Development of Allergic Airway Disease in Mice following Antibiotic Therapy and Fungal Microbiota Increase: Role of Host Genetics, Antigen, and Interleukin-13 (original) (raw)
Abstract
Lending support to the hygiene hypothesis, epidemiological studies have demonstrated that allergic disease correlates with widespread use of antibiotics and alterations in fecal microbiota (“microflora”). Antibiotics also lead to overgrowth of the yeast Candida albicans, which can secrete potent prostaglandin-like immune response modulators, from the microbiota. We have recently developed a mouse model of antibiotic-induced gastrointestinal microbiota disruption that is characterized by stable increases in levels of gastrointestinal enteric bacteria and Candida. Using this model, we have previously demonstrated that microbiota disruption can drive the development of a CD4 T-cell-mediated airway allergic response to mold spore challenge in immunocompetent C57BL/6 mice without previous systemic antigen priming. The studies presented here address important questions concerning the universality of the model. To investigate the role of host genetics, we tested BALB/c mice. As with C57BL/6 mice, microbiota disruption promoted the development of an allergic response in the lungs of BALB/c mice upon subsequent challenge with mold spores. In addition, this allergic response required interleukin-13 (IL-13) (the response was absent in IL-13−/− mice). To investigate the role of antigen, we subjected mice with disrupted microbiota to intranasal challenge with ovalbumin (OVA). In the absence of systemic priming, only mice with altered microbiota developed airway allergic responses to OVA. The studies presented here demonstrate that the effects of microbiota disruption are largely independent of host genetics and the nature of the antigen and that IL-13 is required for the airway allergic response that follows microbiota disruption.
Animal models of allergic airway disease have been extremely useful in dissecting the manifestation of Th2 responses in the airways and the long-term airway remodeling that accompanies chronic inflammation. However, current animal models of allergic airway disease do not focus on the question, “What are the possible initiating events that lead to sensitization to allergens?”
The vast majority of murine models of allergic airway disease use systemic immunization prior to airway exposure. One of the most widely studied animal models of allergic airway disease uses airway challenge of mice previously sensitized with ovalbumin (OVA) (see, for example, references 5, 15, 34, 36, 39, and 43). In this model, normal mice are sensitized by immunization with OVA prior to airway challenge. One variation of this model does not require prior sensitization but uses mice (A/J) that are genetically deficient in C5 production (15, 39). Other models, using allergens such as cockroach antigen (4), dust mite antigen (42), or Aspergillus antigen (11, 24), also use an immunization step prior to airway challenge. A purified Aspergillus allergen protein that retains protease activity can stimulate airway Th2 responses without an adjuvant (16). However, humans are not exposed to this protease alone; rather, they are exposed to conidia or hyphal elements (18). Neither conidia nor hyphal elements can induce allergic airway disease without systemic priming (11, 19). Thus, in murine allergic airway disease models that use biochemically disparate allergenic compounds such as OVA and Aspergillus conidia, a common feature is the need to immunize systemically prior to challenge.
In humans, allergies do not arise from immunization with an allergen. They develop following exposure to the allergen (25-27). However, both animal and human studies have demonstrated that repeated airway exposure to an antigen leads to tolerance rather than sensitization (1, 2, 12, 35-38). Thus, there have been no animal models with which to study how airway exposure to allergens without prior systemic sensitization in immunocompetent individuals can lead to sensitization.
Noverr et al. have developed a mouse model of allergic airway disease that results from antibiotic therapy and microbiota disruption (30). This model includes stable increases in the levels of gastrointestinal (GI) entericbactera and Candida, with no introduction of microbes into the lungs. Mice (C57BL/6) are treated for 5 days with cefoperazone in the drinking water, followed by a single oral gavage of Candida albicans. This procedure results in an increase in the fungal microbiota and an alteration of the GI bacterial microbiota for at least 3 weeks. If these mice are subsequently exposed intranasally to mold spores (Aspergillus fumigatus conidia) without previous systemic priming, they develop a CD4 T-cell-mediated allergic airway response to the spores. Increased levels of eosinophils, mast cells, interleukin 5 (IL-5), IL-13, gamma interferon (IFN-γ), immunoglobulin E (IgE), and mucus-secreting cells characterize the allergic response in the lungs. In the absence of microbiota disruption, mice exposed to Aspergillus conidia do not develop an allergic response in the airways. This model demonstrates experimentally that it is feasible that a clinically common scenario (antibiotic therapy leading to bacterial and fungal microbiota changes) could predispose a host to allergic airway disease. It also opens up the possibility that afferent events in allergic sensitization may occur outside the lungs and involve host-microbiota communication. The objectives of the studies described here were to determine whether the effects of microbiota disruption were largely independent of host genetics and the nature of the antigen and to determine whether IL-13 was required for the airway allergic response that follows microbiota disruption.
MATERIALS AND METHODS
Mice.
Female BALB/c mice (18 ± 2 g) (Jackson Laboratories, Indianapolis, Ind.) or IL-13−/− mice (breeding colony, University of Michigan Unit for Laboratory Animal Medicine) were housed under specific-pathogen-free conditions in enclosed filter-top cages. Food and sterile water were given ad libitum. The mice were maintained by the Unit for Laboratory Animal Medicine at the University of Michigan (Ann Arbor, Mich.), and protocols were approved by an animal institutional review board.
Antibiotic treatment.
Cefoperazone (0.5 mg/ml) (Sigma-Aldrich, St. Louis, Mo.) was administered orally to mice ad libitum in drinking water. Antibiotic treatment was continued for 5 days to allow for C. albicans colonization. After 5 days, drinking water was replaced with sterile water. Fecal cultures were analyzed at the termination of antibiotic therapy to confirm the efficacy of the antibiotic in decreasing GI microbiota populations. To ensure consistent microbiota recolonization, an untreated mouse was always housed with the antibiotic-treated mice.
