Activation of caspases-8 and -10 by FLIPL (original) (raw)
Abstract
The first step in caspase activation is transition of the latent zymogen to an active form. For the initiator caspases, this occurs through dimerization of monomeric zymogens at an activating complex. Recent studies have suggested that FLIPL [FLICE-like inhibitory protein, long form; FLICE is FADD (Fas-associated death domain protein)-like interleukin-1β-converting enzyme], previously thought to act solely as an inhibitor of caspase-8 activation, can under certain circumstances function to enhance caspase activation. Using an in vitro induced-proximity assay, we demonstrate that activation of caspases-8 and -10 occurs independently of cleavage of either the caspase or FLIPL. FLIPL activates caspase-8 by forming heterodimeric enzyme molecules with substrate specificity and catalytic activity indistinguishable from those of caspase-8 homodimers. Significantly, the barrier for heterodimer formation is lower than that for homodimer formation, suggesting that FLIPL is a more potent activator of caspase-8 than is caspase-8 itself.
Keywords: apoptosis, caspase-8, FLIPL (FLICE-like inhibitory protein), protease, zymogen
Abbreviations: Ac-IETD-AFC, acetyl-Ile-Glu-Thr-Asp-7-amido-4-fluoromethylcoumarin; DED, death effector domain; DISC, death-inducing signalling complex; FADD, Fas-associated death domain protein; FLICE, FADD-like interleukin-1β-converting enzyme; FLIP, FLICE-like inhibitory protein; FLIPL, FLIP long form; FLIPS, FLIP short form; MMP, matrix metalloprotease; MT-MMP, membrane-type matrix metalloprotease; NFκB, nuclear factor κB; TIMP, tissue inhibitor of metalloproteases; Z-VAD-FMK, benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone
INTRODUCTION
Apoptotic cell death is carried out through the concerted action of caspases, a family of cysteine-dependent aspartate-specific proteases. The caspases exist within the cell as inactive zymogens and are activated via a proteolytic cascade, the first phase of which is initiator caspase activation. This is achieved via one of two pathways, classified on the basis of the origin of the death stimulus: the extrinsic (death receptor) pathway and the intrinsic (mitochondrial) pathway. Upon receipt of a death signal, initiator caspases are recruited to a multiprotein activating complex – the DISC (death-inducing signalling complex) for the extrinsic initiator caspases-8 and -10, and the apoptosome for the intrinsic initiator caspase-9. Both death pathways culminate with activation of the executioner caspases-3 and -7.
The mechanism by which initiator caspases are activated has been the subject of intense investigation. It was initially thought that all caspases are activated by limited proteolysis, with an activating complex serving to bring initiator caspase zymogens into such close proximity as to allow them to utilize a low level of activity to trans-process their neighbouring zymogen. While this model, espoused as the induced proximity hypothesis, offered a viable explanation given the data at the time, a unified model for apical caspase activation was recently proposed to encompass new findings in the field.
This unified model is based on two main findings: (i) apical caspase zymogens are monomeric, and (ii) proteolytic cleavage is dispensable for apical caspase activation. The unified model postulates that homodimerization of monomeric zymogens is necessary and sufficient for apical caspase activation. Dimerization allows structural rearrangements to order the catalytic site into an active conformation, independent of any proteolytic cleavage. In contrast with the apical caspases, proteolytic cleavage within the catalytic unit of the executioner caspases (caspases-3, -6 and -7) is absolutely required, primarily to permit translocation of the activation loop and subsequent ordering of the active site to occur. Certain predictions arise from the unified model, and perhaps the most intriguing of these is that heterodimerization functions as a mechanism for activating apical caspases.
FLIPL [FLICE-like inhibitory protein, long form; FLICE is FADD (Fas-associated death domain protein)-like interleukin-1β-converting enzyme] is a caspase-8 homologue, containing two N-terminal DEDs (death effector domains) and a caspase-like domain. Importantly, FLIPL lacks key residues necessary to form a protease active site, and is therefore devoid of any catalytic activity. The role of FLIPL in apoptosis has been hotly debated. The prevailing hypothesis was that FLIPL functions solely as an inhibitor of apoptosis, preventing caspase-8 activation at the DISC through competition for binding sites. While this model certainly holds true, under certain conditions where cellular levels of FLIPL are high, recent work has shown that FLIPL can act as an activator of caspase-8 [1,2]. In this paper we investigate in detail the mechanism by which FLIPL activates caspases-8 and -10, and we test the hypothesis that this activation is a natural consequence of dimerization predicted by the unified model.
EXPERIMENTAL
Recombinant protein expression and purification
ΔDED FLIPL (lacking the fist 186 residues) was subcloned into the _Nde_I and _Xho_I sites of a pET-15b derivative plasmid, providing an N-terminal His6 tag. The ΔDED FLIPL D297A mutant (residue numbering follows the caspase-1 convention [3]) was generated by Asp→Ala substitution at the underlined residue LEVD. The ΔDED FLIPL Q390D mutant was generated by a Gln→Ala mutation at VWLQ. ΔDED caspase-10 (lacking the first 202 residues) was subcloned into the _Nde_I and _Bam_HI sites of pET-15b, providing an N-terminal His5 tag. ΔDED non-cleavable caspase-10 mutant was generated by Asp→Ala substitution at IEAD297. ΔDED caspase-10 C285A mutant was generated by a Cys→Ala mutation of the conserved catalytic cysteine. ΔDED caspase-8, ΔDED non-cleavable caspase-8, ΔDED caspase-8 C285A, non-cleavable caspase-9 and caspase-9 C285A mutants were cloned as described in [4]. All mutations were generated by overlap PCR and confirmed by sequencing.
