Direct p53 Transcriptional Repression: In Vivo Analysis of CCAAT-Containing G2/M Promoters (original) (raw)
Abstract
In response to DNA damage, p53 activates G1/S blocking and apoptotic genes through sequence-specific binding. p53 also represses genes with no target site, such as those for Cdc2 and cyclin B, key regulators of the G2/M transition. Like most G2/M promoters, they rely on multiple CCAAT boxes activated by NF-Y, whose binding to DNA is temporally regulated during the cell cycle. NF-Y associates with p53 in vitro and in vivo through the αC helix of NF-YC (a subunit of NF-Y) and a region close to the tetramerization domain of p53. Chromatin immunoprecipitation experiments indicated that p53 is associated with cyclin B2, CDC25C, and Cdc2 promoters in vivo before and after DNA damage, requiring DNA-bound NF-Y. Following DNA damage, p53 is rapidly acetylated at K320 and K373 to K382, histones are deacetylated, and the release of PCAF and p300 correlates with the recruitment of histone deacetylases (HDACs)—HDAC1 before HDAC4 and HDAC5—and promoter repression. HDAC recruitment requires intact NF-Y binding sites. In transfection assays, PCAF represses cyclin B2, and a nonacetylated p53 mutant shows a complete loss of repression potential, despite its abilities to bind NF-Y and to be recruited on G2/M promoters. These data (i) detail a strategy of direct p53 repression through associations with multiple NF-Y trimers that is independent of sequence-specific binding of p53 and that requires C-terminal acetylation, (ii) suggest that p53 is a DNA damage sentinel of the G2/M transition, and (iii) delineate a new role for PCAF in cell cycle control.
Promoters and enhancers are a combinatorial puzzle of DNA elements recognized by sequence-specific regulators that act in a chromatin context. The CCAAT box is a common promoter element, usually positioned in either orientation between −60 and −100. A combination of electrophoretic mobility shift assays (EMSAs) and transfections with highly diagnostic dominant-negative vectors implicated NF-Y as the CCAAT activator (33). NF-Y is composed of three subunits, NF-YA, NF-YB, and NF-YC, all necessary for DNA binding. NF-YB and NF-YC contain histone fold motifs (HFMs) common to all core histones; dimerization is essential for NF-YA association and sequence-specific DNA binding (reference 43 and references therein). A recent bioinformatic analysis of cell cycle promoters showed a remarkable and specific abundance of CCAAT boxes in promoters regulated during the G2/M phase (9). Key regulators, such as CDC25B, CDC25C, cyclin B1, cyclin B2, Cdc2, and topoisomerase IIα, all contain multiple CCAAT boxes, invariably shown to be crucial for proper regulation (4, 7, 11, 12, 32, 45, 47, 59, 60). Chromatin immunoprecipitation (ChIP) experiments determined that NF-Y is dynamically bound in the different phases of the cell cycle (6, 45, 47). Most importantly, recent experiments determined that an NF-YA mutant defective in Cdk2 phosphorylation behaves in a dominant-negative manner, blocking cell cycle progression in G1 and G2 (7).
An important aspect of the regulation of G2/M promoters is represented by their capacity, upon DNA damage, to be repressed through the activity of p53 (reviewed in reference 50). p53 controls the activation of a variety of genes whose products are critical for the regulation of the cell cycle and for the induction of apoptosis (1). DNA-damaging agents induce a wealth of posttranslational modifications—such as phosphorylation, sumoylation, and acetylation—that “activate” p53 transcriptionally. In particular, two histone acetyltransferase (HAT) enzymes acetylate different residues in the C-terminal domain: p300 targets lysines 372, 373, 381, and 382 (15, 18, 44), while PCAF/hGCN5 acetylates lysine 320 (29). In general, acetylation increases the affinity of recombinant p53 for its DNA target in in vitro EMSA experiments (15). Further reports have shown that activator-coactivator interactions are positively affected by the p300-mediated acetylation of p53 (3). However, nonacetylated p53 is bound to certain targets prior to DNA damage (22).
The mechanisms of repression by p53 are less clear. Promoters of genes critical for the G2/M transition are strongly down-modulated by treatment of cells with DNA-damaging agents or by p53 overexpression, without any recognizable binding element (17, 27, 32, 51, 52). Such experiments are incapable of discriminating a direct effect on promoter function from a secondary one due to the activation of G1/S-blocking genes, such as the p21 gene (30, 59, 60). We and others have reported that the negative activity of p53 on G2/M promoters depends upon NF-Y binding (21, 27, 32, 35, 59). This conclusion relied upon the observations that the elimination of CCAAT boxes and the coexpression of dominant-negative NF-YA or of dominant-negative p53 led to inhibition of the negative effects of DNA-damaging agents. To better understand the mechanisms of this phenomenon and to ascertain whether p53 acts directly on G2/M promoters, we used in vivo ChIP, transfection, and protein-protein and DNA-protein interaction assays.
MATERIALS AND METHODS
Protein production and purification.
Production and purification of NF-Y were described previously (13). Glutathione _S_-transferase (GST)-p53 and His-p53 fusion proteins were purified from soluble fractions. NF-YC αC mutants were produced in the backbone of the His-YC5 mutant (43).
Cell cultures and treatments.
NIH 3T3 mouse fibroblasts were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS). Cell line H1299, derived from a human large-cell lung carcinoma, was cultured in RPMI medium supplemented with 10% FCS.
Adriamycin was added at 0.5 μg/ml and incubated for 20 h (see Fig. 1 and 6B), for 8 h (see Fig. 4B), or for various times (see Fig. 6A, C, and D, 7A, and 9D). Cells were collected, and DNA distribution analysis of propidum iodide-stained cells was performed with an Epics cytofluorometer (Coulter).
FIG. 1.
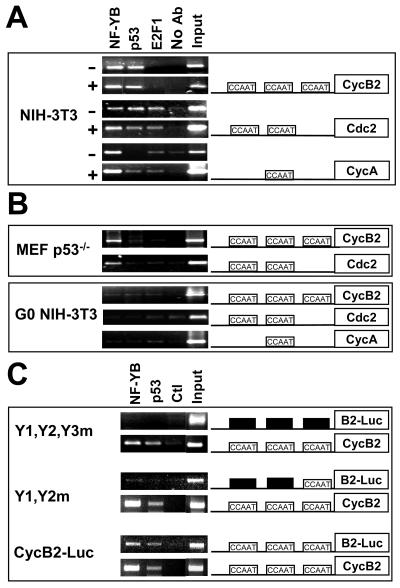
ChIP analysis of NF-Y and p53 on G2/M promoters. (A) ChIP was performed with NIH 3T3 mouse fibroblasts not treated (−) or treated (+) with adriamycin and the indicated antibodies (Ab). (B) (Upper panel) p53−/− mouse embryo fibroblasts (MEF) were used in a similar ChIP analysis with the indicated antibodies. (Lower panel) NIH 3T3 cells arrested in G0 by serum withdrawal were used in a ChIP analysis (6). (C) ChIP with NIH 3T3 cells stably transfected with a reporter construct containing the wild-type cyclin B2 promoter, a mutant lacking two CCAAT boxes (Y1-Y2m), or a mutant lacking all three CCAAT boxes (Y1-Y2-Y3m). Immunoprecipitated DNAs were amplified with oligonucleotides, revealing the luciferase (Luc) transgene as well as the respective endogenous cyclin B2 promoter. Ctl, control.
FIG. 6.
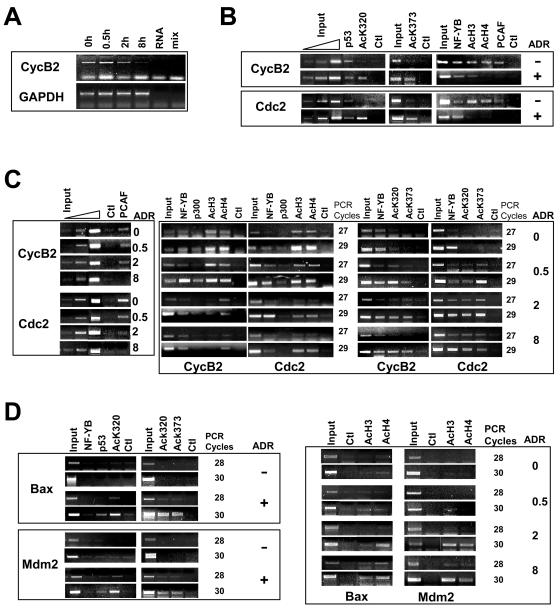
ChIP analysis of p53, H3, and H4 acetylation and HAT association on G2/M promoters. (A) RT-PCR analysis of cyclin B2 and control GAPDH mRNAs following DNA damage by adryamicin for the indicated times. (B) (Left panels) ChIP analysis of NIH 3T3 cells not treated (−) or treated with adriamycin (ADR) for 20 h (+) with the indicated antibodies against nonacetylated p53, Ac-p53 K320, and Ac-p53 K373-382. (Middle and right panels) Anti-YB antibodies used as a positive control, together with anti-Ac-H3, anti-Ac-H4, and anti-PCAF antibodies. Titration of the input DNAs is shown. (C) NIH 3T3 cells were treated with adriamycin for the indicated times (in hours), and ChIP analysis of the cyclin B and Cdc2 promoters was performed with the indicated antibodies as described above. (Left panels) PCAF and control antibodies. Titration of the input DNAs is shown. (Right panels) Representative cycles of semiquantitative PCR. (D) (Left panels) ChIP analysis of the Bax and Mdm2 promoters. (Right panels) Time course for PCR amplification of the Mdm2 and Bax promoters.
FIG. 4.