C. albicans GI inoculation.
C. albicans strain CHN1 (a human pulmonary clinical isolate) was grown to stationary phase (72 h) at 37°C in Sabouraud dextrose broth (1% neopeptone, 2% dextrose [Difco, Detroit, Mich.]) with shaking. For infection, the cultures were washed in sterile nonpyrogenic saline, counted by using a hemocytometer, and diluted to 2 × 108 CFU/ml in sterile nonpyrogenic saline. Mice were inoculated with C. albicans (107 CFU in 50 μl) by oral administration using a 24-gauge feeding needle attached to a 1-ml syringe. The syringe containing C. albicans was mounted on a Stepper repetitive pipette (Tridak, Brookfield, Conn.) to deliver an equivalent amount of inoculum to each mouse. Aliquots of the inoculum were analyzed for the number of CFU to monitor the amount delivered.
Intranasal inoculation with A. fumigatus conidia.
A. fumigatus (ATCC 13073) was grown on Sabouraud dextrose agar (SDA; Difco) for 14 days. Conidia were harvested by washing plates with sterile 0.1% Tween 80. The resulting fungal suspension was then filtered through two layers of sterile gauze to remove hyphae. For infection, the conidia were washed in nonpyrogenic saline (Abbott Laboratories, Chicago, Ill.), counted by using a hemocytometer, and diluted to 109/ml in sterile nonpyrogenic saline in order to administer 107 conidia/mouse in a 10-μl volume. Prior to intranasal inoculation, mice were anesthetized by intraperitoneal injection with a ketamine-xylazine solution (2.5 mg of ketamine [Fort Dodge Animal Health, Fort Dodge, Iowa]/mouse plus 0.1 g of xylazine [Lloyd Laboratories, Shenandoah, Iowa]/mouse). Mice were given a 5-μl bolus of A. fumigatus in each nostril followed by a 10-μl flush with sterile saline in each nostril.
Intranasal challenge with OVA.
OVA (Sigma-Aldrich) was resuspended in nonpyrogenic saline (Abbott Laboratories) to a final concentration of 10 mg/ml. Prior to intranasal inoculation, mice were anesthetized by intraperitoneal injection with a ketamine-xylazine solution (2.5 mg of ketamine [Fort Dodge Animal Health]/mouse plus 0.1 g of xylazine [Lloyd Laboratories]/mouse). Mice were given a 5-μl bolus of OVA in each nostril (total ovalbumin, 50 μg) followed by a 10-μl flush with sterile saline in each nostril.
Tissue CFU assay.
Murine tissues were excised, placed in tubes containing 10 ml of sterile water, weighed, and homogenized mechanically by using a Tissue-tearor (Biospec Products, Bartlesville, Okla.). Aliquots of the homogenates were plated out in 10-fold dilutions. Several types of media were used to select for bacteria or yeast (SDA for yeast [Difco], violet red bile agar [VRBA; Difco] for facultatively anaerobic coliform bacteria, and Trypticase soy agar with 5% sheep blood [TSA II; BD Biosciences, Franklin Lakes, N.J.] for total bacteria). SDA and VRBA plates were incubated aerobically at 37°C, and TSA II plates were incubated under anaerobic conditions (85% N2, 10% CO2, and 5% H2) at 37°C. Colonies were counted 24 h later.
Lung leukocyte culture and cytokine ELISA.
Mice were euthanized by CO2. Lungs were excised, minced, and enzymatically digested as described previously (32a). Cell concentrations were determined by counting cells diluted in trypan blue by using a hemocytometer. Isolated leukocytes from individual mice were standardized to 5 × 106 cells/ml and cultured in complete medium without additional stimulation at 37°C under 5% CO2. Supernatants were harvested at 24 h and assayed for cytokine production by an enzyme-linked immunosorbent assay (ELISA) (OptEIA; Pharmingen, San Diego, Calif.).
Cell staining.
Leukocyte differentials (neutrophils, eosinophils, macrophages, and moncytes/lymphocytes) were visually counted after Wright-Giemsa staining of lung leukocyte samples cytospun onto glass slides (Shandon Cytospin, Pittsburgh, Pa.). The percentage of a leukocyte subset was multiplied by the total number of leukocytes to yield the absolute number of that leukocyte subset.
RESULTS
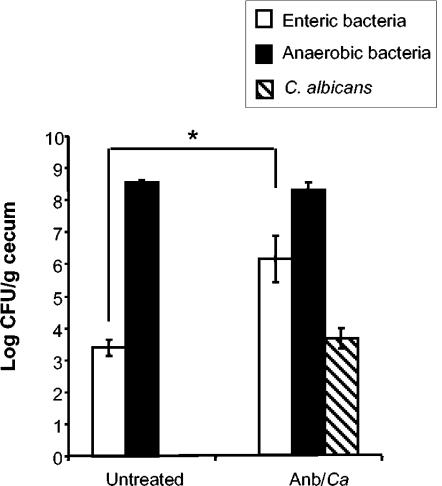
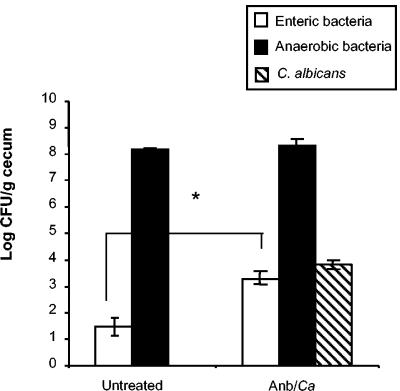
To establish a model of altered GI microbiota that included reproducible yeast overgrowth, C57BL/6 mice were first treated with a short course of a broad-spectrum antibiotic (cefoperazone for 4 days) in the drinking water to decrease total GI bacterial microbiota, followed by a single oral dose of C. albicans to establish a reproducible, low-level elevation of yeast numbers in the microbiota (Fig. 1). This protocol reduced levels of anaerobic and enteric bacteria in the gut by 99.99% at day 4 of antibiotic treatment and resulted in elevated C. albicans levels in the GI tract (30). When antibiotic treatment was discontinued (day 0), the numbers of both anaerobic and enteric bacteria increased (30). However, recolonization by enteric bacteria following termination of antibiotics resulted in enteric bacterial levels that were 10- to 100-fold higher at day 12 post-antibiotic treatment than those in untreated mice (Fig. 2). Levels of C. albicans in the GI tract decreased during regrowth of the bacterial microbiota but remained elevated at day 12 post-antibiotic treatment (data not shown and Fig. 2). C. albicans did not cause overt disease symptoms and did not disseminate from the GI tract (data not shown). Thus, 12 days after antibiotic treatment, the GI microbiota of these mice was still disrupted and was characterized by an elevated ratio of enteric to anaerobic bacteria combined with low-level persistence of C. albicans.