Expression and purification of caspases was carried out as described previously [5]. Non-cleavable caspase-8 contains a trace amount (∼1%) of fully active dimer. As the presence of this fully active enzyme complicated zymogen activation studies, it was removed by incubation with 10 μM Z-VAD-FMK (benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone) for 10 min at 37 °C. Unbound inhibitor was then removed by dialysis into 20 mM Tris (pH 8.0)/50 mM NaCl. The expression and purification of all ΔDED FLIPL proteins was carried out exactly as described for the caspases, with the exception that purification was carried out at pH 7.4 rather than pH 8.0. Purified ΔDED FLIPL proteins were dialysed into 20 mM Tris/HCl (pH 7.3)/200 mM NaCl and stored at −80 °C until the time of assay.
Tissue culture and preparation of cytosolic extracts
Jurkat cells were cultured in RPMI-1640 with 10% (v/v) fetal bovine serum, 100 units/ml penicillin and 2 mM L-glutamine. Jurkat cell cytosolic extracts were prepared as previously described [6].
Immunoblotting
Samples were resolved on an 8–18% (w/v) acrylamide gradient gel, then transferred to a PVDF membrane in 10 mM Caps, pH 11, and 10% (v/v) methanol at a constant current of 400 mA for 45 min. Following passivation in 10% (w/v) dried milk powder, the resulting blots were immunoblotted using the monoclonal anti-caspase-8 antibody C15 raised against an epitope of the large subunit (a kind gift from Markus Peter, University of Chicago, IL, U.S.A.) at 1:250 dilution overnight. Horseradish peroxidase-conjugated secondary antibody (1:7000; APBiotech, Piscataway, NJ, U.S.A.) was detected using SuperSignal detection reagents (Pierce Chemical Co., Rockford, IL, U.S.A.).
Immunoprecipitation
A 15 μl portion of Jurkat cytosolic extract containing 150 μg of total protein was used for each immunoprecipitation. Non-cleavable caspase-8 (15 nM) was prepared simultaneously in 15 μl of lysis buffer. This concentration of recombinant caspase-8 had been determined previously by quantitative Western blot to approximate that of endogenous caspase-8 in a cytosolic extract. Polyclonal anti-caspase-8 antiserum raised against recombinant caspase-8 was a gift from Dr Stan Krajewski (The Burnham Institute). This antiserum was coupled covalently to Protein A/G beads for immunoprecipitation experiments as described in [7]. Immunoprecipitates were diluted to 100 μl in mRIPA (50 mM Tris, pH 7.4, 100 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS) and 10 μl of coupled beads was added. Following a 16 h incubation at 4 °C, immunoprecipitates were washed three times with mRIPA. Samples were resolved on SDS/PAGE gels and immunoblotting was performed as described above. Samples destined for enzymic analysis were resuspended in mRIPA and a portion assayed as described below.
Gel filtration
Experiments were done using a Pharmacia AKTA purifier system equipped with a Superdex 200 HR 10/30 column with a flow rate of 0.5 ml/min (Amersham Pharmacia Biotech), and 0.5 ml fractions were collected for analysis. For separation of monomeric and dimeric ΔDED caspase-8, 20 mM Tris (pH 8.0)/100 mM NaCl was used, and leading and trailing fractions from protein peaks were collected respectively. The column was calibrated using gel filtration standards from Bio-Rad.
Enzymic assays
The indicated concentrations of recombinant caspases and ΔDED FLIPL mutants were preincubated in the presence of the indicated buffer for 20 min at 37 °C. The composition of the low-salt buffer was 20 mM Tris and 100 mM NaCl, pH 7.4. The kosmotrope (high salt) buffer composition was 0.7 M sodium citrate, 20 mM Tris, pH 7.4, and 100 mM NaCl. The indicated fluorogenic substrate was then added in a 10% volume of the appropriate low- or high-salt buffer to give a final concentration of 100 μM. Amidolytic activity was measured on an f-max Molecular Device spectrofluorimeter at 37 °C (excitation 405 nm, emission 510 nm). The active concentrations of all enzymes were determined by active-site titration using Z-VAD-FMK as described [8]. Prior to active-site titration, enzymes were preincubated with the indicated amount of ΔDED FLIPL or buffer control in the presence of low-salt buffer or kosmotrope buffer for 20 min at 37 °C. For kinetic assays on immunoprecipitated proteins, the equivalent of 5 μl of beads was added to the wells of a 96-well plate in 20 μl of mRIPA. FLIP or buffer control was added, and then low-salt or kosmotrope buffer to a final concentration of 0.7 M sodium citrate. Following a 20 min incubation at 37 °C, Ac-IETD-AFC (acetyl-Ile-Glu-Thr-Asp-7-amido-4-fluoromethylcoumarin) was added and activity measured as described above.
N-terminal sequencing of protein samples
Protein samples were resolved by SDS/PAGE and transferred to a PVDF membrane by electroblotting, and the membrane was briefly stained in Coomassie Brilliant Blue R250. Appropriate bands were excised and sequenced by Edman degradation at the University of Georgia Sequencing and Synthesis Facility.