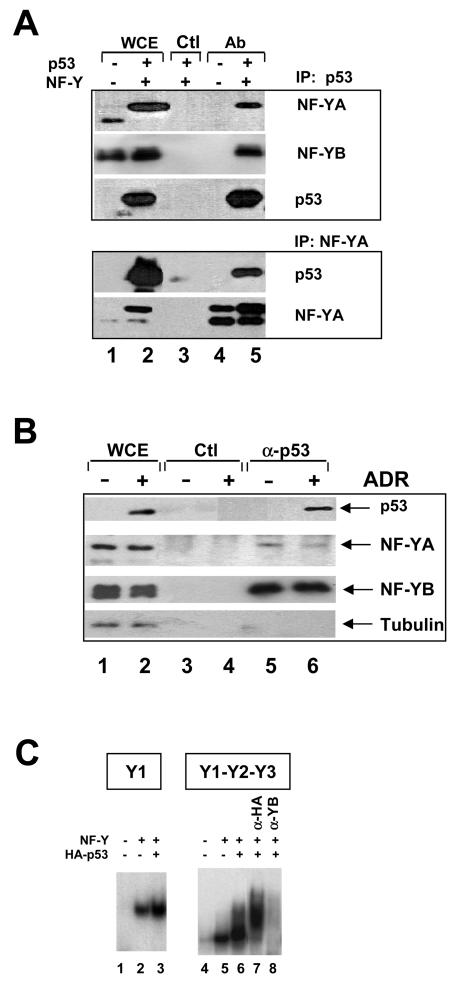
Binding of p53 to NF-Y in vivo. (A) NF-Y-p53 interactions in vivo in H1299 cells. Immunoprecipitation (IP) with anti-p53 or anti-YA (lanes 4 and 5), and control (Ctl) (lane 3) antibodies (Ab) was followed by Western blot analysis with the indicated antibodies. In lanes 1 and 2, extracts were tested directly in Western blots. NF-Y and p53 were overexpressed in the extracts used in lanes 2, 3, and 5. The different sizes of NF-YA are due to the prevalence of the “short” splicing isoform in lane 1 and overexpression of the “long” isoform in lanes 2, 3, and 5. WCE, whole-cell extracts. (B) Evaluation of endogenous NF-Y-p53 interactions. NIH 3T3 cells were not treated (lanes 1, 3, and 5) or were treated with adriamycin (ADR) for 8 h (lanes 2, 4, and 6). Extracts were analyzed directly in Western blots (lanes 1 and 2) or immunoprecipitated with control (lanes 3 and 4) or anti-p53 (lanes 5 and 6) antibodies. Western blot analysis of eluates with the indicated antibodies is shown in lanes 3 to 6. (C) EMSAs of in vivo-produced NF-Y and p53. The Y1 CCAAT oligonucleotide was used in lanes 1 to 3; a cyclin B2 fragment (−129 to +48) was used in lanes 4 to 8. In lanes 1 and 4, 1 μl of mock-transfected Saos2 cell extracts was used; in lanes 2, 3, and 5 to 8, equivalent amounts of extracts from cells transfected with NF-Y expression vectors were used, together with HA-p53 in lanes 3 and 6 to 8. Supershift with anti-HA antibodies and inhibition with anti-YB antibodies are shown in lanes 7 and 8, respectively.
FIG. 7.
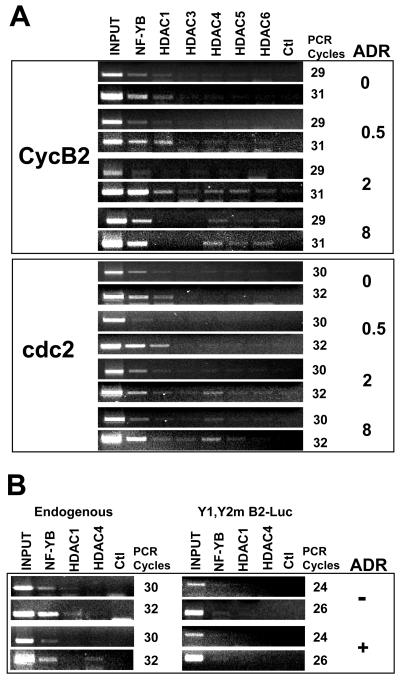
HDAC recruitment to G2/M promoters upon DNA damage. (A) NIH 3T3 cells were treated for the indicated times with adriamycin (ADR), and ChIP analysis was performed with antibodies against the indicated HDACs. Two sets of representative PCR cycles are shown for each time point. Ctl, control. (B) Chromatin from the Y1-Y2m-containing stable clones (Fig. 1) was used for ChIP analysis with the indicated antibodies. Immunoprecipitated DNAs were PCR amplified to show endogenous cyclin B2 (left panels) and the luciferase reporter construct mutated in two of the three CCAAT boxes (right panels).
FIG. 9.
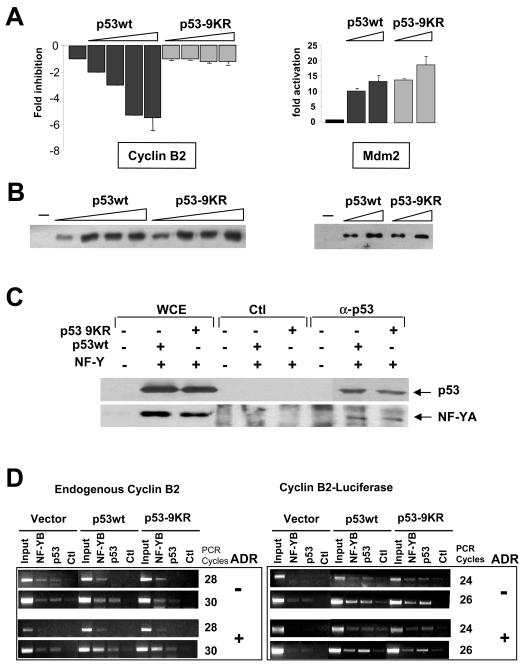
Effect of a nonacetylated p53 mutant on transcriptional repression. (A) Dose-response analysis of wild-type p53 (p53wt) and mutant p53-9KR in Saos2 cells with the cyclin B2 and Mdm2 reporters. Error bars indicate standard deviations. (B) Levels of expression of wild-type p53 and p53-9KR in panel A are shown in Western blots. (C) Immunoprecipitation of overexpressed wild-type p53 and mutant p53-9KR from transfected H1299 cells. Western blot analysis was performed with anti-p53 and anti-YA antibodies. WCE, whole-cell extracts; Ctl, control. (D) ChIP analysis of NF-Y and p53 in NIH 3T3 cells transfected with wild-type p53, mutant p53-9KR, and control vectors, together with the cyclin B2-luciferase construct, before and after adriamycin (ADR) treatment for 8 h. PCRs with the transfected templates are shown in the right panels, and those with the control endogenous cyclin B2 gene are shown in the left panels. Two sets of PCRs are shown. Note that fewer PCR cycles are required for the transfected templates.
Adenovirus vectors for expression of NF-YA or dominant-negative mutant YAm29 were generated by using an AdEasy system. The NF-YA coding regions were excised with HindIII and XbaI from the corresponding pcDNA3-based vectors and introduced into the same sites of shuttle vector pAdTrack-CMV. This plasmid was allowed to recombine with vector pAdEasy1, followed by treatment with PacI and transfection into an E1-complementing cell line. Viruses were amplified, and titers were determined. We infected 107 exponentially growing NIH 3T3 cells for 7 h in the absence of serum. FCS then was added, and the cells were incubated for 48 h.
EMSAs and footprinting.
For electrophoretic mobility shift assays (EMSAs), we used 32P-labeled fragments containing either the complete cyclin B2 promoter (−129 to +48) obtained by PCR or an oligonucleotide with the Y1 CCAAT box (4, 45). For supershift experiments, we used 300 ng of anti-NF-YB, anti-p53 (DO1), antihemagglutinin (HA), and anti-GST (control) antibodies. For some EMSAs (see Fig. 4), Saos2 cells were transfected with 100 ng of expression vectors coding for the three NF-Y subunits and with or without 200 ng of HA-p53. After 24 h, extracts were prepared by resuspending cells in 100 μl of a buffer containing 0.5% NP-40, 50 mM Tris-HCl (pH 8), 120 mM NaCl, 1 mM dithiothreitol, and protease inhibitors. Mock-transfected extracts were used as controls. Samples of 1 to 2 μl of extracts were used in EMSAs with 50 ng of poly(dI-dC) (Sigma).
Immunoprecipitation analyses.
In vitro immunoprecipitation was performed as described previously (13). Recombinant proteins (50 to 100 ng) were incubated in 100 μl of NDB100 (100 mM KCl, 20 mM HEPES [pH 7.9], 0.1% NP-40, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) with rotation for 2 h at 4°C. The samples then were added to 10 μl of protein G-Sepharose to which 5 μg of anti-NF-YB, anti-NF-YA (Mab7), or anti-HA antibodies had been bound. Incubation was carried out for 2 h at 4°C, unbound material was recovered after centrifugation, and the beads were washed with NDB100. Sodium dodecyl sulfate (SDS) buffer was added, and the samples were boiled at 90°C for 5 min and loaded on SDS gels. Western blotting was performed according to standard procedures with the appropriate primary antibodies (DO1 for p53).
For overexpression, H1299 cells were transfected with 10 μg of the eukaryotic expression vector containing the human wild-type cDNA for the NF-YA subunit under the control of the simian virus 40 promoter, the pcDNA3-p53 vector driven by the cytomegalovirus early promoter-enhancer (kindly provided by G. Blandino, Istituto Regina Elena, Rome, Italy), or the vector alone. The cells were harvested at 36 h after transfection in lysis buffer 1 (50 mM HEPES [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 5 mM KCl, 5 mM MnCl2, 5 mM EGTA, 2 mM EDTA, 20 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 1 mM dithiothreitol, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride, 20 mM NaF, 0.1% NP-40) and homogenized. After 30 min on ice, lysates were sonicated for 10 s and centrifuged for 10 min at 14,000 rpm (Eppendorf; Microfuge), and supernatants were used for further studies.
Immunoprecipitation was performed by incubating 1.5 mg of whole-cell extract with 50 μl of anti-NF-YA antibody, 5 μl of anti-p53 sheep serum (Ab-7; Calbiochem), or mouse control serum, with protein A-agarose (Pierce), and with 0.05% bovine serum albumin (BSA) at 4°C for 2 h. The beads were washed twice with phosphate-buffered saline-0.05% BSA plus protease inhibitors, eluted in SDS buffer, and loaded on SDS-12% polyacrylamide gels. Western blotting was performed with anti-p53 mouse monoclonal antibody DO1, anti-NF-YA rabbit polyclonal antibody, anti-NF-YB rabbit polyclonal antibody, and antitubulin mouse monoclonal antibody (Sigma). Immunostained bands were detected with a chemiluminescence system (Amersham Pharmacia).
ChIP analyses.