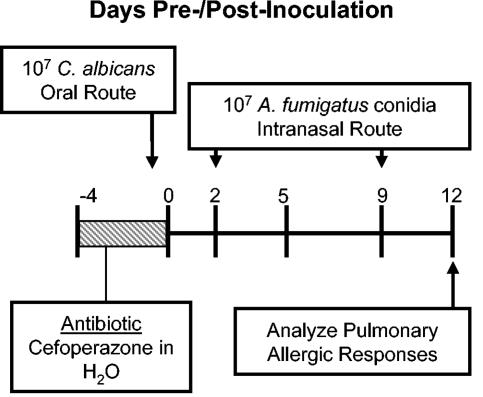
FIG. 1.
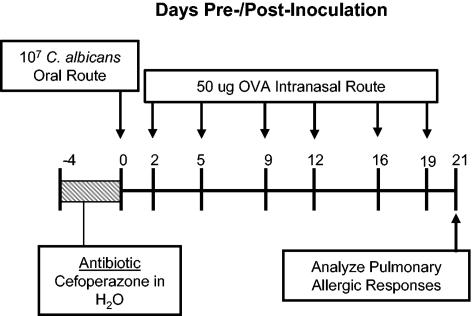
Experimental timeline for conidium challenge model of pulmonary hypersensitivity. BALB/C or IL-13−/− mice were given oral cefoperazone (0.5 mg/ml) for 5 days (day −4 through day 0). At day 0, C. albicans (107 CFU) was administered orally, and mice were challenged (days 2 and 9) by intranasal exposure to A. fumigatus (107 conidia/mouse). Mice were analyzed at day 12 post-C. albicans inoculation.
FIG. 2.
Effect of antibiotic treatment on microbiota populations. At day 12 post-C. albicans inoculation, murine ceca were harvested, homogenized in sterile water, and plated on the following media for enumeration of cecal bacterial populations: VRBA for enteric bacteria, TSA II blood agar for total anaerobic bacteria, and SDA for yeast. Groups are as follows: untreated (conidium challenged, no antibiotic, no C. albicans inoculation) and Anb/Ca (conidium challenged, antibiotic treated, C. albicans oral inoculation). n = 6 to 7 mice per time point pooled from two separate experiments.
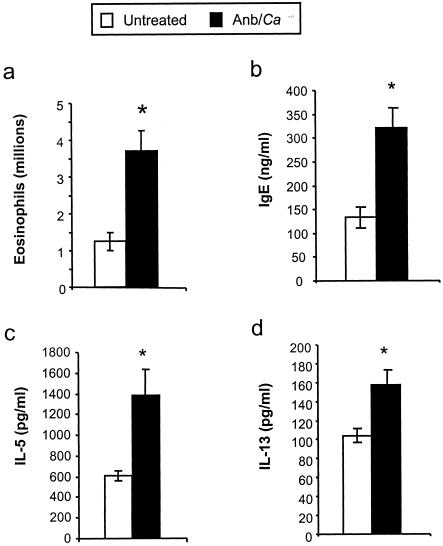
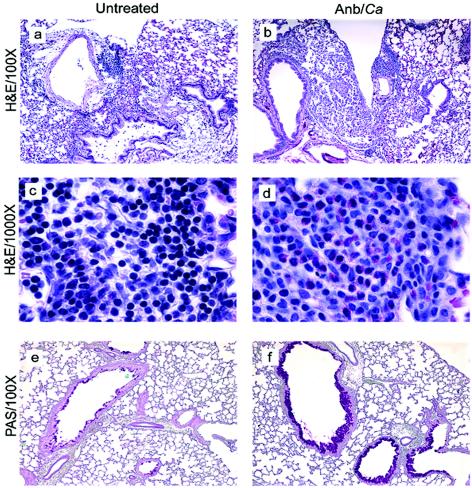
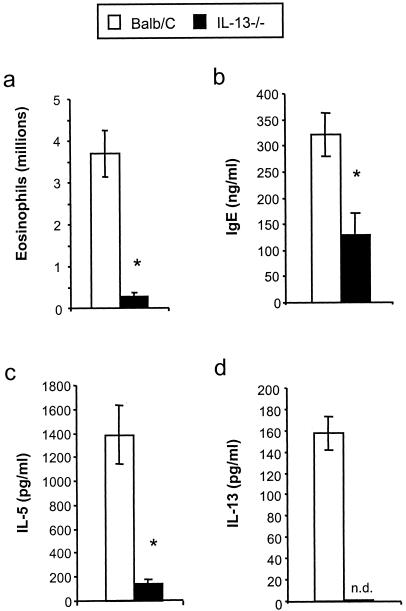
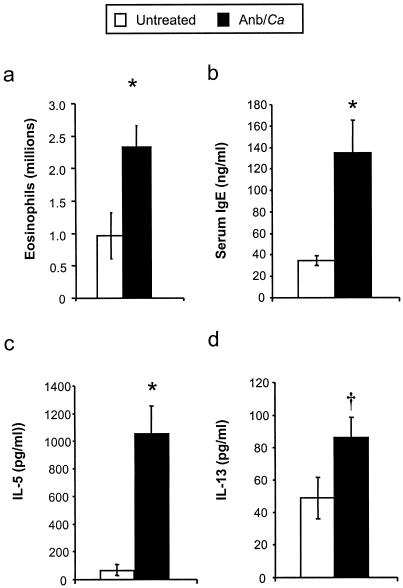
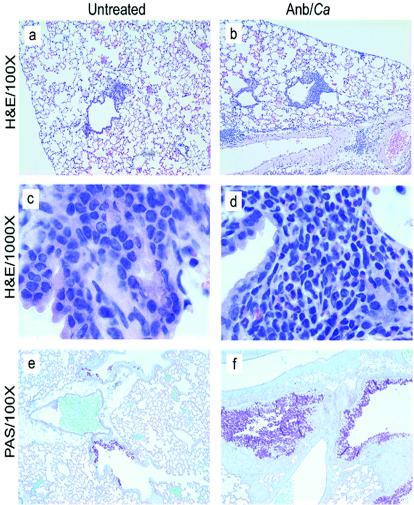
We exposed BALB/c mice intransally to mold spores (A. fumigatus conidia) at days 2 and 9 post-antibiotic treatment (Fig. 1) to determine whether the allergic response observed in microbiota-disrupted mice was restricted to the C57BL/6 host genetic background. The pulmonary immune response following conidial challenge in Anb/Ca (antibiotic-treated, with persistent GI Candida growth) BALB/c mice was significantly different from that in untreated BALB/c mice. Conidial challenge of Anb/Ca mice produced (i) a significant increase in the number of eosinophils in the lungs, (ii) high serum IgE levels, and (iii) elevated IL-5 and IL-13 production (Fig. 3). The eosinophilic nature of the inflammatory response was also evident by histological analysis of the lungs (Fig. 4b and d). In addition, the high IL-13 levels (Fig. 3d) were accompanied by increased goblet cell metaplasia, as indicated by the increased number of cells staining with periodic acid-Schiff stain (PAS) in the airways (Fig. 4f). We were unable to demonstrate that the lung leukocytes produced IL-4, but the increase in serum IgE levels indirectly indicated that IL-4 was induced in these mice (Fig. 3b). In the absence of microbiota disruption, the response in the airways consisted of a few eosinophils; low-level production of IL-5, IL-13, and serum IgE; and minimal goblet cell metaplasia (Fig. 3 and 4a, c, and e). IFN-γ levels and neutrophil numbers (data not shown) were slightly elevated in all conidium-challenged mice, regardless of antibiotic treatment. Control mice not challenged with mold spores exhibited no pulmonary hypersensitivity responses regardless of microbiota manipulation (data not shown), indicating that the microbiota manipulation alone did not induce a hypersensitivity response. The airway response to microbiota disruption in BALB/c mice was virtually identical to that in C57BL/6 mice (30). Thus, these results demonstrate that genetics does not play a major role in the allergic airway disease that accompanies microbiota disruption in this model.