RESULTS
Conventions and definitions
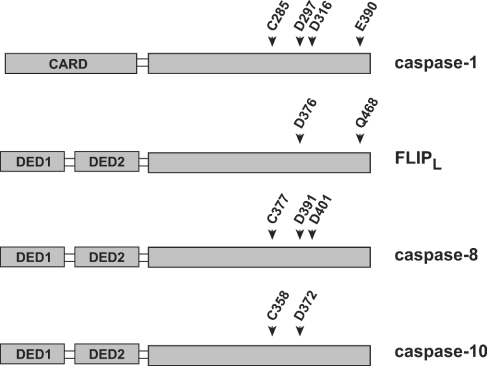
Throughout this paper the caspase-1 numbering convention is used as recommended to describe important residues in caspase-8, caspase-10 and FLIPL [3]. This allows easy translation of conserved structural elements across the caspase family members. Figure 1 provides a reference to the transition from caspase-1 numbering to the actual amino acids of the other proteins. As the presence of DEDs complicates expression of proteins in Escherichia coli, they were deleted from recombinant FLIPL, caspase-8 and caspase-10 mutants. For simplicity, the ΔDED prefix is omitted from the descriptions of these proteins throughout the remaining text.
Figure 1. Schematic representation of conserved caspase residues.
Conserved residues of the caspase family are shown. As the caspase-1 numbering system is used throughout, this Figure allows translation of conserved residues across the various members of the caspase family. For caspases-8 and -10, the numbering system describing isoform a is used.
Caspases frequently occur in two forms – cleaved (two chains) and uncleaved (single chain). This cleavage occurs at the interdomain linker and separates the large and small subunits. A caspase catalytic unit is composed of one large and one small subunit. We define a caspase containing one catalytic unit as a monomer, and one containing two catalytic units as a dimer. Regardless of cleavage status, an initiator caspase monomer is a zymogen, with the dimer being the active form.
FLIPL activates procaspase-8
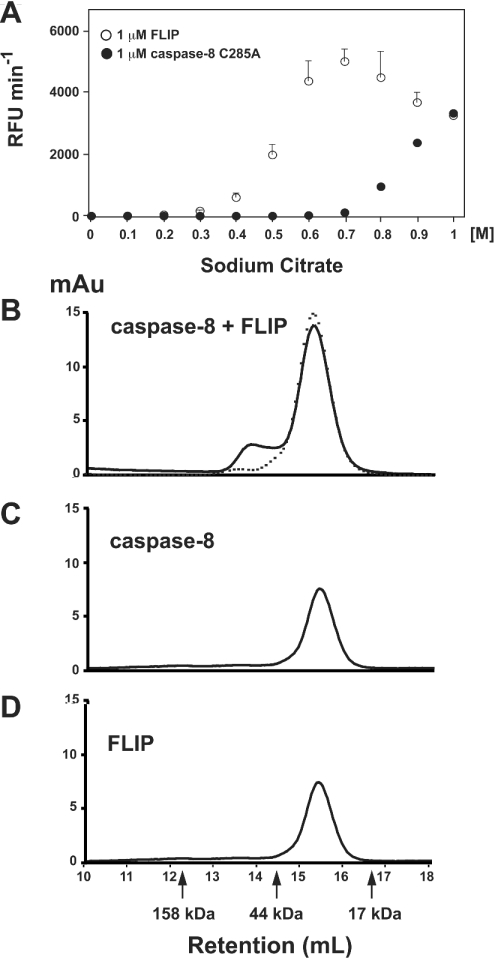
High concentrations of kosmotropic salts have long been a tool of researchers to study protein–protein interactions, as they are capable of salting out proteins from solution, creating local areas of high protein concentrations [9]. Previous work from our laboratory established that high concentrations of kosmotropic salts can induce dimerization of initiator caspases, providing a tool to recapitulate the events of induced proximity in vitro [4]. Therefore we tested the hypothesis that FLIPL can activate caspase-8 by observing caspase activity in the presence of FLIPL and kosmotrope. We used a non-cleavable form of caspase-8 (residues 297 and 316 mutated to Ala) to simulate the endogenous zymogen form, and asked whether it could be activated in the presence of kosmotrope by FLIPL. This enzyme contains 1% fully active dimer, and was treated with Z-VAD-FMK to remove this trace amount of fully active enzyme, as described in the Experimental section. As illustrated in Figure 2(A), FLIPL can indeed activate caspase-8, with a maximum activation at around 0.7 M sodium citrate under the conditions employed.
Figure 2. FLIPL activates caspase-8 by heterodimerization.
(A) Non-cleavable caspase-8 (50 nM) was incubated with either 1 μM FLIPL or 1 μM caspase-8 C285A at the indicated concentrations of sodium citrate at 37 °C, followed 20 min later by the addition of Ac-IETD-AFC. Substrate hydrolysis was followed at 37 °C, and is expressed as relative fluorescence units (RFU)·min−1. (B) Both non-cleavable caspase-8 and FLIPL (each at 7.5 μM) were incubated for 45 min at 37 °C in the presence of 10 mM dithiothreitol and 10 μM Z-VAD-FMK. Following incubation, samples were analysed by size exclusion chromatography on a Superdex 200 column. The broken line indicates samples prepared in the absence of Z-VAD-FMK. (C) Same as (B) with the exclusion of FLIPL; (D) same as (B) with the exclusion of caspase.
Postulating that FLIPL activation of caspase-8 resulted in a heterodimeric enzyme with one active site per heterodimer, we tested whether a caspase catalytic mutant would be able to activate its respective caspase pro-form in a FLIPL-like manner. Figure 2(A) compares the ability of caspase-8 C285A and FLIPL to activate non-cleavable caspase-8 in a kosmotrope-dependent fashion. Notably, FLIPL-mediated hetero-activation required much less kosmotrope than caspase-8-mediated homo-activation. The decrease in activity at high kosmotrope concentrations seen with FLIPL could be due to competition for homodimerization of either the caspase or FLIPL. Whatever the cause, the decrease was repeatable, as demonstrated by the low margin of error. A sodium citrate concentration of 0.7 M provided the largest difference between hetero-activation and homo-activation. Therefore throughout the subsequent studies this concentration of kosmotrope was used to study the kinetics of caspase–FLIPL interactions, as it provides the lowest background activity due to homo-activation.