Formaldehyde cross-linking and ChIP were performed as described previously (6). NIH 3T3 cells (0.5 × 108 to 1 × 108) were washed with phosphate-buffered saline and incubated for 10 min with 1% formaldehyde. After the reaction was quenched with 0.1 M glycine, the cells were sonicated into chromatin fragments with an average length of 500 to 800 bp. Chromatin was kept at −80°C. Immunoprecipitation was performed with protein G-Sepharose (Kierkegaard & Perry Laboratories) and 3 to 5 μg of antibodies to the following: NF-YB (purified rabbit polyclonal antibody) (6), p53 (Ab7; Oncogene Science), acetyl (Ac)-p53 K320 (06-915; Upstate Biotechnology), Ac-p53 K373 to K382 (Ac-p53 K373-382) (06-758; Upstate), E2F1 (sc-193; Santa Cruz Biotechnology), Ac-histone H3 (06-609; Upstate), Ac-histone H4 (06-866; Upstate), p300 (sc-585x; Santa Cruz), PCAF (rabbit serum from Y. Nakatani, Harvard University) (6), histone deacetylase 1 (HDAC1) (H3284; Sigma), and HDAC3 to HDAC6 (active motif) (6). The chromatin solution was precleared by incubation with protein G-Sepharose for 2 h at 4°C, divided into aliquots, and incubated with the antibodies overnight at 4°C. Before use, protein G-Sepharose was blocked twice at 4°C with salmon sperm DNA (1 μg/μl) that had been sheared to a 500-bp length and BSA (1 μg/μl) for 2 h and overnight.
PCRs were performed with previously described Cdc2, cyclin B2, and cyclin A primers (6). The oligonucleotides were as follows: luciferase, 5′-TTGCTCTCCAGCGGTTCCAT; CDC25C, 5′-GGCGAGAGAATTTAGTACAAGGA and 5′-CTCCGGAGATGGCCTGAAGGC; MDM2, 5′-GCCGGGATGCGGCTTCCCGG and 5′-TCCGGTCGGTCTCCCGCTCG; BAX, 5′-CTTACTTAATGGTGCAGCTTGG and 5′-GATGCCCAGAGTTGGTTGTTTC; and p21, 5′-GAGGATACCTTGCAAGGCTGGA and 3′-GCACACCATTGCACGTGAATGT.
Plasmids and transfections.
Stable transfections were performed with the wild-type cyclin B2-luciferase vector or the Y1-Y2m and Y1-Y2-Y3m mutants (3, 45), together with a plasmid encoding hygromycin resistance. A total of 30 to 40 clones were pooled and further grown in selective media. Luciferase activities were normalized for protein concentrations as measured by the Bradford assay. Chromatin was prepared as described above. For transient transfections, 2.5 × 104 COS cells (NIH 3T3 cells in some experiments [see Fig. 8E]) were transfected by using Lipofectamine (Gibco-BRL) with 0.1 μg of cyclin B2-luciferase vector, 0.5 μg of Mdm2-CAT vector, 50 ng of NβGAL vector, and carrier plasmid to keep the total DNA concentration constant at 800 ng. Cells were recovered at 24 or 36 h after transfection and resuspended in 150 mM NaCl-40 mM Tris-HCl (pH 7.4) for measurement of chloramphenicol acetyltransferase activities or lysis buffer 2 (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA) for measurement of luciferase activities. β-Galactosidase was assayed to control for transfection efficiencies. Three to eight independent transfections were performed in duplicate. p53 K320Q and p53 K320R were obtained from M. L. Avvantaggiati and T. Halazonetis (Wistar, Philadelphia, Pa.), respectively; p53-9KR was a kind gift from S. McMahon (Wistar).
FIG. 8.
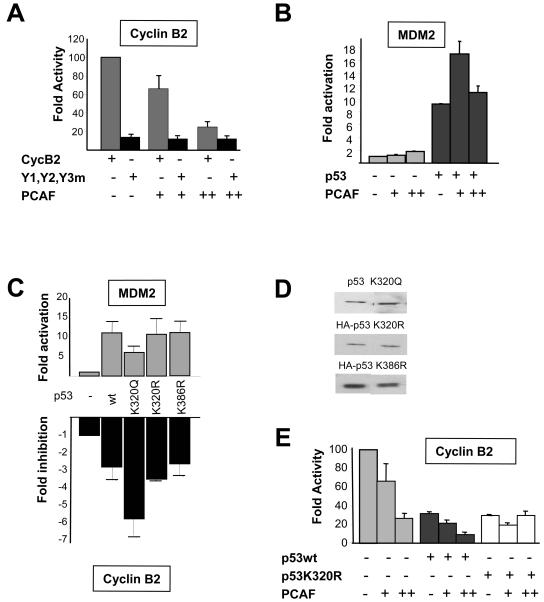
Repression of cyclin B2 by p53 and PCAF. (A) PCAF (100 and 300 ng) was cotransfected in COS cells with 100 ng of p53, with wild-type cyclin B2, and with Y1-Y2-Y3m. Error bars indicate standard deviations. (B) Same as panel A, except that PCAF (100 and 300 ng) was transfected alone or with p53, together with the Mdm2 reporter. (C) Transcriptional analysis of Mdm2 and cyclin B2 reporters in p53 −/− Saos2 cells with 100 ng of wild-type (wt) p53 and the indicated mutants of p53. (D) Representative levels of the p53 proteins in panel C were checked in Western blots. (E) Same as panel A, except that wild-type p53 or p53 K320R (100 ng) was transfected with PCAF and assayed on the cyclin B2 promoter in NIH 3T3 cells.
Some of the ChIP analyses (see Fig. 9D) were carried out with NIH 3T3 cells (15-cm plates; 6 × 106 cells) transfected with the p53 plasmids (10 μg) and the cyclin B2-luciferase vector (10 μg). After 24 h, one plate was treated with adriamycin for 8 h. ChIP analyses then were performed as described above.
RT-PCR analysis.
RNA was extracted by using an RNeasy kit (Qiagen, Hilden, Germany) according to the manuifacturer's protocol from NIH 3T3 cells not treated or treated for various times with adriamycin as described for the ChIP experiments. For cDNA synthesis, 4 μg of RNA was used with a Moloney murine leukemia virus reverse transcriptase (RT) kit (Invitrogen). Semiquantitative PCR was performed with oligonucleotides CycB2ex6 (5′-ACTGGTGTAAGCATTATCTG) and CycB2ex3 (5′-CTGTGAAACCAGTGCAGATG). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control RT-PCR was performed with standard oligonucleotides.
RESULTS
Multiple CCAAT boxes are important for the transcriptional response induced by DNA damage.
Previous experiments suggested that the binding of NF-Y is important not only for the activation of G2/M promoters but also for their repression following DNA damage (17, 27, 32). To investigate the importance of the multiplicity of CCAAT boxes in repression, we stably transfected into NIH 3T3 cells the wild-type cyclin B2-luciferase construct and Y1-Y2m, a mutant in which the distal and middle but not the proximal CCAAT boxes are altered (4, 45). The mutation in the proximal CCAAT box is functionally equivalent to the mutations in the other two CCAAT boxes (45). Pools of several clones for each transgene were harvested, expanded, and assayed for luciferase activities; as expected, the mutant promoter showed a severe decrease in activity (Table 1). Upon treatment with adriamycin for 8 h, the wild-type promoter but not the mutant promoter showed a decrease in luciferase activity. Because the enzyme is short-lived in mammalian cells, these data indicate (i) that the presence of only one CCAAT box is insufficient to render the promoter responsive to a DNA-damaging stimulus and (ii) that repression mechanisms are relatively quick, being observed by 8 h following treatment.
TABLE 1.
Luciferase activitiesa
| Construct | Adriamycin | Luciferase activity (RLU/μg of extract) | SD | Fold repression |
|---|---|---|---|---|
| Cyclin B2-luciferase | − | 5,715 | 437 | |
| Cyclin B2-luciferase | + | 2,264 | 898 | 2.6 |
| CycB-Y1-Y2m | − | 162 | 3 | |
| CycB-Y1-Y2m | + | 135 | 21 | 0.2 |
p53 is bound to promoters with multiple CCAAT boxes in vivo.
The in vivo sequence-specific binding of p53 has been documented by ChIP (3, 22, 36, 48). We investigated whether G2/M promoters are bound by p53 following adriamycin-induced DNA damage by using anti-p53, anti-NF-YB, and anti-E2F1 control antibodies. NIH 3T3 cells were fixed before or after treatment with adriamycin, which induces cell cycle arrest in late G1 and late G2 (data not shown). PCR amplification of various promoters is shown in Fig. 1A; cyclin B2, Cdc2, and cyclin A were bound by NF-Y before and after adriamycin addition. In accordance with published data, E2F1 was bound to cyclin A and Cdc2 but not to cyclin B2 (6, 49, 56). Cyclin B2 and Cdc2 but not cyclin A were positive for p53, even in the absence of DNA damage. PCR amplification of the Mdm2 and Bax promoters determined that p53 but not NF-Y was bound after DNA damage (see below). These data suggest that p53 is bound to G2/M promoters prior to DNA damage. The specificity of our p53 ChIP assays was further checked with p53 knockout mouse embryo fibroblasts; NF-Y was present on cyclin B2 and Cdc2, as expected, whereas p53 was not (Fig. 1B, upper panel). Finally, it was recently shown that NF-Y is not bound to G2/M promoters in G0-arrested NIH 3T3 cells (6). G0 cells were assayed for p53 binding; indeed, NF-Y was not bound, and p53 was also absent (Fig. 1B, lower panel).
To verify the importance of the CCAAT boxes for p53 association, we used NIH 3T3 clones stably transfected as mentioned above; in addition, we also derived a cyclin B2 triple-CCAAT mutant linked to the luciferase transgene (4, 45). We used the same chromatin to PCR amplify the transgene as well as the endogenous cyclin B2 gene; all endogenous targets were bound by NF-YB and p53, as expected (Fig. 1C). On the other hand, only wild-type cyclin B2 and not the Y1-Y2-Y3m or the Y1-Y2m mutant was associated with either NF-Y or p53. We conclude that under no conditions tested was p53 associated with G2/M promoters in the absence of a CCAAT box and of NF-Y bound to it.