FIG. 3.
Effects of microbiota perturbation on pulmonary eosinophil recruitment, Th2 cytokine production by lung leukocytes, and serum IgE production in conidium-challenged mice. Both groups of mice were challenged with conidia. Mice were treated as diagrammed Fig. 1. Leukocytes were isolated from lungs by enzymatic digestion and mechanical dispersion. (a) Eosinophils were phenotyped by Wright-Giemsa staining of cytospun samples. Results are expressed as the mean number of leukocytes per mouse ± standard error of the mean. (b) Serum IgE concentrations were measured by ELISA. (c and d) For Th2 cytokine measurements, leukocytes were isolated from whole lungs and cultured for 24 h without additional stimulation. Supernatants were collected and assayed for IL-5 (c) and IL-13 (d) by ELISA. Results are expressed as the mean ± standard error of the mean. n = 7 to 8 mice pooled from two separate experiments. Groups are as follows: untreated (conidium challenged, no antibiotic, no C. albicans inoculation) and Anb/Ca (conidium challenged, antibiotic treated, C. albicans oral inoculation). *, P < 0.05 as determined by Student's t test.
FIG. 4.
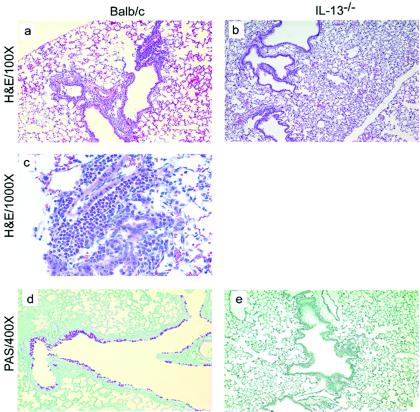
Histological analysis of lungs of mice exposed to conidia. Mice were treated as outlined in Fig. 1. At day 12 post-C. albicans inoculation, lungs were harvested, fixed, sectioned, and stained with hematoxylin and eosin (H&E), which differentially stains leukocytes (a-d). Lung sections were also stained with PAS-hematoxylin, which stains mucus pink (e, f). Groups are as follows: untreated (conidium challenged, no antibiotic, no C. albicans inoculation) and Anb/Ca (conidium challenged, antibiotic treated, C. albicans oral inoculation).
Our next objective was to test whether IL-13 was required for manifestation of the allergic airway response. High-level production of IL-13 is both necessary and sufficient to induce the features of allergic disease, including goblet cell metaplasia (21-23, 40). BALB/c IL-13+/+ and IL-13−/− mice were treated with antibiotics and allowed to establish low-grade Candida persistence prior to intranasal conidial challenge (as outlined in Fig. 1). In IL-13−/− Anb/Ca mice, pulmonary eosinophil numbers, lung IL-5 and IL-13 production, serum IgE levels, and goblet cell metaplasia did not increase following conidial challenge (Fig. 5 and 6b and e). In contrast, all of these allergic response indices were elevated in IL-13+/+ (wild-type) Anb/Ca mice (Fig. 5 and 6a, c, and d). Therefore, the inflammatory response in the lungs of mice with disrupted microbiota following mold spore exposure requires CD4+ T cells and IL-13, well-described hallmarks of allergic airway disease in mice and humans.