Previous studies have shown that it can be difficult to capture the dimerized form of non-cleavable caspase-8 due to its rapid dissociation into monomers [4,10]. We therefore employed Z-VAD-FMK to trap the active caspase in a form suitable for analysis by size exclusion chromatography. As shown in Figure 2(B), co-incubation of FLIPL and non-cleavable caspase-8 in the presence of Z-VAD-FMK resulted in the formation of a complex that was eluted with the predicted size of a heterodimer (66 kDa). The final heterodimer/monomer ratio under these conditions was approx. 1:6. Omission of Z-VAD-FMK from the incubation abrogated the ability of the proteins to form heterodimers that were stable to gel filtration conditions. Incubation of either protein alone under identical conditions resulted in the protein eluting as a monomer [31 kDa for non-cleavable caspase-8 (Figure 2C) and 35 kDa for FLIPL (Figure 2D)], despite the fact that Z-VAD-FMK was present in these samples, indicating that homodimerization does not occur as readily as heterodimerization.
FLIPL preferentially activates the zymogen form of caspase-8
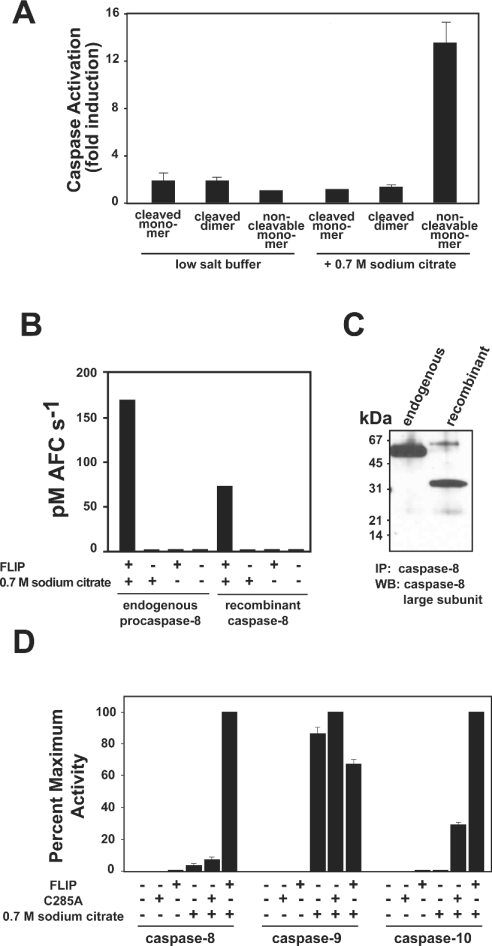
During overexpression in E. coli, high concentrations of proteins occur. In the case of caspase-8, this induces dimerization and subsequent autoprocessing [4,10]. Following purification, wild-type caspase-8 is found to exist as two distinct species, an inactive monomer and an active dimer, separable by size exclusion chromatography [4,10]. Given that cleavage of caspase-8 most often occurs as a post-activation event, we postulated that FLIPL would preferentially activate an uncleaved monomeric caspase-8, as this most closely approximates the zymogen form. In order to assess the ability of FLIPL to activate a zymogen-like form of caspase-8, we again utilized non-cleavable caspase-8. This protein remains unprocessed during expression and purification and is approx. 99% monomeric, with the remainder being a fully active uncleaved dimer [4]. This trace quantity of active enzyme was removed from subsequent zymogen activation studies by treatment with Z-VAD-FMK as described in the Experimental section. Incubation with an equimolar amount of FLIPL led to a modest increase in the activity of all forms of caspase-8. However, in the presence of 0.7 M sodium citrate only the non-cleavable form of caspase-8 was activated substantially by FLIPL, indicating that FLIPL preferentially activates the uncleaved form of caspase-8, which most accurately simulates the endogenous zymogen (Figure 3A).
Figure 3. Non-cleavable caspase-8 is activated most efficiently by FLIPL and closely approximates endogenous procaspase-8.
(A) Caspase-8 cleaved monomer, cleaved dimer or non-cleavable monomer at 75 nM was incubated with an equimolar amount of FLIPL in the presence of 20 mM Tris, pH 7.4, 100 mM NaCl (low-salt buffer) or kosmotrope buffer (containing 0.7 M sodium citrate). After 20 min at 37 °C, Ac-IETD-AFC substrate was added at 100 μM and substrate hydrolysis monitored at 37 °C. Results are expressed as fold increase over caspase alone under the respective buffer conditions. (B, C) Endogenous procaspase-8 from a Jurkat cytosolic extract or non-cleavable caspase-8 prepared in hypotonic buffer at a comparable concentration (15 nM) was immunoprecipitated using polyclonal antisera raised against recombinant caspase-8. (B) Immunoprecipitates were assayed for their ability to hydrolyse Ac-IETD-AFC substrate in the presence or absence of 1 μM FLIPL in either low-salt buffer or kosmotrope buffer as indicated. (C) Western blot of immunoprecipitates using monoclonal anti-caspase-8 antiserum raised against an epitope of the large subunit. (D) Non-cleavable caspase-8, -9 or -10 (500 nM) was added to buffer control, 1 μM of their respective C285A mutant or 1 μM FLIPL in either low-salt buffer or kosmotrope buffer for 20 min at 37 °C before the addition of substrate (Ac-IETD-AFC for caspase-8, Ac-LEHD-AFC for caspase-9, and Ac-DEVD-AFC for caspase-10).