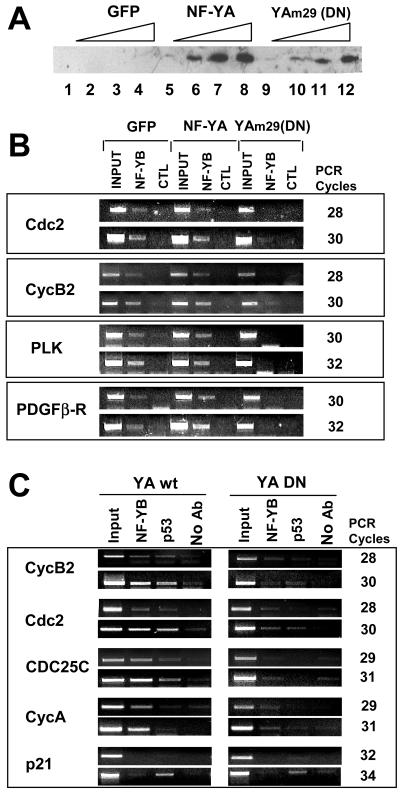
To further clarify the role of NF-Y in p53 recruitment on G2/M genes, we prepared adenovirus vectors expressing wild-type NF-YA and the well-characterized mutant YAm29 (reference 33 and references therein); this mutant, crippled in the DNA binding subdomain, is able to associate with the HFM NF-YB-NF-YC dimer, rendering the complex incapable of binding to a CCAAT box. Infection of NIH 3T3 cells with increasing doses of the two viruses but not with an empty control yielded increasing amounts of NF-YA proteins (Fig. 2A, upper panel). ChIP analysis of the infected cells with anti-YB and control antibodies is shown in Fig. 2B; with NF-Y targets (Cdc2, cyclin B2, platelet-derived growth factor β receptor [PDGFβ-R], and PLK), the control and wild-type NF-YA infections had negligible effects on NF-Y binding in vivo. On the contrary, the YAm29 mutant substantially removed the trimer from promoters.
FIG. 2.
Overexpression of a dominant-negative (DN) NF-YA mutant prevents p53 association with CCAAT promoters. (A) NIH 3T3 cells were infected for 7 h with green fluorescent protein (GFP) control adenovirus (lanes 1 to 4), wild-type NF-YA adenovirus (lanes 5 to 8), or YAm29 adenovirus (lanes 9 to 12). Western blot analysis of overexpressed NF-YA is shown. (B) ChIP analysis with anti-NF-YB antibodies of chromatin derived from cells infected with the three viruses. The indicated bona fide NF-Y targets were PCR amplified. For each promoter, we show two amplifications at different PCR cycles. CTL, control; PLK, Polo-like kinase. (C) Same as panel B, except that wild-type NF-YA chromatin (YA wt) and YAm29 chromatin (YA DN) were analyzed with anti-NF-YB and anti-p53 antibodies (Ab). We included as controls the cyclin A promoter (single CCAAT box) and the p21 promoter (containing a bona fide p53 binding element but not CCAAT boxes).
We next analyzed the binding of p53; with Cdc2, cyclin B2, and CDC25C (another triple-CCAAT promoter) (63) promoters, p53 was present only in wild-type NF-YA-infected cells and was absent in those expressing the Yam29 mutant. As a control, we used the cyclin A single-CCAAT promoter; only NF-Y and not p53 was present with the wild-type vector but not with the mutant vector (Fig. 2C). Finally, to verify whether NF-YA overexpression affects sequence-specific DNA binding of p53, we PCR amplified the p53 binding site in the p21 promoter, because it was shown that p53 is constitutively associated with this region in the absence of DNA damage (22); indeed, p53 was present in nonstress conditions, and little effect was observed whether the control, the wild type, or the mutant was overexpressed (Fig. 2C, lower panel). These data indicate that the removal of p53 from double- or triple-CCAAT promoters by dominant-negative NF-YA is specific for CCAAT boxes and is not due to general sequestration of p53.
Taken together, these data indicate that (i) p53 is bound in vivo to promoters lacking consensus p53 binding sites but harboring at least two CCAAT boxes, (ii) NF-Y association is absolutely required for p53 to access these sites, (iii) NF-Y overexpression has no effect on sequence-specific p53 binding, and (iv) DNA damage has a minimal effect on NF-Y and p53 binding, ruling out the possibility that repression of these promoters might be due to removal of the DNA-bound activator.
NF-Y and p53 interact in vitro and in vivo.
The ChIP data suggested that direct interactions between NF-Y and p53 might be possible. To investigate this point, recombinant NF-Y and p53 were used in in vitro immunoprecipitation assays. Using an anti-NF-YA monoclonal antibody (Mab7) and an anti-HA antibody, we were able to immunoprecipitate HA-p53 and NF-YB with both antibodies. The latter result was expected, as the trimer is associated under these conditions, whereas the former result indicated interactions between NF-Y and p53 (Fig. 3A, upper panel). When incubated in the absence of HA-p53, the anti-HA antibody did not immunoprecipitate the NF-Y trimer (Fig. 3A, lower panel). Similar results were obtained with a nitrilotriacetic acid affinity assay with His-tagged proteins (data not shown).
FIG. 3.
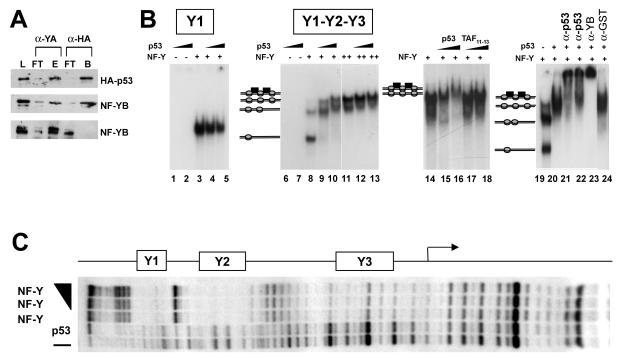
Binding of p53 to NF-Y in solution and on DNA in vitro. (A) Purified recombinant His-tagged NF-Y trimer and HA-p53 were immunoprecipitated with anti-NF-YA (Mab7) and anti-HA antibodies. Load (L), flowthrough (FT), and bound (B) fractions were assayed in Western blots with anti-HA and anti-NF-YB antibodies. In the lowest panel, immunoprecipitation was performed without HA-p53. (B) EMSAs with recombinant NF-Y and p53 and with the cyclin B2 Y1 high-affinity NF-Y binding site (lanes 1 to 5) or a cyclin B2 fragment (−129 to +48) (lanes 6 to 24). p53 was used at 50 ng in lanes 1, 4, 6, 9, 12, 15, and 20 to 24 and at 150 ng in lanes 2, 5, 7, 10, 13, and 16; NF-Y was used at 1 ng in lanes 3 to 5 and 8 to 10, at 2 ng in lanes 19 to 24, and at 5 ng in lanes 11 to 18; TAF11-TAF13 dimer was used at 200 and 600 ng in lanes 15 and 16, respectively. For the supershift EMSAs in lanes 19 to 24, anti-p53 DO1 was incubated with recombinant p53 before (lane 21) or after (lane 22) NF-Y and DNA were added; anti-NF-YB and anti-GST controls were added after NF-Y, p53, and DNA were preincubated. (C) Footprint analysis of NF-Y and p53 on a cyclin B2 fragment (−129 to +48). In the upper rows, 20 ng of NF-Y was incubated alone or with 100 and 300 ng of p53. The lower rows contained unbound DNA and p53 (300 ng) alone. The positions of the Y1, Y2, and proximal Y3 CCAAT boxes are indicated.
We next extended these observations to complexes formed in the presence of DNA. NF-Y binds to the cyclin B2 triple-CCAAT core promoter with a relative affinity of Y1 > Y2 > Y3 (4, 45). We used EMSAs with NF-Y and p53 on the high-affinity Y1 binding site and on a fragment containing the three CCAAT sequences (Y1, Y2, and Y3); upon the addition of increasing doses of p53, no binding for the two probes was observed in the absence of NF-Y (Fig. 3B, lanes 1, 2, 6, and 7). The addition of an anti-p53 antibody and of Ac-p53 did not induce p53 binding, a result that was expected, since there are no consensus p53 binding sites (data not shown). Increasing concentrations of NF-Y generated a single complex with Y1 and multiple complexes with Y1-Y2-Y3 (Fig. 3B, lanes 3, 8, and 11), corresponding to the occupation of the Y1, Y2 and, finally, Y3 boxes (see below). The addition of p53 resulted in the formation of additional, slower-migrating complexes with Y1-Y2-Y3 (Fig. 3B, lanes 9, 10, 12, and 13) but not with Y1 (lanes 3 to 5). Controls were performed to ascertain the nature of these upper complexes. First, we used the HFM TAF11-TAF13, a dimer that interacts with NF-Y in immunoprecipitation and nitrilotriacetic acid affinity assays similar to those shown here (13); high concentrations of TAF11-TAF13 did not modify the NF-Y-CCAAT complexes on Y1-Y2-Y3 (Fig. 3B, lanes 14 to 18). Next, we incubated the NF-Y-p53 complexes with anti-p53, anti-YB, and irrelevant antibodies; the former two types of antibodies further supershifted the complexes, while the latter had no effect (Fig. 3B, lanes 19 to 24).
We next wished to know whether the association of p53 with NF-Y might lead to the protection of additional DNA sequences on the cyclin B2 promoter in a footprinting analysis. Figure 3C indicates that NF-Y efficiently protects the Y1 and Y2 CCAAT elements and, to a lesser degree, the Y3 CCAAT element (45); p53 alone, even at high concentrations, is incapable of either binding NF-Y or modifying the NF-Y pattern. These data are a further indication that the association of p53 is independent from contact with DNA.
These in vitro observations were extended with in vivo assays. We overexpressed p53 with NF-Y in H1299 cells and performed immuprecipitation experiments with anti-p53 and anti-NF-Y antibodies. As shown in Fig. 4A, NF-YA and NF-YB were coimmunoprecipitated from extracts in overexpressing cells with anti-p53 antibodies but not with control antibodies. The reciprocal was also true; that is, p53 was present in anti-NF-Y antibody immunoprecipitates. We next assayed whether endogenous p53 and NF-Y interacted in NIH 3T3 cells before or after adriamycin addition. Figure 4B shows that NF-Y subunits were coimmunoprecipitated in the absence of DNA damage with anti-p53 antibodies but not with control antibodies. As expected, p53 expression was vastly increased in whole-cell extracts after DNA damage. Interestingly, NF-Y-p53 interactions could be detected from the small amounts of p53 present in undamaged cells, but there was no increase in NF-Y-p53 associations following DNA damage; these data suggested that the interactions did not require posttranslational modifications of p53, in keeping with the in vitro data and with the in vivo ChIP data. The additional non-NF-Y-bound p53 present after damage was most likely devoted to sequence-specific binding and activation.