FIG. 5.
Effects of microbiota perturbation on pulmonary eosinophil recruitment, Th2 cytokine production by lung leukocytes, and serum IgE production in conidium-challenged IL-13−/− mice. Mice were treated as outlined in Fig. 1. Leukocytes were isolated from lungs by enzymatic digestion and mechanical dispersion. (a) Eosinophils were phenotyped by Wright-Giemsa staining of cytospun samples. Results are expressed as the mean number of leukocytes per mouse ± standard error of the mean. (b) Serum IgE concentrations were measured by ELISA. (c and d) For Th2 cytokine measurements, leukocytes were isolated from whole lungs and cultured for 24 h without additional stimulation. Supernatants were collected and assayed for IL-5 (c) and IL-13 (d) by ELISA. Results are expressed as means ± standard errors of the means. n = 7 to 8 mice pooled from two separate experiments. Both BALB/c and IL-13−/− mice were challenged with A. fumigatus, treated with an antibiotic, and orally inoculated with C. albicans. n.d., not detected. *, P < 0.05 as determined by Student's t test.
FIG. 6.
Histological analysis of lungs of IL-13−/− mice exposed to A. fumigatus. Mice were treated as outlined in Fig. 1. At day 12 post-C. albicans inoculation, lungs were harvested, fixed, sectioned, and stained with hematoxylin and eosin (H&E), which differentially stains leukocytes (a-c). Lung sections were also stained with PAS-methyl green, which stains mucus pink (d, e). Both BALB/c and IL-13−/− mice were challenged with A. fumigatus, treated with an antibiotic, and orally inoculated with C. albicans.
In addition to mold spores, a number of aerosolized allergens, including pollens, dander, dust mite particles, and cockroach feces, can elicit an allergic response in the airways. Our objective was to demonstrate that the allergic airway disease following microbiota disruption was not restricted to mold spores, especially since the presence of the fungus C. albicans in the GI tract during priming appears to be required. Thus, we tested Sigma fraction V OVA as an allergen because it is not a fungal product, it is commonly used in animal models of allergic airway disease (5, 15, 34, 36, 39, 43), and allergic responses to this “antigen” require some form of systemic priming before inhalation exposure (6).
A vigorous allergic airway disease also developed in BALB/c mice with disrupted microbiota following repeated intranasal OVA challenge. Despite the use of a different type of intranasal antigenic challenge (Fig. 7), the microbiota disruption protocol created the same general changes in the GI microbiota of OVA-challenged BALB/c mice as in mold spore-challenged BALB/c mice (Fig. 2 and 8). The pulmonary immune response following OVA exposure in Anb/Ca BALB/c mice was significantly different from that in untreated BALB/c mice. Exposure of Anb/Ca mice to OVA produced (i) a significant increase in the number of eosinophils in the lungs, (ii) high serum IgE levels, and (iii) elevated IL-5 and IL-13 production (Fig. 9). The eosinophilic nature of the inflammatory response was also evident in histological analysis of the lungs (Fig. 10b and d). Again, we were unable to demonstrate that the lung leukocytes produced IL-4, but the increase in serum IgE levels indirectly indicated that IL-4 was induced in these mice (Fig. 9b). In the absence of microbiota disruption, the response in the airways consisted of a few eosinophils and low-level production of IL-5, IL-13, and serum IgE (Fig. 9 and 10a and c). IFN-γ levels and neutrophil numbers (data not shown) were slightly elevated in all mice exposed to fraction V OVA, regardless of antibiotic treatment. The most striking change following microbiota disruption was the development of a widespread goblet cell metaplasia (Fig. 10f) that was even more pronounced than that in mice challenged with Aspergillus conidia (Fig. 4f and 6d). Overall, these experiments demonstrate that the allergic airway disease following microbiota disruption is not restricted to mold spores and can be stimulated by the “model allergen” Sigma fraction V OVA.
FIG. 7.
Experimental timeline for OVA challenge model of pulmonary hypersensitivity. BALB/c mice were given oral cefoperazone (0.5 mg/ml) for 5 days (day −4 through day 0). At day 0, C. albicans (107 CFU) was administered orally. Mice were challenged (days 2, 5, 9, 12, 16, and 19) with intranasal ovalbumin (50 μg/mouse). Mice were analyzed at day 21 post-C. albicans inoculation.
FIG. 8.
Effect of antibiotic treatment on cecal microbiota populations. At day 21, ceca were harvested and plated on the following media to enumerate cecal bacterial populations: VRBA for enteric bacteria, TSA II blood agar for total anaerobic bacteria, and SDA for yeast. n = 7 mice per time point pooled from two separate experiments. Groups are as follows: untreated (OVA challenged, no antibiotic, no C. albicans inoculation) and Anb/Ca (OVA challenged, antibiotic treated, C. albicans oral inoculation). *, P < 0.05 as determined by Student's t test.
FIG. 9.