Endogenous procaspase-8 can be activated by FLIPL
Non-cleavable caspase-8 resembles endogenous procaspase-8 in that both are uncleaved monomers. However, the recombinant proenzyme lacks the N-terminal DEDs and the ability to be cleaved during activation. To validate non-cleavable caspase-8 as a model for studying the activation of endogenous procaspase-8, we compared the ability of the two proteins to be activated by FLIPL in our in vitro induced-proximity assay. Endogenous procaspase-8 immunoprecipitated from a Jurkat cytosolic extract required the presence of 0.7 M sodium citrate and 1 μM FLIPL to achieve catalytic activity (Figure 3B). This result was similar to that obtained using an equivalent amount of non-cleavable caspase-8 that had been subject to identical immunoprecipitation conditions. Importantly, the activity achieved in the presence of kosmotrope and FLIPL manifested itself at comparable levels with the two enzymes. Figure 3(C) demonstrates that the enzymes were present in the assay at roughly comparable levels, as assessed by Western blot using monoclonal antisera. Taken together, these results validate our in vitro initiator caspase activation assay and demonstrate that endogenous full-length procaspase-8 is activated in a FLIPL-dependent manner.
Although our results suggest that the presence of DEDs on endogenous caspase-8 do not inhibit its activation by ΔDED FLIPL in the presence of kosmotrope, activation of caspase-8 by full-length FLIPL awaits further characterization. One possibility is that the DEDs serve to prevent adventitious activation by blocking the dimerization interface of the zymogens. Binding of the DEDs at the DISC would then relieve this block by freeing the dimerization interface. However, it is unlikely that this type of block is necessary, as our in vitro data suggest that the barrier for both hetero- and homo-dimerization is sufficiently high as to prevent unsolicited activation in the absence of an induced-proximity event (i.e. kosmotrope or binding at the DISC).
To determine if FLIPL can activate other initiator caspases, we tested its ability to activate the recombinant pro-forms of caspases-9 and -10. FLIPL activated non-cleavable caspases-8 and -10 in the presence and absence of kosmotrope, but under no conditions was non-cleavable caspase-9 activated by FLIPL (Figure 3D). Therefore FLIPL is a specific activator of initiator caspase zymogens of the extrinsic pathway. In the present study we focused on delineating the role of FLIPL in the modulation of apoptotic pathways, and thus restricted our studies to the canonical death caspases. More attention was given to caspases-8 and -10 as they, like FLIPL, contain N-terminal DEDs and are known to be recruited to the DISC. However, the formal possibility remains that FLIPL might influence the activity of other initiator caspase zymogens, such as caspases-1, -2 and -5, and this will probably depend on the complementarity of the dimer interface(s).
Cleavage of FLIPL is not required for the activation of procaspases-8 and -10
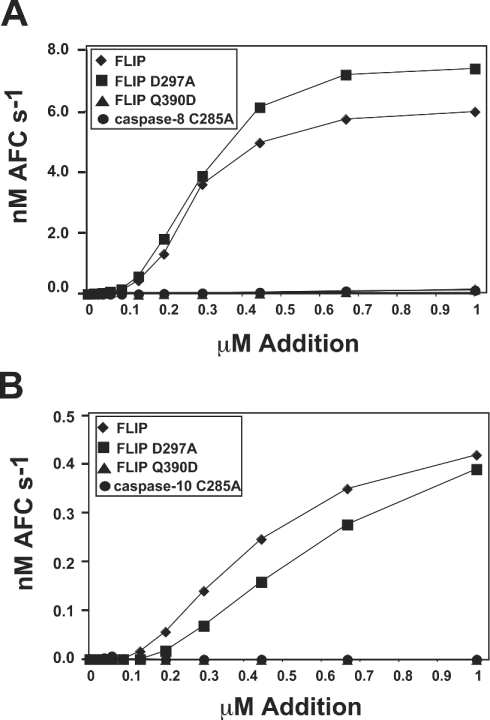
Several reports indicate that FLIPL is cleaved at the DISC by caspase-8 [2,11–13]. In our in vitro activation experiments, we found that a small proportion of FLIPL was cleaved at Asp297 during its activation of non-cleavable caspase-8, as confirmed by N-terminal sequencing (results not shown). To investigate the importance of this cleavage event for the activation process, we mutated this site to Ala, producing FLIPL D297A. We found this mutant to be as efficient in activating non-cleavable caspases-8 and -10 as wild-type FLIPL (Figures 4A and 4B), despite the fact that no detectable cleavage of FLIPL occurred during the course of the experiment (results not shown). Indeed, FLIPL D297A seemed to enhance activity of non-cleavable caspase-8 somewhat more efficiently than wild-type FLIPL.
Figure 4. Cleavage of FLIPL is not required for activation of caspase-8, but an ability to form heterodimers is.
(A) Non-cleavable caspase-8 (50 nM) was incubated with increasing concentrations of the indicated protein in the presence of 0.7 M sodium citrate for 20 min at 37 °C. Following the activation period, activity was monitored by Ac-IETD-AFC (AFC) hydrolysis. (B) ΔDED caspase-10 (50 nM) was treated as in (A). Following the incubation, activity was monitored by hydrolysis of Ac-DEVD-AFC. Symbols represent FLIP (♦), non-cleavable FLIP D287A (▪), dimer blocking FLIP Q390D (▴), and a catalytic mutant of caspase-8 or -10 (•).
Requirement of heterodimerization for activation of caspase-8 by FLIPL
To investigate the role of heterodimerization in the activation of procaspase-8 by FLIPL, a radical mutation was introduced at the predicted heterodimer interface of FLIPL, producing FLIPL Q390D [2]. This single point mutation is expected to prevent heterodimerization, and completely abrogated the ability of FLIPL to activate non-cleavable caspase-8 or -10 (Figures 4A and 4B).