Finally, we overexpressed the NF-Y trimer, with or without p53, and assayed the resulting extracts in EMSAs with cyclin B2 probes. CCAAT binding activity was vastly increased upon cotransfection of the NF-Y trimer (Fig. 4C, lanes 1, 2, 4, and 5); this result was expected and strictly depended upon the use of all three NF-Y expression vectors (C. Imbriano and R. Mantovani, unpublished data). With the Y1 CCAAT probe, cotransfection of p53 had no effect on the NF-Y complex (Fig. 4C, lanes 2 and 3), whereas slower-migrating complexes were observed with the triple-CCAAT probe (lane 6). These complexes were supershifted by anti-HA antibodies recognizing transfected p53 and were inhibited by anti-NF-YB antibodies, as has been commonly observed for the latter reagent. These results are in full agreement with the EMSA results shown in Fig. 3. Therefore, the data in Fig. 3 and 4 demonstrate that (i) p53 can associate with NF-Y in vitro and in vivo, (ii) the interaction is observed on DNA and requires multiple CCAAT boxes, (iii) p53 binding is independent from the recognition of a specific DNA sequence, and (iv) p53 does not impair NF-Y binding.
Mapping of the domains required for p53-NF-Y interactions.
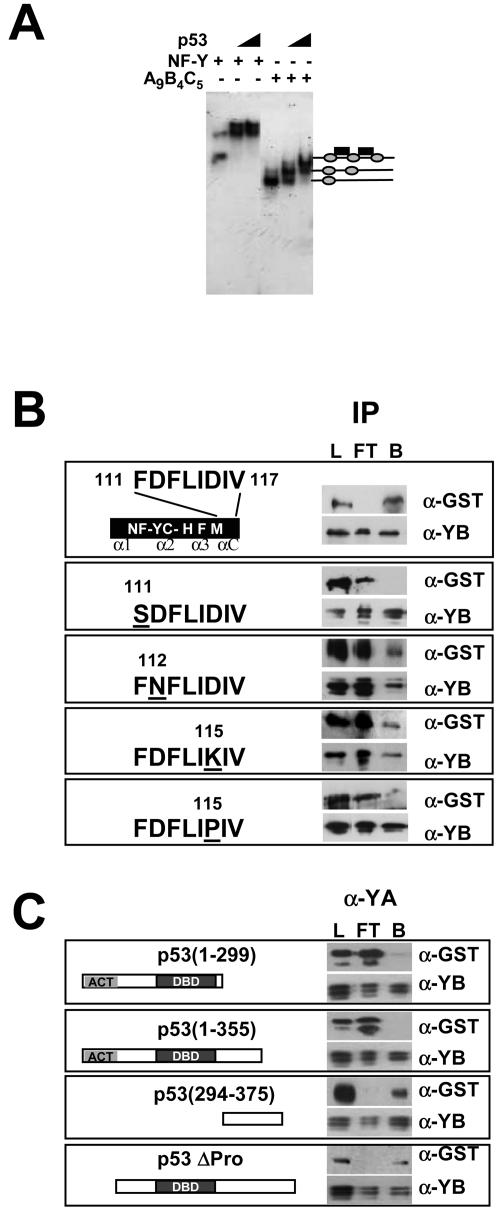
To locate the NF-Y domain required for p53 binding, we incubated various combinations of deletion mutants of the three NF-Y subunits; all DNA binding variants formed p53 complexes on the triple-CCAAT construct. This result was exemplified by the smallest mutant still being capable of binding DNA containing the homology domains of the three subunits, YA9, YB4, and YC5 (references 13 and 43 and references therein). The fast-migrating complex was still shifted by the addition of p53 (Fig. 5A, compare lanes 1 to 3 and lanes 4 to 6). It has been shown that the HFM αC helix of NF-YC is required for both NF-YA association and MYC binding (19, 24). We next assayed by immunoprecipitation the single-amino-acid mutants described previously (43). Figure 5B shows that among the mutants that retained DNA binding, YC5-F111S was diminished in p53 complex formation. In contrast, YC5-D112N and YC5-L115K still retained the capacity to bind p53. The YC5-I115P mutant, which does not bind DNA, presumably because of an alteration in the αC conformation, was also inefficient in p53 binding (Fig. 5B, lowest panel). Therefore, the integrity of the conserved αC helix of NF-YC, particularly F111, is important for p53 association. Finally, to map the region of p53 involved in NF-Y binding, we performed similar immunoprecipitation assays with the GST-tagged p53 mutants shown in Fig. 5C. p53 constructs 1 to 298 and 1 to 355 were negative (Fig. 5C, upper panels), whereas 294 to 375 and ΔPro, lacking the N-terminal activation domain, did interact with the trimer (lower panels). We conclude that a region at the C terminus of p53, in particular, between amino acids 355 and 375, is required for NF-Y interactions.
FIG. 5.
Mapping of the NF-Y-p53 association domains. (A) EMSA of wild-type NF-Y (left panel) and HFM NF-YC-NF-YB dimer with a 56-amino-acid NF-YA mutant (YA9) sufficient for DNA binding (right panel) alone and with increasing doses of p53. (B) Immunoprecipitation (IP) of GST-p53 mutants with the indicated NF-YC mutations and of full-length NF-YB with anti-NF-YB antibodies. Flowthrough (FT) and bound (B) fractions were assayed in Western blots with the indicated antibodies. L, load. (C) Immunoprecipitation of full-length NF-Y trimer with the indicated GST-p53 mutations with antibody Mab7. Western blots are shown.
p53 is rapidly acetylated on the repressed cyclin B2 and Cdc2 promoters in vivo after DNA damage.
Although many studies have described a decrease in cyclin B expression induced by p53, none have reported the rapidity of such a decrease. It is therefore important to establish the kinetics of mRNA decay. We performed RT-PCR analysis of cyclin B2 mRNA following adriamycin addition. Fig. 6A shows that cyclin B2 expression was slightly decreased at 2 h and virtually absent at 8 h after damage. No change was observed with the GAPDH control. The results of this experiment, which measured steady-state mRNA levels, were consistent with the rapid inactivation of the promoter and a decrease in the half-life of mitotic cyclin mRNAs observed after DNA damage (31), paralleling the promoter repression assay results shown in Table 1.
p53 is acetylated at K320 and K373-382 (3, 15, 18, 29, 44). To determine the acetylation status of p53 on G2/M promoters, we used specific anti-Ac-p53 antibodies directed against Ac-p53 K320 and Ac-p53 K373-382 in ChIP experiments; these antibodies recognize only p53 acetylated in vitro by recombinant PCAF and p300, respectively (data not shown). ChIP experiments with untreated and adriamycin-treated NIH 3T3 cells are shown in Fig. 6B; the anti-Ac-p53 antibodies were positive for cyclin B2 and Cdc2 after DNA damage, whereas the anti-p53 control antibodies were positive before and after (left panels). In parallel, we verified the presence of PCAF and the acetylation status of neighboring histones; PCAF was bound before but not after DNA damage. H4 and H3 were highly acetylated before adriamycin treatment but less so after adriamycin treatment. NF-Y bound to these promoters both before and after DNA damage. The same immunoprecipitated materials were also used to amplify two bona fide targets of p53, Mdm2 and Bax. Figure 6D clearly shows that neither promoter was bound by p53 in the absence of adriamycin, in agreement with previous results (3, 48). Both anti-Ac-p53 antibodies were positive after damage. As expected, NF-Y did not bind to the two CCAAT-less promoters. These data fully confirm and extend to K320 previous observations made with antibodies against Ac-p53 K373-382 (3). Thus, under DNA damage conditions, Ac-p53 is bound to repressed as well as to activated targets.
Next, we determined the kinetics of p53 acetylation in ChIP experiments. As shown in Fig. 6C (right panels), the acetylation of p53 was already observed at 30 min after adriamycin addition and was maximal by 2 h. Histone tails were deacetylated rapidly, a reduction already being evident at 30 min. However, with longer times, cyclin B2 showed minimal levels of histone acetylation, whereas the levels were reduced but not abolished on Cdc2 (Fig. 6C, compare H3-H4 with input DNA at time zero and at 8 h). As shown before (6), PCAF and p300 were present on both promoters in cycling cells; the former remained bound at 2 h and disappeared by 8 h after adriamycin addition (Fig. 6C, left and middle panels), and the latter disappeared at an earlier time. In general, the dynamics of association of both HATs are consistent with rapid p53 acetylation. In parallel experiments, we controlled the H3 and H4 acetylation status at Mdm2 and Bax loci; as expected, little acetylation was scored before adriamycin addition, and a clear increase was observed after 2 to 8 h of treatment (Fig. 6D, right panels). These kinetics are in line with the observed dynamics of p53 binding and activation of p53-activated genes (48).
Taken together, these data indicate that p53 is not acetylated under nonstress conditions on G2/M promoters; following DNA damage, p53 is rapidly modified at lysines known to be modified by p300 and PCAF, histone tails become deacetylated, and p300 and PCAF are released at later time points.
HDAC recruitment on G2/M promoters following DNA damage.
The results shown in Fig. 6 indicate that histone tails become rapidly deacetylated upon damage, suggesting that HDACs are concomitantly recruited. To substantiate this point, we performed ChIP experiments with anti-HDAC antibodies under the same experimental conditions. Figure 7A shows that under growing conditions, HDAC1 was present at relatively low levels. This result was anticipated, based on our NIH 3T3 cell cycle analysis of Cdc2 and cyclin B2 promoters (6), and it was most likely due to cells in G1 in our asyncronous cell population. Upon adriamycin addition, HDAC1 binding increased and was still observed at 2 h on cyclin B2 but not on Cdc2 (Fig. 7A, upper panels). At this time point, the class II HDACs (HDAC4, HDAC5, and HDAC6) were associated with and indeed were the only HDACs bound at 8 h to cyclin B2. Only HDAC4 was observed on Cdc2. We were not able to detail the binding of HDAC3. To ascertain whether HDAC recruitment was dependent upon the presence of CCAAT boxes and hence of NF-Y-p53 complexes, we used chromatin from the Y1-Y2m clones shown in Fig. 1. As expected, the endogenous control cyclin B2 was positive for NF-Y and, to a much lesser extent, for HDAC1 under growing conditions, while HDAC4 was recruited after 8 h of adriamycin treatment (Fig. 7B, left panels). On the other hand, the corresponding mutated Y1-Y2m templates were negative for all of these factors (Fig. 7B, right panels). Overall, these data (i) are well in line with the kinetics of deacetylation of histones, (ii) correlate the more pronounced deacetylation of cyclin B2 than of Cdc2 with the greater extent of association with HDACs, (iii) suggest differential temporal roles of class I and class II HDACs in promoter repression, and (iv) establish an essential role of CCAAT-NF-Y-p53 complexes in HDAC recruitment.