Effects of microbiota perturbation on pulmonary eosinophil recruitment, Th2 cytokine production by lung leukocytes, and serum IgE production in OVA-challenged mice. Both groups of mice were challenged with OVA. Mice were treated as diagrammed in Fig. 7. Leukocytes were isolated from lungs by enzymatic digestion and mechanical dispersion. (a) Eosinophils were phenotyped by Wright-Giemsa staining of cytospun samples. Results are expressed as the mean number of leukocytes per mouse ± standard error of the mean. (b) Serum IgE concentrations were measured by ELISA. (c and d) For Th2 cytokine measurements, leukocytes were isolated from whole lungs and cultured for 24 h without additional stimulation. Supernatants were collected and assayed for IL-5 (c) and IL-13 (d) by ELISA. Results are means ± standard errors of the means. n = 7 mice pooled from two separate experiments. Groups are as follows: untreated (OVA challenged, no antibiotic, no C. albicans inoculation) and Anb/Ca (OVA challenged, antibiotic treated, C. albicans oral inoculation). *, P < 0.05 as determined by Student's t test; †, P < 0.05 as determined by the Mann-Whitney test.
FIG. 10.
Histological analysis of lungs of OVA-challenged mice. Mice were treated to disrupt the microbiota and were challenged intranasally with OVA as outlined in the experimental design (Fig. 7). At day 21 post-C. albicans inoculation, lungs were harvested, fixed, sectioned, and stained with hematoxylin and eosin (H&E), which differentially stains leukocytes (a-c). Lung sections were also stained with PAS-methyl green, which stains mucus pink (d, e). Groups are as follows: untreated (OVA challenged, no antibiotic, no C. albicans inoculation) and Anb/Ca (OVA challenged, antibiotic treated, C. albicans oral inoculation).
DISCUSSION
The rates of allergy and asthma in Westernized countries have increased significantly over the past 4 decades, and increased antibiotic use and altered fecal microbiota (microflora) both correlate with these increased rates of allergies and asthma (reviewed in references 29 and 41). Germfree animals also display numerous defects in the regulation of immune responses (20), raising the possibility that changes in the microbiota are a predisposing factor for development of allergic responses in the airways. One of the most common changes in the microbiota following antibiotic therapy is increased numbers of fungal microbiota, notably the yeast C. albicans, a member of the normal microbiota in humans (3). Control of C. albicans numbers by the normal bacterial microbiota is very important. C. albicans (and many other fungi) secretes prostaglandin-like oxylipin molecules de novo or via conversion of exogenous arachidonic acid (31, 32). Oxylipins are potent immunomodulatory molecules, providing a potential mechanism by which growth of fungal microbiota in a mucosal site could potentiate or alter immune responses on the mucosa (28). The animal model described in this report utilizes antibiotic therapy and low-grade GI Candida persistence to model a common clinical scenario. Our objective was to use this model to address whether microbiota disruption could predispose an individual to allergic airway disease via a mechanism that is independent of genetics or the type of antigen (fungal versus nonfungal).
In this report, we have demonstrated that microbiota disruption can alter immune regulation in the airways, leading to upregulation of Th2 responses to both mold spore and OVA exposure in BALB/c mice. A similar response to mold spore challenge in C57BL/6 mice with disrupted microbiota has been reported previously (30). The most notable feature of this model is that systemic priming is not necessary to generate an immune response in the airways to antigen exposure. Intranasal antigen exposure of mice with disrupted microbiota will elicit a Th2 immune response rather than a tolerogenic response (1, 2, 12, 35-38). Thus, antibiotic therapy and increased fungal microbiota persistence can disrupt normal airway tolerance, regardless of the genetic background of the host (BALB/c versus C57BL/6) or the biochemical nature of the antigen (A. fumigatus conidia versus OVA) to which the host is exposed.
We chose to compare the responses in BALB/c and C57BL/6 mice because these two inbred mouse strains with disparate major histocompatibility complex classes have been reported to differ significantly in the polarization of their T-cell responses to a number of antigens both systemically and in the airways (7, 10, 13, 14, 17, 33). These two mouse strains have been strongly suggested to have genetic predispositions to polarize in opposite directions toward either Th1 or Th2 responses, depending on the stimulus (7, 10, 13, 14, 17, 33). Thus, testing the microbiota disruption model on these two mouse strains provides a proof in principle that genetics does not play a major role in this process.
We have also shown that the airway allergic response in mice with disrupted microbiota is driven by IL-13. This Th2 cytokine is functionally similar to IL-4; the IL-13 receptor complex is composed of a low-affinity IL-13α chain and the IL-4 receptor α subunit (9). IL-13 is produced not only by both CD4+ and CD8+ T cells but also by granulocytic cells involved in allergic responses (eosinophils, basophils, mast cells) (22). Both IL-4 and IL-13 are involved in perpetuating Th2 responses; however, IL-13 is a key regulator of allergy and asthma and is both necessary and sufficient for induction of all features of allergic disease (21-23, 40). It has been shown previously that CD4+ T cells are required for the response (30). In addition to the Th2 nature of the response, IFN-γ was also produced by lung leukocytes during the inflammatory response. The production of IFN-γ and Th2 cytokines in the lungs is consistent with an allergic airway response (8).
The afferent events in this model of allergic disease remain to be dissected but appear to be strongly influenced by the microbiota of the host. As hypothesized in more detail in other reports, there are striking correlates between oral tolerance, airway tolerance, intact microbiota, and the development of regulatory T cells (29, 30). Thus, one possibility is that the microbiota plays a role in promoting the development of regulatory T cells that downregulate Th2 responses in the airways. Future studies will address this potential mechanism. Altogether, the immunologic studies in this and our previous work indicate that intranasal antigen challenge of mice with disrupted microbiota will produce a classic allergic-type response in the airways that is absent if the microbiota is intact. The present studies demonstrate that the allergic response can be generated in genetically disparate inbred mouse strains by using both fungal and nonfungal antigens.
Acknowledgments
We thank Galen Toews for scholarly contributions and Rachael Noggle for technical support.
This work was supported by a New Investigator Award in Molecular Pathogenic Mycology from the Burroughs-Wellcome Fund (to G.B.H.) and grants RO1-HL65912 and RO1-AI059201 (to G.B.H.) from the National Institutes of Health. M.C.N. was supported by NIH-NHLBI training grant 2T32HL007749-11.