In the interests of characterizing this novel heterodimeric protease, the catalytic parameters of FLIPL-activated non-cleavable caspase-8 were determined and compared with those of homodimeric wild-type caspase-8. As the titration experiment in Figure 4 illustrates, 50 nM caspase-8 was maximally activated by 1 μM FLIPL. Table 1 summarizes the findings of experiments using this ratio of enzyme to activator where appropriate. Incubation in 0.7 M sodium citrate improved the _K_m and _k_cat values for homodimeric wild-type caspase-8, probably due to stabilization of the active site. Incubation of the heterodimer in 0.7 M sodium citrate yielded a robust enzyme with _K_m and _k_cat values very similar to those of the wild-type enzyme. Notably, non-cleavable caspase-8 in 0.7 M sodium citrate did not possess measurable catalytic activity in the absence of FLIPL, indicating that the reported values are attributable entirely to heteroactivation.
Table 1. Kinetic comparison of homodimeric caspase-8 and heterodimeric caspase-8/FLIPL.
Values for enzyme concentration were obtained using active-site-titrated enzymes, as described in the Experimental section.
| Enzyme | Sodium citrate | [Enzyme] (nM) | _K_m (μM) | _k_cat (s−1) | 10−4×_k_cat/_K_m (M−1·s−1) |
|---|---|---|---|---|---|
| Cleaved caspase-8 | − | 58.0 | 55.2±2.0 | 0.65±0.1 | 1.2±0.2 |
| Cleaved caspase-8 | + | 29.0 | 20.6±0.3 | 1.25±0.2 | 6.1±1.1 |
| Non-cleavable caspase-8 | + | 50.0* | No measurable activity | ||
| Non-cleavable caspase-8+1 μM FLIP | + | 20.9 | 31.3±0.6 | 0.98±0.1 | 3.1±0.4 |
| Non-cleavable caspase-8+1 μM FLIP D297A | + | 20.9 | 29.3±0.7 | 0.78±0.1 | 2.7±0.4 |
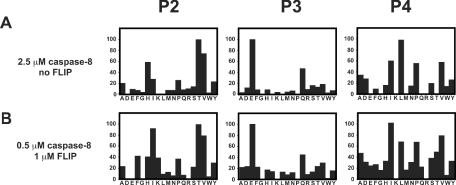
To characterize further the catalytic parameters of the homodimer and the heterodimer, a comparison of subsite specificity was performed using a positional scanning library with a fixed P1 aspartic acid. As illustrated in Figure 5, the subsite specificities of the homo- and hetero-dimers were very similar. Of notable importance is the finding that, in the absence of FLIP, non-cleavable caspase-8 exhibited such a low level of activity that in order to perform the experiment under these conditions 2.5 μM caspase was required.
Figure 5. Substrate specificity of the caspase-8 homodimer and the caspase-8/FLIPL heterodimer.
Caspase-8 was incubated in 0.7 M sodium citrate in the presence (A) or absence (B) of FLIPL and assayed with a positional scanning substrate library with P1 fixed as aspartic acid. The _y_-axis is the rate of hydrolysis presented as a percentage of the maximal rate observed. The _x_-axis provides the positionally defined L-amino acid (single-letter code).
DISCUSSION
The extrinsic apoptosis pathway is used to eliminate unwanted cells during development, immune system education, and immunosurveillance. It is initiated by ligation of a trans-membrane death receptor of the tumour necrosis factor receptor type 1 superfamily, which acts as a conduit for the transfer of apoptotic signals into a cell (reviewed in [14]). Upon ligation, death receptors form aggregates at the cell surface providing a platform, the DISC, for the intracellular recruitment of the extrinsic apoptotic machinery. In its simplest form the DISC is composed of Fas, the adapter FADD and caspase-8, and together these constitute the minimal caspase-8 or -10 activator. The fundamental activation event is dimerization of caspase-8 monomers, induced by proximity of the zymogen monomers at the cytosolic face of the DISC. The original induced-proximity model postulated that a low level of activity is intrinsic to initiator caspase zymogens, allowing them to trans-process their neighbouring zymogens upon recruitment to the DISC [15]. However, recent studies have determined that initiator caspase zymogens are in fact completely inactive monomers with an absolute requirement for dimerization for activation, and that clustering at oligomeric activation platforms lowers the barrier to dimer formation and concomitant activation (reviewed in [16]). Following dimerization to the catalytically active form, the N-terminal DEDs of caspase-8 are frequently removed, presumably allowing the activated caspase to be released into the cytosol [17].
FLIPL as a caspase activator
In many early studies, FLIPL was seen as an inhibitor of DISC formation because it acts as a competitor of caspase-8 or -10 recruitment [18–21]. This is seen most often with the short form of FLIP, FLIPS, which contains only the DEDs and is analogous to the DED-only viral FLIPs [22]. However, some of the initial reports on the full-length protein indicated that it could also be pro-apoptotic [13,23–25]. More recently this has been confirmed by others and the apparent conflict rationalized on the grounds that FLIPL may be able to form heterocomplexes with procaspase-8 that are proteolytically active [1,2]. Thus short forms of FLIP, lacking the caspase homology domain (c-FLIPS and viral FLIP) function solely as inhibitors, but FLIPL may be an inhibitor or activator of caspase-8, depending on the local concentrations at the DISC. In this scenario, high FLIP concentrations, found in many cancer cells, block caspase activation by saturating FADD molecules during DISC formation, preventing caspase-8 or -10 recruitment [1]. Lower concentrations favour caspase-8 or -10 activation [1], presumably by the heterodimerization mechanism described here.