Acetylation of p53 is important for repression.
We and others previously showed that cotransfection of p300 activated the cyclin B2 promoter, provided that the integrity of the CCAAT sequences and their correct spacing were respected (45, 54). The ChIP data showing the rapid removal of p300 after damage are in agreement with a positive function of this HAT. However, PCAF removal is effective between 2 and 8 h, a timing that follows HDAC association, histone deacetylation, and promoter inactivation. We therefore wished to determine the effect of PCAF in this system. Upon cotransfection of COS cells with a PCAF vector, the wild-type cyclin B2 promoter was repressed in a dose-dependent way; this effect was absent when the triple-CCAAT mutant was used (Fig. 8A). In line with previous reports, a positive effect of equivalent doses of PCAF was observed on Mdm2, either alone or with p53 (Fig. 8B). The same results were obtained with NIH 3T3 cells. Thus, contrary to that of p300, PCAF overexpression has a negative effect on cyclin B2 promoter function.
To assess the role of K320 acetylation in repression, we transfected p53−/− Saos2 cells with cyclin B2 together with p53 vectors carrying single-amino-acid mutations at K320. In parallel experiments, the same mutants were assayed on the Mdm2 promoter. The results of such experiments are shown in Fig. 8C. As expected, p53 reduced transcription while activating Mdm2. The K320R mutant showed minor differences compared to wild-type p53, both in activation and in repression. Liu et al. (29) also reported that the reduction in the activation of a p21-thymidine kinase construct was modest (15%); this result might have been due to the presence of two flanking lysines (K319 and K321), one of which might be an optional binding site for PCAF (see Fig. 3 in reference 29). On the other hand, the K320Q mutant was less active in activating Mdm2 while being significantly more potent as a repressor. We also assayed p53 mutated in the residue (K386) modified by sumoylation; K386R showed no differences in the assays. A comparison of the amounts of p53 proteins expressed indicated comparable levels (Fig. 8D). Therefore, a mutant with a mutation of lysine 320 acetylated by PCAF to glutamine, potentially mimicking acetylaton, has enhanced repression capacity. In cotransfection assays with PCAF in NIH 3T3 cells, wild-type p53 had a clearly additive effect, whereas the K320R mutant revealed only the repression function of PCAF and no further effect of p53 (Fig. 8E). Together, these results indicate that, unlike that of p300, PCAF overexpression has a negative effect on cyclin B2 promoter function that is additive with that of wild-type p53 but not mutant p53 and that K320 is not the only determinant of repression.
Activation versus repression of acetylated p53.
The repression observed with the K320R mutant and the fact that p300-targeted residues were also rapidly acetylated on G2/M promoters suggested that lysines other than K320 might be important in repression. We used a p53 mutant (p53-9KR) in which all lysines of the C terminus were mutated to arginines. Dose-response repression assays on cyclin B2 in Saos2 cells clearly indicated that p53-9KR had completely lost repression capacity, even at high protein concentrations (Fig. 9A). In parallel, we transfected the Mdm2 reporter on the same dose scales as wild-type p53 and p53-9KR; we obtained robust activation, as reported previously (2). The levels of the p53 proteins were determined by Western blot analysis and shown to be equivalent (Fig. 9B). The loss of repression capacity prompted us to investigate the responsible mechanisms, in particular, by verifying whether the mutant was capable of interacting with NF-Y in in vivo overexpression assays. Indeed, immunoprecipitation showed little effect of the K-R mutations at the C terminus of p53 compared to wild-type p53, in terms of NF-Y association (Fig. 9C). We next performed ChIP experiments by cotransfecting a wild-type cyclin B2-luciferase reporter and the p53 vectors. We noted that both wild-type p53 and p53-9KR were similarly capable of associating with the transfected reporter compared to the results obtained in control ChIP experiments, in which an empty vector was cotransfected (Fig. 9D, right panels; compare the vector with wild-type p53 and p53-9KR); little effect was observed at 8 h after adriamycin addition. When the endogenous cyclin B2 promoter was PCR amplified, a modest increase in the binding of NF-Y and p53 was observed (Fig. 9D, left panel; compare the vector with wild-type p53 and p53-9KR). Interestingly, although we only cotransfected p53 and not NF-Y vectors, an increase in NF-Y binding was evident on the cyclin B2-luciferase reporter when p53 vectors were used (Fig. 9D, right panel; compare NF-Y levels in vector, wild-type p53, and p53-9KR lanes); this result suggested that p53 overexpression might actually favor NF-Y binding or stabilize it on transfected templates. We conclude that p53 C-terminal lysines that are acetylated in vivo do play a critical role in repression but that they are collectively expendable for NF-Y association and G2/M promoter recruitment.
DISCUSSION
In this report, we investigated the mechanisms of p53-mediated repression of promoters active in the G2/M phase of the cell cycle. We came to the following relevant conclusions. (i) Under normal conditions, NF-Y and p53 are coresident on selected classes of promoters containing multiple CCAAT boxes and lacking p53 binding sequences. (ii) A direct association between the two factors requires the evolutionarily conserved αC helix of NF-YC and a region within the C-terminal part of p53 adjacent to the tetramerization domain. (iii) After DNA damage, NF-Y and p53 remain bound, p53 K320 and p53 K373-382 become rapidly acetylated, HDACs are recruited, histones are deacetylated, and PCAF and p300 are released from the promoters. These events coincide with rapid promoter repression. (iv) An intact CCAAT-NF-Y-p53 complex is required but not sufficient for HDAC recruitment and repression. (v) Acetylated p53 residues are collectively important for repression.
NF-Y and p53.
We have catalogued 580 promoters regulated by NF-Y. CCAAT boxes are found in genes expressed in a tissue-specific way and in genes rapidly induced by environmental stimuli or growth conditions but are essentially absent in housekeeping genes (F. Romani, A. Testa, and R. Mantovani, unpublished data). Promoters of genes with key roles in the G2/M transition have multiple CCAAT boxes (9). A common feature of these promoters is the conservation of the distance between two CCAAT boxes, 32 or 33 bp; this alignment is important for function through p300-mediated coactivation (45). It is clear that multiple CCAAT boxes are important for p53-mediated repression as well. The data in Fig. 1 and 2 show that promoters containing double but not single CCAAT boxes can be bound by p53 in vivo and that in the absence of NF-Y, p53 does not bind. Given its histone-like structure and DNA bending capacity (reference 43 and references therein), NF-Y is likely to have a predisposition for a peculiar local architecture for p53 association. The p53 docking area on NF-YC is distant from the DNA binding contacts of the HFM subunits; indeed, DNA footprinting shows no further protection of p53 with respect to NF-Y. p53 association might require the severely distorted DNA configuration resulting from multiple NF-Y complexes. This scenario likely reflects the tetrameric status of p53 and the need for multiple points to stably associate with NF-Y. The p53 stretch required for NF-Y association (amino acids 355 to 375) lies next to the tetramerization domain, between the PCAF and p300 acetylated sites. In fact, mutations in the C-terminal flanking lysines do not affect NF-Y binding (Fig. 9). On the NF-Y side, the binding of p53 requires the αC helix of the H2A-like subunit NF-YC. This part is clearly divergent with respect to core histones (43), and its integrity is required for NF-YA association; mutations that grossly alter it, such as the introduction of prolines at residues 115 and 117, abolish binding to p53 and to NF-YA (24, 43). p53 requires the phenylalanine at position 111 in the center of a highly hydrophobic core that is formed by several residues in the α2, α3, and αC helices of NF-YC and in the α2 helix of NF-YB. All amino acids involved in this structure, particularly F111, are conspicuously conserved in evolution. Whether other residues in NF-YB and NF-YA are also required remains to be tested. It is notable that the αC domain of NF-YC was shown to be required for the association of MYC, another transcription factor known to serve a dual role in transcription. Repression is observed on PDGFβ-R, which lacks E boxes but does contain CCAAT boxes (19). It is tempting to speculate that MYC recruitment on such promoters is accomplished through NF-Y binding sites by mechanisms similar to those described here for p53.
Direct versus indirect p53 repression.
p53 induces cell cycle arrest, DNA repair, and apoptosis in response to a variety of environmental stresses (1). In contrast to p53-mediated G1 arrest, which is relatively well understood, the G2 block is less clear, particularly concerning the regulation of genes controlling the G2/M phase. A large body of evidence indicates that p53 utilizes multiple strategies to perform such a function (50). It targets Cdc2, the crucial cyclin-dependent kinase, by inducing p21. Indeed, p21 is necessary for p53-mediated repression of several genes, and overexpressed p21 is even sufficient to repress a subset of these genes in the absence of p53 (30). Through the reduction of the phosphorylation of Rb family proteins, the repression of E2F-responsive promoters can be expected. In fact, several G2/M promoters emerged in studies coupling E2F4 ChIP analysis with microarray analysis of genomic targets (42, 55); evidence that repressive E2F factors bind to G2/M promoters in G0 has been presented, including from our laboratory (6, 49, 56). The sequence(s) bound by E2F factors is unclear, but the likeliest targets are the CDE-CHR elements that are important for repression during the cell cycle (52, 63).
p53 also represses the transcription of the cyclin B, CDC25C, topoisomerase II, Chk2, and Cdc2 genes; the fundamental question is whether repression is direct on promoters, indirect (through p21), or both. Our in vivo data prove that a direct mechanism is operational, as the rapidity of local p53 activation (through acetylation) correlates with promoter repression (Fig. 6). In particular, our data indicate that p53 accounts for short-term repression activity. Previous experiments are in keeping with this line of reasoning. (i) Dissociation of activation from the repressive functions of p53 was reported (25, 26). (ii) Functional and ChIP analyses of MAP4 and survivin indicate direct p53 repression, exerted through sequence-specific binding to DNA (16, 37); however, these results are somewhat controversial, as the elimination of the p53 binding element in the survivin promoter does not impair repression (30, 36). (iii) The cyclin B1 core promoter is still repressed in p21−/− mouse embryo fibroblasts (39). The data shown in Fig. 6 for Mdm2 and Bax are in perfect agreement with the kinetics of p53 binding and mRNA accumulation for responsive genes (48), and it is clear that the decreased cyclin B2 mRNA accumulation is not slower than CDKI accumulation. Does this mean that indirect effects are irrelevant? Not necessarily. Cdk2 phosphorylates NF-YA at serines 320 and 326, suggesting that the inhibition of Cdk2 by p21 may have a direct negative effect on NF-Y function (60). Most importantly, a Ser-to-Ala mutant acts in a dominant-negative way on Cdc2 transcription, blocking cells in G1 and G2 (7), clearly proving that NF-YA is a crucial Cdk2 target for cell cycle progression. Therefore, an indirect role of p21 would be in blocking a phosphorylation event on NF-YA, hampering activation by the trimer.