REFERENCES
- 1.Akbari, O., R. H. DeKruyff, and D. T. Umetsu. 2001. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat. Immunol. 2**:**725-731. [DOI] [PubMed] [Google Scholar]
- 2.Andrew, D. K., R. R. Schellenberg, J. C. Hogg, C. J. Hanna, and P. D. Pare. 1984. Physiological and immunological effects of chronic antigen exposure in immunized guinea pigs. Int. Arch. Allergy Appl. Immunol. 75**:**208-213. [DOI] [PubMed] [Google Scholar]
- 3.Calderone, R. A. (ed.) 2001. Candida and candidiasis. ASM Press, Washington, D.C.
- 4.Campbell, E. M., I. F. Charo, S. L. Kunkel, R. M. Strieter, L. Boring, J. Gosling, and N. W. Lukacs. 1999. Monocyte chemoattractant protein-1 mediates cockroach allergen-induced bronchial hyperreactivity in normal but not CCR2−/− mice: the role of mast cells. J. Immunol. 163**:**2160-2167. [PubMed] [Google Scholar]
- 5.Corry, D. B., H. G. Folkesson, M. L. Warnock, D. J. Erle, M. A. Matthay, J. P. Wiener-Kronish, and R. M. Locksley. 1996. Interleukin 4, but not interleukin 5 or eosinophils, is required in a murine model of acute airway hyperreactivity. J. Exp. Med. 183**:**109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deurloo, D. T., B. C. van Esch, C. L. Hofstra, F. P. Nijkamp, and A. J. van Oosterhout. 2001. CTLA4-IgG reverses asthma manifestations in a mild but not in a more “severe” ongoing murine model. Am. J. Respir. Cell Mol. Biol. 25**:**751-760. [DOI] [PubMed] [Google Scholar]
- 7.Guery, J. C., F. Galbiati, S. Smiroldo, and L. Adorini. 1996. Selective development of T helper (Th)2 cells induced by continuous administration of low dose soluble proteins to normal and β2-microglobulin-deficient BALB/c mice. J. Exp. Med. 183**:**485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, G., G. Berry, R. H. DeKruyff, and D. T. Umetsu. 1999. Allergen-specific Th1 cells fail to counterbalance Th2 cell-induced airway hyperreactivity but cause severe airway inflammation. J. Clin. Investig. 103**:**175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilton, D. J., J. G. Zhang, D. Metcalf, W. S. Alexander, N. A. Nicola, and T. A. Willson. 1996. Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proc. Natl. Acad. Sci. USA 93**:**497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoag, K. A., N. E. Street, G. B. Huffnagle, and M. F. Lipscomb. 1995. Early cytokine production in pulmonary Cryptococcus neoformans infections distinguishes susceptible and resistant mice. Am. J. Respir. Cell Mol. Biol. 13**:**487-495. [DOI] [PubMed] [Google Scholar]
- 11.Hogaboam, C. M., K. Blease, B. Mehrad, M. L. Steinhauser, T. J. Standiford, S. L. Kunkel, and N. W. Lukacs. 2000. Chronic airway hyperreactivity, goblet cell hyperplasia, and peribronchial fibrosis during allergic airway disease induced by Aspergillus fumigatus. Am. J. Pathol. 156**:**723-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt, P. G., and C. McMenamin. 1989. Defence against allergic sensitization in the healthy lung: the role of inhalation tolerance. Clin. Exp. Allergy 19**:**255-262. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh, C. S., S. E. Macatonia, A. O'Garra, and K. M. Murphy. 1995. T cell genetic background determines default T helper phenotype development in vitro. J. Exp. Med. 181**:**713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huffnagle, G. B., M. B. Boyd, N. E. Street, and M. F. Lipscomb. 1998. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible mice (C57BL/6). J. Immunol. 160**:**2393-2400. [PubMed] [Google Scholar]
- 15.Karp, C. L., A. Grupe, E. Schadt, S. L. Ewart, M. Keane-Moore, P. J. Cuomo, J. Kohl, L. Wahl, D. Kuperman, S. Germer, D. Aud, G. Peltz, and M. Wills-Karp. 2000. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat. Immunol. 1**:**221-226. [DOI] [PubMed] [Google Scholar]
- 16.Kheradmand, F., A. Kiss, J. Xu, S. H. Lee, P. E. Kolattukudy, and D. B. Corry. 2002. A protease-activated pathway underlying Th cell type 2 activation and allergic lung disease. J. Immunol. 169**:**5904-5911. [DOI] [PubMed] [Google Scholar]
- 17.Kuroda, E., T. Sugiura, K. Zeki, Y. Yoshida, and U. Yamashita. 2000. Sensitivity difference to the suppressive effect of prostaglandin E2 among mouse strains: a possible mechanism to polarize Th2 type response in BALB/c mice. J. Immunol. 164**:**2386-2395. [DOI] [PubMed] [Google Scholar]
- 18.Kurup, V., H. Shen, and B. Banerjee. 2000. Respiratory fungal allergy. Microbes Infect. 2**:**1101-1110. [DOI] [PubMed] [Google Scholar]
- 19.Kurup, V. P., S. Mauze, H. Choi, B. W. Seymour, and R. L. Coffman. 1992. A murine model of allergic bronchopulmonary aspergillosis with elevated eosinophils and IgE. J. Immunol. 148**:**3783-3788. [PubMed] [Google Scholar]
- 20.Macpherson, A. J., and N. L. Harris. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4**:**478-485. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie, A. N., J. A. Culpepper, R. de Waal Malefyt, F. Briere, J. Punnonen, G. Aversa, A. Sato, W. Dang, B. G. Cocks, S. Menon, et al. 1993. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc. Natl. Acad. Sci. USA 90**:**3735-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenzie, G. J., A. Bancroft, R. K. Grencis, and A. N. McKenzie. 1998. A distinct role for interleukin-13 in Th2-cell-mediated immune responses. Curr. Biol. 8**:**339-342. [DOI] [PubMed] [Google Scholar]
