMIS Double Mutants
Figure Sup_4. Sequence Analysis of V(D)J Recombination Coding Joints