Jung and coworkers suggested that the mechanism of G2/M promoter repression is related to the inhibition of NF-Y DNA binding capacity by p53 (21). Elimination of DNA binding in vitro (Fig. 3 and 4) or in vivo (Fig. 1, 6, 7, and 9) by p53 clearly was not observed here. Both NF-Y and p53 remained bound to promoters after damage. Thus, we propose that local changes in p53 (and possibly NF-Y) and not the elimination of NF-Y binding are instrumental in the release of HATs and the recruitment of HDACs under genotoxic conditions. A repressive role for NF-Y has been described in other systems. In one study, GH receptor function was negatively controlled by upstream CCAAT boxes; upon the release of NF-Y binding, histone tails became acetylated and the promoter became derepressed (14). For the von Willebrand factor promoter, NF-Y was shown to serve the dual role of recruiting HATs under activating conditions and HDACs under repressing ones; indeed, this study documented a direct association of NF-Y with HDAC1 and HDAC2 (40). Multiple studies indicated that HDAC inhibitors, such as TSA and SAHA, activate promoters through NF-Y binding sites, most likely by relieving repression (references 20, 38, and 62 and references therein). Similarly, p53 was also shown to bind to HDAC1 through the C-terminal domain (28). HDAC4 takes part in genotoxic responses through interactions with p53BP1, a p53 binding protein, becoming associated with DNA repair foci after DNA damage (23). In general, the differential timing of HDAC promoter association (and of HDAC1 displacement) is peculiar and may reflect a relocalization of class II HDACs, largely found in the cytoplasmic compartment of most cell types, into the nucleus upon DNA damage. Whether this is the case and whether other subunits of the class I HDAC complexes follow the fate of HDAC1 remain to be tested.
Role of p53 acetylation in repression.
p53 posttranslational modifications, in particular, acetylation, are clearly essential for the activation of the protein (reviewed in reference 41). Although they increase DNA binding affinity, in vitro and in vivo studies documented the binding of nonacetylated p53 to DNA (10, 22), suggesting that there may be a hierarchy of p53-activated targets, some bound before activation of the protein. Indeed, in NIH 3T3 cells, we did find p53 binding to a site in the p21 promoter before adriamycin addition (Fig. 2), while Mdm2 and BAX were bound only after (Fig. 6). We have started to discriminate the opposing functions of p53 through the use of the p53-9KR mutant; it was previously shown that this mutant is defective in recruiting the TRRAP coactivator while retaining the capacity to increase Mdm2 function to wild-type p53 levels (2). We confirmed these results and determined that mutation of acetylated lysines abolished the repression capacity. Interestingly, this effect did not occur through a lack of NF-Y binding or a loss of promoter association (Fig. 9), indicating that acetylated lysines are instrumental in the recruitment of repressor complexes. In keeping with these findings, our ChIP analysis indicated that p53 is acetylated on repressed promoters. In particular, this is the first demonstration that PCAF-targeted K320 is acetylated on target promoters in vivo. Kinetic analysis suggested that local p53 acetylation on Cdc2 and cyclin B2 promoters occurs no later than and even precedes that on activated promoters. Irrespective of the residue modified, it is possible that p53 activation induces conformational changes. This action is clearly not required for NF-Y association, as demonstrated by protein-protein interactions (in vitro with recombinant proteins and in vivo under nonstress conditions), by EMSAs with nonacetylated p53, and by ChIP assays under growing conditions. Nevertheless, CCAAT boxes are essential for HDAC recruitment, helping to explain the above-mentioned data on the dual function of NF-Y (and p53) in repression and activation.
The binding of NF-Y, PCAF, and p300 to G2/M promoters is regulated during the cell cycle with a precise hierarchy (6). Despite the coresidency of two HATs, p53 is locally not acetylated under normal conditions. Therefore, the crucial question is how HAT activities are locally directed to histones tails but not to p53. The overexpression of p300 leads to the activation of cyclin B (45, 54), while PCAF represses in such assays (Fig. 8). This action is not necessarily contradictory to the presence of PCAF while cell cycle promoters are being transcribed during the cell cycle (6). Indeed, we noted that PCAF association is antecedent to that of p300 and, in some situations, to that of NF-Y. PCAF appears to associate with some promoters while they are still repressed (6). Most importantly, Yamagoe and collaborators detailed the interactions of PCAF/hGCN5 and class I HDACs in a poorly characterized complex, apparently distinct from other HDAC and HAT complexes (57). Indeed, it is possible that PCAF acts as a switch to help recruit HDACs. Regulation is most likely conferred by associated polypeptides in the PCAF complex (also known as TFTC or STAGA) which is formed by 15 or more proteins (5, 34; reviewed in reference 46). Connections between NF-Y and some of these proteins emerged. PCAF/hGCN5 interacts directly with HFM subunits (8, 20, 38). NF-Y contacts HFM-containing TAF12, possibly a partner of H2A-like STAF42 (34); other H3- or H4-like proteins (the presumed TAF6b-TAF9 dimer) may also interact with NF-Y (13). Moreover, TFTC is recruited to UV-damaged DNA (5), and one of the subunits, SAP130, is similar to the xeroderma pigmentosum group E protein (34). Other subunits of the complex are important for p53 function; the TRRAP/PAF400 subunit is a member of the ATM/ATR family of protein kinases targeting p53 (2), and hADA3 binding to phosphorylated p53 is important for function (53). The role of the PCAF complex as a DNA damage sensor, as suggested here, would be entirely consistent with the tumor suppressor function proposed by Schiltz and Nakatani (46).
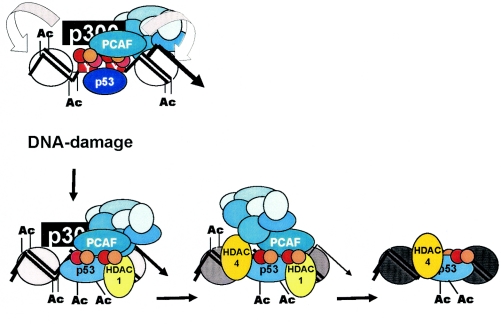
In summary, the following scenario can be proposed (Fig. 10). DNA damage is sensed by one of the PCAF complex subunits, thus allowing rapid redirection of PCAF (and p300) HAT activities, targeting promoter-bound p53. This step is followed by the recruitment of HDACs, first class I and then class II, causing a decrease in the acetylation of nearby nucleosomes. HATs are released, first p300 and then PCAF. The deacetylation of histones occurs while HATs are still on the promoter, and PCAF, modified p53, and NF-Y may recruit HDACs. The fact that NF-Y and modified p53 are still bound may indicate that the promoter is in an idle, “standby” position, waiting for the damage to be cleared to resume growth. Later events (not tested here) may be important in sustaining the block; these include inhibition of Cdk2 by p21, further impairing NF-Y transcriptional activity, repressive E2F binding to CDE-CHR elements, and additional repressor associations (58). This model is a working framework to be pursued with the in vivo assays used here in order to verify the behavior of the numerous subunits of the PCAF and HDAC complexes. Moreover, it is clearly crucial to understand (i) the role of NF-Y posttranslational modifications in the process and (ii) how p53 acetylation, phosphorylation, sumoylation, and other modifications in general, such as the recently described Pin1-mediated prolyl isomerization (61), are transmitted to the structure of the protein, resulting locally in opposite transcriptional outcomes. Finally, the parallel use of wild-type p53 and p53-9KR in overexpression assays may help to reveal the genes which are specifically and directly repressed by p53.
FIG. 10.
Short-term events on G2/M promoters following DNA damage.
Acknowledgments
We thank Y. Nakatani for the gift of PCAF antibodies and T. Halazonetis, S. Berger, M. L. Avvantaggiati, and S. McMahon for the p53 reagents. We thank B. Amati for many helpful discussions and S. Soddu for helpful comments on the article. R.M. thanks V. Rossi for inspiration.