- 23.McKenzie, G. J., C. L. Emson, S. E. Bell, S. Anderson, P. Fallon, G. Zurawski, R. Murray, R. Grencis, and A. N. McKenzie. 1998. Impaired development of Th2 cells in IL-13-deficient mice. Immunity 9**:**423-432. [DOI] [PubMed] [Google Scholar]
- 24.Murali, P. S., G. Dai, A. Kumar, J. N. Fink, and V. P. Kurup. 1992. Aspergillus antigen-induced eosinophil differentiation in a murine model. Infect. Immun. 60**:**1952-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, C. S., A. Woodcock, and A. Custovic. 2001. The role of indoor allergen exposure in the development of sensitization and asthma. Curr. Opin. Allergy Clin. Immunol. 1**:**407-412. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen, G. D., J. S. Hansen, R. M. Lund, M. Bergqvist, S. T. Larsen, S. K. Clausen, P. Thygesen, and O. M. Poulsen. 2002. IgE-mediated asthma and rhinitis I: a role of allergen exposure? Pharmacol. Toxicol. 90**:**231-242. [DOI] [PubMed] [Google Scholar]
- 27.Nolte, H., V. Backer, and C. Porsbjerg. 2001. Environmental factors as a cause for the increase in allergic disease. Ann. Allergy Asthma Immunol. 87**:**7-11. [DOI] [PubMed] [Google Scholar]
- 28.Noverr, M. C., J. R. Erb-Downward, and G. B. Huffnagle. 2003. Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clin. Microbiol. Rev. 16**:**517-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noverr, M. C., and G. B. Huffnagle. 2004. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 12**:**562-568. [DOI] [PubMed] [Google Scholar]
- 30.Noverr, M. C., R. M. Noggle, G. B. Toews, and G. B. Huffnagle. 2004. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 72**:**4996-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noverr, M. C., S. M. Phare, G. B. Toews, M. J. Coffey, and G. B. Huffnagle. 2001. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 69**:**2957-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noverr, M. C., D. D. Williams, G. B. Toews, and G. B. Huffnagle. 2002. Production of prostaglandins and leukotrienes by pathogenic fungi. Infect. Immun. 70**:**400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Noverr, M. C., G. M. Cox, J. R. Perfect, and G. B. Huffnagle. 2003. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect. Immun. 71**:**1538-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiner, S. L., and R. M. Locksley. 1995. The regulation of immunity to Leishmania major. Annu. Rev. Immunol. 13**:**151-177. [DOI] [PubMed] [Google Scholar]
- 34.Russo, M., M. A. Nahori, J. Lefort, E. Gomes, A. de Castro Keller, D. Rodriguez, O. G. Ribeiro, S. Adriouch, V. Gallois, A. M. de Faria, and B. B. Vargaftig. 2001. Suppression of asthma-like responses in different mouse strains by oral tolerance. Am. J. Respir. Cell Mol. Biol. 24**:**518-526. [DOI] [PubMed] [Google Scholar]
- 35.Sakai, K., A. Yokoyama, N. Kohno, H. Hamada, and K. Hiwada. 2001. Prolonged antigen exposure ameliorates airway inflammation but not remodeling in a mouse model of bronchial asthma. Int. Arch. Allergy Immunol. 126**:**126-134. [DOI] [PubMed] [Google Scholar]
- 36.Seymour, B. W., L. J. Gershwin, and R. L. Coffman. 1998. Aerosol-induced immunoglobulin (Ig)-E unresponsiveness to ovalbumin does not require CD8+ or T cell receptor (TCR)-γ/δ+ T cells or interferon (IFN)-γ in a murine model of allergen sensitization. J. Exp. Med. 187**:**721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsitoura, D. C., R. L. Blumenthal, G. Berry, R. H. Dekruyff, and D. T. Umetsu. 2000. Mechanisms preventing allergen-induced airway hyperreactivity: role of tolerance and immune deviation. J. Allergy Clin. Immunol. 106**:**239-246. [DOI] [PubMed] [Google Scholar]
- 38.Tsitoura, D. C., S. Kim, K. Dabbagh, G. Berry, D. B. Lewis, and D. T. Umetsu. 2000. Respiratory infection with influenza A virus interferes with the induction of tolerance to aeroallergens. J. Immunol. 165**:**3484-3491. [DOI] [PubMed] [Google Scholar]
- 39.Wills-Karp, M., and S. L. Ewart. 1997. The genetics of allergen-induced airway hyperresponsiveness in mice. Am. J. Respir. Crit. Care Med. 156**:**S89-S96. [DOI] [PubMed] [Google Scholar]
- 40.Wills-Karp, M., J. Luyimbazi, X. Xu, B. Schofield, T. Y. Neben, C. L. Karp, and D. D. Donaldson. 1998. Interleukin-13: central mediator of allergic asthma. Science 282**:**2258-2261. [DOI] [PubMed] [Google Scholar]
- 41.Wills-Karp, M., J. Santeliz, and C. L. Karp. 2001. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat. Rev. Immunol. 1**:**69-75. [DOI] [PubMed] [Google Scholar]
- 42.Yu, C. K., S. C. Lee, J. Y. Wang, T. R. Hsiue, and H. Y. Lei. 1996. Early-type hypersensitivity-associated airway inflammation and eosinophilia induced by Dermatophagoides farinae in sensitized mice. J. Immunol. 156**:**1923-1930. [PubMed] [Google Scholar]
- 43.Zhang, Y., W. J. Lamm, R. K. Albert, E. Y. Chi, W. R. Henderson, Jr., and D. B. Lewis. 1997. Influence of the route of allergen administration and genetic background on the murine allergic pulmonary response. Am. J. Respir. Crit. Care Med. 155**:**661-669. [DOI] [PubMed] [Google Scholar]