Our data support this hypothesis, demonstrating for the first time proteolytic activity within the caspase-8/FLIPL heterodimer. We hypothesize that kosmotropic salts duplicate the situation in vivo by lowering the barrier to dimer formation of initiator caspases [4], and in this environment the kosmotrope-induced dimers simulate the activity of natural caspases-8 and -10 in their active forms. The mechanism of heterodimerization is readily understood, since FLIPL should in principle be conformationally similar to procaspases-8 and -10 [2] and therefore undergo similar protein interactions. Significantly, hetero-activation requires less kosmotrope than homo-activation, suggesting that heterodimerization of caspase-8 with FLIPL has a lower kinetic barrier than homodimerization of caspase-8.
The reasons for the lowered barrier are unknown. Probably the most important structures governing activation are loops that influence translation of the 340 activation loop through the central cavity of caspase dimers. Insertions into this cavity are seen in the zymogens of caspase-7, where they prevent translation of the 340 activation loop to its catalytic conformation [26,27]. In caspase-9 the 240 loop occludes the central cavity and favours translation of the analogous 340 activation loop [28]. The structure of FLIPL is unknown, but we note that it may contain insertions around the interface symmetry residue (390) with respect to other caspases, and also insertions in the interdomain linker (downstream of residue 283). We speculate that these insertions may provide a barrier to the integration of the interdomain linker into the central cavity at the dimer interface, thereby lowering the barrier to the translation of the all-important 340 activation loop of the caspase-8 unit of the heterodimer.
Our data imply that at the DISC, in the presence of equimolar FLIPL and caspase, a molecule of caspase-8 zymogen is more likely to form FLIP heterodimers than caspase homodimers. The stoichiometry of the DISC is unknown, and likely to vary based on the origin of the signal, the available concentrations of components, and the number of DISCs formed. The concentration of FLIPL within the cell has been estimated to be approx. 1% of that of caspase-8, while at the DISC there is an approximate 1:5 ratio of FLIPL to caspase-8, presumably due to differential preferences of the DEDs of the protein for binding sites of FADD [1].
There are some interesting similarities between FLIPL-mediated caspase-8 activation and activation of MMP-2 (matrix metalloprotease-2) mediated by its inhibitor, TIMP-2 (tissue inhibitor of metalloproteases-2). In the extracellular milieu, low or intermediate TIMP-2 concentrations drive the activation of MMP-2 by MT1-MMP (membrane-type 1 MMP), with TIMP-2 acting to recruit proMMP-2 to cell-surface-bound MT1-MMP [29]. High concentrations of TIMP-2, however, rapidly inhibit activated MMP-2 [30], and so the local concentrations of the inhibitor TIMP-2 regulate the activation of the enzyme MMP-2. Thus the concept of an inhibitor regulating ‘activate/don't activate’ signals is not unique, although the mechanisms of caspase-8/FLIPL and MMP-2/TIMP-2/MT1-MMP are radically different.
Role of FLIPL/caspase-8 heterodimers
Abundant evidence suggests that caspase-8 participates in specific cellular proliferative events, in addition to its broader role in initiating apoptotic events. For example, caspase-8 is required for T-cell proliferation, at least in humans [31]. A recent study suggests that cleavage of FLIPL by caspase-8 is required for FLIPL-mediated propagation of pro-life signals through the NFκB (nuclear factor κB) pathway [11]. Our findings clearly demonstrate that cleavage of FLIPL by caspase-8 does not influence the activity of the heterodimer. However, cleavage may function to reveal neo-epitopes in FLIPL that mediate signalling through the NFκB pathway. This demonstrates the absolute importance of being able to distinguish between proteolytic activation events and those that may have another role in modulating the function of the heterodimer.
We were not able to detect significant kinetic or specificity differences between the homodimers and the heterodimers by using tetrapeptide synthetic substrates, so there seem to be no intrinsic differences based on whether caspase-8 contains one active site or two, and whether it is activated by homodimerization or heterodimerization. However, this does not preclude differences in cellular targets of the two enzymes in vivo. Moreover, a number of studies have demonstrated that the DEDs of caspase-8 are removed during its activation at the DISC. However, removal of the DEDs of FLIPL has not been detected, probably due to the fact that it lacks the conserved cleavage site between the DEDs and the large subunit. Therefore the heterodimer presumably remains sequestered at the DISC and hence may be confined to cleaving local substrates, such as the kinase RIP (receptor-interacting kinase) [2]. Thus the cellular location of the active enzyme, i.e. DISC-bound or released from the DISC, may confer differential substrate utilization. Another intriguing possibility is that heterodimeric caspase-8 may have a decreased ability to process dimeric substrates when compared with homodimeric caspase-8. Given that the executioner caspase zymogens are dimeric, this could potentially explain proposed pro-life signalling events mediated by caspase-8/FLIP heterodimers, compared with pro-death signalling believed to be mediated by caspase-8. Further studies are required to address this issue, but this proposal could account in part for the differential roles of caspase-8 in cell death pathways and in cell proliferation pathways that may be modulated by FLIP.
Acknowledgments
We thank Scott Snipas and Annamarie Price for expert technical assistance, Markus Peter and Stan Krajewski for supplying caspase-8 antisera, and Pablo Fuentes-Prior for helpful discussions. This work was supported by California Breast Cancer Research Program Fellowship 8GB-0137 (K.M.B.) and NIH grant CA69381.