C.I. and S.D.A. are the recipients of a FIRC fellowship; A.G. is the recipient of a MIUR-FIRB contract; R.M., G.P., and G.D.S. are supported by grants from AIRC; R.M. and G.D.S. are supported by MIUR-COFIN; R.M. and G.P. are supported by FIRB and Ministero della Sanitá (R.F. 02/184); and R.M. is supported by Fondazione Cariplo. M.D. acknowledges support from Wilhelm Sander Stiftung.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268**:**2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Ard, P. G., C. Chatterjee, S. Kunjibettu, L. R. Adside, L. E. Gralinski, and S. B. McMahon. 2002. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol. Cell. Biol. 22**:**5650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8**:**1243-1254. [DOI] [PubMed] [Google Scholar]
- 4.Bolognese, F., M. Wasner, C. Lange-zu Dohna, A. Gurtner, A. Ronchi, H. Muller, I. Manni, J. Mossner, G. Piaggio, R. Mantovani, and K. Engeland. 1999. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene 18**:**1845-1853. [DOI] [PubMed] [Google Scholar]
- 5.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20**:**3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caretti, G., V. Salsi, C. Vecchi, C. Imbriano, and R. Mantovani. 2003. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 278**:**30435-30440. [DOI] [PubMed] [Google Scholar]
- 7.Chae, H. D., J. Yun, Y. J. Bang, and D. Y. Shin. 2004. Cdk2-dependent phosphorylation of the NF-Y transcription factor is essential for the expression of the cell cycle-regulatory genes and cell cycle G1/S and G2/M transitions. Oncogene 23**:**4084-4088. [DOI] [PubMed] [Google Scholar]
- 8.Currie, R. A. 1998. Biochemical characterization of the NF-Y transcription factor complex during B lymphocyte development. J. Biol. Chem. 273**:**1430-1434. [DOI] [PubMed] [Google Scholar]
- 9.Elkon, R., C. Linhart, R. Sharan, R. Shamir, and Y. Shiloh. 2003. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 13**:**773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinosa, J. M., and B. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8**:**57-69. [DOI] [PubMed] [Google Scholar]
- 11.Farina, A., I. Manni, G. Fontemaggi, M. Tiainen, C. Cenciarelli, M. Bellorini, R. Mantovani, A. Sacchi, and G. Piaggio. 1999. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene 18**:**2818-2827. [DOI] [PubMed] [Google Scholar]
- 12.Flatt, P. M., L. J. Tang, C. D. Scatena, S. T. Szak, and J. A. Pietenpol. 2000. p53 regulation of G2 checkpoint is retinoblastoma protein dependent. Mol. Cell. Biol. 20**:**4210-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frontini, M., C. Imbriano, A. diSilvio, B. Bell, A. Bogni, C. Romier, D. Moras, L. Tora, I. Davidson, and R. Mantovani. 2002. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 277**:**5841-5848. [DOI] [PubMed] [Google Scholar]
- 14.Gowri, P. M., J. H. Yu, A. Shaufl, M. A. Sperling, and R. K. Menon. 2003. Recruitment of a repressosome complex at the growth hormone receptor promoter and its potential role in diabetic nephropathy. Mol. Cell. Biol. 23**:**815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90**:**605-606. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman, W. H., S. Biade, J. T. Zilfou, J. Chen, and M. Murphy. 2002. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 277**:**3248-3257. [DOI] [PubMed] [Google Scholar]
- 17.Innocente, S. A., J. L. Abrahamson, J. P. Cogswell, and J. M. Lee. 1999. p53 regulates a G2 checkpoint through cyclin B1. Proc. Natl. Acad. Sci. USA 96**:**2148-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20**:**1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumi, H., C. Molander, L. Z. Penn, A. Ishisaki, K. Kohno, and K. Funa. 2001. Mechanism for the transcriptional repression by c-Myc on PDGF beta-receptor. J. Cell Sci. 114**:**1533-1544. [DOI] [PubMed] [Google Scholar]
- 20.Jin, S., and K. W. Scotto. 1998. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol. Cell. Biol. 18**:**4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, M. S., J. Yun, H. D. Chae, J. M. Kim, S. C. Kim, T. S. Choi, and D. Y. Shin. 2001. p53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene 20**:**5818-5825. [DOI] [PubMed] [Google Scholar]
- 22.Kaeser, M. D., and R. D. Iggo. 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. USA 99**:**95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao, G. D., W. G. McKenna, M. G. Guenther, R. J. Muschel, M. A. Lazar, and T. J. Yen. 2003. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 160**:**1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, I. S., S. Sinha, B. de Crombrugghe, and S. N. Maity. 1996. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 16**:**4003-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kokontis, J. M., A. J. Wagner, M. O'Leary, S. Liao, and N. Hay. 2001. A transcriptional activation function of p53 is dispensable for and inhibitory of its apoptotic function. Oncogene 20**:**660-668. [DOI] [PubMed] [Google Scholar]
- 26.Koumenis, C., R. Alarcon, E. Hammond, P. Sutphin, W. Hoffman, M. Murphy, J. Derr, Y. Taya, S. W. Lowe, M. Kastan, and A. Giaccia. 2001. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 21**:**1297-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krause, K., M. Wasner, W. Reinhard, U. Haugwitz, C. L. Dohna, J. Mossner, and K. Engeland. 2000. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 28**:**4410-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275**:**20436-20443. [DOI] [PubMed] [Google Scholar]
- 29.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19**:**1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohr, K., C. Moritz, A. Contente, and M. Dobbelstein. 2003. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 278**:**32507-32516. [DOI] [PubMed] [Google Scholar]
- 31.Maity, A., W. G. McKenna, and R. J. Muschel. 1995. Evidence for post-transcriptional regulation of cyclin B1 mRNA in the cell cycle and following irradiation in HeLa cells. EMBO J. 14**:**603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manni, I., G. Mazzero, A. Gurtner, R. Mantovani, U. Haugwitz, K. Krause, K. Engeland, A. Sacchi, S. Soddu, and G. Piaggio. 2001. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and Cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 276**:**5570-5576. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239**:**15-27. [DOI] [PubMed] [Google Scholar]
- 34.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 20**:**6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsui, T., Y. Katsuno, T. Inoue, F. Fujita, T. Joh, H. Niida, H. Murakami, M. Itoh, and M. Nakanishi. 2004. Negative regulation of Chk2 expression by p53 is dependent on the CCAAT-binding transcription factor NF-Y. J. Biol. Chem. 279**:**25093-25100. [DOI] [PubMed] [Google Scholar]
- 36.Mirza, A., M. McGuirk, T. N. Hockenberry, Q. Wu, H. Ashar, S. Black, S. F. Wen, L. Wang, P. Kirschmeier, W. R. Bishop, L. L. Nielsen, C. B. Pickett, and S. Liu. 2002. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21**:**2613-2622. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13**:**2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S. H., S. R. Lee, B. C. Kim, E. A. Cho, S. P. Patel, H. B. Kang, E. A. Sausville, O. Nakanishi, J. B. Trepel, B. I. Lee, and S. J. Kim. 2002. Transcriptional regulation of the transforming growth factor beta type II receptor gene by histone acetyltransferase and deacetylase is mediated by NF-Y in human breast cancer cells. J. Biol. Chem. 277**:**5168-5174. [DOI] [PubMed] [Google Scholar]
- 39.Passalaris, T. M., J. A. Benanti, L. Gewin, T. Kiyono, and D. A. Galloway. 1999. The G2 checkpoint is maintained by redundant pathways. Mol. Cell. Biol. 19**:**5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, Y., and N. Jahroudi. 2003. The NFY transcription factor inhibits von Willebrand factor promoter activation in non-endothelial cells through recruitment of histone deacetylases. J. Biol. Chem. 278**:**8385-8394. [DOI] [PubMed] [Google Scholar]
- 41.Prives, C., and J. L. Manley. 2002. Why is p53 acetylated? Cell 107**:**815-818. [DOI] [PubMed] [Google Scholar]
- 42.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, and R. A. Young., and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication and G(2)/M checkpoints. Genes Dev. 16**:**245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romier, C., F. Cocchiarella, R. Mantovani, and D. Moras. 2003. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278**:**1336-1345. [DOI] [PubMed] [Google Scholar]
- 44.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12**:**2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salsi, S., G. Caretti, M. Wasner, W. Reinhard, U. Haugwitz, K. Engeland, and R. Mantovani. 2003. Interactions between p300 and multiple NF-Y trimers govern cyclin B2 promoter function. J. Biol. Chem. 278**:**6642-6650. [DOI] [PubMed] [Google Scholar]
- 46.Schiltz, R. L., and Y. Nakatani. 2000. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta 1480**:**M37-M53. [DOI] [PubMed] [Google Scholar]
- 47.Sciortino, S., A. Gurtner, I. Manni, G. Fontemaggi, A. Dey, A. Sacchi, K. Ozato, and G. Piaggio. 2001. The cyclin B1 gene is actively transcribed during mitosis in HeLa cells. EMBO Rep. 2**:**1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szak, S. T., D. Mays, and J. A. Pietenpol. 2001. Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 21**:**3375-3386.11313463 [Google Scholar]
- 49.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14**:**804-816. [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20**:**1803-1815. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, W. R., S. E. DePrimo, A. Agarwal, M. L. Agarwal, A. H. Schonthal, K. S. Katula, and G. R. Stark. 1999. Mechanisms of G2 arrest in response to overexpression of p53. Mol. Biol. Cell 19**:**3607-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor, W. R., A. H. Schonthal, J. Galante, and G. R. Stark. 2001. p130/E2F4 binds to and represses the Cdc2 promoter in response to p53. J. Biol. Chem. 276**:**1998-2006. [DOI] [PubMed] [Google Scholar]
- 53.Wang, T., T. Kobayashi, R. Takimoto, A. E. Denes, E. L. Snyder, W. S. el-Deiry, and R. K. Brachmann. 2001. hADA3 is required for p53 activity. EMBO J. 20**:**6404-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wasner, M., K. Tschop, K. Spiesbach, U. Haugwitz, C. Johne, J. Mossner, R. Mantovani, and K. Engeland. 2003. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 536**:**66-70. [DOI] [PubMed] [Google Scholar]
- 55.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16**:**235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20**:**5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamagoe, S., T. Kanno, Y. Kanno, S. Sasaki, R. M. Siegel, M. J. Lenardo, G. Humphrey, Y. Wang, Y. Nakatani, B. H. Howard, and K. Ozato. 2003. Interaction of histone acetylases and deacetylases in vivo. Mol. Cell. Biol. 23**:**1025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon, H. S., and V. W. Yang. 2004. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J. Biol. Chem. 279**:**5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yun, J., H. D. Chae, H. E. Choy, J. Chung, H. S. Yoo, M. H. Han, and D. Y. Shin. 1999. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 274**:**29677-29682. [DOI] [PubMed] [Google Scholar]
- 60.Yun, J., H. D. Chae, T. S. Choi, E. H. Kim, Y. J. Bang, J. Chung, K. S. Choi, R. Mantovani, and D. Y. Shin. 2003. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J. Biol. Chem. 278**:**36966-36972. [DOI] [PubMed] [Google Scholar]
- 61.Zacchi, P., M. Gostissa, T. Uchida, C. Salvagno, F. Avorio, S. Volinia, Z. Ronai, G. Blandino, C. Schneider, and G. Del Sal. 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419**:**853-857. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X., W. Wharton, Z. Yuan, S. C. Tsai, N. Olashaw, and E. Seto. 2004. Activation of the growth differentiation factor 11 gene by the histone deacetylase (HDAC) inhibitor trichostatin A and repression by HDAC3. Mol. Cell. Biol. 24**:**5106-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zwicker, J., F. C. Lucibello, L. A. Wolfraim, C. Gross, M. Truss, K. Engeland, and R. Muller. 1995. Cell cycle regulation of the cyclin A, Cdc25C and Cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 14**:**4514-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]