References
- 1.Chang D. W., Xing Z., Pan Y., Algeciras-Schimnich A., Barnhart B. C., Yaish-Ohad S., Peter M. E., Yang X. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micheau O., Thome M., Schneider P., Holler N., Tschopp J., Nicholson D. W., Briand C., Grutter M. G. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E. S., Livingston D. J., Nicholson D. W., Salvesen G., Thornberry N. A., Wong W. W., Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I., Ricci J.-E., Edris W. A., Sutherlin D. P., Green D. R., Salvesen G. S. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 5.Stennicke H. R., Salvesen G. S. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- 6.Deveraux Q., Takahashi R., Salvesen G. S., Reed J. C. X-linked IAP is a direct inhibitor of cell death proteases. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 7.Simanis V., Lane D. P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 8.Stennicke H. R., Salvesen G. S. Caspase assays. Methods Enzymol. 2000;322:91–100. doi: 10.1016/s0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt U., Darke P. L. Dimerization and activation of the herpes simplex virus type 1 protease. J. Biol. Chem. 1997;272:7732–7735. doi: 10.1074/jbc.272.12.7732. [DOI] [PubMed] [Google Scholar]
- 10.Donepudi M., MacSweeney A., Briand C., Gruetter M. G. Insights into the regulatory mechanism for caspase-8 activation. Mol. Cell. 2003;11:543–549. doi: 10.1016/s1097-2765(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka T., Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol. Cell. Biol. 2004;24:2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scaffidi C., Schmitz I., Krammer P. H., Peter M. E. The role of c-FLIP in modulation of CD95-induced apoptosis. J. Biol. Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 13.Shu H. B., Halpin D. R., Goeddel D. V. Casper is a FADD- and caspase-related inducer of apoptosis. Immunity. 1997;6:751–763. doi: 10.1016/s1074-7613(00)80450-1. [DOI] [PubMed] [Google Scholar]
- 14.Ashkenazi A., Dixit V. M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 15.Salvesen G. S., Dixit V. M. Caspase activation: The induced-proximity model. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boatright K. M., Salvesen G. S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Chang D. W., Xing Z., Capacio V. L., Peter M. E., Yang X. Inter-dimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irmler M., Thome M., Hahne M., Schneider P., Hofmann K., Steiner V., Bodmer J.-L., Schroter M., Burns K., Mattmann C., et al. Inhibition of death receptors signals by cellular FLIP. Nature (London) 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 19.Hu S., Vincenz C., Ni J., Gentz R., Dixit V. M. I-FLICE, a novel inhibitor of tumor necrosis factor receptor-1- and CD-95-induced apoptosis. J. Biol. Chem. 1997;272:17255–17257. doi: 10.1074/jbc.272.28.17255. [DOI] [PubMed] [Google Scholar]
- 20.Rasper D. M., Vaillancourt J. P., Hadano S., Houtzager V. M., Seiden I., Keen S. L., Tawa P., Xanthoudakis S., Nasir J., Martindale D., et al. Cell death attenuation by ‘Usurpin’, a mammalian DED-caspase homologue that precludes caspase-8 recruitment and activation by the CD-95 (Fas, APO-1) receptor complex. Cell Death Differ. 1998;5:271–288. doi: 10.1038/sj.cdd.4400370. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasula S. M., Ahmad M., Ottile S., Bullrich F., Banks S., Wang Y., Fernandes-Alnemri T., Croce C. M., Litwack G., Tomaselli K. J., et al. FLAME-1, a novel FADD-like anti-apoptotic molecule that regulates Fas/TNFR1-induced apoptosis. J. Biol. Chem. 1997;272:18542–18545. doi: 10.1074/jbc.272.30.18542. [DOI] [PubMed] [Google Scholar]
- 22.Thome M., Schneider P., Hofmann K., Fickenscher H., Meinl E., Neipel F., Mattmann C., Burns K., Bodmer J.-L., Schroter M., et al. Viral FLICE-inhibitory proteins (FLIPs) prevents apoptosis induced by death receptors. Nature (London) 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 23.Goltsev Y. V., Kovalenko A. V., Arnold E., Varfolomeev E. E., Brodianskii V. M., Wallach D. CASH, a novel caspase homologue with death effector domains. J. Biol. Chem. 1997;272:19641–19644. doi: 10.1074/jbc.272.32.19641. [DOI] [PubMed] [Google Scholar]
- 24.Han D. K. M., Chaudhary P. M., Wright M. E., Friedman C., Trask B. J., Riedel R. T., Baskin D. G., Schwartz S. M., Hood L. MRIT, a novel death-effector domain-containing protein, interacts with caspases and Bcl-XL and initiates cell death. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11333–11338. doi: 10.1073/pnas.94.21.11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inohara N., Koseki T., Hu Y., Chen S., Nunez G. CLARP, a death effector domain containing protein interacts with caspase-8 and regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10717–10722. doi: 10.1073/pnas.94.20.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl S. J., Fuentes-Prior P., Renatus M., Kairies N., Krapp R., Huber R., Salvesen G. S., Bode W. Structural basis for the activation of human procaspase-7. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chai J., Wu Q., Shiozaki E., Srinivasula S. M., Alnemri E. S., Shi Y. Crystal structure of a procaspase-7 zymogen. Mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 28.Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seiki M., Mori H., Kajita M., Uekita T., Itoh Y. Membrane-type 1 matrix metalloproteinase and cell migration. Biochem. Soc. Symp. 2003;70:253–262. doi: 10.1042/bss0700253. [DOI] [PubMed] [Google Scholar]
- 30.Howard E. W., Bullen E. C., Banda M. J. Regulation of the autoactivation of human 72-kDa progelatinase by tissue inhibitor of metalloproteinases-2. J. Biol. Chem. 1991;266:13064–13069. [PubMed] [Google Scholar]
- 31.Chun H. J., Zheng L., Ahmad M., Wang J., Speirs C. K., Siegel R. M., Dale J. K., Puck J., Davis J., Hall C. G., et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature (London) 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]